Abstract
In the infected cell, HIV-1 protease (PR) is initially synthesized as part of the GagPol polyprotein. PR autoprocessing is a virus-specific process by which the PR domain embedded in the precursor catalyzes proteolytic reactions responsible for liberation of free mature PRs, which then recognize and cleave at least ten different peptide sequences in the Gag and GagPol polyproteins. Despite extensive structure and function studies of the mature PRs as well as the successful development of ten US FDA-approved catalytic-site inhibitors, the precursor autoprocessing mechanism remains an intriguing yet-to-be-solved puzzle. This article discusses current understanding of the autoprocessing mechanism, in an effort to prompt the development of novel anti-HIV drugs that selectively target precursor autoprocessing.
HIV-1 is the causative pathogen of AIDS. The HIV-1 protease (PR) is a virus-encoded proteolytic enzyme that is initially synthesized as part of the GagPol polyprotein. PR autoprocessing is a regulated process by which the precursor PR catalyzes the cleavage reactions leading to liberation of the free mature PR upon or shortly after progeny virion is released from the infected cell. HIV-1 PR is an aspartic protease with the catalytic site mapped to residue D25; alterations of D25 to A, Y, H or N abolish its enzymatic activity [1–4]. In the human genome, aspartic proteases are the smallest class with only 15 members found in two clans, clan AA and clan AD [5]. Clan AA has A1 and A2 families. The A1 family contains classical aspartyl proteases, such as pepsin A/C, cathepsin D/E, BACE1/2. The HIV-1 PR is a member of the A2 family. Clan AD contains proteases, such as the presenilins and signal peptide peptidase that cleave transmembrane peptides within the lipid bilayer [5].
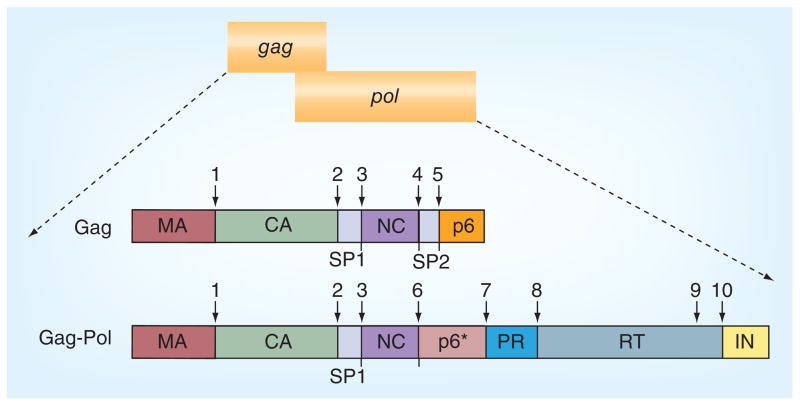
In the HIV infected cell, the unspliced genomic RNA also serves as mRNA directing synthesis of the Gag and GagPol polyproteins. Both Gag and GagPol polyproteins have the same N-termini [6,7]; approximately 5% of translation undergoes a −1 ribosomal frameshift, resulting in production of the GagPol precursor [8–10]. Within the GagPol polyprotein, the HIV PR is flanked by a transframe region, namely TFR or p6*, at the N-terminus and by the reverse transcriptase at the C-terminus (Figure 1) [2,11]. At least two proteolytic reactions are required to release the mature PR, one at the N-terminal and other at the C-terminal of the PR (sites 7 and 8, respectively, in Figure 1). These reactions are catalyzed by the GagPol polyprotein itself – a process referred to as PR precursor autoprocessing – in which the GagPol precursor serves as both the enzyme and substrate at the same time.
Figure 1. HIV-1 proviral genome and the protease cleavage sites in the Gag and GagPol.
CA: Capsid; MA: Matrix; NC: Nucleocapsid; SP: Spacer peptide.
The released mature PR recognizes and cleaves at least ten sites in the Gag and GagPol polyproteins (Figure 1 & Table 1). The substrate residues are usually numbered P1, P2, P3 and P1′, P2′, P3′, beginning from each side of the scissile bond [12]. The HIV-1 PR accepts Y, F, L, M and N in P1 site and has a slight preference for P over A, M, F, L and Y in P1′ position (Table 1) [13–16]. Many cleavage sites are highly conserved among HIV-1 viruses except for some polymorphisms that emerge in drug-resistant strains at the p2-nucleocapsid (p2-NC) and p1-p6 sites [17–20]. However, there is no single consensus sequence that can be extrapolated, suggesting that HIV-1 PR can process a wide variety of substrates. Accurate and precise PR processing of these sites is absolutely required for the production of infectious progeny virions [21–27]. Because of its critical role in viral replication, HIV-1 PR has been a major target for anti- AIDS drug development. In fact, unprecedented efforts from academic and industrial laboratories have made the mature HIV-1 PR one of the best characterized enzymes as documented by a series of excellent reviews published over last 20 years [2,5,13,28–33]. As a result, multiple US FDA-approved HIV-1 PR inhibitors have been developed to treat HIV-1-positive patients [34,35].
Table 1.
Common HIV-1 protease cleavage sites.
| Site | Location | Substrate peptide sequence (P4-P1 ↓ P1′-P4′) |
|---|---|---|
| 1 | MA-CA | AQNY ↓ PIVQ |
| 2 | CA-sp1 | ARVL ↓ AEAM |
| 3 | sp1-NC | ATIM ↓ MQRG† |
| 4 | NC-sp2 | RQAN ↓ FLGK |
| 5 | sp2-p6 | PGNF ↓ LQSR† |
| 6 | NC-p6* | RQAN ↓ FLRE† |
| 7 | p6*-PR | SFNF ↓ PQIT |
| 8 | PR-RT | TLNF ↓ PISP |
| 9 | Within RT | AETP ↓ YVDG |
| 10 | PR-IN | RKIL ↓ FLDG |
The sequences are from the HXB2 strain; these sites display sequence polymorphism among different strains.
CA: Capsid; MA: Matrix; NC: Nucleocapsid.
In contrast to extensive investigation of HIV-1 mature PR structure and function, the molecular and cellular mechanisms of precursor autoprocessing are largely undefined and many fundamental questions remain to be answered. For example, autoprocessing of the newly synthesized GagPol precursor needs to be suppressed until Gag assembly and virion release are nearly completed. Whether and how precursor PR is suppressed and later activated are completely unknown. Also, it remains unclear whether the initial cleavage is intra- or inter-molecular. This article will first briefly describe the mature HIV-1 PR structure, activity and its inhibitors to set a stage for comparison with the precursor PR. A focus of this article is to give an overview of the methods that have been developed for PR autoprocessing characterization and the current concept of PR autoprocessing mechanism. Additionally, this article proposes an off the beaten path theory that attempts to reconcile the exiting data. This will hopefully provide a platform to inspire pioneering research with novel approaches to further advance our understanding of autoprocessing mechanisms, which will lay the groundwork for the development of novel autoprocessing inhibitors.
Mature HIV-1 PR structure, activity & inhibitors
Many aspartic proteases share a general fold with three distinct domains. For single-chained proteases, such as pepsins, the N- and C-terminal domains, each contributing one catalytic aspartic acid residue, are connected by an antiparallel β-sheet. The active site is located at the bottom of a cleft formed by the two domains and is covered by two β-hairpin structures or ‘flaps’ that are highly flexible to allow binding and release of the substrates. Two aspartic amino acids working cooperatively at the optimal pH in the presence of a water molecule essential for amide bond hydrolysis [36]. The β-sheet domain plays a critical role in stabilizing the overall structure and comprises β-strands from discontinuous regions of the polypeptide [37]. In the case of mature cathepsin D, which is normally found in the lysosomes of higher eukaryotes, a two-chain form is predominant due to a post-translational autocleavage between residues 98 and 106 resulting in the noncovalent association of light and heavy chains [38]. HIV-1 mature PR is a distinguished aspartic PR by two structural features. First, mature PR is a homodimer of 99-amino acid subunits [2,11,39] and is the smallest aspartic PR, in terms of size, that has been identified to date. Second, the ‘interdomain’ β-sheet is drastically shorter with fewer β-strands. This region is highly conserved in amino-acid composition and contributes approximately 75% of the dimer stabilization energy [40]. There are over 500 mature HIV-1 PR structures that have been reported in the literature with most of them being complexes of PR with different inhibitors [41–49]. The mature PR dimer is extremely stable with a dissociation constant Kd of <5 nM [50], suggesting that once the mature PR dimer is formed, reversible dissociation to the individual monomer is less probable.
A fluorometric assay has been routinely used to quantify the mature PR activity in vitro using purified recombinant PR and synthetic substrate peptides derived from various cleavage sites found in Gag and/or GagPol polyproteins. For example, a hexapeptide substrate derived from the capsid (CA)-sp1 cleavage site (site 2 in Figure 1), Ac-Thr-Ile-Nle-Nle-Gln-Arg-NH2, incorporation of 2-aminobenzoic acid, in place of the acetyl group as the donor, and p-NO2-Phe at the P1′ position, as the acceptor, intramolecularly quenches fluorogenic substrate. Peptide cleavage by mature HIV PR releases the fluorescent N-terminal tripeptide from its quenching nitro-benzyl group, resulting in an increase in fluorescence. Subsequently, fluorescence intensity can be measured to reflect enzymatic activity [51]. Diverse commercial kits use synthetic oligopeptides derived from different cleavage sites. Purified recombinant HIV-1 PR revealed several 100-fold variations in catalytic kinetics among different peptide sequences [52]. A similar scale of variation was also observed with HIV GagPol as substrate in an in vitro assay [15,53,54]. Out of the five HIV-1 Gag-processing sites, the sp1-NC site is processed at the very early stage and cleavage of the CA-sp1 site appears much slower in vitro [15]. Interestingly, it was recently demonstrated that not only the substrate sequence but also the flanking domains can differentially modulate mature PR kinetics [55,56] suggesting contribution of complex substrate interactions beyond the active site to PR activity. Accessibility of these substrates might be another mechanism to regulate processing of each site [18,20]. These varied processing rates might be critical to regulate the order of different sites that are cleaved by HIV-1 PR. It remains to be illustrated whether these sites are cleaved in a defined sequence during production of infectious virions.
There are currently nine FDA-approved PR inhibitors (amprenavir was discontinued and replaced by its prodrug fosamprenavir in 2004) that are used by themselves or in combination with other antiretroviral agents (reverse transcriptase inhibitors, fusion inhibitors and integrase inhibitors and so forth) to treat HIV-1 positive patients (Figure 2). These PR inhibitors are noncleavable substrate analogs targeting the catalytic site of the mature PR with high binding affinities. Unfortunately, this single mode of inhibition is insufficient at completely blocking HIV replication as various drug-resistant HIV strains often emerge in patients treated with these PR inhibitors. These variants usually carry a single or a combination of point mutations in PR [34], and the resulting mutant PRs become insensitive to the inhibitors but are still able to support viral replication even though some PRs may be appreciably compromised by 10–20-fold in kcat/Km as measured in vitro [57]. Such structural plasticity seems intrinsic to HIV-1 PR as it is made to recognize and process a variety of substrates with diverse amino acid sequences [41]. Substrate sequence co-evolution also contributes to the development of drug-resistant strains [19]. The constant emergence of drug-resistant HIV strains in patients undergoing these therapies and the long-term side effects associated with many of these drugs underscore the urgent need for the development of new PR inhibitors with novel action mechanisms. In this regard, PR precursor autoprocessing could be a promising target as it is responsible for liberation of the mature PR and is a virus specific process that has not yet been extensively exploited for anti-HIV drug discovery.
Figure 2. Ten US FDA-approved HIV-1 protease inhibitors with the year of initial FDA approval in parentheses.
Fosamprenavir is a prodrug of amprenavir; production of amprenavir was discontinued by the manufacturer in 2004. Tipranavir is a non-peptidomemetic inhibitor while the others are peptidomemtic inhibitors.
Definitions of precursor PR & precursor autoprocessing
The GagPol polyprotein is the full-length PR precursor that is initially synthesized in the infected cell. Additionally, various constructs containing the PR domain along with different flanking sequences are often used in autoprocessing studies. These constructs exhibit diverse proteolytic activities when expressed in vitro [3,58–61], in Escherichia coli [1,11,50,62,63] or in mammalian cells [22,63–66]. The only common component in these constructs is the PR coding sequence (PR) along with its upstream p6* sequence, which has defined the p6*-PR as a miniprecursor PR [11,42,50,63,67,68].
PR autoprocessing refers to a process by which the precursor catalyzes cleavage reactions leading to liberation of the free mature PR. Extensive investigation confirms that the PR domain is essential and sufficient for PR autoprocessing; alterations of the catalytic aspartic acid to other amino acids abolish precursor autoprocessing. Two cleavage reactions are necessary to set free the mature PR from its precursor (sites 7 and 8 in Figure 1). Mutations that block the C-terminal cleavage site lead to production of PR-RT fusion enzymes that are capable of making viable and infectious virions with approximately 20-fold reduction in infectivity [69], indicating that the C-terminal extension has little impact on PR activity. A transient intermediate consisting of the mature PR and its C-terminal native flanking sequence (19 residues of the RT) demonstrated proteolytic kinetics similar to the free mature PR, suggesting again that the C-terminal sequence does not significantly affect the full catalytic activity [70]. In addition, fusion of fluorescent proteins such as CFP and YFP to the C-terminus demonstrated no obvious impact on virus infectivity [71]. PR precursors with small epitopes fused to the C-terminus also effectively autoprocessed in transfected mammalian cells [63,65]. Collectively, these studies suggest that PR activity is not significantly affected by its downstream sequences. With that being said, it should be noted that the downstream RT might function to modulate precursor PR dimerization in the context of GagPol polyprotein [72]. On the other hand, blocking the N-terminal cleavage (at site 7) led to production of the p6*-PR miniprecursor that was unable to process most of the cleavage sites in the Gag polyprotein and the resulting virions were noninfectious [73,74]. Furthermore, the mature PR activity was only observed upon removal of the upstream p6* in an in vitro assay [50]. These studies have defined the cleavage reaction at site 7 as a critical step of precursor autoprocessing. In this article, a precursor PR is referred to as any construct that carries the PR sequence plus part or all of the upstream p6*; precursor autoprocessing is primarily referred to as the cleavage reaction at site 7.
It is intuitive to assume that the precursor PR and mature PR are enzymatically similar as the amino acid sequence in the PR region is identical. However, a body of evidence demonstrates that these two forms of PR are quite different in their catalytic properties. For example, the precursor PR is only capable of processing a limited number of cleavage sites, such as sites 3, 6, 8 and 10 (Figure 1), whereas the mature PR recognizes and processes all the cleavage sites [14,74]. The authors [64,66] and others [50,57,60] have demonstrated that the precursor PR is considerably less sensitive than the mature PR to inhibition by the catalytic site inhibitors. The contemporary concept assumes that both precursor and mature PRs share the same catalytic site but the N-terminal extension peptide (p6*) significantly destabilizes the dimeric precursor such that the precursor PR forms the catalytic site only transiently. This idea is mainly inferred from structure analyses of in vitro purified p6*-PR model precursors PR [50,75,76] and thermodynamic simulation of folding kinetics [77–79]. Particularly, a recombinant inactive precursor carrying four amino acids extended from the N-terminus and lacking the last four amino acids of the mature PR forms highly transient dimeric complexes (3–5% of population in solution) that resemble the mature protease configuration. In the atomic structure of a complex formed between the pseudo-wildtype PR with four residues extended from the N-terminus and darunavir (a potent PR inhibitor), the N-terminal end of one monomer is found to bends away from the stabilizing anti-parallel β-sheet, suggesting that precursor PR dimer is much less stable compared with the mature protease and that the N-terminal end might go into the catalytic site to be cleaved intra-molecularly. According to this model, one should expect little or no proteolytic activity from the precursor PR especially when it exists at low concentrations. However, the in vitro transcribed/translated GagPol precursor (estimated up to ~0.2 nM) effectively processes sites 3, 6 and 8. Another emerging hypothesis is that the precursor PR is different from the mature PR in their catalytic site conformations and that allosteric regulations are attributed to the different proteolysis activity between the precursor and mature PRs. This hypothesis also implies structural flexibility of the precursor PR in response to different conditions and conformational changes from the precursor to the mature PR. This allosteric regulation theory is able to reconcile most autoprocessing data obtained from different models but the direct evidence remains to be established.
Model systems for precursor autoprocessing study
Autoproteolysis of precursor PRs imposes a technical challenge to isolation and purification of the wild-type unprocessed precursors for autoprocessing characterization, which partially underlies the fact that the molecular mechanism of precursor autoprocessing is poorly understood. No detailed (wild-type) precursor PR structure (not complexed with protease inhibitors) is currently available. This section summarizes model systems that have been developed for autoprocessing study to date. It is worth noting that different systems utilize different kinds of precursor PRs, which might have complicated data interpretation and reconciliation.
In vitro purified PR miniprecursors
When expressed in E. coli, the mature PR is predominantly associated with the inclusion bodies, from which large quantities of PR can be purified under denaturing conditions. The purified recombinant mature PR displays proteolytic activity upon in vitro refolding. This provides an easy way to obtain purified mature PR at decent amounts. In fact, most mature PR structures are solved using samples that are prepared this way. The wild-type mature PR tends to undergo rapid self-degradation as there are several sites that are highly sensitive to PR autoproteolysis [80,81]. To circumvent these hurdles, several laboratories modified PR sequences and demonstrated that substitutions Q7K, L33I and L63I significantly improve the stability [80,81]. Also, C67A and C95A were generated to restrict cysteine thiol oxidation-mediated PR aggregation in vitro [11,50,68]. The resulting PR bearing these mutations displayed kinetic properties similar to the wild-type PR as measured by the standard in vitro fluorometric assay [80,81]. Consequently, a pseudo-wild-type PR bearing these five mutations has been utilized in many biochemical and structural analyses [11,42,50,63,67,68,82].
Most active precursor PRs undergo effective autoprocessing when expressed in E. coli, making it impractical to isolate and purify recombinant precursor PR. Interestingly, a miniprecursor consisting of the pseudo wild-type PR and an additional R8Q mutation led to approximately equal amount of precursor and mature PR in the inclusion bodies [50]. The mature R8Q PR displayed enzymatic kinetics similar to the pseudo wild-type PR. This has enabled in vitro characterization of precursor autoprocessing using the R8Q miniprecursor [11,50,57,68,75,76,83]. Upon refolding at low pH conditions, the purified R8Q precursor undergoes autoprocessing releasing the mature PR. Extensive enzyme kinetics and PR structure analyses have been carried out with this in vitro system and the results of these studies are nicely summarized in recent reviews [11,68]. One intrinsic limit of this approach is that the in vitro refolding procedure might induce the conformation that is most favorable and stable under the conditions and, hence, other functional conformations might be missed by this approach. Inhibitor titration experiment also suggested that not all of the proteins refold into the mature PR-like conformation [63]. Therefore, this model system is not applicable to examine and evaluate the allosteric regulation theory. Additionally, this system is anticipated to be less productive for the development of precursor specific inhibitors as the in vitro folding conditions are designed to favor the folding of structures that are similar to the mature protease.
In vitro transcribed & translated precursors
Another system utilizes in vitro transcription coupled translation of precursors in cell-free lysates such as wheat-germ lysates or rabbit reticulocyte lysates. Pulse-chase analysis of the [35S] methionine-labeled Gag-PR precursors reveal several self-cleaved products that could be detected by radiography based on their sizes and [35S] methionine content [3]. The full-length GagPol precursor also demonstrated similar proteolytic activity and yielded several processing products [14,60]. In this system, the sp1-NC (site 3 in Figure 1) site appears to be the first one that is processed followed by the cleavage that separates the NC from the p6* peptide (site 6). The latter site varies among different HIV-1 strains probably due to sequence polymorphism, that is, F/L in the HXB isolate [60] and N/F in the BH10 isolate [3]. Mutations that block these early sites lead to alternative cleavages at neighboring sites [60], suggesting that the PR precursor is able to process non-conventional substrates under certain conditions. Processing at these two sites was relatively rapid (stable after 15 min), but no additional cleavage of the other GagPol sites was observed after few hours of incubation. These in vitro transcribed/translated Gag or GagPol precursors can be processed effectively with purified mature PR added in trans [14,84–86], suggesting that the cleavage sites in these proteins are at least accessible to the mature PR. These analyses confirmed intrinsic catalytic activity of the precursor PR but liberation of the mature PR was not observed in this system.
It is worth noting that the estimated GagPol precursor concentrations with an optimized rabbit reticulocyte lysates system are only up to 0.2 nM [14,15], which is lower than the dissociation constant of the mature PR (i.e., ~5 nM). This raises a fundamental question of how precursor PR forms a mature PR-like dimer at such low concentration if dimerization is required for proteolytic activity. Additionally, the fact that no detectable mature PR is released argues that this system lacks essential elements for precursor autoprocessing although precursor PR activity is readily manifested. This system alone is also too complicated to be used for functional screening of precursor PR inhibitors in a high-throughput format.
Proviral constructs
Mammalian cells transfected with proviral constructs have also been used to study precursor autoprocessing. The transfected cell mimics the infected cell by expressing all the viral encoded components that mediate assembly and release of virion particles into the culture medium. This allows for an examination of both cell and virion associated proteins by standard western blotting. Another advantage of the provirus system is that it allows for the determination of how mutations affect virus infectivity and fitness, the biologically relevant parameters that cannot be evaluated with other model systems. However, direct detection of the autoprocessing intermediates and the final product (mature PR) in transfected cells and virions has been less successful. This is partially due to a lack of highly sensitive antibodies specific to the PR and that many PR-containing fragments only exist transiently; for example, the mature PR undergoes rapid self-degradation. Instead, quantification of Gag processing has served as an indirect measurement to reflect autoprocessing efficiency [65]. In particular, the amount of CA relative to the other CA-containing proteins is normally used to determine autoprocessing efficiency as the cleavage at the CA-sp1 site absolutely requires the autoprocessing product, the mature PR; the precursor PR is unable to process this site. One limit of this system is that some Gag cleavage sites can be processed by either the precursor or mature PR and, hence, data interpretation is complicated by not knowing the exact contributing players in the system.
Another provirus-based assay was recently reported to study precursor dimerization [71,87,88]. In brief, a CFP or YFP is inserted between PR and RT with a polyalaline linker (non-cleavable) connecting PR and CFP (or YFP). The C-terminus of CFP (or YFP) is connected to RT through phenylalanine and proline (cleavable) so that the PR–CFP fusion can be cleaved from RT. Cos-7 cells co-transfected with the CFP/YFP proviral plasmid pairs were able to produce viral particles that contained CFP (or YFP)-tags PR and were capable of infecting CD4+ MT-4 cells, confirming that the C-terminal sequences do not significantly impact PR activity. The rationale of this design is that PR dimerization brings CFP and YFP into a close proximity and, thus, mediates fluorescence (Förster) resonance energy transfer (FRET) in the co-transfected cell. To quantify FRET efficiency, fluorescence intensity of the donor (CFP) after and before photobleaching is individually measured and used to calculate the A/B ratio (i.e., after intensity/before intensity). The average CFPA/B ratio of the wild-type control is 1.24 ± 0.11. Single amino acid substitutions that are expected to disrupt mature PR dimerization also demonstrated reduced CFPA/B ratios in the transfected cell and the resulting viruses are replication deficient. Therefore, this assay provides another useful measurement of PR dimerization in the context of transfected cells. However, one technical limitation of this assay is wide distribution of CFPA/B ratios due to variations in signal intensity and transfection efficiency among different cells. In addition, according to the current concept, the majority of precursor PR should be inactive (i.e., not dimerized yet) in the transfected cell as precursor dimerization is mostly suppressed until or shortly after virion release. Subsequently, positive FRET resulted from precursor dimerization is expected to come from a very small percentage of the whole precursor population. Data interpretation of this assay is further complicated by the fact that the mature PR forms stable dimers as a result of effective precursor autoprocessing. Collectively, it is practically challenging to distinguish precursor dimerization-mediated FRET from false-positive signals (from mature PR dimers) and background noises. Therefore, this assay needs further development before it can be widely used for precursor dimerization and autoprocessing studies. For the purpose of drug development, both provirus-based models are very low-throughput considering the complicated protocols that are involved. Subsequently, they are more suitable for hit compound validation as secondary or tertiary assays, but neither is an appropriate primary assay for high-throughput screening of precursor inhibitors.
Fusion precursors expressed in cells
Many constructs containing PR precursor sequences fused to non-viral components are used for autoprocessing study as well. GST is an example of the fusion tags. GST is a well-characterized homodimer with the dissociation constants for dimeric GST ranging from the low micromolar to the low nanomolar, depending on assay conditions [89,90]. Owing to the tight dimer association and slow exchange kinetics [91], the GST moiety is expected to facilitate dimerization of the fusion precursor and subsequent autoprocessing. Additionally, small peptide epitopes, such as HA, Myc and Flag, can be engineered into different locations of the fusion precursor to facilitate direct detection of the autoprocessing intermediates and mature PR. A typical fusion precursor contains the miniprecursor (p6*-PR) sandwiched between GST and HA tags. Expression of GST fused precursor PR leads to effective autoprocessing and production of mature HIV-1 PR in cells (E. coli or mammalians) [63–65,92]. Fusion precursors carrying other N-terminal tags, including GFP, hsp70 and GCN4, are also autoprocessing competent when expressed in the transfected HEK293T cells [66]. This has provided an easy assay to specifically study the autoprocessing reactions inside cells, which provides a more biologically relevant environment than in a test tube at low pH conditions. This system allows simultaneous examination of drug effects on the mature PR and precursor PR, thus providing side-by-side comparison of the two under the same conditions. Importantly, this assay also enables cell-based functional screens of cell-permeable compounds that impede precursor autoprocessing. Such screens might identify hit compounds that suppress precursor autoprocessing via various action modes. Further characterization of these compounds will not only shed light on autoprocessing mechanism but also help to identify novel autoprocessing inhibitors that target regions outside the active site of the mature PR.
MBP is another fusion tag that is widely used to increase solubility of target protein. Unlike GST that exists as stable dimers, MBP is generally considered as a monomeric protein at concentrations up to 1 mM. In the first reported MBP fusion precursor, a polypeptide containing the PR domain plus 12 and 19 amino acids extended from its N- and C-terminus, respectively, is fused to the C-terminus of MBP [82,93]. Interestingly, when expressed in E. coli, the resulting fusion is soluble but autoprocessing deficient [82,93]. The purified MBP fusion precursor releases the mature PR after going through an in vitro denature-and-refold cycle at 0.1–50 μM concentrations. One could argue that MBP fusion precursor might exist as a monomer thus suppressing the dimerization essential for the formation of the active site. However, size-exclusion column chromatography analysis indicated that the fusion precursor forms oligomeric complexes with an apparent molecular mass of 1.1 × 106 Da. Even more intriguingly, the authors recently observed that several MBP fusion precursors are effectively autoprocessed in the expressing mammalian cells [Huang L et al., Unpublished Data], suggesting a host-dependent (E. coli vs mammalian cells) regulation of precursor autoprocessing. This indirectly supports the allosteric regulation theory in that the precursor PR activity can be regulated by a tag (MBP) in different host cells. Therefore, additional characterizations would be necessary to provide insights into the underlying mechanism.
Another advantage of this model system is that it can be used to identify small-molecule inhibitors that selectively suppress precursor autoprocessing via functional screening. With this assay, precursor autoprocessing efficiency inversely correlates with the amount of the full-length fusion precursor, which can be quantified by amplified luminescent proximity homogeneous assay ELISA (AlphaLISA) technology [94–97] using crude cell lysates. Our preliminary results demonstrated highly sensitive detection (up to 30 pM) [Chen C et al., Unpublished Data], suggesting the exciting potential of this assay for its application to drug discovery. This is the first high-throughput compatible assay that enables identification of autoprocessing-specific inhibitors while bypassing the lack of knowledge about the underlying enzymatic mechanisms. Further characterization of the hit compounds will not only provide powerful probes for the study of autoprocessing mechanism but will also lead to the development of a new class of drugs that complement the currently available protease inhibitors by targeting regions/areas beyond the catalytic site of the mature protease. These new drugs will offer alternative treatments to combat HIV-1 strains that are resistant to the current protease inhibitors.
Mechanism & regulation of precursor autoprocessing
It has been a long-standing mystery how autoprocessing is initiated and regulated in the infected cell. A fascinating feature of PR autoprocessing is that the autoproteolysis does not occur immediately after the synthesis of the GagPol precursor; rather, it is temporally coordinated with the late virion-release event. The underlying suppression and activation mechanisms are currently largely unknown. According to the mature PR structure, it is widely accepted that precursor PR dimerization is required for precursor autoprocessing, although definitive evidence demonstrating that the precursor dimers form an active site similar to that observed with the mature PR is absent. Therefore, it remains debatable how precursor PR dimerizes whereas the upstream p6* peptide functions to destabilize precursor dimerization. In addition, it is worth noting that the current autoprocessing concept is primarily built upon studies utilizing a recombinant model precursor that is purified from E. coli inclusion body under denaturing conditions and then refolded in vitro (see above). This approach eliminates other viral and/or cellular factors, if any, that may be involved in regulating the precursor PR activity. This section gives an overview of the recent advances in our understanding of HIV-1 PR autoprocessing while highlighting the outstanding questions that remain to be addressed.
Role of the p6* peptide in HIV PR autoprocessing
The p6* peptide is immediately upstream of the mature PR and makes up the N-terminal portion of the miniprecursor. The p6* peptide by itself is predicted to be intrinsically disordered; synthetic p6* alone does not display any defined structure. Accumulating data suggest that it plays an inhibitory role on precursor dimerization and precursor activity although the underlying mechanisms are still unclear. The recombinant p6*-PR precursor demonstrated an apparent dissociation constant of approximately 700 nM, which is >100-fold higher than the mature PR dissociation constant (Kd < 5 nM) [50]. Structural and biochemical analyses also demonstrated that an N-terminal extension of as few as four amino acids disrupts the β-sheet that is critical in stabilizing PR dimers [75,98]. Subsequently, one putative model is that the p6* peptide simply functions as a disordered N-terminal extension that prevents precursor dimerization. Consistent with this speculation, the majority of the peptide seems dispensable for virus replication. A previous study demonstrated that the central region of p6 (S14-I31), also corresponding to the central region of p6* (R20-D39), is dispensable for virus replication in cultured cells and smaller deletion can be tolerated in vivo as well [99]. It was recently demonstrated that approximately 65% of p6* region can be deleted without affecting PR autoprocessing and Gag processing after genetic uncoupling of this region from the overlapping p6 coding sequence [100]. The authors recently reported that truncation of the majority of the p6* region does not affect autoprocessing of GST fused precursor PRs in transfected mammalian cells [64]. These observations further support the idea that the N-terminal extension rather than the specific sequence hinders precursor dimerization and removal of the extension renders the full mature PR activity. Consequently, the N-terminal cleavage site seems to be an interesting target on its own, as blocking this cleavage would prevent release of the mature PR. In this regards, a peptide (P27) consisting of the N-terminal cleavage site along with the flanking sequence demonstrated encouraging protection in acutely infected MT-2 [67]. However, this peptide also inhibited virion production for an unknown reason. Therefore, additional investigation of the N-terminal cleavage site conformation and cleavage mechanism is needed for the further development of inhibitors specifically targeting this site.
A puzzling paradox is how p6*-PR precursor exerts proteolytic activity if dimerization is required to form the catalytic site as observed with the mature PR whereas the p6* peptide prevents dimerization. The in vitro transcribed/translated GagPol precursor is estimated to be at subnanomolar concentrations (up to 0.2 nM) that are at least tenfold lower than the mature PR dissociation constant (<5 nM) and approximately 3500-fold lower than the precursor PR dissociation constant (~700 nM). Consequently, GagPol precursor dimerization seems unfavorable under this condition, yet this GagPol precursor effectively cleaves the sp1-NC and NC-p6* sites. Assuming that dimerization is a must for catalytic activity, one could argue that the other domains (e.g., RT) flanking the p6*-PR precursor provide enough force to increase the local concentration to facilitate p6*-PR dimerization. This argument also infers that virion assembly at the plasma membrane leads to an increase in local concentrations of GagPol precursor to trigger precursor autoprocessing. However, this model fails to explain the result observed with the proviral system where the N-terminal cleavage site is mutated to non-cleavable [74]. The mutant virus contains normal amounts of RT and IN that are processed by the p6*-PR precursor whereas Gag polyprotein processing was severely impaired; only the cleavage at sp1- NC site was observed. It is estimated that in the released virus particles the p6*-PR precursor could reach up to approximately 220 mM [101], which is approximately 300-fold higher than the dissociation constant reported for the recombinant p6*-PR precursor [50]. If concentration is the only driving force for dimerization of the p6*-PR precursor, some mature PR activity would be expected in the released virions. The complete lack of Gag processing at sites other than sp1-NC indicated that regulation of the p6*-PR precursor activity appears more complicated than a simple concentration driven process. The allosteric regulation theory offers a possible solution to this apparent controversy. According to this theory, the precursor PR and the mature PR are two related but different enzymes and the mutation preventing the N-terminal cleavage also blocks the conformational change from the precursor to mature PR. As a result, only the precursor activity was manifested and no detectable mature PR activity could be observed. Therefore, it is fundamental to determine precursor structures in order to better understand autoprocessing mechanism and to develop precursor PR-specific inhibitors.
Role of the residues at the dimer interfaces on HIV-1 PR activity
Due to the fact that detailed structure information of precursor PR is not available, residues that are determined to be important for dimerization are all based on the mature PR structures. For HIV-1 mature PR, the interface interactions contribute close to 75% of the free energy of dimer stabilization [102,103]. Therefore, interfering with the residues contributing dimer formation is expected to decrease dimer stability and hence reduce PR activity. For example, deletions of the N- and C-terminal residues completely abolished dimer formation; the resulting PR5–95 was found to exist as monomers [42,98]. The catalytic site mutation D25N alone increased the equilibrium dimer dissociation constant by a factor >100-fold (1.3 ± 0.09 μM) relative to the wild-type mature PR [47]. Other residues including D25, I49, G50 at the interface were also exploited to promote the formation of heterodimers between the wild-type PR and the inactive mutant [104,105]. Interestingly, when introduced into the provirus and mixed with the wild-type provirus DNA at ratio of 2:1, the resulting mutant reduced virus infectivity by 82%, providing a proof-of-concept to inhibit HIV replication via manipulating PR dimer interface. Unlike the development of catalytic site inhibitors, the field of interfacial binding inhibitors for fighting against HIV is still in its infancy; new techniques are being developed that will hopefully provide some guidelines for the design of novel drug candidates [87,106,107].
Role of H69 & the surrounding residues in HIV-1 precursor autoprocessing
More than two decades ago, Loeb et al. individually altered each amino acid of HIV PR to different missense mutations and analyzed the resulting mutants for their autoprocessing activities in E. coli [1]. Results of this analysis have provided a landscape to study functionally important regions. For example, V82 polymorphism was observed in a long stretch of conserved residues (T74-L90) and V82 variants were later identified in many drug-resistant HIV strains. The highly conserved G86 was found to play a critical role in catalytic activity but have minimal influence on dimerization [48]. Functions of many other amino acids in PR activity and autoprocessing regulation remain to be revealed.
Chen and co-workers recently made an intriguing observation suggesting that H69 is involved in modulation of precursor structure and the subsequent autoprocessing [63,65]. Based on the mature PR structure, H69 is a surface residue that is approximately 25 Å away from the catalytic site and is located close to the stabilizing anti-parallel sheet PR. Alterations of H69 to neutral or positively charged amino acids have no obvious impact on precursor autoprocessing but changes to negatively charged amino acids (e.g., E or D), impede precursor autoprocessing. This phenomenon was reproducibly observed with three different model systems. Interesting enough, the recombinant mature PR carrying H69E mutation only demonstrated moderate reduction in catalytic activity. The authors’ data demonstrate that an acidic amino acid at position 69 does not drastically impair the mature PR activity, but prevents productive precursor autoprocessing. It was inferred that the local conformation of H69 residue in the precursor PR is different from that in the mature PR, supporting the idea that the precursor PR and the mature PR are not identical in their detailed structures.
H69 residue is also in a close proximity of the two highly conserved cysteines (C67 and C95) in the mature PR structure. The regulatory function of C67 and C95 was first noticed because copper salts inhibited the mature PR activity and oxidation of cysteine residues was essential for the inhibition [108,109] because precursors carrying C67A/C95A mutation are not inhibited by copper salts. Interestingly, column binding analysis revealed that H69, not C67, is responsible for reversible high affinity binding to the immobilized Cu2+ [110]. Biochemical analyses using in vitro purified mature PR demonstrated that glutathione modification at C95 abolished enzymatic activity by disrupting dimerization and dithiothreitol or cellular thioltransferase reversed the inhibition [111,112]. These studies suggested reversible oxidative modification of these cysteines as a regulation mechanism of precursor autoprocessing [113], which was further confirmed using the in vitro transcription/ translation system [61]. It was also observed that the H69E mutation severely impairs precursor autoprocessing in the context of cysteine-null background (C67A/C95A) but inhibits precursor autoprocessing to a lesser extend in the context of C67/C95 backbone [65]. Collectively, this general region containing residues C67, H69 and C95 seems flexible in its local conformation but plays an important role in regulating the precursor activity. Chemicals disturbing the function of this region might hold promise for the development of novel autoprocessing inhibitions. In this regard, Chang MW et al. recently identified one compound that inhibits the wild-type mature PR, as well as a drug-resistant PR with six point mutations. Docking analysis suggests a binding of the compound to the groove that also contains C67 and H69 residues via an allosteric non-competitive mechanism [114]. Such a mechanism has been underappreciated probably due to the approach that is used for structural analysis of the mature PR; the in vitro refolding procedure might force the PR to fold into the most stable conformation under the conditions. Therefore, it is essential to develop new approaches that would allow characterization of precursor conformations under physiological conditions.
Temporal correlation of HIV-1 PR autoprocessing with virion production
It is long believed that precursor autoprocessing has to be suppressed until the point when virion assembly is almost completed; early PR activation is detrimental to virion production and infectivity [23,72]. However, the link establishing this temporal correlation is still missing. One speculation is through the late domain that is located in the C-terminal region of the Gag polyprotein, responsible for recruiting the cellular ESCRT complexes to facilitate virions release [115–118]. In line with this idea, it was reported almost three decades ago that HIV late domain mutation inhibited HIV production in the presence of active PR; double mutation of the late domain and D25A rescued virion production [4]. The players mediating this interplay are yet to be identified. If additional viral and/or cellular components are indeed required for regulation of precursor autoprocessing (suppression during early assembly stage and activation upon virion release) the conventional crystallographic approach would be less fruitful without knowing the contributing factors. Therefore, identification and characterization of these factors that are associated with the precursor and/or autoprocessing intermediates would be critical for advancing our understanding of autoprocessing temporal regulation.
Precursor autoprocessing in the presence of catalytic site inhibitors
The currently available catalytic site inhibitors bind to the mature PR with high affinities, ranging from high pM to low nM, under conditions in which peptide substrates demonstrate high μM affinities [101,119]. These inhibitors are very effective at suppressing the mature PR activity due to their high-binding affinities with low nanomolar IC50 values. However, they are drastically insufficient at inhibiting the precursor PR activity. For example, 1 μM of ritonavir is required to inhibit proteolytic activity of the in vitro transcribed/translated GagPol precursor (estimated to be up to ~0.2 nM) [60]. Similarly, it has been repeatedly reported that μM concentrations of PR inhibitors are needed to precursor PR activity in various model systems [50,57,60,64,66]. The underlying mechanism of this phenomenon has been another long-standing mystery. Based on the contemporary model, one should that expect less PR inhibitors are required to suppress precursor PR activity given that only 3–5% of precursor would fold into mature PR-like catalytic site [75]. Once again, the allosteric regulation theory, proposed in this review, offers a sounding interpretation that the precursor PR is induced to fold into a mature PR-like structure in the presence of PR inhibitors. Such a process appears less efficient and thus requires high concentrations of PR inhibitors. Of course, experimental data supporting this idea are vital for justification.
Future perspective
The HIV-1 PR is a proven target for anti-AIDS drug development because of its critical role in virus infectivity. Extensive research efforts have primarily focused on the mature PR in the past three decades; whereas investigation of the precursor PR structure and function along with its autoprocessing mechanism has been mostly neglected. This is, in part, due to the widespread perception that the precursor and the mature PR are very similar in their structure and proteolytic activity although the definitive evidence is lacking. Accumulating evidence suggests that the precursor PR is enzymatically different from the well-characterized mature PR; the currently available FDA-approved PR inhibitors (targeting the catalytic site of the mature PR) are significantly less effective at suppressing precursor autoprocessing. Therefore, it is more critical than ever to better understand the precursor PR biochemistry and its mediated autoprocessing reactions. Extensive research efforts are vital to address outstanding questions, such as how different (enzymatically and structurally) is the precursor PR from the mature PR, how is the precursor PR activity suppressed during virion assembly and later activated upon virion release. Nonconventional approaches that pay special attention to better preserving precursor PR conformations under physiologically relevant conditions are also essential to examine and evaluate the proposed allosteric regulation theory. Results of these studies will hopefully identify regions that are outside the catalytic site of the mature PR but are essential for precursor autoprocessing. Chemicals targeting these regions will interfere with precursor autoprocessing and form a new class of anti-AIDS inhibitors. A combined use of new autoprocessing inhibitors with the existing catalytic site inhibitors would significantly increase the genetic barrier for the evolution of HIV-1 strains resistant PR to both classes of inhibitors. Although such compounds remain to be identified, this approach, if successful, will offer new promise to the campaign of achieving an AIDS-free generation.
Executive summary.
HIV-1 protease (PR) is a proven target for anti-HIV drug development. It has been increasingly appreciated that HIV-1 protease exists in at least two enzymatically different forms – the precursor and mature HIV-1 proteases. This unconventional theory would provide conceptual guidance for novel therapeutic development, although it remains to be further defined and evaluated.
Mechanistic characterization of precursor autoprocessing requires new approaches and methods that take biologically relevant conditions into consideration, as HIV-1 precursor proteases are expected to be flexible and capable of adapting into different conformations in response to various environments according to the proposed theory.
The currently available US FDA-approved protease inhibitors primarily target the catalytic site of the mature protease based on the known structure. These inhibitors are significantly less effective at suppressing the precursor-mediated autoprocessing reaction; no specific precursor protease inhibitor is available.
Emergence of drug-resistant HIV-1 strains is a constant concern and novel inhibitors with distinct action modes and/or different target regions are in urgent need. The precursor protease is a promising target given that it is enzymatically different from the mature protease.
Structure-based drug design is not practical for the development of precursor-specific inhibitors due to absence of detailed information of its conformation under physiologically relevant conditions. The current structural information is derived from recombinant protease purified under denaturing conditions followed by in vitro refolding.
The described cell-based assay allows for quantification of precursor autoprocessing, which provides a platform for high-throughput screening of autoprocessing inhibitors using AlphaLISA™ technology.
Acknowledgments
The authors would like to thank CJ Counts for help editing the manuscript.
Footnotes
For reprint orders, please contact reprints@future-science.com
Financial & competing interests disclosure
The authors were in part supported by an NIH/NIAID R21 A1080351 grant to C Chen. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
- 1.Loeb DD, Swanstrom R, Everitt L, et al. Complete mutagenesis of the HIV-1 protease. Nature. 1989;340(6232):397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- 2.Oroszlan S, Luftig RB. Retroviral proteinases. Curr Top Microbiol Immunol. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- 3.Partin K, Zybarth G, Ehrlich L, et al. Deletion of sequences upstream of the proteinase improves the proteolytic processing of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1991;88(11):4776–4780. doi: 10.1073/pnas.88.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eder J, Hommel U, Cumin F, Martoglio B, Gerhartz B. Aspartic proteases in drug discovery. Curr Pharm Des. 2007;13(3):271–285. doi: 10.2174/138161207779313560. [DOI] [PubMed] [Google Scholar]
- 6.Barre-Sinoussi F, Chermann JC, Rey F, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 7.Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; NY, USA: 1997. [PubMed] [Google Scholar]
- 8.Parkin NT, Chamorro M, Varmus HE. Human immunodeficiency virus type 1 GagPol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J Virol. 1992;66(8):5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung M, Patel P, Davis S, Green SR. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J Virol. 1998;72(6):4819–4824. doi: 10.1128/jvi.72.6.4819-4824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Montelaro RC. Characterization of RNA elements that regulate GagPol ribosomal frameshifting in equine infectious anemia virus. J Virol. 2003;77(19):10280–10287. doi: 10.1128/JVI.77.19.10280-10287.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis JM, Weber IT, Tozser J, Clore GM, Gronenborn AM. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv Pharmacol. 2000;49:111–146. doi: 10.1016/s1054-3589(00)49025-3. [DOI] [PubMed] [Google Scholar]
- 12.Schechter I, Berger A. On the size of the active site in proteases. I Papain Biochem Biophys Res Commun. 1967;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 13.Prejdova J, Soucek M, Konvalinka J. Determining and overcoming resistance to HIV protease inhibitors. Curr Drug Targets Infect Disord. 2004;4(2):137–152. doi: 10.2174/1568005043340984. [DOI] [PubMed] [Google Scholar]
- 14.Pettit SC, Clemente JC, Jeung JA, Dunn BM, Kaplan AH. Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease. J Virol. 2005;79(16):10601–10607. doi: 10.1128/JVI.79.16.10601-10607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettit SC, Lindquist JN, Kaplan AH, Swanstrom R. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology. 2005;2:66. doi: 10.1186/1742-4690-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paulus C, Ludwig C, Wagner R. Contribution of the Gag Pol transframe domain p6* and its coding sequence to morphogenesis and replication of human immunodeficiency virus type 1. Virology. 2004;330(1):271. doi: 10.1016/j.virol.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Feher A, Weber IT, Bagossi P, et al. Effect of sequence polymorphism and drug resistance on two HIV-1 Gag processing sites. Eur J Biochem. 2002;269(16):4114–4120. doi: 10.1046/j.1432-1033.2002.03105.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, Chen CH, Aiken C. Human immunodeficiency virus type 1 resistance to the small molecule maturation inhibitor 3-O-(3′,3′-dimethylsuccinyl)-betulinic acid is conferred by a variety of single amino acid substitutions at the CA-SP1 cleavage site in Gag. J Virol. 2006;80(24):12095–12101. doi: 10.1128/JVI.01626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nijhuis M, van Maarseveen NM, Lastere S, et al. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 2007;4(1):e36. doi: 10.1371/journal.pmed.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adamson CS, Sakalian M, Salzwedel K, Freed EO. Polymorphisms in Gag spacer peptide 1 confer varying levels of resistance to the HIV-1 maturation inhibitor bevirimat. Retrovirology. 2010;7:36. doi: 10.1186/1742-4690-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohl NE, Emini EA, Schleif WA, et al. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85(13):4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karacostas V, Wolffe EJ, Nagashima K, Gonda MA, Moss B. Overexpression of the HIV-1 Gag Pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193(2):661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 23.Krausslich HG. Human immunodeficiency virus proteinase dimer as component of the viral polyprotein prevents particle assembly and viral infectivity. Proc Natl Acad Sci USA. 1991;88(8):3213–3217. doi: 10.1073/pnas.88.8.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan AH, Zack JA, Knigge M, et al. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J Virol. 1993;67(7):4050–4055. doi: 10.1128/jvi.67.7.4050-4055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiegers K, Rutter G, Kottler H, et al. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72(4):2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F, Goila-Gaur R, Salzwedel K, et al. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci USA. 2003;100(23):13555–13560. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dulude D, Berchiche YA, Gendron K, Brakier-Gingras L, Heveker N. Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology. 2006;345(1):127–136. doi: 10.1016/j.virol.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 28.Skalka AM. Retroviral proteases: first glimpses at the anatomy of a processing machine. Cell. 1989;56(6):911–913. doi: 10.1016/0092-8674(89)90621-1. [DOI] [PubMed] [Google Scholar]
- 29.Darke PL, Huff JR. HIV protease as an inhibitor target for the treatment of AIDS. Adv Pharmacol. 1994;25:399–454. doi: 10.1016/s1054-3589(08)60438-x. [DOI] [PubMed] [Google Scholar]
- 30.Wlodawer A, Vondrasek J. Inhibitors of HIV-1 protease: a major success of structure-assisted drug design. Annu Rev Biophys Biomol Struct. 1998;27:249–284. doi: 10.1146/annurev.biophys.27.1.249. [DOI] [PubMed] [Google Scholar]
- 31.Wlodawer A, Gustchina A. Structural and biochemical studies of retroviral proteases. Biochim Biophys Acta. 2000;1477:16–34. doi: 10.1016/s0167-4838(99)00267-8. [DOI] [PubMed] [Google Scholar]
- 32.Dunn BM, Goodenow MM, Gustchina A, Wlodawer A. Retroviral proteases. Genome Biol. 2002;3(4):reviews3006–reviews3006.7. doi: 10.1186/gb-2002-3-4-reviews3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vondrasek J, Wlodawer A. HIVdb: a database of the structures of human immunodeficiency virus protease. Proteins. 2002;49(4):429–431. doi: 10.1002/prot.10246. [DOI] [PubMed] [Google Scholar]
- 34.King JR, Acosta EP. Tipranavir: a novel nonpeptidic protease inhibitor of HIV. Clin Pharmacokinet. 2006;45(7):665–682. doi: 10.2165/00003088-200645070-00003. [DOI] [PubMed] [Google Scholar]
- 35.Wensing AM, Van Maarseveen NM, Nijhuis M. Fifteen years of HIV protease inhibitors: raising the barrier to resistance. Antiviral Res. 2010;85(1):59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006;5(9):785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 37.Sielecki AR, Fedorov AA, Boodhoo A, Andreeva NS, James MN. Molecular and crystal structures of monoclinic porcine pepsin refined at 1.8 A resolution. J Mol Biol. 1990;214(1):143–170. doi: 10.1016/0022-2836(90)90153-D. [DOI] [PubMed] [Google Scholar]
- 38.Baldwin ET, Bhat TN, Gulnik S, et al. Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design. Proc Natl Acad Sci USA. 1993;90(14):6796–6800. doi: 10.1073/pnas.90.14.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearl LH, Taylor WR. A structural model for the retroviral proteases. Nature. 1987;329(6137):351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- 40.Bannwarth L, Reboud-Ravaux M. An alternative strategy for inhibiting multidrug-resistant mutants of the dimeric HIV-1 protease by targeting the subunit interface. Biochem Soc Trans. 2007;35(Pt 3):551–554. doi: 10.1042/BST0350551. [DOI] [PubMed] [Google Scholar]
- 41.Heaslet H, Rosenfeld R, Giffin M, et al. Conformational flexibility in the flap domains of ligand-free HIV protease. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 8):866–875. doi: 10.1107/S0907444907029125. [DOI] [PubMed] [Google Scholar]
- 42.Ishima R, Ghirlando R, Tozser J, et al. Folded monomer of HIV-1 protease. J Biol Chem. 2001;276(52):49110–49116. doi: 10.1074/jbc.M108136200. [DOI] [PubMed] [Google Scholar]
- 43.Ishima R, Torchia DA, Lynch SM, Gronenborn AM, Louis JM. Solution structure of the mature HIV-1 protease monomer: insight into the tertiary fold and stability of a precursor. J Biol Chem. 2003;278(44):43311–43319. doi: 10.1074/jbc.M307549200. [DOI] [PubMed] [Google Scholar]
- 44.Tie Y, Boross PI, Wang YF, et al. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multidrug-resistant clinical strains. J Mol Biol. 2004;338(2):341–352. doi: 10.1016/j.jmb.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 45.Tie Y, Boross PI, Wang YF, et al. Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs. FEBS J. 2005;272(20):5265–5277. doi: 10.1111/j.1742-4658.2005.04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prabu-Jeyabalan M, Nalivaika EA, Romano K, Schiffer CA. Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate. J Virol. 2006;80(7):3607–3616. doi: 10.1128/JVI.80.7.3607-3616.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sayer JM, Liu F, Ishima R, Weber IT, Louis JM. Effect of the active site D25N mutation on the structure, stability, and ligand binding of the mature HIV-1 protease. J Biol Chem. 2008;283(19):13459–13470. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishima R, Gong Q, Tie Y, Weber IT, Louis JM. Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease. Proteins. 2009;78(4):1015–1025. doi: 10.1002/prot.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mittl PR, Grutter MG. Opportunities for structure-based design of protease-directed drugs. Curr Opin Struct Biol. 2006;16(6):769–775. doi: 10.1016/j.sbi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 50.Louis JM, Clore GM, Gronenborn AM. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat Struct Biol. 1999;6:868–875. doi: 10.1038/12327. [DOI] [PubMed] [Google Scholar]
- 51.Toth MV, Marshall GR. A simple, continuous fluorometric assay for HIV protease. Int J Pept Protein Res. 1990;36(6):544–550. doi: 10.1111/j.1399-3011.1990.tb00994.x. [DOI] [PubMed] [Google Scholar]
- 52.Tozser J, Blaha I, Copeland TD, Wondrak EM, Oroszlan S. Comparison of the HIV-1 and HIV-2 proteinases using oligopeptide substrates representing cleavage sites in Gag and GagPol polyproteins. FEBS Lett. 1991;281(1–2):77–80. doi: 10.1016/0014-5793(91)80362-7. [DOI] [PubMed] [Google Scholar]
- 53.Pettit SC, Moody MD, Wehbie RS, et al. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68(12):8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettit SC, Henderson GJ, Schiffer CA, Swanstrom R. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J Virol. 2002;76(20):10226–10233. doi: 10.1128/JVI.76.20.10226-10233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55▪.Lee SK, Potempa M, Kolli M, et al. Context surrounding processing sites is crucial in determining cleavage rate of a subset of processing sites in HIV-1 Gag and Gag-Pro-Pol polyprotein precursors by viral protease. J Biol Chem. 2012;287(16):13279–13290. doi: 10.1074/jbc.M112.339374. Demonstrates that HIV-1 protease activity is modulated by complex interactions beyond the catalytic site and the substrate peptide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Breuer S, Sepulveda H, Chen Y, Trotter J, Torbett BE. A cleavage enzyme-cytometric bead array provides biochemical profiling of resistance mutations in HIV-1 Gag and protease. Biochemistry. 2011;50(20):4371–4381. doi: 10.1021/bi200031m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57▪.Louis JM, Aniana A, Weber IT, Sayer JM. Inhibition of autoprocessing of natural variants and multidrug resistant mutant precursors of HIV-1 protease by clinical inhibitors. Proc Natl Acad Sci USA. 2011;108(22):9072–9077. doi: 10.1073/pnas.1102278108. Reveals that precursor proteases are less responsive than the mature proteases to clinical inhibtors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shehu-Xhilaga M, Kraeusslich HG, Pettit S, et al. Proteolytic processing of the P2/ nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J Virol. 2001;75(19):9156–9164. doi: 10.1128/JVI.75.19.9156-9164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettit SC, Gulnik S, Everitt L, Kaplan AH. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. J Virol. 2003;77(1):366–374. doi: 10.1128/JVI.77.1.366-374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J Virol. 2004;78(16):8477–8485. doi: 10.1128/JVI.78.16.8477-8485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daniels SI, Davis DA, Soule EE, et al. The initial step in human immunodeficiency virus type 1 GagProPol processing can be regulated by reversible oxidation. PLoS ONE. 2010;5(10):e13595. doi: 10.1371/journal.pone.0013595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Debouck C, Gorniak JG, Strickler JE, et al. Human immunodeficiency virus protease expressed in Escherichia coli exhibits autoprocessing and specific maturation of the gag precursor. Proc Natl Acad Sci USA. 1987;84(24):8903–8906. doi: 10.1073/pnas.84.24.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63▪.Huang L, Sayer JM, Swinford M, Louis JM, Chen C. Modulation of human immunodeficiency virus type 1 protease autoprocessing by charge properties of surface residue 69. J Virol. 2009;83(15):7789–7793. doi: 10.1128/JVI.00473-09. Reports on the role of H69 in regulating precursor processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang L, Chen C. Autoprocessing of human immunodeficiency virus type 1 protease miniprecursor fusions in mammalian cells. AIDS Res Ther. 2010;7(1):27. doi: 10.1186/1742-6405-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65▪.Huang L, Hall A, Chen C. Cysteine 95 and other residues influence the regulatory effects of histidine 69 mutations on human immunodeficiency virus type 1 protease autoprocessing. Retrovirology. 2010;7:24. doi: 10.1186/1742-4690-7-24. Describes a novel cell-based precursor autoprocessing assay that recaptulates many previously observed phenomena. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang L, Li Y, Chen C. Flexible catalytic site conformations implicated in modulation of HIV-1 protease autoprocessing reactions. Retrovirology. 2011;8(1):79. doi: 10.1186/1742-4690-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davis DA, Brown CA, Singer KE, et al. Inhibition of HIV-1 replication by a peptide dimerization inhibitor of HIV-1 protease. Antiviral Res. 2006;72(2):89–99. doi: 10.1016/j.antiviral.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 68.Louis JM, Ishima R, Torchia DA, Weber IT. HIV-1 protease: structure, dynamics, and inhibition. Adv Pharmacol. 2007;55:261–298. doi: 10.1016/S1054-3589(07)55008-8. [DOI] [PubMed] [Google Scholar]
- 69.Cherry E, Liang C, Rong L, et al. Characterization of human immunodeficiency virus type-1 (HIV-1) particles that express protease-reverse transcriptase fusion proteins. J Mol Biol. 1998;284(1):43–56. doi: 10.1006/jmbi.1998.1968. [DOI] [PubMed] [Google Scholar]
- 70.Wondrak EM, Nashed NT, Haber MT, Jerina DM, Louis JM. A transient precursor of the HIV-1 protease. Isolation, characterization, and kinetics of maturation. J Biol Chem. 1996;271(8):4477–4481. doi: 10.1074/jbc.271.8.4477. [DOI] [PubMed] [Google Scholar]
- 71.Koh Y, Matsumi S, Das D, et al. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. J Biol Chem. 2007;282(39):28709–28720. doi: 10.1074/jbc.M703938200. [DOI] [PubMed] [Google Scholar]
- 72▪.Pan YY, Wang SM, Huang KJ, Chiang CC, Wang CT. Placement of leucine zipper motifs at the carboxyl terminus of HIV-1 protease significantly reduces virion production. PLoS ONE. 2012;7(3):e32845. doi: 10.1371/journal.pone.0032845. Reports a fluorescence resonance energy transfer-based method for quantification of precursor dimerization in co-transfected mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tessmer U, Krausslich HG. Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity. J Virol. 1998;72(4):3459–3463. doi: 10.1128/jvi.72.4.3459-3463.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ludwig C, Leiherer A, Wagner R. Importance of protease cleavage sites within and flanking human immunodeficiency virus type 1 transframe protein p6* for spatiotemporal regulation of protease activation. J Virol. 2008;82(9):4573–4584. doi: 10.1128/JVI.02353-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang C, Louis JM, Aniana A, Suh JY, Clore GM. Visualizing transient events in aminoterminal autoprocessing of HIV-1 protease. Nature. 2008;455(7213):693–696. doi: 10.1038/nature07342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76▪.Agniswamy J, Sayer JM, Weber IT, Louis JM. Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease. Biochemistry. 2012;51(5):1041–1050. doi: 10.1021/bi201809s. Structural analysis of complexes formed between a model precursor HIV-protease and darunavir illustrating the terminal interface conformations that destablize precursor dimers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chatterjee A, Mridula P, Mishra RK, Mittal R, Hosur RV. Folding regulates autoprocessing of HIV-1 protease precursor. J Biol Chem. 2005;280(12):11369–11378. doi: 10.1074/jbc.M412603200. [DOI] [PubMed] [Google Scholar]
- 78.Burnley BT, Afonine PV, Adams PD, Gros P. Modelling dynamics in protein crystal structures by ensemble refinement. eLife. 2012;1:e00311. doi: 10.7554/eLife.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sadiq SK, Noe F, De Fabritiis G. Kinetic characterization of the critical step in HIV-1 protease maturation. Proc Natl Acad Sci USA. 2012;109(50):20449–20454. doi: 10.1073/pnas.1210983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rose JR, Salto R, Craik CS. Regulation of autoproteolysis of the HIV-1 and HIV-2 proteases with engineered amino acid substitutions. J Biol Chem. 1993;268(16):11939–11945. [PubMed] [Google Scholar]
- 81.Mildner AM, Rothrock DJ, Leone JW, et al. The HIV-1 protease as enzyme and substrate: mutagenesis of autolysis sites and generation of a stable mutant with retained kinetic properties. Biochemistry. 1994;33(32):9405–9413. doi: 10.1021/bi00198a005. [DOI] [PubMed] [Google Scholar]
- 82.Louis JM, Nashed NT, Parris KD, Kimmel AR, Jerina DM. Kinetics and mechanism of autoprocessing of human immunodeficiency virus type 1 protease from an analog of the GagPol polyprotein. Proc Natl Acad Sci USA. 1994;91(17):7970–7974. doi: 10.1073/pnas.91.17.7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Agniswamy J, Shen CH, Aniana A, et al. HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements. Biochemistry. 2012;51(13):2819–2828. doi: 10.1021/bi2018317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Erickson-Viitanen S, Manfredi J, Viitanen P, et al. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res Hum Retroviruses. 1989;5(6):577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 85.Krausslich HG, Schneider H, Zybarth G, Carter CA, Wimmer E. Processing of in vitro-synthesized gag precursor proteins of human immunodeficiency virus (HIV) type 1 by HIV proteinase generated in Escherichia coli. J Virol. 1988;62(11):4393–4397. doi: 10.1128/jvi.62.11.4393-4397.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krausslich HG, Ingraham RH, Skoog MT, et al. Activity of purified biosynthetic proteinase of human immunodeficiency virus on natural substrates and synthetic peptides. Proc Natl Acad Sci USA. 1989;86(3):807–811. doi: 10.1073/pnas.86.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koh Y, Aoki M, Danish ML, et al. Loss of protease dimerization inhibition activity of darunavir is associated with the acquisition of resistance to darunavir by HIV-1. J Virol. 2011;85(19):10079–10089. doi: 10.1128/JVI.05121-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aoki M, Danish ML, Aoki-Ogata H, et al. Loss of the protease dimerization inhibition activity of tipranavir (TPV) and its association with the acquisition of resistance to TPV by HIV-1. J Virol. 2012;86(24):13384–13396. doi: 10.1128/JVI.07234-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang YC, Misquitta S, Blond SY, Adams E, Colman RF. Catalytically active monomer of glutathione S-transferase pi and key residues involved in the electrostatic interaction between subunits. J Biol Chem. 2008;283(47):32880–32888. doi: 10.1074/jbc.M805484200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fabrini R, De Luca A, Stella L, et al. Monomer–dimer equilibrium in glutathione transferases: a critical re-examination. Biochemistry. 2009;48(43):10473–10482. doi: 10.1021/bi901238t. [DOI] [PubMed] [Google Scholar]
- 91.Kaplan W, Husler P, Klump H, et al. Conformational stability of pGEX-expressed Schistosoma japonicum glutathione S-transferase: a detoxification enzyme and fusion-protein affinity tag. Protein Sci. 1997;6(2):399–406. doi: 10.1002/pro.5560060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wan M, Takagi M, Loh BN, Xu XZ, Imanaka T. Autoprocessing: an essential step for the activation of HIV-1 protease. Biochem J. 1996;316(Pt 2):569–573. doi: 10.1042/bj3160569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Louis JM, McDonald RA, Nashed NT, et al. Autoprocessing of the HIV-1 protease using purified wild-type and mutated fusion proteins expressed at high levels in Escherichia coli. Eur J Biochem. 1991;199(2):361–369. doi: 10.1111/j.1432-1033.1991.tb16132.x. [DOI] [PubMed] [Google Scholar]
- 94.Ullman EF, Kirakossian H, Singh S, et al. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc Natl Acad Sci USA. 1994;91(12):5426–5430. doi: 10.1073/pnas.91.12.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ullman EF, Kirakossian H, Switchenko AC, et al. Luminescent oxygen channeling assay (LOCI): sensitive, broadly applicable homogeneous immunoassay method. Clin Chem. 1996;42(9):1518–1526. [PubMed] [Google Scholar]
- 96.Mcgiven JA, Sawyer J, Perrett LL, et al. A new homogeneous assay for high throughput serological diagnosis of brucellosis in ruminants. J Immunol Methods. 2008;337(1):7–15. doi: 10.1016/j.jim.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 97.Eglen RM, Reisine T, Roby P, et al. The use of AlphaScreen technology in HTS: current status. Curr Chem Genomics. 2008;1:2–10. doi: 10.2174/1875397300801010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ishima R, Torchia DA, Louis JM. Mutational and structural studies aimed at characterizing the monomer of HIV-1 protease and its precursor. J Biol Chem. 2007;282(23):17190–17199. doi: 10.1074/jbc.M701304200. [DOI] [PubMed] [Google Scholar]
- 99.Bleiber G, Peters S, Martinez R, et al. The central region of human immunodeficiency virus type 1 p6 protein (Gag residues S14-I31) is dispensable for the virus in vitro. J Gen Virol. 2004;85(Pt 4):921–927. doi: 10.1099/vir.0.19576-0. [DOI] [PubMed] [Google Scholar]
- 100.Leiherer A, Ludwig C, Wagner R. Uncoupling human immunodeficiency virus type 1 Gag and Pol reading frames: role of the transframe protein p6* in viral replication. J Virol. 2009;83(14):7210–7220. doi: 10.1128/JVI.02603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee SK, Potempa M, Swanstrom R. The choreography of HIV-1 proteolytic processing and virion assembly. J Biol Chem. 2012;287(49):40867–40874. doi: 10.1074/jbc.R112.399444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Todd MJ, Semo N, Freire E. The structural stability of the HIV-1 protease. J Mol Biol. 1998;283(2):475–488. doi: 10.1006/jmbi.1998.2090. [DOI] [PubMed] [Google Scholar]
- 103.Hamacher K. Relating sequence evolution of HIV1-protease to its underlying molecular mechanics. Gene. 2008;422(1–2):30–36. doi: 10.1016/j.gene.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 104.Rozzelle JE, Dauber DS, Todd S, Kelley R, Craik CS. Macromolecular inhibitors of HIV-1 protease. Characterization of designed heterodimers. J Biol Chem. 2000;275(10):7080–7086. doi: 10.1074/jbc.275.10.7080. [DOI] [PubMed] [Google Scholar]
- 105.Miklossy G, Tozser J, Kadas J, et al. Novel macromolecular inhibitors of human immunodeficiency virus-1 protease. Protein Eng Des Sel. 2008;21(7):453–461. doi: 10.1093/protein/gzn022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Camarasa MJ, Velazquez S, San-Felix A, Perez-Perez MJ, Gago F. Dimerization inhibitors of HIV-1 reverse transcriptase, protease and integrase: a single mode of inhibition for the three HIV enzymes? Antiviral Res. 2006;71(2–3):260–267. doi: 10.1016/j.antiviral.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 107.Davis DA, Tebbs IR, Daniels SI, et al. Analysis and characterization of dimerization inhibition of a multidrug-resistant human immunodeficiency virus type 1 protease using a novel size-exclusion chromatographic approach. Biochem J. 2009;419(2):497–506. doi: 10.1042/BJ20082068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Karlstrom AR, Levine RL. Copper inhibits the protease from human immunodeficiency virus 1 by both cysteine-dependent and cysteine-independent mechanisms. Proc Natl Acad Sci USA. 1991;88(13):5552–5556. doi: 10.1073/pnas.88.13.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Karlstrom AR, Shames BD, Levine RL. Reactivity of cysteine residues in the protease from human immunodeficiency virus: identification of a surface-exposed region which affects enzyme function. Arch Biochem Biophys. 1993;304(1):163–169. doi: 10.1006/abbi.1993.1334. [DOI] [PubMed] [Google Scholar]
- 110.Danielson H, Lindgren MT, Markgren PO, Nillroth U. Investigation of an allosteric site of HIV-1 proteinase involved in inhibition by Cu2+ Adv Exp Med Biol. 1998;436:99–103. doi: 10.1007/978-1-4615-5373-1_13. [DOI] [PubMed] [Google Scholar]
- 111.Davis DA, Dorsey K, Wingfield PT, et al. Regulation of HIV-1 protease activity through cysteine modification. Biochemistry. 1996;35(7):2482–2488. doi: 10.1021/bi951525k. [DOI] [PubMed] [Google Scholar]
- 112.Davis DA, Newcomb FM, Starke DW, et al. Thioltransferase (glutaredoxin) is detected within HIV-1 and can regulate the activity of glutathionylated HIV-1 protease in vitro. J Biol Chem. 1997;272(41):25935–25940. doi: 10.1074/jbc.272.41.25935. [DOI] [PubMed] [Google Scholar]
- 113.Davis DA, Brown CA, Newcomb FM, et al. Reversible oxidative modification as a mechanism for regulating retroviral protease dimerization and activation. J Virol. 2003;77(5):3319–3325. doi: 10.1128/JVI.77.5.3319-3325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chang MW, Giffin MJ, Muller R, et al. Identification of broad-based HIV-1 protease inhibitors from combinatorial libraries. Biochem J. 2010;429(3):527–532. doi: 10.1042/BJ20091645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Freed EO. Viral late domains. J Virol. 2002;76(10):4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Freed EO. HIV-1 and the host cell: an intimate association. Trends Microbiol. 2004;12(4):170–177. doi: 10.1016/j.tim.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 117.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 118.Goff SP. Host factors exploited by retroviruses. Nat Rev Microbiol. 2007;5(4):253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 119.Anderson J, Schiffer C, Lee SK, Swanstrom R. Viral protease inhibitors. Handbook Exp Pharmacol. 2009;189(189):85–110. doi: 10.1007/978-3-540-79086-0_4. [DOI] [PMC free article] [PubMed] [Google Scholar]