Abstract
Populations on continental islands are often distinguishable from mainland conspecifics with respect to body size, appearance, behaviour or life history, and this is often congruent with genetic patterns. It is commonly assumed that such differences developed following the complete isolation of populations by sea-level rise following the Last Glacial Maximum (LGM). However, population divergence may predate the LGM, or marine dispersal and colonization of islands may have occurred more recently; in both cases, populations may have also diverged despite ongoing gene flow. Here, we test these alternative hypotheses for the divergence between wedge-tailed eagles from mainland Australia (Aquila audax audax) and the threatened Tasmanian subspecies (Aquila audax fleayi), based on variation at 20 microsatellite loci and mtDNA. Coalescent analyses indicate that population divergence appreciably postdates the severance of terrestrial habitat continuity and occurred without any subsequent gene flow. We infer a recent colonization of Tasmania by marine dispersal and cannot discount founder effects as the cause of differences in body size and life history. We call into question the general assumption of post-LGM marine transgression as the initiator of divergence of terrestrial lineages on continental islands and adjacent mainland, and highlight the range of alternative scenarios that should be considered.
Keywords: sea level, Last Glacial Maximum, marine dispersal, vicariance, subspecies, inbreeding depression
1. Introduction
The comparison of animal populations on islands with their close relatives on continents has provided striking examples of the potential for rapid divergence between lineages in morphological [1,2], behavioural [3–6] and life-history [7] traits. With respect to examples involving oceanic islands—those emerging from oceanic crust without prior terrestrial connection to other land masses—these divergences most probably occurred under conditions of complete genetic isolation, of duration constrained by the age of island emergence [8]. However, most island biotas are not found on oceanic islands, but rather continental islands—islands that experienced a repeated history of marine isolation and terrestrial connectivity with the adjacent mainland during the Pleistocene, owing to their shared position on a continental plate [8]. Therefore, the circumstances under which the majority of island biotas diverged from that on the adjacent mainland may differ from those inferred for oceanic islands.
Continental islands that are presently separated from adjacent mainland by seaways shallower than 100 m would have experienced multiple cycles of connection via continuous terrestrial habitat, and subsequent isolation via marine transgression, during the last 3 Myr [9], assuming that any tectonic uplift or subsistence in the region was minor. The most recent isolation event is represented by the marine transgression following the Last Glacial Maximum (LGM, 26.5–19.0 kya), during which sea levels were up to 120 m lower than the present [10]. Three or possibly four marine transgressions of similar magnitude have occurred during the last 0.5 Myr, and additional transgressions of less certain magnitudes occurred earlier during the Pleistocene [11].
Despite the number and significance of continental island biotas (e.g. Taiwan, Indonesia, New Guinea, Sri Lanka, Britain), their divergence from those on the adjacent mainland has received somewhat less attention than those of oceanic islands. Divergences of continental island biotas are often assumed to reflect vicariant isolation from the adjacent mainland during the marine transgression that followed the LGM (figure 1a), without full consideration of alternative hypotheses [12]. Even if divergence was initiated by the most recent marine transgression, divergence may have continued under gene flow in taxa with marine dispersal capability ([13]; figure 1b). Similarly, divergence may have been initiated by an earlier event (not necessarily a marine transgression), and proceeded through subsequent periods of terrestrial connectivity, either with or without gene flow ([14]; figure 1c,d). Finally, marine dispersal, rather than terrestrial vicariance, may have initiated the divergence of lineages through the establishment of the island (or potentially mainland) lineage, analogous to the default situation invoked for geologically recent oceanic islands, and again, divergence could have proceeded either with or without subsequent gene flow ([15,16], figure 1e,f). Knowledge of which biogeographic scenarios have operated is important to achieve a greater understanding of the processes that have led to the distinctiveness of lineages on continental islands compared with their mainland relatives (e.g. potential for local adaptation, phenotypic plasticity or founder effect divergence), and can now be tested under analytical frameworks that estimate the age of population divergence, subsequent rates of gene flow and population sizes [17–22].
Figure 1.
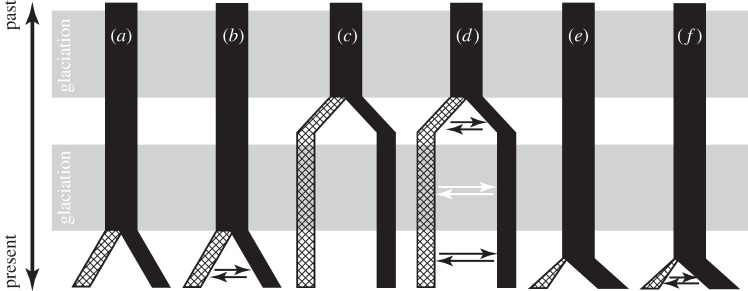
Potential divergence scenarios of lineages from a continental island and adjacent mainland. Black bars represent lineages occupying the mainland, while crosshatched bars are lineages occupying islands (in these examples, the mainland is assumed to be the ancestral distribution, but the opposite is also possible). Arrows represent gene flow between lineages (here illustrated as bidirectional, but may also be unidirectional). Grey shading indicates periods of glaciation. (a,b) Vicariant divergence mediated by the most recent marine transgression. (c,d) An earlier divergence, with lineages maintaining their distinction through subsequent low sea stands, either with or without gene flow between lineages during glacial or interglacial periods. (e,f) Divergence initiated by marine dispersal, either with or without subsequent gene flow.
The island of Tasmania provides an excellent framework to investigate these alternative lineage divergence scenarios for continental island and mainland taxa. Tasmania is presently separated from mainland Australia by an expansive (approx. 240 km) but shallow (less than 70 m; figure 2; [23]) seaway (Bass Strait), with high tectonic stability, such that repeated terrestrial connections existed during the Pleistocene, most recently during the LGM [10]. Tasmanian and mainland populations of several species have also undergone trait divergence with respect to morphology, physical appearance, behaviour and life-history characters [24–26]. In particular, a high proportion of the Tasmanian terrestrial avifauna (20 out of 145 species) exhibit divergence from mainland relatives at a level considered worthy of subspecies recognition [27].
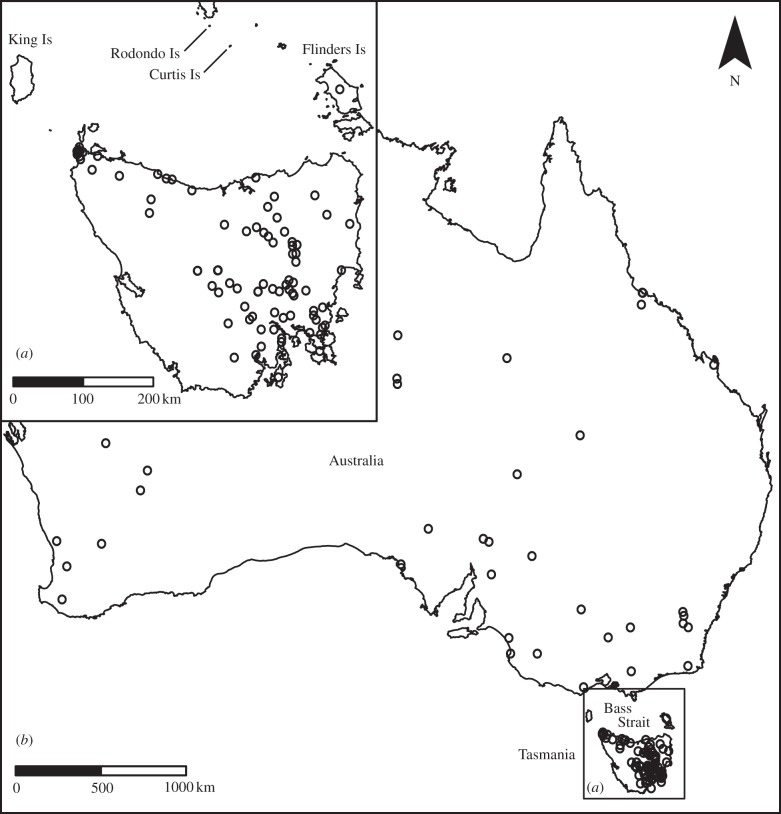
Figure 2.
Distribution of A. audax samples employed for genetic analysis from (a) Tasmania (A. a. fleayi, n = 175) and (b) mainland Australia (A. a. audax, n = 49). Note that two mainland and 60 Tasmanian individuals are not illustrated owing to the absence of fine-scale collection locality information.
The wedge-tailed eagle (Accipitridae: Aquila audax) is Australia's largest bird of prey (wingspan 1.9–2.3 m, mass 3.5–5.5 kg) [28,29] and represents an ideal species to test alternative scenarios for the divergence of continental island and mainland lineages. Firstly, A. audax is presently widespread across Tasmania, mainland Australia and parts of New Guinea, and is found in almost every habitat available, including deserts, grasslands, forests and alpine herbfields [28]. This suggests an ability to occupy the landbridge between Tasmania and the mainland when it was exposed, enabling vicariance-mediated divergence. Secondly, the species exhibits long-distance terrestrial dispersal by non-breeding juveniles of up to 800 km [30]. Thus marine dispersal, either subsequent to the initiation of population divergence (i.e. gene flow), or as the initiator of divergence itself (i.e. colonization), can also be realistically entertained. Suggestions of a vagrant on King Island, at the western margin of Bass Strait, may indicate marine dispersal capability assuming that it was correctly identified [31]. Thirdly, there is reputedly trait divergence between Tasmanian and mainland populations; Tasmanian adults are darker in colour [28,32,33], larger in body size [34,35], roost only in trees and produce smaller clutches (usually one egg, versus usually two on the mainland) [29,36]. Consequently, mainland and Tasmanian populations have been recognized as distinct subspecies (mainland: A. a. audax; Tasmania: A. a. fleayi) [32], with the latter listed as endangered under the Tasmanian Threatened Species Protection Act 1995 and the Environment Protection and Biodiversity Conservation Act 1999 [37]. The wedge-tailed eagle feeds on both live prey and carrion, comprising reptiles, birds and mammals, particularly rabbits and hares, and appears to breed in solitary and monogamous pairs, with a longevity of up to 20 years in the wild [28,38].
The aim of this study was to test the alternative biogeographic scenarios under which Tasmanian and mainland populations of A. audax diverged, specifically, the duration of their divergence with respect to the dynamic history of terrestrial connectivity between these regions and the amount gene flow that may have occurred between these populations while undergoing divergence. We surveyed genetic variation in mitochondrial DNA (control region and the ATPase 6/8 genes), and at 20 nuclear microsatellite loci, and used the ‘Isolation with Migration’ framework of Hey & Nielsen [17] to address these questions, along with more traditional population genetics tests for population isolation.
2. Material and methods
(a). Sampling
A total of 224 individuals from across Australia (mainland = 49, Tasmania = 175) were sampled for genetic material (figure 2), obtained from existing museum specimens (see electronic supplementary material), captive individuals, shed feathers and a Tasmanian Government frozen collection of deceased individuals accumulated during 1996–2012. Tissue samples were obtained from feathers in the case of live and museum specimens, or additionally from breast muscle using a 6 mm biopsy punch in the case of frozen specimens. DNA extraction and purification employed the QIAGEN DNeasy Blood & Tissue kit, following manufacturer's instructions, and DNA concentrations were standardized to approximately 20 ng µl−1 following quantification via spectrophotometry.
(b). Mitochondrial DNA
Mitochondrial control region and the ATPase6/8 gene sequences were obtained from 70 individuals (40 of mainland and 30 of Tasmania). An approximately 250 bp fragment of the mitochondrial control region was amplified using primers Eagle D loop F (CCCCGTATGTATTATTGTACAT) and Eagle D loop R (CAAGTTATGACCTGCTACC), and an approximately 1000 bp fragment spanning the entire ATPase6/8 genes was amplified using CO2GQL (GGA CAA TGC TCA GAA ATC TGC GG) and CO3HMH (ATG GGC TGG GGT CRA CTA TGT G), or in two smaller, overlapping fragments using primers Eagle CO3 F1 (GGG GCT AAC CAC AGC TAC AT) and Eagle ATPase R1 (CCT AGT AAG TTG ATT GTT AGT AGG AGA), and ATPase F1 (ACC CCT AAA TAA AGG AGG ACA) and Eagle CO2 R1 (ATG GGC TGG GGT CGA CTA T). Each PCR (25 μl final volume) contained 1× Hotmaster buffer, 0.2 mM each dNTP, 0.2 μM of each primer and 0.5 units of Hotmaster Taq DNA Polymerase (5 PRIME). Thermocycling comprised an initial denaturation and enzyme activation at 94°C for 2 min, followed by 35 cycles of denaturing at 94°C for 20 s, primer annealing at 55°C for 10 s, and elongation at 65°C for 1 min. Samples then underwent final elongation and adenylization for 10 min at 65°C. Positive PCRs were purified using AMPure (Agencourt) following manufacturer's instructions, and both strands were sequenced using BigDye 3.1 Sanger sequencing chemistry and an Applied Biosystems 3130XL DNA Analyzer.
Mitochondrial DNA sequences were aligned, and homogeneity of haplotype frequencies among Tasmanian and mainland populations was examined using exact tests in Arlequin 3.5 [39], which was also used to quantify population structuring via FST based on haplotype frequencies alone. A minimum spanning network was constructed using Arlequin.
(c). Microsatellites
Individuals were genotyped for 20 polymorphic microsatellite loci (electronic supplementary material). Allele scoring was performed using Genemapper 4.0 (Applied Biosystems). Allele frequencies in each population were tested for concordance with Hardy–Weinberg equilibrium and genotypic disequilibrium using Genepop [40]. Tests of Hardy–Weinberg equilibrium were performed using an ‘Exact H-W test’, with complete enumeration of p-values whenever there were less than five alleles [41], otherwise estimation of p-values via 1000 Markov chain batches [42]. Tests of genotypic disequilibrium used the log-likelihood ratio statistic and 1000 Markov chain batches [40]. Sequential Bonferroni correction was applied for multiple tests [43]. Allele frequency homogeneity between Tasmania and the mainland was examined using G tests in Genepop, with 1000 Markov chain batches, and population genetic structuring (GST and 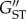 ) was quantified using GenoDive [44].
) was quantified using GenoDive [44].
Bayesian clustering was employed to test for the presence of individuals that may have recently dispersed across Bass Strait, or their descendants, or population structuring within Tasmania or the mainland that may require specific accommodation during subsequent coalescent analyses of population divergence. Bayesian clustering employed STRUCTURE 2.3.3 [45] with 100 000 Markov chain batches under the population admixture model with correlated allele frequencies, and potential values of K (number of population clusters) between one and five. Selection of the optimum number of clusters followed Evanno et al. [46], based on 20 replicate analyses for each value of K, assessed using STRUCTURE HARVESTER [47].
Levels of genetic variation between Tasmanian and mainland populations (number of alleles, Ho and He at each locus) were compared using a one-tailed Mann Whitney U-test, under the expectation that the island population would exhibit lower variation [48]. The effective inbreeding coefficient, 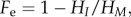 where HI is the expected heterozygosity of the Tasmanian population, and HM is the corresponding value for the mainland population, was calculated to place into the context of values observed for other island–mainland comparisons and populations suffering from inbreeding depression [48].
where HI is the expected heterozygosity of the Tasmanian population, and HM is the corresponding value for the mainland population, was calculated to place into the context of values observed for other island–mainland comparisons and populations suffering from inbreeding depression [48].
(d). Coalescence analysis
The age of divergence between mainland and Tasmanian populations and the level of gene flow during divergence was assessed under the ‘Isolation with Migration’ framework of Hey & Nielsen [17], employing IMa2 [49]. Mitochondrial and microsatellite markers were analysed concurrently to estimate the six parameters of the model (two contemporary and one ancestral population size, divergence time of populations and rates of post-isolation gene flow between populations in each direction). The HKY mutation model was employed for mtDNA sequence data, while the stepwise mutation model (SMM) was employed for microsatellites. Two microsatellite loci were excluded from the analysis as they exhibited alleles inconsistent with SMM (Aau348, Aau89; allele size differences that suggest changes other than the repeat number of the microsatellite motif). To expedite the analysis, a random subsample of 30 Tasmanian and 30 mainland individuals were selected for analysis (containing data for mtDNA and all microsatellite loci).
Information on generation time and mutation rate is required to convert the output of IMa2 into units of years and individuals. Good life table data for A. audax are lacking. Sexual maturity is thought to be attained at 4–6 years, but age of first reproduction also depends on the procurement of a breeding territory and a mate and is thought to take 6–7 years [28]. Mortality is high during early life, but longevity is thought to extend to 20–25 years [38]. Therefore, we employed 10 and 15 years as estimates for the average age of successfully reproducing individuals. Mitochondrial mutation rates were employed in the analysis, against which mutation rates at the nuclear loci would be scaled. Estimates of rates for birds vary widely and also appear to vary in a time-dependent manner [50,51]. Here, we started with an initial value of 2.1% sequence divergence per million years [52], but then explored the consequences of using a faster rate that may be more applicable to reconstructing demographic history over recent (less than 100 000 kyr) timescales [53]. Failure to entertain time-dependent rates of molecular change will lead to overestimation of divergence time and population sizes, and underestimation of gene flow [53].
Prior distributions were constructed following the guidelines in the IMa2 manual. Uniform priors were employed for population size and divergence time parameters, while exponential priors were employed for gene flow, given an expectation that low rates of gene flow were likely. Upper limits on uniform priors of θ ( = 4Neµ) for contemporary populations were initially five times the largest population geometric mean of θ among microsatellite loci and mtDNA, and that for ancestral θ was the sum of the two. For microsatellite loci, θhom was calculated using the SMM method of Ohta & Kimura [54], and θπ was calculated for mtDNA, both using Arlequin. The upper limit of the uniform population divergence time prior was set in a manner to encompass the last approximately 80 kyr when accommodating a rapid mtDNA mutation rate (0.1/site/lineage/Myr). This accommodates population divergence scenarios that occurred coincidentally (vicariant divergence), after, and substantially before the most recent inundation of Bass Strait. Exponential priors for gene flow were set around a value approximating a mean of 1 individual per generation.
An initial run was conducted until stationarity was achieved, employing MCMC sampling with 80 chains, a geometric chain heating scheme with first and second heating parameters of 0.95 and 0.5, respectively, and eight chain swap attempts per step. This was then used to seed 24 independent 24-h runs each with a short (4 h) burn-in, but otherwise using the same parameters as above. These 24 runs produced a total of 334 814 genealogies for analysis of model parameters.
3. Results
(a). Genetic variation
Variation was observed at only 12 nucleotide positions within the 1119 bp of the mitochondrial genome sequenced (168 bp control region, 951 bp ATPase6/8), defining 15 haplotypes. The Tasmanian individuals were fixed for a haplotype shared with mainland Australia (figure 3).
Figure 3.
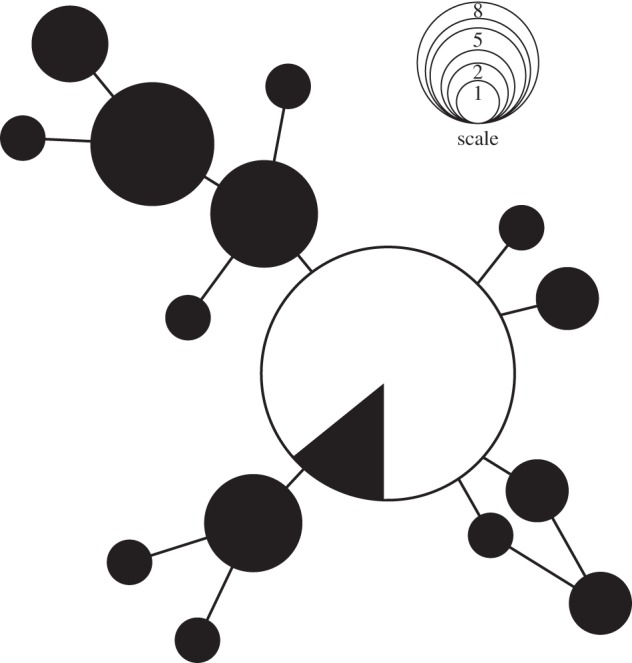
Minimum spanning network among mitochondrial haplotypes in A. audax. Circles represent haplotypes, with areas proportional to numbers of observations (smallest circles represent 1 individual, largest circle represents 35, others defined by the scale). Black shading represents mainland individuals (30 observations, slice of central haplotype corresponds to five individuals), white shading represents Tasmanian individuals. Lines linking haplotypes represent single mutation steps.
Following Bonferroni correction, only locus Aau252 from the mainland population deviated from Hardy–Weinberg equilibrium. Only genotypic disequilibrium was found for the three possible comparisons among Aau190, Aau299, and Aau89. Levels of genetic variation were significantly lower in the Tasmanian population relative to the mainland population with respect to number of alleles per locus, Ho and He (U0.05(1),20,20 = 288, 291 and 293, respectively, p < 0.01; table 1). In particular, three loci were monomorphic in the Tasmanian population, in comparison to none in the mainland population, despite almost three times as many Tasmanian individuals in the dataset. The effective inbreeding coefficient for the Tasmanian population relative to the mainland population was Fe = 0.41.
Table 1.
Genetic variation in Tasmanian and mainland Australian populations of A. audax based on 20 microsatellite markers and 1119 bp of mitochondrial DNA. n represents sample size, Ho and He are observed and expected heterozygosity, respectively, h is haplotype diversity and π is nucleotide diversity.
| microsatellite variation (20 loci) |
mitochondrial variation |
|||||||
|---|---|---|---|---|---|---|---|---|
| n | no. alleles | Ho | He | n | no. haplotypes | h | π | |
| mean (s.e.) | mean (s.e.) | mean (s.e.) | mean (s.d.) | mean (s.d.) | ||||
| Tasmania | 175 | 3.000 (0.324) | 0.325 (0.063) | 0.333 (0.063) | 30 | 1 | 0 | 0 |
| mainland | 49 | 5.785 (0.786) | 0.550 (0.059) | 0.568 (0.058) | 40 | 15 | 0.91 ± 0.02 | 0.0019 (0.0012) |
(b). Population structuring
Bayesian clustering of individuals based on microsatellite genotypes without prior population collection information suggested two populations, corresponding to mainland and Tasmanian individuals (mean Ln Pr(X|K) = −5883.355) (figure 4). The degree of genetic structuring between the two populations was quantified as FST = 0.234 for mtDNA, and GST = 0.118 and 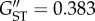 for microsatellites, and all were significantly greater than zero (p < 0.001). Exact tests of population differentiation were also significant for both mtDNA (p < 0.01) and microsatellites (p < 0.01 for 19 loci, all of which were significant following Bonferroni correction).
for microsatellites, and all were significantly greater than zero (p < 0.001). Exact tests of population differentiation were also significant for both mtDNA (p < 0.01) and microsatellites (p < 0.01 for 19 loci, all of which were significant following Bonferroni correction).
Figure 4.

Estimated population structure from STRUCTURE analysis. Each individual is represented by a vertical bar proportional to its estimated membership to the mainland (black) or Tasmanian (white) population, with the actual source of the individual demarcated along the top of the figure.
(c). Coalescent analysis
The posterior distributions from the coalescent analysis are indicated in figure 5. Posterior probabilities of θ for the mainland population never returned to zero at large values, but replicate analysis with different upper bounds for this parameter did not appear to influence the posterior distribution for any of the other parameters. Posterior distributions for gene flow were exponentially distributed but differed from that of the prior. Conversion of parameters to demographic scales suggests a much smaller effective size for the Tasmanian population and a very recent divergence from the mainland population, in the order of 3–95 years ago (table 2).
Figure 5.
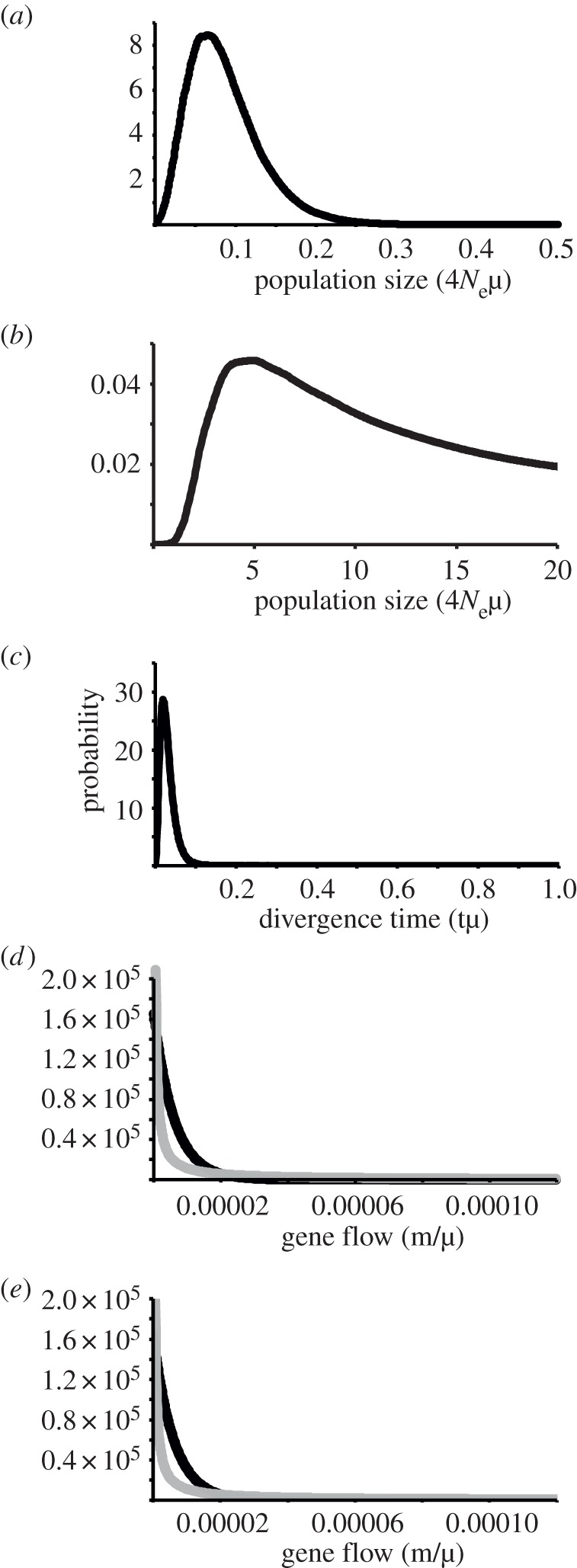
Posterior distributions from analysis under the ‘Isolation with Migration’ model, indicated with black curves. Gene flow prior distributions (grey curves) are illustrated to demonstrate that the posterior distributions differ from them. (a) θ Tasmania, (b) θ mainland, (c) divergence time, (d) gene flow, mainland to Tasmania, going backwards in time, (e) gene flow, Tasmania to mainland, going backwards in time. Ne is effective population size, µ is mutation rate per generation, t is time since population splitting, and m is migration rate (gene flow).
Table 2.
Posterior estimates of parameters from ‘Isolation with Migration’ analysis, and conversion to demographic scales based on a mean generation time of 10 years. Median values from posteriors are reported, along with 95% highest posterior densities (HPD, in parentheses). Analyses with faster mutation rates (e.g. 10% divergence/Myr) produced much smaller estimates of effective population sizes and divergence times, while increasing the generation time to 15 years reduced effective population size estimates slightly.
| mtDNA divergence rate = 2.1%/Myr | mtDNA divergence rate = 10.0%/Myr | |
|---|---|---|
| Ne mainland | 120 (HPD indefinable) | 25 (HPD indefinable) |
| Ne Tasmania | 3 (0–10) | 1 (0–2) |
| Ne ancestral | 70 (49–104) | 15 (10–22) |
| gene flow (Tas→mainland) | 0 (less than 1 × 10−10) | 0 (less than 1 × 10−10) |
| gene flow (mainland→Tas) | 0 (less than 1 × 10−10) | 0 (less than 1 × 10−10) |
| divergence time (years since) | 22 (3–95) | 5 (1–20) |
4. Discussion
The Tasmanian population of A. audax became isolated very recently from the mainland population, contrary to the paradigm of marine transgression-induced vicariance of continental island populations [12,37,55]. Coalescent estimates employing ‘conventional’ avian mitochondrial substitution rates suggest divergence 3–95 years ago. While these divergence dates must be considered an underestimate because the first European sighting of A. audax in Tasmania was documented in 1836 [56], this likely reflects the lack of unique molecular variation in the Tasmanian population, which confidently refutes divergence times compatible with divergence accompanying the flooding of Bass Strait greater than 13 kya [10]. Furthermore, under time-dependent molecular rates [50,51,53], such an ancient divergence is even less likely. Despite the inferred recency of founding, there was no evidence for subsequent or ongoing gene flow, from coalescent analysis, Bayesian clustering or tests of allele frequency homogeneity. These results suggest that the Tasmanian population was founded appreciably later than the marine transgression that flooded the Bassian plain and severed terrestrial connection to mainland Australia approximately 13 kya [10].
(a). Colonization via unassisted marine dispersal
The inferred age of lineage divergence and differences in levels of genetic variation appear compatible with marine dispersal by A. audax across a flooded Bass Strait, from the mainland to Tasmania, and the founding of a new population. The record of a vagrant on King Island [31], if correct, provides evidence of marine dispersal over short time scales, and a single genotype obtained from Flinders Island was also very similar to those in Tasmania, despite the long duration of intervening marine isolation. Individuals on Curtis and Rodondo Islands [57,58] in Bass Strait are also likely examples of marine dispersal. Examples of marine dispersal of similar or greater magnitude by other raptors (some migratory) are also documented both from direct observations [59,60], palaeontological evidence ([61], e.g. the swamp harrier, Circus approximans, colonizing New Zealand probably less than 1000 years ago; [62]), and reconstructions of phylogenetic relationships [63–65].
While human-mediated dispersal of accipitrids, and raptors more generally, has occurred in the past for various reasons, for example control of pest populations [66], it appears unlikely to explain our observations for A. audax. Although A. audax have been trained and kept for hunting [33], documented sightings from different parts of Tasmania in 1836 [56] would appear to be too early after English colonization in 1803 to be explained by undocumented translocation by Europeans, given a generation time of about 10 years [28], and low fecundity (no greater than two chicks per annum per pair) [29,36]. Furthermore, Tasmanian aboriginal names, for example ‘Nairana’, may apply to A. audax, suggesting an earlier presence in Tasmania, although there is some doubt as to whether this name was ascribed specifically to A. audax or other raptors as well [67].
(b). The diversity of divergence scenarios observed for continental island and mainland lineages
Our results highlight the possibility that the divergence of terrestrial lineages on a continental island and its adjacent mainland postdate the cessation of the most recent terrestrial connection between these landmasses. Similarly, two of the three other bird species which have been compared genetically between Tasmania and the mainland also exhibit divergences compatible with post-LGM colonization of Tasmania or gene flow with the mainland [15,16]; divergence dates for populations of the third species may be explained by recent colonization, but are also compatible with vicariance induced by post-LGM marine transgression [68]. Examples are also emerging in support for the other possible biogeographic scenarios highlighted in figure 1. Li et al. [14] suggest that divergence of Taiwanese and mainland hwamei (Leucodioptron spp., passerines) was initiated 3.47 Ma, and gene flow occurred between lineages up until 0.52 Ma, after which there was no further genetic exchange. In this instance, the maintenance of genetic isolation through subsequent low sea stands may owe to unfavourable intervening terrestrial habitat, as the lineages have been observed to interbreed following the artificial establishment of sympatric populations. Physical isolation of black-throated laughing thrush (Garrulax chinensis) on Hainan and adjacent mainland China has also been suggested to persist through low sea stands owing to unfavourable intervening habitats, and here the initial divergence occurred in the absence of gene flow [20]. However, in other systems, the most recent marine transgression does indeed provide a good explanation for the isolation of lineages on continental islands, such as among Skyros wall lizards (Podarcis gaigeae) on Greek islands, even though dispersal capabilities demonstrated for other lizards would appear to favour recent gene flow over these marine geographical scales [19].
Given the results of this study, and others that have applied similar tests when studying related lineages on continental islands and adjacent continents, it appears that the divergence of such lineages cannot be safely assumed to be the product of post-LGM marine transgression and testing of this and alternative hypotheses are required in each instance. It also cautions the assumption of divergence accompanying the post-LGM marine transgression for the calibration of molecular clocks [69].
(c). Colonization or recolonization?
As we have rejected the hypothesis of vicariance for the divergence of continental island and mainland lineages, an obvious question arises whether A. audax could have been present in Tasmania previously, and the observed divergence from the mainland represents a recolonization. A close phylogenetic relationship is observed between A. audax and Aquila gurneyi from New Guinea and adjacent islands [70,71], and if this represents a proxy for the existence of A. audax in Australia, then the amount of cytochrome b sequence divergence between these lineages (average p distance 3.7%; [70]) rejects the possibility that A. audax is too young to have colonized Tasmania before the LGM. Whereas changes in climate across the LGM have been implicated for extirpation and recolonization of mid-latitude populations in the Northern Hemisphere [72], they appear to have been less influential on species distributions at similar latitudes in the Southern Hemisphere [73,74]. Other animals presently distributed in Tasmania and mainland Australia, including prey species of A. audax, exhibit genetic divergences compatible with occupation of Tasmania during glacial periods [24,75–77]. Therefore, the hypothesis that we have reconstructed the first arrival in Tasmania of A. audax can only be rejected by fossil or subfossil evidence for an earlier occupation, of which we are unaware.
(d). Taxonomy and conservation
As with many biota on continental islands, the Tasmanian A. audax differs in morphology from that of the adjacent mainland and has been afforded subspecific status. The Tasmanian population also has endangered species status given extinction concerns owing to its small size (1000–1500 birds in 426 breeding territories), loss and disturbance of breeding habitat and high human-induced mortality [29,78]. However, when describing the subspecies Condon & Amadon [32] noted that they ‘found no conclusive evidence in favour of’ the size difference suggested by Fleay [33] and suggestions of coloration differences were employed for taxonomic distinction even though only three Tasmanian individuals (all female) were examined. Analysis of 159 and 116 specimens from Tasmania and the mainland, respectively, revealed significant size differences, with Tasmanian individuals indeed larger [34,35]. However, occasional mainland individuals are as large as those in Tasmania and rapid morphological divergence may also accompany founder events [19,79,80]. It has been recommended that the presence of substantial pre- and/or postzygotic isolation be used when defining taxa for conservation purposes [81], rather than the Taxonomic Species Concept as presently applied to A. a. audax and A. a. fleayi.
While we call into question the present taxonomic distinction of these Aquila lineages, the ecological significance of the Tasmanian population should be quantified before judgements are made regarding its conservation priority, particularly as the time and effort to re-establish such a population using mainland individuals would be onerous. Concomitantly, it is plausible that the Tasmanian population may be suffering from inbreeding depression, and therefore the introduction of mainland individuals to ameliorate any such effects is a potential management action [82]. Inbreeding depression has been previously implicated for the reduced fitness of island relative to mainland populations [83,84]. There has been significant attention given to fitness of the Tasmanian population given its elevated conservation concern, with observations suggesting lower fledgling success and greater sensitivity to anthropogenic disturbance than mainland populations [85]. The effective inbreeding coefficient for Tasmanian A. audax (Fe = 0.41) is approaching the point at which increases in F generated during laboratory experiments produced elevated extinction probabilities in benign environments [48], and inbreeding depression in a wild population is even more likely.
Acknowledgements
We thank Doug Black, James Harris, Nicki de Preu, Chris Thompson, Tony Zidarich, Kyong Nahm, Skye Wassens, Doreen Krumbiegel, Kimiko Naemi Krumbiegel, Gregory Wright, Robert Madden, Steve Murphy, Lisa Harris, Michelle Olsson, Bourne's Bird Museum, Western Australian Museum, Museum Victoria, South Australian Museum, Australian National Wildlife Collection (CSIRO Ecosystem Sciences) and the Department of Primary Industries, Parks, Water and the Environment (Tasmania) for providing samples. Peter Unmack provided the template for figure 2.
Data accessibility
DNA sequences are deposited in GenBank (KF683172-91; and mitochondrial DNA sequences, microsatellite genotypes and museum specimen details are available in Dryad, http://dx.doi.org/10.5061/dryad.tc88k.
Funding statement
J.J.A. is supported by an ARC Future Fellowship. This research was funded by the School of Earth and Environmental Sciences, University of Adelaide (to J.J.A., J.W. and L.O.), the School of Zoology, University of Tasmania (to C.P.B. and D.L.N.), and Roaring 40's (to C.P.B.) and NP Power (to J.J.A.).
References
- 1.Keogh JS, Scott IAW, Hayes C. 2005. Rapid and repeated origin of insular gigantism and dwarfism in Australian tiger snakes. Evolution 59, 226–233 (doi:10.1554/04-310) [PubMed] [Google Scholar]
- 2.Clegg SM, Frentiu FD, Kikkawa J, Tavecchia G, Owens IPF. 2008. 4000 years of phenotypic change in an island bird: heterogeneity of selection over three microevolutionary timescales. Evolution 62, 2393–2410 (doi:10.1111/j.1558-5646.2008.00437.x) [DOI] [PubMed] [Google Scholar]
- 3.Myers S, Brown G, Kleindorfer S. 2010. Divergence in New Holland Honeyeaters (Phylidonyris novaehollandiae): evidence from morphology and feeding behavior. J. Ornithol. 151, 287–296 (doi:10.1007/s10336-009-0454-7) [Google Scholar]
- 4.Castilla AM, Herrel A, Gosa A. 2008. Mainland versus island differences in behaviour of Podarcis lizards confronted with dangerous prey: the scorpion Buthus occitanus. J. Nat. Hist. 42, 2331–2342 (doi:10.1080/00222930802254763) [Google Scholar]
- 5.Schlotfeldt BE, Kleindorfer S. 2006. Adaptive divergence in the Superb Fairy-wren (Malurus cyaneus): a mainland versus island comparison of morphology and foraging behaviour. Emu 106, 309–319 (doi:10.1071/mu06004) [Google Scholar]
- 6.Pafilis P, Foufopoulos J, Poulakakis N, Lymberakis P, Valakos ED. 2009. Tail shedding in island lizards Lacertidae, Reptilia: decline of antipredator defenses in relaxed predation environments. Evolution 63, 1262–1278 (doi:10.1111/j.1558-5646.2009.00635.x) [DOI] [PubMed] [Google Scholar]
- 7.Johansson F, Lind MI, Ingvarsson PK, Bokma F. 2012. Evolution of the G-matrix in life history traits in the common frog during a recent colonisation of an island system. Evol. Ecol. 26, 863–878 (doi:10.1007/s10682-011-9542-2) [Google Scholar]
- 8.Whittaker RJ, Fernández-Palacios JM. 2007. Island biogeography. Ecology, evolution and conservation, 2nd edn Oxford, UK: Oxford University Press [Google Scholar]
- 9.Lambeck K, Esat TM, Potter E-K. 2002. Links between climate and sea levels for the past three million years. Nature 419, 199–206 (doi:10.1038/nature01089) [DOI] [PubMed] [Google Scholar]
- 10.Lambeck K, Chappell J. 2001. Sea level change through the last glacial cycle. Science 292, 679–686 (doi:10.1126/science.1059549) [DOI] [PubMed] [Google Scholar]
- 11.Rohling EJ, Fenton M, Jorissen FJ, Bertrand P, Ganssen G, Caulet JP. 1998. Magnitudes of sea-level lowstands of the past 500,000 years. Nature 394, 162–165 (doi:10.1038/28134) [Google Scholar]
- 12.Velo-Anton G, Zamudio KR, Cordero-Rivera A. 2012. Genetic drift and rapid evolution of viviparity in insular fire salamanders (Salamandra salamandra). Heredity 108, 410–418 (doi:10.1038/hdy.2011.91) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nosil P. 2008. Speciation with gene flow could be common. Mol. Ecol. 17, 2103–2106 (doi:10.1111/j.1365-294X.2008.03715.x) [DOI] [PubMed] [Google Scholar]
- 14.Li J-W, et al. 2010. Rejecting strictly allopatric speciation on a continental island: prolonged postdivergence gene flow between Taiwan (Leucodioptron taewanus, Passeriformes Timaliidae) and Chinese (L. canorum canorum) hwameis. Mol. Ecol. 19, 494–507 (doi:10.1111/j.1365-294X.2009.04494.x) [DOI] [PubMed] [Google Scholar]
- 15.Murphy SA, Joseph L, Burbidge AH, Austin J. 2011. A cryptic and critically endangered species revealed by mitochondrial DNA analyses: the Western Ground Parrot. Conserv. Genet. 12, 595–600 (doi:10.1007/s10592-010-0161-1) [Google Scholar]
- 16.Shephard JM, Hughes JM, Catterall CP, Olsen PD. 2005. Conservation status of the white-bellied sea-eagle Haliaeetus leucogaster in Australia determined using mtDNA control region sequence data. Conserv. Genet. 6, 413–429 (doi:10.1007/s10592-005-4987-x) [Google Scholar]
- 17.Hey J, Nielsen R. 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167, 747–760 (doi:10.1534/genetics.103.024182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hey J. 2005. On the number of New World founders: A population genetic portrait of the peopling of the Americas. PLoS Biol. 3, 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Runemark A, Hey J, Hansson B, Svensson EI. 2012. Vicariance divergence and gene flow among islet populations of an endemic lizard. Mol. Ecol. 21, 117–129 (doi:10.1111/j.1365-294X.2011.05377.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Huang J, Zhang M, Luo S, Zhang Y, Lei F, Sheldon FH, Zou F. 2012. Genetic divergence and population demography of the Hainan endemic black-throated Laughingthrush (Ayes: Timaliidae, Garrulax chinensis monachus) and adjacent mainland subspecies. Mol. Phylogenet. Evol. 65, 482–489 (doi:10.1016/j.ympev.2012.07.005) [DOI] [PubMed] [Google Scholar]
- 21.Lee Y-H, Lin C-P. 2012. Pleistocene speciation with and without gene flow in Euphaea damselflies of subtropical and tropical East Asian islands. Mol. Ecol. 21, 3739–3756 (doi:10.1111/j.1365-294X.2012.05654.x) [DOI] [PubMed] [Google Scholar]
- 22.Burridge CP. 2012. Divergence of island biotas when they were not always islands. Front. Biogeogr. 3.4, 125–126 [Google Scholar]
- 23.Harris P, Heap A, Passlow V, Sbaffi L, Fellows M, Porter-Smith R, Buchanan C, Daniell J. 2005. Geomorphic features of the continental margin of Australia. Canberra, Australia: Geoscience Australia [Google Scholar]
- 24.Gongora J, et al. 2012. Genetic structure and phylogeography of platypuses revealed by mitochondrial DNA. J. Zool. 286, 110–119 (doi:10.1111/j.1469-7998.2011.00854.x) [Google Scholar]
- 25.Firestone KB, Elphinstone MS, Sherwin WB, Houlden BA. 1999. Phylogeographical population structure of tiger quolls Dasyurus maculatus (Dasyuridae : Marsupialia), an endangered carnivorous marsupial. Mol. Ecol. 8, 1613–1625 (doi:10.1046/j.1365-294x.1999.00745.x) [DOI] [PubMed] [Google Scholar]
- 26.Morrow G, Nicol SC. 2009. Cool Sex? Hibernation and reproduction overlap in the Echidna. PLoS ONE 4, e6070 (doi:10.1371/journal.pone.0006070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morcombe M. 2000. Field guide to Australian birds. Archerfield, Australia: Steve Parish Publishing [Google Scholar]
- 28.Marchant S, Higgins PJ. 1993. Handbook of Australian, New Zealand and Antarctic Birds. Volume two—raptors to lapwings. Melbourne, Australia: Oxford University Press [Google Scholar]
- 29.Bell P, Mooney N. 1998. National recovery plan for the wedge-tailed eagle 1998–2003. Hobart, Australia: Department of Primary Industries, Water, and Environment [Google Scholar]
- 30.Ridpath MG, Brooker MG. 1986. Age, movements and the management of the wedge-tailed eagle, Aquila audax, in arid Western Australia. Aust. Wildl. Res. 13, 245–260 (doi:10.1071/WR9860245) [Google Scholar]
- 31.Green RH. 1993. Birds of Tasmania. Launceston, Australia: Potoroo Publishing [Google Scholar]
- 32.Condon HT, Amadon D. 1954. Taxonomic notes on Australian Hawks. Rec. S. Aust. Mus. 11, 189–246 [Google Scholar]
- 33.Fleay D. 1952. With a wedge-tailed eagle at the nest. Emu 52, 1–16 (doi:10.1071/MU952001) [Google Scholar]
- 34.Nankervis DL. 2010. Spatial patterns of genetic and morphological variation in the wedge-tailed eagle (Aquila audax). Hobart, Australia: University of Tasmania [Google Scholar]
- 35.Wadley J. 2009. Phylogeography and conservation genetics of Wedge-tailed Eagles (Aquila audax). Adelaide, Australia: University of Adelaide [Google Scholar]
- 36.Olsen P, Marples TG. 1993. Geographic-variation in egg size, clutch size and date of laying of Australian raptors (Falconiformes and Strigiformes). Emu 93, 167–179 (doi:10.1071/MU9930167) [Google Scholar]
- 37.Bekessy SA, et al. 2009. Modelling human impacts on the Tasmanian wedge-tailed eagle (Aquila audax fleayi). Biol. Conserv. 142, 2438–2448 (doi:10.1016/j.biocon.2009.05.010) [Google Scholar]
- 38.Gaffney RF, Mooney NJ. 1992. The wedge-tailed eagle recovery plan: management phase. Hobart, Australia: Department of Parks, Wildlife and Heritage [Google Scholar]
- 39.Excoffier L, Laval G, Schneider S. 2005. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 [PMC free article] [PubMed] [Google Scholar]
- 40.Raymond M, Rousset F. 1995. Genepop (version-1.2) - population-genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249 [Google Scholar]
- 41.Louis EJ, Dempster ER. 1987. An exact test for Hardy–Weinberg and multiple alleles. Biometrics 43, 805–811 (doi:10.2307/2531534) [PubMed] [Google Scholar]
- 42.Guo SW, Thompson EA. 1992. Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 48, 361–372 (doi:10.2307/2532296) [PubMed] [Google Scholar]
- 43.Rice TK, Schork NJ, Rao DC, Rao DC, Gu CC. 2008. Methods for handling multiple testing. In Advances in genetics (eds Rao DC, Gu CC.), pp. 293–308 New York, NY: Academic Press; [DOI] [PubMed] [Google Scholar]
- 44.Meirmans PG, Van Tienderen PH. 2004. genotype and genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 4, 792–794 (doi:10.1111/j.1471-8286.2004.00770.x) [Google Scholar]
- 45.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620 (doi:10.1111/j.1365-294X.2005.02553.x) [DOI] [PubMed] [Google Scholar]
- 47.Earl D, vonHoldt B. 2012. Structure Harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361 (doi:10.1007/s12686-011-9548-7) [Google Scholar]
- 48.Frankham R. 1998. Inbreeding and extinction: island populations. Conserv. Biol. 12, 665–674 (doi:10.1046/j.1523-1739.1998.96456.x) [Google Scholar]
- 49.Hey J. 2010. Isolation with migration models for more than two populations. Mol. Biol. Evol. 27, 905–920 (doi:10.1093/molbev/msp296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho SYW, Phillips MJ, Cooper A, Drummond AJ. 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22, 1561–1568 (doi:10.1093/molbev/msi145) [DOI] [PubMed] [Google Scholar]
- 51.Ho SYW, Shapiro B, Phillips MJ, Cooper A, Drummond AJ. 2007. Evidence for time dependency of molecular rate estimates. Syst. Biol. 56, 515–522 (doi:10.1088/10635150701435401) [DOI] [PubMed] [Google Scholar]
- 52.Weir JT, Schluter D. 2008. Calibrating the avian molecular clock. Mol. Ecol. 17, 2321–2328 (doi:10.1111/j.1365-294X.2008.03742.x) [DOI] [PubMed] [Google Scholar]
- 53.Burridge CP, Craw D, Fletcher D, Waters JM. 2008. Geological dates and molecular rates: fish DNA sheds light on time dependency. Mol. Biol. Evol. 25, 624–633 (doi:10.1093/molbev/msm271) [DOI] [PubMed] [Google Scholar]
- 54.Ohta T, Kimura M. 1973. Model of mutation appropriate to estimate number of electrophoretically detectable alleles in a finite population. Genet. Res. 22, 201–204 (doi:10.1017/S0016672300012994) [DOI] [PubMed] [Google Scholar]
- 55.Wright NA, Steadman DW. 2012. Insular avian adaptations on two Neotropical continental islands. J. Biogeogr. 39, 1891–1899 (doi:10.1111/j.1365-2699.2012.02754.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis WE. 2009. Early Tasmanian ornithology: the correspondence of Ronald Campbell Gunn and James Grant, 1836–1838. In Memoirs of the Nuttall Ornithological Club (ed. Boles WE.), pp. 263 Cambridge, MA: Nuttall Ornithological Club [Google Scholar]
- 57.Brothers N. 1985. Unusual observations of raptors in Bass Strait. Australas. Raptor Assoc. News 6, 15 [Google Scholar]
- 58.Brothers N, Pemberton D, Pryor H, Halley V. 2001. Tasmania's offshore islands: seabirds and other natural features, 1st edn, 643 p; Hobart, Australia: Tasmanian Museum and Art Gallery [Google Scholar]
- 59.Carter M. 2012. Nankeen Kestrels colonising remote oceanic islands. Boobook 30, 46 [Google Scholar]
- 60.Meyburg B-U, Graszynski K, Langgemach T, Sömmer P, Bergmanis U. 2008. Cainism, nestling management in Germany in 2004–2007 and satellite tracking of juveniles in the Lesser Spotted Eagle (Aquila pomarina). Slovak Raptor J. 2, 53–72 [Google Scholar]
- 61.Worthy TH, Holdaway RN. 2002. Lost World of the Moa: prehistoric life of New Zealand. Bloomington, IN: Indiana University Press [Google Scholar]
- 62.Holdaway RN, Worthy TH, Tennyson AJD. 2001. A working list of breeding bird species of the New Zealand region at first human contact. N. Z. J. Zool. 28, 119–187 (doi:10.1080/03014223.2001.9518262) [Google Scholar]
- 63.Bunce M, Szulkin M, Lerner HRL, Barnes I, Shapiro B, Cooper A, Holdaway RN. 2005. Ancient DNA provides new insights into the evolutionary history of New Zealand's extinct giant eagle. PLoS Biol. 3, e9 (doi:10.1371/journal.pbio.0030009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hull JM, Savage WK, Bollmer JL, Kimball RT, Parker PG, Whiteman NK, Ernest HB. 2008. On the origin of the Galapagos hawk: an examination of phenotypic differentiation and mitochondrial paraphyly. Biol. J. Linnean Soc. 95, 779–789 (doi:10.1111/j.1095-8312.2008.01082.x) [Google Scholar]
- 65.Fuchs J, et al. 2008. Tracing the colonization history of the Indian Ocean scops-owls (Strigiformes: Otus) with further insight into the spatio-temporal origin of the Malagasy avifauna. BMC Evol. Biol. 8, 197 (doi:10.1186/1471-2148-8-197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hogan FE, Campbell C, Harrison KA, Milledge D, Cooke R. 2013. Molecular data contradicts historical records and cautions translocation of the Lord Howe Island masked owl. Biol. Conserv. 159, 313–320 (doi:10.1016/j.biocon.2012.12.006) [Google Scholar]
- 67.Plomley NJB. 1976. A word-list of the Tasmanian Aboriginal languages. Launceston, Australia: Published by the author in association with the Government of Tasmania [Google Scholar]
- 68.Toon A, Mather PB, Baker AM, Durrant KL, Hughes JM. 2007. Pleistocene refugia in an arid landscape: Analysis of a widely distributed Australian passerine. Mol. Ecol. 16, 2525–2541 (doi:10.1111/j.1365-294X.2007.03289.x) [DOI] [PubMed] [Google Scholar]
- 69.Beerli P, Hotz H, Uzzell T. 1996. Geologically dated sea barriers calibrate a protein clock for Aegean water frogs. Evolution 50, 1676–1687 (doi:10.2307/2410903) [DOI] [PubMed] [Google Scholar]
- 70.Haring E, Kvaløy K, Gjershaug JO, Røv N, Gamauf A. 2007. Convergent evolution and paraphyly of the hawk-eagles of the genus Spizaetus (Aves, Accipitridae) – phylogenetic analyses based on mitochondrial markers. J. Zool. Syst. Evol. Res. 45, 353–365 (doi:10.1111/j.1439-0469.2007.00410.x) [Google Scholar]
- 71.Lerner HRL, Mindell DP. 2005. Phylogeny of eagles, Old World vultures, and other Accipitridae based on nuclear and mitochondrial DNA. Mol. Phylogenet. Evol. 37, 327–346 (doi:10.1016/j.ympev.2005.04.010) [DOI] [PubMed] [Google Scholar]
- 72.Hewitt GM. 2004. Genetic consequences of climatic oscillations in the quaternary. Phil. Trans. R. Soc. Lond. B 359, 183–195 (doi:10.1098/rstb.2003.1388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wallis GP, Trewick SA. 2009. New Zealand phylogeography: evolution on a small continent. Mol. Ecol. 18, 3548–3580 (doi:10.1111/j.1365-294X.2009.04294.x) [DOI] [PubMed] [Google Scholar]
- 74.Sérsic AN, Cosacov A, Cocucci AA, Johnson LA, Pozner R, Avila LJ, Sites JW, Jr, Morando M. 2011. Emerging phylogeographical patterns of plants and terrestrial vertebrates from Patagonia. Biol. J. Linnean Soc. 103, 475–494 (doi:10.1111/j.1095-8312.2011.01656.x) [Google Scholar]
- 75.Frankham GJ, Handasyde KA, Eldridge MDB. 2012. Novel insights into the phylogenetic relationships of the endangered marsupial genus Potorous. Mol. Phylogenet. Evol. 64, 592–602 (doi:10.1016/j.ympev.2012.05.013) [DOI] [PubMed] [Google Scholar]
- 76.Dubey S, Shine R. 2010. Evolutionary diversification of the lizard genus Bassiana (Scincidae) across Southern Australia. PLoS ONE 5, e12982 (doi:10.1371/journal.pone.0012982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Symula R, Keogh JS, Cannatella DC. 2008. Ancient phylogeographic divergence in southeastern Australia among populations of the widespread common froglet, Crinia signifera. Mol. Phylogenet. Evol. 47, 569–580 (doi:10.1016/j.ympev.2008.01.011) [DOI] [PubMed] [Google Scholar]
- 78.Threatened Species Section 2006. Threatened Tasmanian eagles recovery plan: 2006–2010. Hobart, Australia: Department of Primary Industries & Water, Tasmanian Government [Google Scholar]
- 79.Grant PR, Grant BR, Petren K. 2001. A population founded by a single pair of individuals: establishment, expansion, and evolution. Genetica 112, 359–382 (doi:10.1023/a:1013363032724) [PubMed] [Google Scholar]
- 80.Louette M, Herremans M, Nagy ZT, de Roland LAR, Jordaens K, Van Houdt J, Sonet G, Breman FC. 2011. Frances's sparrowhawk Accipiter francesiae (Aves: Accipitridae) radiation on the Comoro Islands. Bonn. Zool. Monogr. 57, 133–143 [Google Scholar]
- 81.Frankham R, et al. 2012. Implications of different species concepts for conserving biodiversity. Biol. Conserv. 153, 25–31 (doi:10.1016/j.biocon.2012.04.034) [Google Scholar]
- 82.Frankham R, Ballou JD, Eldridge MDB, Lacy RC, Ralls K, Dudash MR, Fenster CB. 2011. Predicting the probability of outbreeding depression. Conserv. Biol. 25, 465–475 (doi:10.1111/j.1523-1739.2011.01662.x) [DOI] [PubMed] [Google Scholar]
- 83.Eldridge MDB, King JM, Loupis AK, Spencer PBS, Taylor AC, Pope LC, Hall GP. 1999. Unprecedented low levels of genetic variation and inbreeding depression in an island population of the black-footed rock-wallaby. Conserv. Biol. 13, 531–541 (doi:10.1046/j.1523-1739.1999.98115.x) [Google Scholar]
- 84.Miller EJ, Eldridge MDB, Morris KD, Zenger KR, Herbert CA. 2011. Genetic consequences of isolation: island tammar wallaby (Macropus eugenii) populations and the conservation of threatened species. Conserv. Genet. 12, 1619–1631 (doi:10.1007/s10592-011-0265-2) [Google Scholar]
- 85.Mooney NM, Holdsworth M. 1991. The effects of disturbance on nesting wedge-tailed eagles (Aquila audax fleayi) in Tasmania. Tasforests 3, 15–31 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DNA sequences are deposited in GenBank (KF683172-91; and mitochondrial DNA sequences, microsatellite genotypes and museum specimen details are available in Dryad, http://dx.doi.org/10.5061/dryad.tc88k.