Abstract
Rationale: Previous studies of chronic obstructive pulmonary disease (COPD) have suggested that genetic factors play an important role in the development of disease. However, single-nucleotide polymorphisms that are associated with COPD in genome-wide association studies have been shown to account for only a small percentage of the genetic variance in phenotypes of COPD, such as spirometry and imaging variables. These phenotypes are highly predictive of disease, and family studies have shown that spirometric phenotypes are heritable.
Objectives: To assess the heritability and coheritability of four major COPD-related phenotypes (measurements of FEV1, FEV1/FVC, percent emphysema, and percent gas trapping), and COPD affection status in smokers of non-Hispanic white and African American descent using a population design.
Methods: Single-nucleotide polymorphisms from genome-wide association studies chips were used to calculate the relatedness of pairs of individuals and a mixed model was adopted to estimate genetic variance and covariance.
Measurements and Main Results: In the non-Hispanic whites, estimated heritabilities of FEV1 and FEV1/FVC were both about 37%, consistent with estimates in the literature from family-based studies. For chest computed tomography scan phenotypes, estimated heritabilities were both close to 25%. Heritability of COPD affection status was estimated as 37.7% in both populations.
Conclusions: This study suggests that a large portion of the genetic risk of COPD is yet to be discovered and gives rationale for additional genetic studies of COPD. The estimates of coheritability (genetic covariance) for pairs of the phenotypes suggest considerable overlap of causal genetic loci.
Keywords: missing heritability, pleiotropy, pulmonary function, imaging phenotypes, chromosomal partition
At a Glance Commentary
Scientific Knowledge on the Subject
Novel estimates of heritability for chronic obstructive pulmonary disease and four lung function–related phenotypes are estimated using a population-based dataset and compared with pedigree-derived estimates.
What This Study Adds to the Field
We present heritability estimates of emphysema for the first time. This study suggests that a large portion of the genetic content of chronic obstructive pulmonary disease is yet to be discovered and gives rationale for later genetic studies for chronic obstructive pulmonary disease.
Chronic obstructive pulmonary disease (COPD) is a major cause of disability and has recently risen to become the third leading cause of death in the United States. Although cigarette smoking is the most important risk factor for the development of COPD, only a small proportion of smokers develop clinically significant COPD. In family studies, it has been shown that the risk of COPD is approximately two to three times higher in smokers who have a first-degree relative affected by COPD (1). This suggests that genetic factors play an important role in development of COPD. The best known genetic factor linked to COPD is a deficiency of the serine protease α1-antitrypsin (encoded by SERPINA1), which occurs in 1–3% of patients with COPD (2). Recently, genome-wide association studies (GWAS) have revealed various common variants in several genetic loci that are associated with COPD susceptibility including CHRNA3/CHRNA5/IREB2 (3), HHIP (4), FAM13A (5, 6), and a chromosome 19q region near CYP2A6 (7). However, similar to other common nonmendelian diseases, these genetic variants account for a small percentage of the heritability in COPD-related phenotypes, which has been estimated in previous twin studies and family studies to be 20–40% for spirometric phenotypes, such as FEV1 and FEV1/FVC (8, 9), and even higher, 50–60%, for smoking behavior phenotypes (10, 11). Rare variants may be related to the substantial undetermined heritability because not all rare variants are tagged by GWAS chips. Another explanation is that the heritability is not missing but hiding (12). Recently, Yang and coworkers (12, 13) proposed a method of estimating the total amount of phenotypic variance captured by all single-nucleotide polymorphisms (SNPs) on GWAS commercial genotyping arrays using unrelated individuals. They estimated that about 45% of the phenotypic variance of human height could be explained by all the SNPs genotyped for GWAS. Their approach showed a nearly 10-fold increase from the 5% explained by published and validated individual SNPs in GWAS for height (14–16). Their method can be extended to multiple phenotypes to determine pleiotropy (17) by estimating coheritability between pair of phenotypes. Pleiotropy is said to be present when two or more phenotypes share some causal loci.
By using Yang’s method in the Genetic Epidemiology of COPD (COPDGene) Study population of approximately 10,000 subjects, we report heritability estimates of spirometric phenotypes (post-bronchodilator FEV1 and FEV1/FVC), chest computed tomography (CT) scan phenotypes (percent emphysema and percent gas trapping), and COPD disease status. Because COPD is defined based on reductions in FEV1 and FEV1/FVC (FEV1 <80% of predicted and FEV1/FVC <0.7), heritabilities of these two phenotypes were adjusted for study ascertainment to predict population heritability. Prebronchodilator FEV1 and FEV1/FVC are highly correlated with post-bronchodilator measures and our estimations show that they have very similar heritability estimates. Therefore, we only report post-bronchodilator FEV1 and FEV1/FVC in this paper. Genomic partitioning of the heritability to the separate autosomes shows the distribution of causal variants over the genome, and also enables us to account for any population substructure in the sample (18). We therefore estimated the heritability of two phenotypes (FEV1/FVC and percent emphysema) in non-Hispanic whites (NHW) for each of the autosomes (nonsex chromosomes), the sample size being too small to partition the heritability for the African American (AA) sample. Finally, we estimated the percentage of the overall heritability that can be explained by four previously reported GWAS SNPs (3–6).
Methods
Sample
COPDGene is one of the largest cohorts of well-characterized smokers for respiratory disease research, including 10,192 current and former smokers with airflow obstruction ranging from none to Global Initiative for Chronic Obstructive Lung Disease stage 4 (very severe) COPD. The study design of COPDGene has been reported previously (19). Briefly, both NHW subjects and AA subjects were included between the ages of 45 and 80 with at least a 10 pack-year smoking history. Exclusion criteria included pregnancy, history of other lung disease except asthma, prior lobectomy or lung volume reduction surgery, active cancer undergoing treatment, or known or suspected lung cancer. After data cleaning, 6,678 individuals from the NHW and 3,300 individuals from the AA population with complete genotypic data remained. All of the subjects were not knowingly related to each other. The sample is enriched for COPD subjects by design.
Genotyping was performed on the Illumina OmniExpress platform (Illumina, San Diego, CA). We excluded SNPs that have minor allele frequency less than 0.01 and Hardy-Weinberg equilibrium P value less than 10−8 using PLINK (20). Only SNPs located on the autosomes were used. After filtering, 664,892 and 663,347 autosomal SNPs were retained for analysis in the NHW and AA population, respectively.
Phenotypes
COPDGene subjects underwent extensive phenotypic assessment, including spirometry (pre and post bronchodilator) and chest CT scans (inspiratory and expiratory). Spirometry, chest CT scans, and smoking phenotypes were considered as continuous variables, whereas COPD disease status was a binary outcome in the analysis. Prebronchodilator spirometric phenotypes are highly correlated with post-bronchodilator measures and our estimations show that they have very similar heritability estimates. Therefore, we only report post-bronchodilator FEV1 and FEV1/FVC in this paper.
Spirometric measures of lung function were performed before and after the inhalation of 180 μg (two puffs) of albuterol, according to American Thoracic Society criteria (21). Volumetric chest CT acquisitions were obtained at full inspiration (200 mA), and at the end of normal expiration (50 mA). Quantitative image analysis to calculate percent emphysema and percent gas trapping was performed using 3D SLICER (http://www.slicer.org/). Percent emphysema was defined as the total percentage of both lungs with attenuation values less than −950 Hounsfield units on inspiratory images, and percent gas trapping was defined as the total percentage of both lungs with attenuation values less than −856 Hounsfield units on expiratory images.
Statistical Methods
Heritability and genetics relationship matrix estimation.
We calculated a genetic relationship matrix (GRM) using SNPs from all of the autosomes. The GRM uses SNP data to measure the relatedness between each pair of individuals in our sample. This GRM replaces the known information about relatedness found in pedigrees. Heritabilities for continuous and binary outcomes were estimated using the software package GCTA developed by Yang and coworkers (13). Heritability of disease status was estimated using a liability model as illustrated previously (22, 23).
To determine whether the distribution of genetic variance differs by chromosome, and to account for the effects of population stratification, we first calculated the GRMs from the SNPs separately on each autosome. We then estimated the genetic variances for all 22 autosomes in a joint analysis using a linear mixed model. In this model, the heritability of each autosome is just the ratio of its genetic variance to the overall phenotypic variance. We used one representative spirometric phenotype, FEV1/FVC, and one CT scan phenotype, percent emphysema only in NHW sample because of the small sample size in AA (18).
To quantify the effect of population substructure, we estimated the genetic variance for each autosome in a separate analysis. Using these estimates of the genetic variance, we calculated the autosome-specific heritabilities in two ways: using the jointly estimated genetic variances ( ) and using the separately estimated genetic variances (). The joint estimates, , can be viewed as adjusting for genetic variation on other chromosomes (i.e., the effect of population substructure). The estimated does not take population substructure into account. We then regressed the difference in the two estimates of the genetic variances, jointly and separately, on the length (in mega–base pairs) of the corresponding chromosome. The regression slope can be attributed to population stratification because longer chromosomes are likely to have more ancestry informative markers, assuming that the ancestry informative markers are randomly distributed across the genome (18).
Heritability that can be explained by the four previously reported GWAS SNPs was calculated by comparing the genetic variances estimated with and without using the four SNPs as covariates.
Adjustment for ascertainment.
When the proportion of cases and control subjects are not a random sample from the general population, the sample heritability needs to be adjusted to estimate general population heritability. The adjustment for ascertainment is a straightforward extension of the approach used to estimate heritability from dichotomous data using the threshold model. We first express heritability in the sample as a function of population heritability (h2). Because can be estimated from the data, h2 is therefore determined if the sampling fractions (prevalence in the population and in the sample) and thresholds to define cases are known.
In COPDGene, disease status is defined by two phenotypes: an individual is a case when post-bronchodilator FEV1 is less than 80% predicted and post-bronchodilator FEV1/FVC is less than 0.7. Although slightly more complex, the general approach for ascertainment correction extends readily when disease status is defined using two variables. See the online supplement for a detailed derivation.
Genetic covariance.
The estimation of the genetic covariance between two phenotypes can be seen as an estimation of covariance between multivariate traits within a linear mixed model framework (24, 25). We parameterized covariance structure of Y1 and Y2 to be
where and , , and are the genetic and environmental variances and covariances, with i and j indexing subjects and t indexing traits, and K is the GRM. Each parameter was estimated using a maximum likelihood method (26). The genetic correlation coefficient is defined as follows:
Results
Heritability Estimation for COPD-related Phenotypes
The COPDGene Study enrolled a total of 10,192 smokers. After phenotypic exclusions and genotyping quality control, we started with a total sample of 9,978 individuals with full genotypes including 6,678 NHW subjects and 3,300 AA subjects. All of the individuals within each racial group were used to calculate race-specific genetic relationship coefficients based on previously filtered autosomal SNPs. To avoid inflation caused by closely related individuals, we selectively excluded one of any pair of individuals with an estimated relationship greater than 0.025 (corresponding to the relationships of third or fourth degree of relationship) to maximize the remaining sample size. A total of 6,415 NHW individuals and 2,792 AA individuals remained for genetic relationship coefficient calculations and for various descriptive statistics generation (Table 1). Percent emphysema and percent gas trapping were log-transformed to achieve normality. Phenotypes used for estimation are standardized residuals after adjusting age, sex, age2, age × sex (i.e., interaction between age and sex), and age2 × sex. We also estimated heritability adjusting for smoking by including pack-years and current smoking as covariates. No obvious outliers (larger than ±6 standardized residuals) were found. We found substantial differences in the AA and NHW samples. On average, individuals in the AA sample were younger than in the NHW sample, and the AA sample contained more current smokers but fewer COPD cases. Consistent with the larger fraction of control subjects in the AA sample, the mean values of the spirometric phenotypes in the AA sample were higher than the NHW sample, and the mean of the CT scan emphysema and gas trapping phenotypes was lower. We address this point in comparing the estimated heritabilities from the two racial groups.
TABLE 1.
DESCRIPTIVE STATISTICS OF THE STUDY POPULATION USED FOR HERITABILITY ESTIMATION
| Variable | Non-Hispanic White Mean ± SD (Sample Size) | African American Mean ± SD (Sample Size) |
|---|---|---|
| Age, yr |
61.97 ± 8.79 |
54.45 ± 7.08 |
| (n = 6,415) |
(n = 2,792) |
|
| Male, % |
52.27% |
56.20% |
| (n = 6,415) |
(n = 2,792) |
|
| Body mass index |
28.69 ± 6.09 |
29.09 ± 6.71 |
| (n = 6,415) |
(n = 2,792) |
|
| COPD cases, % |
47.32% |
31.78% |
| (n = 5,354) |
(n = 2,152) |
|
| Current smokers, % |
39.52% |
81.12% |
| (n = 6,415) |
(n = 2,792) |
|
| Pack-years of smoking |
47.59 ± 26.14 |
38.41 ± 21.87 |
| (n = 6,415) |
(n = 2,792) |
|
| Height, cm |
169.90 ± 9.43 |
171.33 ± 9.67 |
| (n = 5,978) |
(n = 2,792) |
|
| FEV1, % predicted (post-bronchodilator) |
72.99 ± 26.03 |
81.57 ± 23.64 |
| (n = 6,398) |
(n = 2,756) |
|
| FEV1/FVC (post-bronchodilator) |
0.64 ± 0.17 |
0.72 ± 0.14 |
| (n = 6,398) |
(n = 2,756) |
|
| % Emphysema |
7.46 ± 10.42 |
3.72 ± 7.44 |
| (n = 5,777) |
(n = 2,514) |
|
| % Gas trapping | 24.51 ± 20.60 |
16.89 ± 17.49 |
| (n = 5,385) | (n = 2,192) |
Definition of abbreviation: COPD = chronic obstructive pulmonary disease.
The results of heritability estimation are displayed in Table 2. We estimated the heritability of FEV1 to be 38.4% in the NHW population and 50.9% in the AA population. For log percent emphysema and log percent gas trapping, the estimated heritabilities are lower and have higher standard errors compared with FEV1 and FEV1/FVC. Among the five phenotypes (COPD disease status, and four COPD-related phenotypes) the ranking of the heritability estimates are the same between two populations (i.e., FEV1 is the most heritable phenotype and percent of gas trapping is the least heritable). Heritability estimates in the AA population were generally higher than the NHW population. FEV1 shows the greatest difference, with an increase from 38.4% in the NHW population to 50.9% in the AA population. To determine whether heritability estimates for well-studied anthropometric characteristics were consistent with other reports, we estimated the heritability of height and body mass index in our cohort. The heritability of height in our cohort was about 60% in NHW, similar to prior estimates (12). The heritability estimates for body mass index were much lower, 18.7% in NHW and 29% in AA, which are also similar to estimates from three combined population-based GWAS (18).
TABLE 2.
HERITABILITY ESTIMATES
| Phenotypes | NHW |
AA |
Wald P Value* | ||
|---|---|---|---|---|---|
| N | Heritability (SE) | N | Heritability (SE) | ||
| FEV1, % predicted† |
6,128 |
38.4% (5.7%)‡ |
2,756 |
50.9% (13.5%)‡ |
0.395 |
| FEV1/FVC† |
6,128 |
37.3% (5.7%)‡ |
2,756 |
46.6% (13.9%)‡ |
0.538 |
| Log of % emphysema |
5,777 |
28.2% (6.0%)‡ |
2,514 |
31.3% (14.5%)‡ |
0.849 |
| Log of % gas trapping |
5,385 |
24.0% (6.4%)‡ |
2,192 |
27.9% (14.9%)‡ |
0.808 |
| COPD disease status† |
4,929 |
37.7% (7.4%)‡ |
2,152 |
37.9% (20.4%) |
0.992 |
| Body mass index |
6,145 |
18.7% (6.0%) |
2,792 |
29.0% (12.8%) |
0.469 |
| Height | 6,145 | 59.4% (5.6%)‡ | 2,792 | 45.5% (13.5%)‡ | 0.341 |
Definition of abbreviations: AA = African American; COPD = chronic obstructive pulmonary disease; NHW = non-Hispanic white.
Wald test for the comparison of heritability estimation between NHW and AA.
After ascertainment adjustment, heritability of FEV1 is still 38.4% and FEV1/FVC is 37.0% in NHW. In the AA population the adjusted heritability of FEV1 is 51.0% and FEV1/FVC is 49.4%.
LRT P value for testing heritability different from zero passed threshold 0.05.
Using the likelihood ratio test, we tested whether each estimated heritability estimate is significantly different from zero. In the NHW population, genetic components of all phenotypes except for body mass index were significantly different from zero; however, in the smaller AA population both body mass index and COPD disease status did not have statistically significant genetic components. Although the heritability estimates were generally higher in the AA, based on a Wald test we found the heritability estimates between NHW and AA were not statistically significantly different from each other.
Our results adjusting for current smoking and pack-years of smoking are shown in Table E1 in the online supplement. The effect of adjustment is negligible for the imaging phenotypes in AA. In both populations, COPD disease status and spirometric phenotypes showed around a 5% drop in heritability after smoking adjustment, although this drop is well within the standard errors. Therefore, for the remainder of the paper we use estimates unadjusted for smoking variables.
Because our sample is enriched for COPD cases, defined by FEV1 less than 80% predicted and FEV1/FVC less than 0.7, we adjusted the estimates of FEV1 and FEV1/FVC for ascertainment. In both populations ascertainment adjustment had little effect on heritability estimates. The heritability of COPD disease status was estimated to be 37.7% under a liability scale assuming 10% prevalence in the NHW population and 37.9% in the AA population assuming the same prevalence, with higher prevalence assumptions leading to higher heritability estimates.
Genome Partitioning of the Heritability
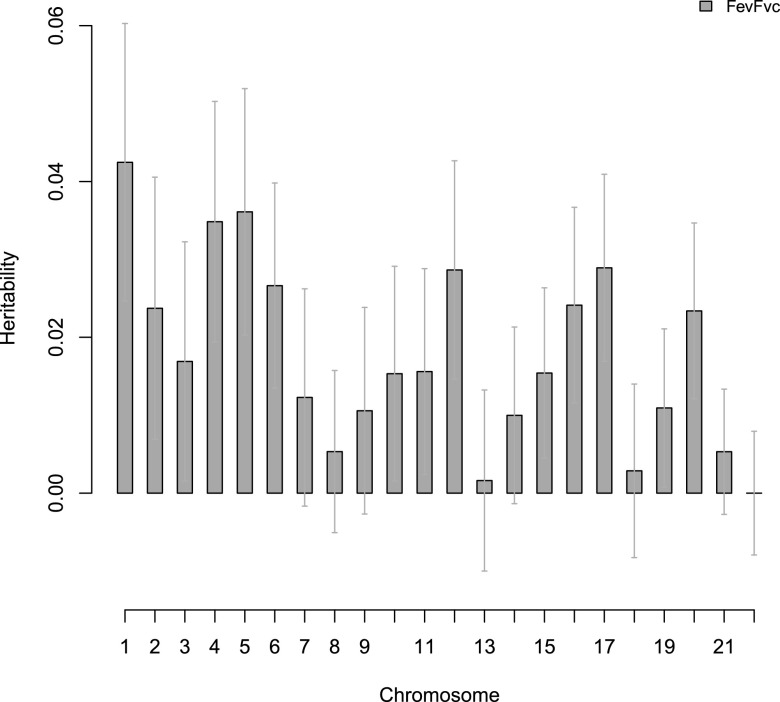
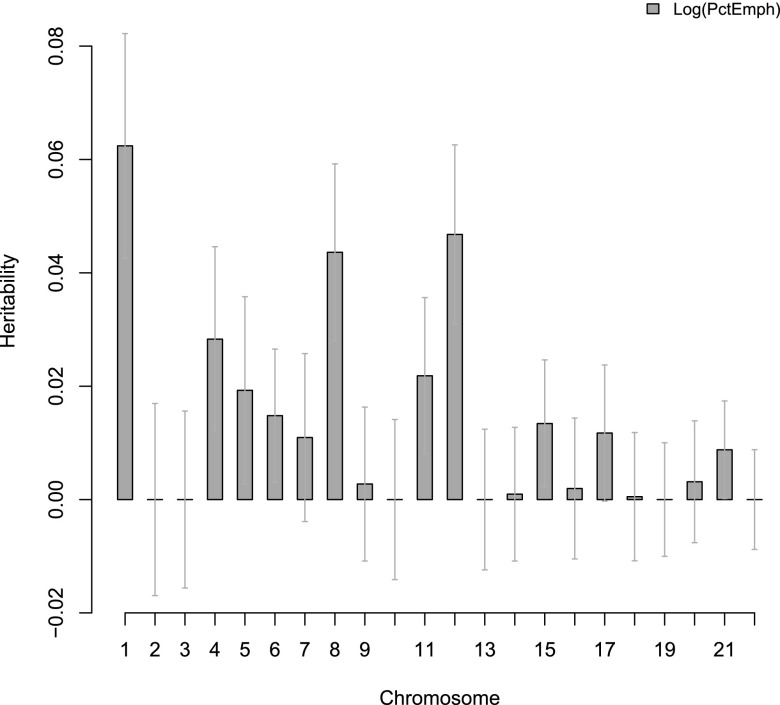
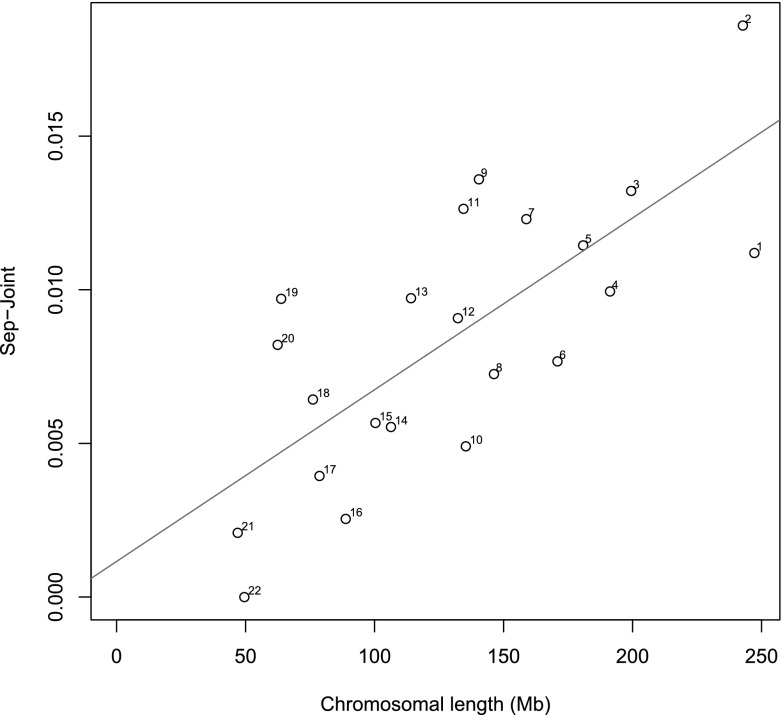
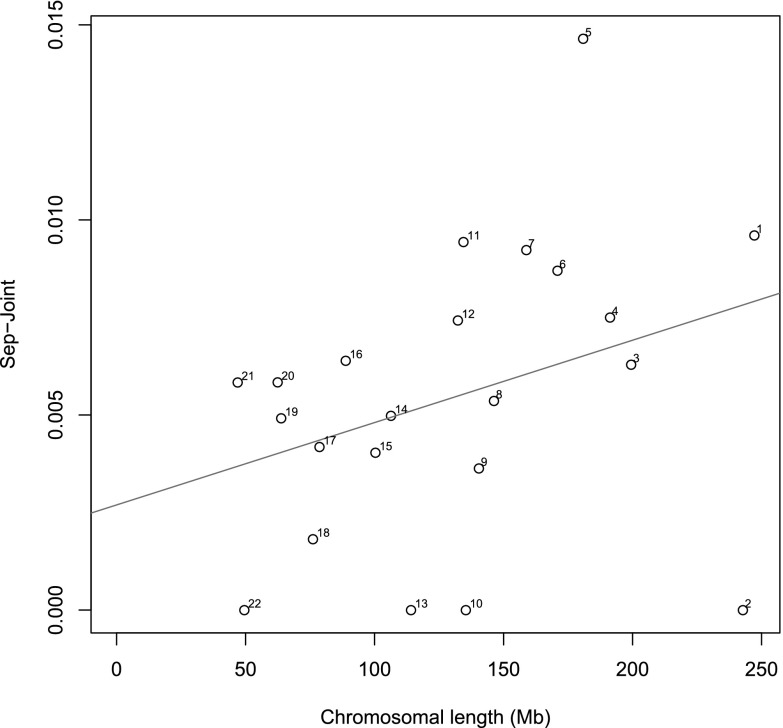
Chromosomal heritabilities (Figures 1 and 2) of both phenotypes significantly correlate with chromosomal length. Although chromosome 1 has the highest heritability for both phenotypes, the standard errors were very large. We found that the proportion of estimated heritability attributed to population structure across the whole genome is minimal: 0.88% and 0.56% for FEV1/FVC and percent emphysema, respectively (Figures 3 and 4).
Figure 1.
Chromosomal partition of phenotypes FEV1/FVC in non-Hispanic white sample. Error bars are the standard error for each estimate.
Figure 2.
Chromosomal partition of percent of emphysema in non-Hispanic white sample. Error bars are the standard error for each estimate.
Figure 3.
Variance caused by cryptic relatedness and population stratification. Shown is the difference between the estimates of variance explained by each chromosome by the separate (Sep) and joint analyses for FEV1/FVC. Numbers next to each dot are the chromosome numbers. Straight line is the regression line (difference between analysis separately and jointly regressed on chromosomal length.)
Figure 4.
Variance caused by cryptic relatedness and population stratification. Shown is the difference between the estimates of variance explained by each chromosome by the separate (Sep) and joint analyses for percent of emphysema. Numbers next to each dot are the chromosome numbers. Straight line is the regression line (difference between analysis separately and jointly regressed on chromosomal length.)
Comparison with Known COPD-associated Variants in NHW Population
To quantify the effect of known COPD-associated variants on the results, we considered four SNPs that have previously been shown to be associated with COPD in NHW: (1) rs7671167 in FAM13A on chromosome 4; (2) rs13180 at the chromosome 15 locus (IREB2/CHRNA3/CHRNA5); (3) rs7937 at the chromosome 19 locus (RAB4B/CYP2A6); and (4) rs7655625 near HHIP on chromosome 4 (r2 = 0.96 with rs13118928). We used these SNPs as covariates in the model when estimating heritability of FEV1. In the NHW population, when compared with the results without including the known associated variants, the overall estimated heritability of FEV1 dropped from 38.4–35.4% indicating 7.8% (3%/38.4%) of the original heritability can be explained by these four SNPs, which is in line with an estimate of approximately 5–10% of variance explained by the previous GWAS (7). With the liability scale, heritability of COPD disease status decreases to 34.2% from 37.7% when the four GWAS variants are included in the model.
Estimated Genetic Correlation Coefficients
In both populations, genetic correlations were all greater than 0.5 (in magnitude) suggesting considerable overlap of the genetic content between the four major COPD-related phenotypes that were analyzed (Table 3). Consistent with intuition, genetic content of lung function and CT scan phenotypes were negatively correlated. Interestingly, although heritability estimates vary between the two racial groups, genetic covariance estimates are similar across populations (Table 4).
TABLE 3.
GENETIC CORRELATION COEFFICIENT ESTIMATES FROM NON-HISPANIC WHITE (ABOVE THE MAIN DIAGONAL) AND AFRICAN AMERICAN (BELOW THE MAIN DIAGONAL) SAMPLES
| FEV1 | FEV1/FVC | Log of % Emphysema | Log of % Gas Trapping | |
|---|---|---|---|---|
| FEV1 |
— |
0.889 |
−0.626 |
−0.844 |
| FEV1/FVC |
0.797 |
— |
−0.818 |
−0.877 |
| Log of % emphysema |
−0.573 |
−0.725 |
— |
0.903 |
| Log of % gas trapping | −0.855 | −0.696 | 0.814 | — |
TABLE 4.
GENETIC COVARIANCE ESTIMATES FROM NON-HISPANIC WHITE (ABOVE THE MAIN DIAGONAL) AND AFRICAN AMERICAN (BELOW THE MAIN DIAGONAL) SAMPLES
| FEV1 | FEV1/FVC | Log of % Emphysema | Log of % Gas Trapping | |
|---|---|---|---|---|
| FEV1 |
— |
0.336 (0.052) |
−0.205 (0.044) |
−0.247 (0.047) |
| FEV1/FVC |
0.394 (0.122) |
— |
−0.264 (0.048) |
−0.261 (0.049) |
| Log of % emphysema |
0.233 (0.106) |
0.260 (0.115) |
— |
0.245 (0.053) |
| Log of % gas trapping | −0.293 (0.109) | −0.253 (0.115) | 0.245 (0.122) | — |
SE is given in parentheses.
Discussion
Issues of missing heritability have been widely discussed in light of the fact that the many genetic variants discovered by GWAS for complex traits can only explain a small portion of the total estimated heritability (27, 28). One of the explanations is that some causal variants have such small effects that they cannot pass the stringent significance threshold used in GWAS. Therefore, variation that is based only on significant SNPs cannot explain all of the heritability. Yang and coworkers (12) developed a method to estimate heritability using all SNPs within a GWAS. In this paper we estimated heritability based on their method in a population of smokers from the COPDGene study, and we also estimated coheritability for COPD status and four major phenotypes. To our knowledge, these are the first estimates of heritability and coheritability for COPD or lung function–related phenotypes using a population-based dataset.
In our GWAS cohort, spirometric phenotypes have higher heritability estimates than chest CT scan phenotypes in NHW and AA samples. There are several possible explanations for the slightly lower heritability estimates of imaging phenotypes compared with spirometric phenotypes. It is possible that the specific imaging phenotypes that we analyzed have higher measurement errors because of technical issues. It is also possible that the imaging phenotypes have lower genetic contributions than spirometric phenotypes. Assessment of additional CT imaging phenotypes is required to resolve this issue (29). Heritability estimates are generally higher in AA sample than in NHW sample, especially spirometry phenotypes, although the differences in the heritability estimates were not statistically significant. Although we cannot rule out the possibility that there is a greater contribution from common genetic variants to these phenotypes in AA, we have low power to detect this because of the small sample size of the AA.
Generally, the previously reported COPD GWAS SNPs account for approximately 8% of the heritability in the NHW population. It was of interest that results for FEV1/FVC were very similar to FEV1 for both populations, because more GWAS associations have been found for FEV1/FVC than for FEV1 in general population samples (30).
Our basic approach to estimation of heritability assumes that the phenotypes are polygenic (i.e., there are numerous causal genetic variants spread throughout the genome). This explains the dramatic difference between our estimates and the heritability attributed to a small number of SNPs identified as statistically significant in GWAS studies. This is likely because many associated SNPs have either smaller effect size or low minor allele frequencies that typical GWAS studies have been unable to detect. The current analysis indicates that there are considerable genetic components for five major COPD-related phenotypes, and between phenotypes there is substantial genetic overlap. Of interest, chest CT scan phenotypes, which have not been explored before, were shown to have a modest, but statistically significant genetic component. Our estimates of heritability for spirometric phenotypes are consistent with results from previous family-based studies in the literature. Further analysis with denser genetic content can be performed to search for the location of causal variants underlying complex traits like COPD.
Acknowledgments
COPDGene Investigators—Core Units:
Administrative Core: James Crapo, M.D. (PI), Edwin Silverman, M.D., Ph.D. (PI), Barry Make, M.D., Elizabeth Regan, M.D., Ph.D., Rochelle Lantz, Lori Stepp, and Sandra Melanson.
Genetic Analysis Core: Terri Beaty, Ph.D., Barbara Klanderman, Ph.D., Nan Laird, Ph.D., Christoph Lange, Ph.D., Michael Cho, M.D., Stephanie Santorico, Ph.D., John Hokanson, M.P.H., Ph.D., Dawn DeMeo, M.D., M.P.H., Nadia Hansel, M.D., M.P.H., Craig Hersh, M.D., M.P.H., Peter Castaldi, M.D., M.Sc., Merry-Lynn McDonald, Ph.D., Jing Zhou, M.D., Ph.D., Manuel Mattheissen, M.D., Ph.D., Emily Wan, M.D., Megan Hardin, M.D., Jacqueline Hetmanski, M.S., Margaret Parker, M.S., and Tanda Murray, M.S.
Imaging Core: David Lynch, M.B., Joyce Schroeder, M.D., John Newell, Jr., M.D., John Reilly, M.D., Harvey Coxson, Ph.D., Philip Judy, Ph.D., Eric Hoffman, Ph.D., George Washko, M.D., Raul San Jose Estepar, Ph.D., James Ross, M.Sc., Mustafa Al Qaisi, M.D., Jordan Zach, Alex Kluiber, Jered Sieren, Tanya Mann, Deanna Richert, Alexander McKenzie, Jaleh Akhavan, and Douglas Stinson.
PFT QA Core, LDS Hospital, Salt Lake City, UT: Robert Jensen, Ph.D.
Biological Repository, Johns Hopkins University, Baltimore, MD: Homayoon Farzadegan, Ph.D., Stacey Meyerer, Shivam Chandan, and Samantha Bragan.
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, Ph.D., Andre Williams, Ph.D., Carla Wilson, M.S., Anna Forssen, M.S., Amber Powell, and Joe Piccoli.
Epidemiology Core, University of Colorado School of Public Health, Denver, CO: John Hokanson, M.P.H., Ph.D., Marci Sontag, Ph.D., Jennifer Black-Shinn, M.P.H., Gregory Kinney, M.P.H., Ph.Dc., and Sharon Lutz, M.P.H., Ph.D.
COPDGene Investigators—Clinical Centers:
Ann Arbor VA, Ann Arbor, MI: Jeffrey Curtis, M.D. and Ella Kazerooni, M.D.
Baylor College of Medicine, Houston, TX: Nicola Hanania, M.D., M.S., Philip Alapat, M.D., Venkata Bandi, M.D., Kalpalatha Guntupalli, M.D., Elizabeth Guy, M.D., Antara Mallampalli, M.D., Charles Trinh, M.D., Mustafa Atik, M.D., Hasan Al-Azzawi, M.D., Marc Willis, D.O., Susan Pinero, M.D., Linda Fahr, M.D., Arun Nachiappan, M.D., Collin Bray, M.D., L. Alexander Frigini, M.D., Carlos Farinas, M.D., David Katz, M.D., Jose Freytes, M.D., and Anne Marie Marciel, M.D.
Brigham and Women’s Hospital, Boston, MA: Dawn DeMeo, M.D., M.P.H., Craig Hersh, M.D., M.P.H., George Washko, M.D., Francine Jacobson, M.D., M.P.H., Hiroto Hatabu, M.D., Ph.D., Peter Clarke, M.D., Ritu Gill, M.D., Andetta Hunsaker, M.D., Beatrice Trotman-Dickenson, M.B.B.S., and Rachna Madan, M.D.
Columbia University, New York, NY: R. Graham Barr, M.D., Dr.P.H., Byron Thomashow, M.D., John Austin, M.D., and Belinda D’Souza, M.D.
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., M.D., Lacey Washington, M.D., and H. Page McAdams, M.D.
Fallon Clinic, Worcester, MA: Richard Rosiello, M.D., Timothy Bresnahan, M.D., Joseph Bradley, M.D., Sharon Kuong, M.D., Steven Meller, M.D., and Suzanne Roland, M.D.
Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, M.D., M.P.H. and Joseph Tashjian, M.D.
Johns Hopkins University, Baltimore, MD: Robert Wise, M.D., Nadia Hansel, M.D., M.P.H., Robert Brown, M.D., Gregory Diette, M.D., and Karen Horton, M.D.
Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Los Angeles, CA: Richard Casaburi, M.D., Janos Porszasz, M.D., Ph.D., Hans Fischer, M.D., Ph.D., Matt Budoff, M.D., and Mehdi Rambod, M.D.
Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, M.D., Charles Trinh, M.D., Hirani Kamal, M.D., Roham Darvishi, M.D., Marc Willis, D.O., Susan Pinero, M.D., Linda Fahr, M.D., Arun Nachiappan, M.D., Collin Bray, M.D., L. Alexander Frigini, M.D., Carlos Farinas, M.D., David Katz, M.D., Jose Freytes, M.D., and Anne Marie Marciel, M.D.
Minneapolis VA, Minneapolis, MN: Dennis Niewoehner, M.D., Quentin Anderson, M.D., Kathryn Rice, M.D., and Audrey Caine, M.D.
Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, M.D., M.S., Gloria Westney, M.D., M.S., and Eugene Berkowitz, M.D., Ph.D.
National Jewish Health, Denver, CO: Russell Bowler, M.D., Ph.D., David Lynch, M.B., Joyce Schroeder, M.D., Valerie Hale, M.D., John Armstrong, II, M.D., Debra Dyer, M.D., Jonathan Chung, M.D., and Christian Cox, M.D.
Temple University, Philadelphia, PA: Gerard Criner, M.D., Victor Kim, M.D., Nathaniel Marchetti, D.O., Aditi Satti, M.D., A. James Mamary, M.D., Robert Steiner, M.D., Chandra Dass, M.D., and Libby Cone, M.D.
University of Alabama, Birmingham, AL: William Bailey, M.D., Mark Dransfield, M.D., Michael Wells, M.D., Surya Bhatt, M.D., Hrudaya Nath, M.D., and Satinder Singh, M.D.
University of California, San Diego, CA: Joe Ramsdell, M.D. and Paul Friedman, M.D.
University of Iowa, Iowa City, IA: Alejandro Cornellas, M.D., John Newell, Jr., M.D., and Edwin JR van Beek, M.D., Ph.D.
University of Michigan, Ann Arbor, MI: Fernando Martinez, M.D., MeiLan Han, M.D., and Ella Kazerooni, M.D.
University of Minnesota, Minneapolis, MN: Christine Wendt, M.D. and Tadashi Allen, M.D.
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, M.D., Joel Weissfeld, M.D., M.P.H., Carl Fuhrman, M.D., Jessica Bon, M.D., and Danielle Hooper, M.D.
University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, M.D., Sandra Adams, M.D., Carlos Orozco, M.D., Mario Ruiz, M.D., Amy Mumbower, M.D., Ariel Kruger, M.D., Carlos Restrepo, M.D., and Michael Lane, M.D.
Footnotes
Supported by Award Numbers U01HL089897, R01HL089897, U01HL089856, R01HL089856, and P01HL105339; K08 HL097029, R01 HL113264 (M.H.C.) from the National Heart, Lung, and Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health. The COPDGene project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, and Sunovion.
Author Contributions: J.J.Z. conducted the analysis and drafted the manuscript. M.H.C., P.J.C., C.P.H., and E.K.S. provided data and revised the manuscript. N.M.L. and E.K.S. supervised the analysis and revised the manuscript.
Originally Published in Press as DOI: 10.1164/rccm.201302-0263OC on August 23, 2013
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O’Donnell WJ, Reilly JJ, Ginns L, Mentzer S, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med. 1998;157:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 2.Stoller JK, Aboussouan LS. Alpha1-antitrypsin deficiency. Lancet. 2005;365:2225–2236. doi: 10.1016/S0140-6736(05)66781-5. [DOI] [PubMed] [Google Scholar]
- 3.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. ICGN Investigators. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, Myers RH, Borecki IB, Silverman EK, Weiss ST, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YM, Chen TH, Barr RG, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, Himes BE, Sylvia JS, Klanderman BJ, Ziniti JP, et al. ICGN Investigators; ECLIPSE Investigators; COPDGene Investigators. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21:947–957. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeMeo DL, Carey VJ, Chapman HA, Reilly JJ, Ginns LC, Speizer FE, Weiss ST, Silverman EK. Familial aggregation of FEF(25-75) and FEF(25-75)/FVC in families with severe, early onset COPD. Thorax. 2004;59:396–400. doi: 10.1136/thx.2003.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb DJ, Wilk JB, Harmon M, Evans JC, Joost O, Levy D, O’Connor GT, Myers RH. Heritability of longitudinal change in lung function. The Framingham Study. Am J Respir Crit Care Med. 2001;164:1655–1659. doi: 10.1164/ajrccm.164.9.2010122. [DOI] [PubMed] [Google Scholar]
- 10.Ho MK, Tyndale RF. Overview of the pharmacogenomics of cigarette smoking. Pharmacogenomics J. 2007;7:81–98. doi: 10.1038/sj.tpj.6500436. [DOI] [PubMed] [Google Scholar]
- 11.Kim DK, Hersh CP, Washko GR, Hokanson JE, Lynch DA, Newell JD, Murphy JR, Crapo JD, Silverman EK COPD Gene Investigators. Epidemiology, radiology, and genetics of nicotine dependence in COPD. Respir Res. 2011;12:9. doi: 10.1186/1465-9921-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 15.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, et al. Diabetes Genetics Initiative; FUSION; KORA; Prostate, Lung Colorectal and Ovarian Cancer Screening Trial; Nurses’ Health Study; SardiNIA. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, et al. Diabetes Genetics Initiative; Wellcome Trust Case Control Consortium; Cambridge GEM Consortium. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stearns FW. One hundred years of pleiotropy: a retrospective. Genetics. 2010;186:767–773. doi: 10.1534/genetics.110.122549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.American Thoracic Society. Standardization of spirometry, 1994 update. Am J Respir Crit Care Med. 1995;152:1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 22.Falconer DS. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann Hum Genet. 1965;29:51–71. [Google Scholar]
- 23.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lange K. New York: Springer; 2002. Mathematical and statistical methods for genetic analysis. [Google Scholar]
- 25.Korte A, Vilhjálmsson BJ, Segura V, Platt A, Long Q, Nordborg M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat Genet. 2012;44:1066–1071. doi: 10.1038/ng.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange K, Papp JC, Sinsheimer JS, Sripracha R, Zhou H, Sobel EM. Mendel: the Swiss army knife of genetic analysis programs. Bioinformatics. 2013;29:1568–1570. doi: 10.1093/bioinformatics/btt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 28.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Sluis S, Verhage M, Posthuma D, Dolan CV. Phenotypic complexity, measurement bias, and poor phenotypic resolution contribute to the missing heritability problem in genetic association studies. PLoS ONE. 2010;5:e13929. doi: 10.1371/journal.pone.0013929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, Zhai G, Zhao JH, Smith AV, Huffman JE, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]