Abstract
The cysteine cathepsins are a family of proteases that play important roles in both normal cellular physiology and many human diseases. In cancer, the activity of many of the cysteine cathepsins is upregulated and can be exploited for tumor imaging. Here we present the design and synthesis of a new class of quenched fluorescent activity-based probes (qABPs) containing a phenoxymethyl ketone (PMK) electrophile. These reagents show enhanced in vivo properties and broad reactivity resulting in dramatically improved labeling and tumor imaging properties compared to previously reported ABPs.
INTRODUCTION
The cysteine cathepsins are a family of proteases that play important roles in health and disease.1 Although their function has mainly been described as being confined to the endosomal pathway, evidence is accumulating they are major regulators of matrix degradation and cell motility, suggesting that they also function in an extracellular context.2 Members of the cysteine cathepsin family have also been shown to be major players in the development and progression of several types of cancer.3, 4 Furthermore, changes in the expression of the endogenous inhibitors of the cathepsins, the cystatins, take place during cancer progression.5 These observations, in combination with the dynamic nature of the intra- and extracellular milieu, stress the importance of tools that allow the direct assessment of the activity of these proteases in the context of a native tumor microenvironment. Activity-based probes (ABPs) are small molecule tools that allow dynamic monitoring of protease activity. These reagents form activity-dependent covalent bonds with protease active site nucleophiles, thereby providing a readout of the levels of active protease in a cell, tissue or even whole organism.6 Several classes of ABPs targeting the cysteine cathepsin family have been reported previously. 7 In particular, fluorescently quenched ABPs (qABPs) have proven to be powerful tools for noninvasive optical imaging of cancer and subsequent characterization of the target cathepsins on a histological, cellular and protein level.8, 9
In this work we set out to develop a qABP with overall improved in vivo tumor imaging properties compared to the existing qABPs. We therefore decided to optimize three major elements of the probe, the quencher, the linker and the electrophilic “warhead”. One of the biggest drawbacks of the cysteine cathepsin qABPs reported to date is their relatively poor aqueous solubility. Therefore we introduced sulfonate groups into the QSY21 quencher10 in order to improve the water solubility and thereby the bio-distribution of the probe. We also varied the length of the spacer tethering the electrophile and the quencher in order to decrease the lipophilicity of the qABP. Finally, we explored a new electrophile in order to increase the range of possible cathepsin targets. Since several members of the cysteine cathepsin family are upregulated in a variety of cancers, mainly but not exclusively due to the infiltration of immune cells with high cysteine cathepsin expression, 3 we expected a brighter fluorescence signal in tumors as the result of probes that target a broad spectrum of cysteine cathepsin activities. In order to obtain a more pan-reactive probe we aimed to decrease the size and increase the reactivity of the electrophile. All qABPs reported to date are based on the acyloxymethyl ketone (AOMK) electrophile,8, 9, 11 with the 2,6-dimethylbenzoic acid derived AOMK being the most optimal in vivo. This is attributed to the steric protection of the ester bond from cleavage by esterases in the serum. We have previously shown that inhibitors armed with a 2,3,5,6-tetrafluoro substituted phenoxymethyl ketone (PMK) electrophile had a greater reactivity for cysteine dipeptidyl aminopeptidases compared to their 2,6-dimethylbenzoic acid derived acyloxymethyl ketone (AOMK) counterparts.12 The introduction of an additional electron withdrawing amide functionality on the para-position of the 2,3,5,6-tetrafluoro PMK to form the tether between the electrophile and the quencher would further increase the reactivity of the warhead. The smaller size of the PMK relative to the previously reported 2,6-dimethylbenzoic acid derived AOMKs could also increase pan-reactivity since the binding grooves of some of the cysteine cathepsins are sterically restricted.8, 13, 14 Furthermore, we expect the phenol ether to be more stable in vivo compared to the AOMK electrophile which contains an ester linkage that can be degraded by esterases.
RESULTS AND DISCUSSION
Probe synthesis
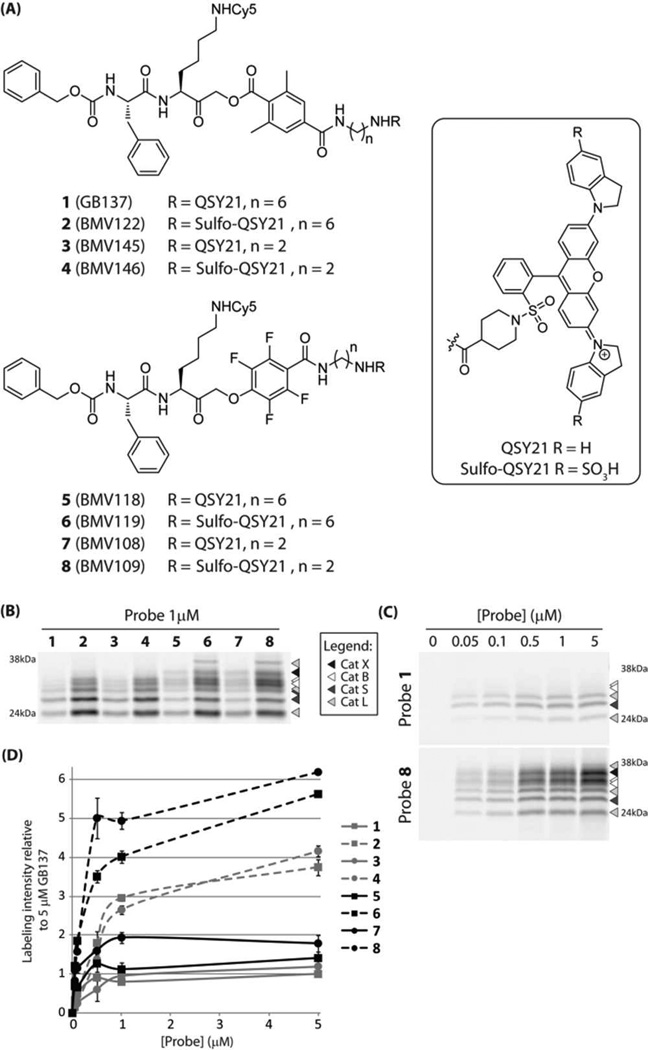
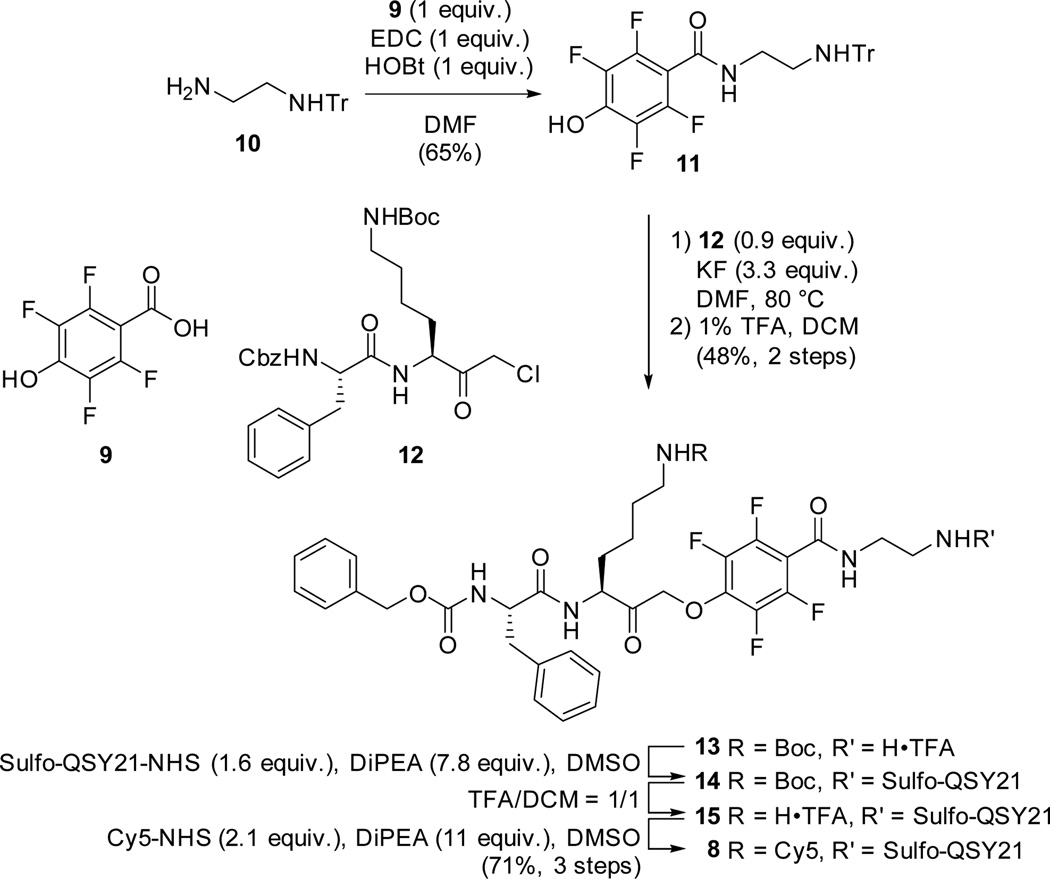
As a starting point for this study we synthesized 7 analogs (2 – 8) of the previously reported qABP GB137 (1)8 (Figure 1a). These compounds represent all combinations of the two electrophiles (AOMK and PMK), two quenchers (QSY21 and sulfo-QSY21) and two linker lengths (hexyl and ethyl). The AOMK probes were synthesized using an optimized, solution chemistry based procedure as described in the Supporting Information (Scheme S1). The PMK probe synthesis commenced with the commercially available 2,3,5,6-tetrafluoro-4-hydroxybenzoic acid (9). The synthesis of qABP 8 is depicted in Scheme 1. After condensation of mono-trityl ethylenediamine (10), phenol 11 was reacted with chloromethyl ketone 1213 at 80 °C under the influence of potassium fluoride in DMF. Heating proved necessary to substitute the chloride with the very deactivated phenolate, since no reaction could be observed at room temperature. Subsequent mild acidic cleavage of the trityl protecting group enabled the coupling of the water soluble quencher sulfo-QSY21 to give intermediate 14. After acidic removal of the Boc protecting group the Cy5 dye could be introduced to give the PMK qABP 8 in a good overall yield.
Figure 1.
(a) Structures of the qABPs based on GB137 containing the AOMK group (1–4) and the new probes containing the PMK group (5 – 8). (b) Labeling profile of probes 1 – 8 in intact, live RAW cells at 1µM. (c) Concentration dependent labeling by probes 1 and 8 in live RAW cells. d) Total cathepsin labeling intensity of probes 1 – 8 in live RAW cells relative to 5µM GB137 (1).
Scheme 1.
Synthesis of PMK qABP 8.
In vitro comparison of qABPs 1–8
We initially tested the specificity and potency of the probes by labeling intact RAW 264.7 cells (mouse leukemic monocyte macrophage cell line) (Figure 1b). We observed several trends in the properties of the probes. All of the sulfo-QSY21 functionalized qABPs (2, 4, 6 and 8) showed stronger overall cathepsin labeling compared to the more hydrophobic QSY21 containing probes (1, 3, 5 and 7). Interestingly, the change in the spacer length from a hexyl to an ethyl linker did not have a dramatic influence on the labeling profile. Perhaps the most striking observation was that the qABPs with the PMK electrophile showed a broader cysteine cathepsin labeling profile compared to their AOMK counterparts. Probes 5 – 8 showed robust cathepsin X labelling. Moreover, Sulfo-QSY21 functionalized PMK probes 6 and 8 were able to label a higher molecular weight proform of cathepsin L. The identities of the fluorescently labeled cathepsins were determined by immunoprecipitation (Figure S1a). Upon performing titration labeling experiments in live RAW cells, we observed several other interesting trends. The most hydrophobic qABPs (1, 3, 5 and 7) showed only low level labeling that reached a maximum intensity at 0.5 – 1µM (Figure 1c,d and Figure S1b,c), suggesting that their reduced water solubility results in precipitation of the probes at the higher concentrations. Now it also became apparent that the shorter spacerlength seems to be beneficial, with all probes carrying the ethyl spacer producing brighter labeling signals at 5 µM compared to their hexyl containing counterparts. When comparing the AOMKs and the PMKs, a clear difference in selectivity was observed. The AOMK qABPs preferentially labeled cathepsins S and L and only at higher concentrations labeled cathepsin B. Surprisingly, the AOMK qABPs 2 – 4 seem to be capable of labeling cathepsin X even though prior studies showed that several other related AOMKs are incapable of labeling this target (Figure S1b).14 The PMK qABPs labeled all target cysteine cathepsins with equal intensity, even at the lower probe concentrations, suggesting that they are more sensitive than their AOMK counterparts. Together, these experiments suggest that increased hydrophilicity improves labeling intensity and that the novel PMK qABPs have a broader, more pan-cysteine cathepsin reactivity.
Further in vitro characterization of qABP 8
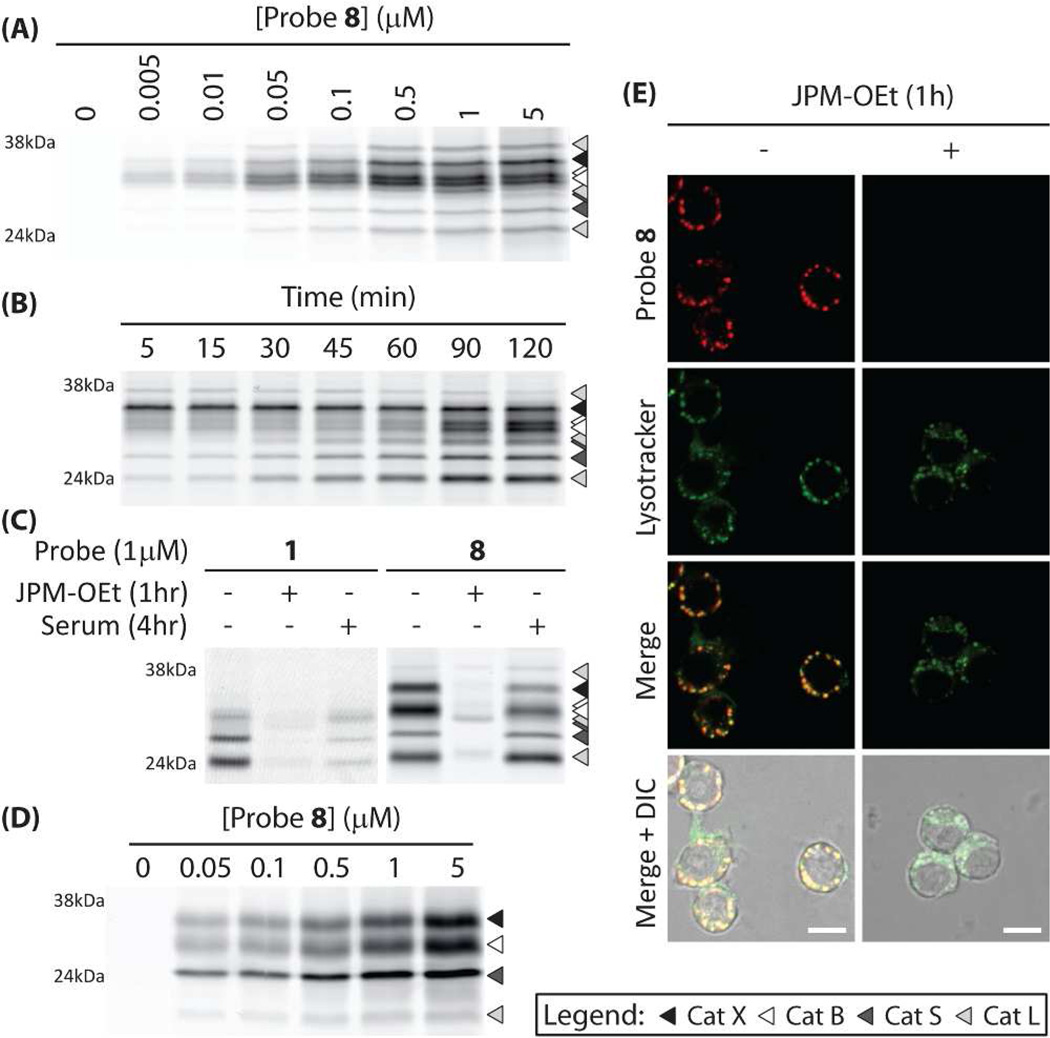
Because the PMK qABP 8 was the most optimal in terms of labeling intensity and broad cathepsin reactivity, we decided to proceed with this probe for further characterization studies. To further define target selectivity, we labeled RAW cell lysates with increasing concentrations of qABP 8 at pH 5.5. These results indicated that the probe has similar potency towards the targeted cysteine cathepsins (B, S, L, X) with labeling observed at concentrations as low as 5 nM. The labeling of all of the targets was saturated by 500 nM of the probe (Figure 2a). When the probe was used for a timecourse labeling of live RAW cells at the set concentration of 500 nM, we observed rapid saturation of cathepsin X followed by more slow labeling of cathepsin S, L and B with cathepsin B labeling signal increasing even up to 120 min (Figure 2b). These data suggest that the probe is likely able to access pools of cathepsin X most rapidly, perhaps due to its localization within or on the surface of the cells. It also suggests that cathepsins B and X may be in alternate locations in the cells, which can be accessed by the probe to different extents. When a similar timecourse labeling experiment of live RAW cells was performed at a probe concentration of 1 µM, labeling intensity close to saturation was reached as fast as 5 minutes after exposure to the probe (Figure S1d). This indicates that probe 8 shows very fast cell internalization and labeling kinetics. In order to test the serum stability of the new PMK probe, we examined the effects of serum exposure on labeling in RAW cells (Figure 2c). Whereas 4 hours of serum pre-exposure to the original AOMK probe 1 resulted in a loss of nearly 70% of target labeling, more than 80% of the labeling was retained for the PMK qABP 8. This may be due to enhanced stability of the PMK over the AOMK towards esterases in the serum. Pre-treatment of the cells with the cysteine cathepsin inhibitor JPM-OEt blocked more than 90% of this labeling. Exposure of primary human monocyte derived macrophages to probe 8 revealed that the pan-reactivity of the PMK qABPs was retained for the human enzymes (Figure 2d). Given the improved labeling properties of the PMK probe, we next performed live cell fluorescence microscopy studies. These results confirmed that the probe produced bright and specific labeling signals and that the majority of the probe labeled cathepsins reside in lysosomes (Figure 2e).
Figure 2.
a) Concentration dependent labeling of RAW cell lysates by probe 8 at pH 5.5. b) Labeling time course of 0.5µM probe 8 in intact, live RAW cells. c) Inhibition of labeling of probes 1 and 8 in living RAW cells by pretreatment with JPM-OEt (50µM) and serum stability. d) Labeling profile of probe 8 in living primary human monocyte derived macrophages. e) Live cell fluorescence microscopy of RAW cells exposed to 1µM probe 8 (Red) and lysotracker (Green, scale bar 10µm).
Noninvasive optical imaging of syngeneic orthotopic mouse breast tumors
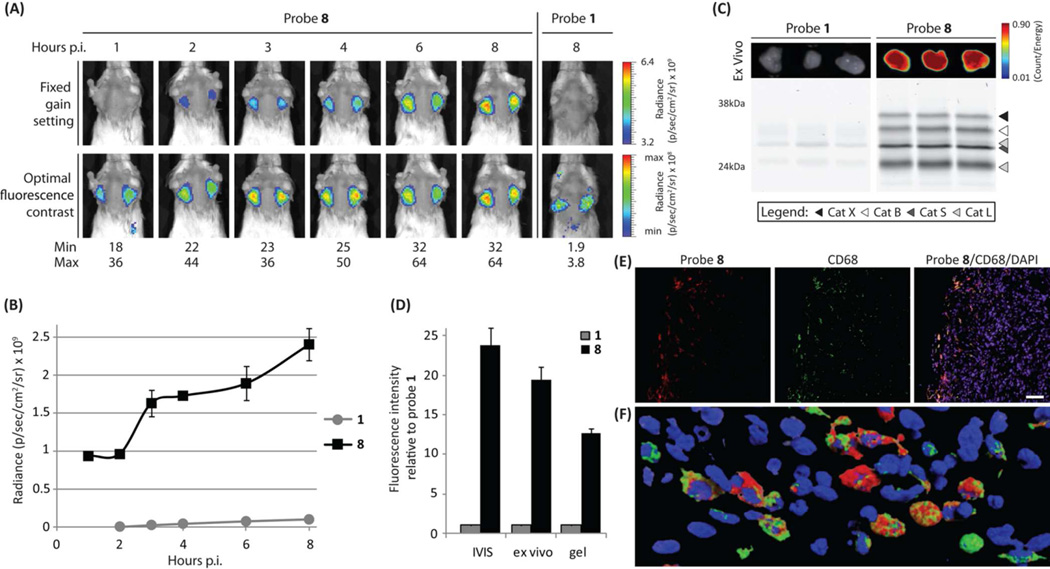
Given the positive live cell labeling properties of the new PMK electrophile, we tested the best performing qABPs 2, 6 and 8 in a syngeneic orthotopic mouse model of breast cancer.15 In addition, we compared these probes to the original AOMK probe 1 (Figure 3 and Figure S2). 4T1 cells were implanted in the number 2 and 7 mammary fat pads of Balb/c mice and tumor growth was monitored. When tumors were established, the mice were injected with equimolar amounts of the qABPs (20 nmol) via tail vein and the Cy5 fluorescence was noninvasively imaged over time (Figure 3 a,b). Again, these results confirmed that the qABP 8 proved to be superior. Robust tumor specific activation of fluorescence with high overall intensity could be observed for probe 8 specifically in the tumor region. Contrary to probe 1, as soon as 1 hour post injection of probe 8, tumor margins were already visible with substantial contrast (Figure 2a and Figure S2c), and signals continued to increase up to the end of the timecourse. Ultimately probe 8 achieved a more than twentyfold enhanced tumor-specific fluorescence signal compared to probe 1. Good tumor-specific signal was also observed for probe 6 and to a lesser extent for probe 2, although both still outcompeted the original AOMK probe by more than tenfold (Figure S2a,b). After the completion of the time course, the tumors were excised and tumor fluorescence was measured ex vivo, followed by homogenization and analysis of the fluorescently labeled proteins by SDS-PAGE (Figure 3c and Figure S2d). The quantification of the ex vivo fluorescence and the total cysteine cathepsin labeling showed a similar trend to that seen in the noninvasive optical imaging studies (Figure 3d and Figure S2e). The labeling profile confirmed that the PMK qABPs are pan-reactive cysteine cathepsin probes in vivo, targeting cathepsin X, B, S and L with similar labeling intensities, contributing to overall brighter tumor fluorescence. We have previously shown that a cathepsin S directed qABP BMV083 mainly targets M2 type tumor-associated macrophages in 4T1 syngeneic orthotopic mouse breast tumors.9 To determine the cellular source of probe 8 fluorescence, we performed immuno-fluorescence staining on tumor tissue sections from probe labeled mice using the macrophage marker CD68 (Figure 3e and Figure S2f). Cy5 fluorescence localized to CD68 positive cells, however, not all CD68 positive cells were probe 8 positive, suggesting different activation states of the tumor-associated macrophages. Probe 8 proves to be able to demarcate the cancer region due to the localization of the probe positive macrophages in the invasive edge of the tumor. More detailed analysis with confocal laser scanning microscopy (CLSM) confirmed that all cells that were positive for probe 8 were CD68 positive (Figure 3f). Taken together, these data confirm that increasing the hydrophilicity of the quencher, shortening of the spacer and the introduction of a more reactive and sterically less restricted nucleophilic trap resulted in a qABP with a broad cysteine cathepsin reactivity and overall improved in vivo properties.
Figure 3.
a) Noninvasive optical imaging time-course of tumor bearing mice injected with probe 8 and 1 (right panels). The lower panels represent the optimal fluorescence contrast at each time point. b) Time-dependent tumor-specific fluorescence (tumor - background) for mice treated with probe 1 or 8 (n = 3; data represent mean values ± standard errors). c) Ex vivo tumor fluorescence (top panel) and in vivo fluorescently labeled proteins after SDS-PAGE visualized by in-gel fluorescence scanning (lower panel). d) Fluorescence intensity at the end point of the noninvasive optical imaging study (shown in a). Ex vivo tumor imaging, and in-gel fluorescence labeling (shown in c). Intensity relative to probe 1 is depicted (n = 3; data represent mean values ± standard errors). e) Fluorescence microscopy of probe 8 (Red) treated tumor tissue section with CD68 immuno-staining (Green) and nuclear staining (DAPI – Blue, scale bar 50µm). f) 3D reconstruction of CLSM of probe 8 (Red) treated tumor tissue section with CD68 immuno-staining (Green) and nuclear staining (DAPI – Blue).
CONCLUSIONS
We have synthesized a novel class of quenched fluorescent activity-based probes bearing a PMK electrophile with greater reactivity and broader selectivity compared to the previously reported AOMK based probes. We have furthermore increased the hydrophilicity of the qABP by introducing a sulfonated quencher and shortening the spacer tethering the electrophile and the quencher. This results in greater aqueous solubility and improved in vivo properties resulting in enhanced specific signal intensity in noninvasive optical imaging of cancer. Although very distinct functions have been described for some of the cysteine cathepsin family members,16 other roles are redundant and alterations in the activity of one cathepsin can influence the activity of others. For example, loss of cathepsin B is compensated by increased activity of cathepsin X17 and upregulation of cathepsin B results in downregulation of cathepsin L.18 Therefore a broad spectrum probe is highly valuable as it facilitates the readout of multiple cysteine cathepsins in one experiment and enables the comparison of the activities of the individual cathepsins with respect to one another. Furthermore, the overall broad reactivity of the probe within the cathepsin family may facilitate diagnostic applications in a wider range of conditions in which cathepsins are known to be key players. In support of this, the usefulness of pan-reactive ABPs has already been demonstrated by the pan-serine hydrolase fluorophosphonate probes19 and the pan-reactive proteasome probe MV151.20 The findings that led to the pan-reactive cysteine cathepsin qABP could aid the development of selective chemical tools to study the cathepsin family. Because the PMK-based qABPs are highly reactive towards cathepsin X, these scaffolds can be used to generate selective qABPs against this still poorly understood cysteine cathepsin.14 The optimization of the qABPs presented in this work is an important step towards the potential clinical translation of these contrast agents. Probe 8 proved to be capable of demarcating tumor margins with substantial contrast as soon as 1 hour after administration. This finding has particular value for applications in fluorescence guided cancer surgery,21 where signal contrast needs to be generated soon after administration of the contrast agent. In addition, topical application of a contrast agent capable of distinguishing the tumor margins would allow further flexibility and utility for contrast agents in tumor surgery. We have previously shown that GB119, an AOMK-based qABP, could be applied topically to aid the resection of tumor tissue during cancer surgery.22 The pan-reactivity that resulted in enhanced fluorescence signal and the ability of probe 8 to penetrate cells and rapidly saturate labeling of target cathepsins are characteristics tailored for a topically applicable contrast agent. Combined these enhanced properties of the PMK containing probes make them important new tools to be added to the current toolbox of optical contrast agents. We are currently working towards the pre-clinical evaluation (i.e. toxicity and GMP production) and testing in human tissues that will be required to initiate clinical studies of the agents in the near future.
Supplementary Material
ACKNOWLEDGMENT
We thank the members of the Bogyo lab for insightful discussions, T. Doyle at the Stanford Small Animal Facility, S. Lynch at the Stanford NMR Facility and A. Chien and T. McLaughlin at the Stanford Mass Spec Facility for their technical assistance. We thank Yaron Carmi for assistance with CLSM.
Funding Sources
This work was supported by NIH grants R01 EB005011 R01 HL116307 (to MB) and the Netherlands Organization for Scientific Research (NWO) Rubicon fellowship (to MV and WL) and NIH Re-entry into Biomedical Sciences Supplement 3R01EB005011-06S1 (to KOB).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Experimental details of the synthesis of probes 1–8 and biological experiments; Cathepsin target identification by immunoprecipitation; All gels; In vivo data for probe 1, 2 and 6; Control images for microscopy. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Reiser J, Adair B, Reinheckel T. J Clin Invest. 2010;120:3421–3431. doi: 10.1172/JCI42918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brömme D, Wilson S. In: Role of Cysteine Cathepsins in Extracellular Proteolysis. Biology of Extracellular Matrix. Parks WC, Mecham RP, editors. Vol. 2. Berlin Heidelberg: Springer; 2011. pp. 23–51. [Google Scholar]
- 3.Mohamed MM, Sloane BF. Nat Rev Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 4.Palermo C, Joyce JA. Trends Pharmacol Sci. 2008;29:22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Cox JL. Front Biosci. 2009;14:463–474. doi: 10.2741/3255. [DOI] [PubMed] [Google Scholar]
- 6.Serim S, Haedke U, Verhelst SH. ChemMedChem. 2012;7:1146–1159. doi: 10.1002/cmdc.201200057. [DOI] [PubMed] [Google Scholar]
- 7.Edgington LE, Verdoes M, Bogyo M. Curr Opin Chem Biol. 2011;15:798–805. doi: 10.1016/j.cbpa.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Nat Chem Biol. 2007;3:668–77. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- 9.Verdoes M, Edgington LE, Scheeren FA, Leyva M, Blum G, Weiskopf K, Bachmann MH, Ellman JA, Bogyo M. Chem Biol. 2012;19:619–628. doi: 10.1016/j.chembiol.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing B, Khanamiryan A, Rao J. J Am Chem Soc. 2005;127:4158–4159. doi: 10.1021/ja042829+. [DOI] [PubMed] [Google Scholar]
- 11.Edgington LE, Verdoes M, Ortega A, Withana NP, Lee J, Syed S, Bachmann MH, Blum G, Bogyo M. J Am Chem Soc. 2013;135:174. doi: 10.1021/ja307083b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deu E, Leyva MJ, Albrow VE, Rice MJ, Ellman JA, Bogyo M. Chem Biol. 2010;17:808–819. doi: 10.1016/j.chembiol.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Nat Chem Biol. 2005;1:203–209. doi: 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- 14.Paulick MG, Bogyo M. ACS Chem Biol. 2011;6:563–572. doi: 10.1021/cb100392r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lelekakis M, Moseley JM, Martin TJ, Hards D, Williams E, Ho P, Lowen D, Javni J, Miller FR, Slavin J, Anderson RL. Clin Exp Metastasis. 1999;17:163–170. doi: 10.1023/a:1006689719505. [DOI] [PubMed] [Google Scholar]
- 16.Conus S, Simon HU. Swiss Med Wkly. 2010;140:w13042. doi: 10.4414/smw.2010.13042. [DOI] [PubMed] [Google Scholar]
- 17.Sevenich L, Schurigt U, Sachse K, Gajda M, Werner F, Müller S, Vasiljeva O, Schwinde A, Klemm N, Deussing J, Peters C, Reinheckel T. Proc Natl Acad Sci U S A. 2010;107:2497–2502. doi: 10.1073/pnas.0907240107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopinathan A, Denicola GM, Frese KK, Cook N, Karreth FA, Mayerle J, Lerch MM, Reinheckel T, Tuveson DA. Gut. 2012;61:877. doi: 10.1136/gutjnl-2011-300850. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Patricelli MP, Cravatt BF. Proc Natl Acad Sci U S A. 1999;96:14694. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verdoes M, Florea BI, Menendez-Benito V, Maynard CJ, Witte MD, van der Linden WA, van den Nieuwendijk AM, Hofmann T, Berkers CR, van Leeuwen FW, Groothuis TA, Leeuwenburgh MA, Ovaa H, Neefjes JJ, Filippov DV, van der Marel GA, Dantuma NP, Overkleeft HS. Chem Biol. 2006;13:1217. doi: 10.1016/j.chembiol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Keereweer S, Hutteman M, Kerrebijn JD, van de Velde CJ, Vahrmeijer AL, Löwik CW. Curr Pharm Biotechnol. 2012;13:498. doi: 10.2174/138920112799436294. [DOI] [PubMed] [Google Scholar]
- 22.Cutter JL, Cohen NT, Wang J, Sloan AE, Cohen AR, Panneerselvam A, Schluchter M, Blum G, Bogyo M, Basilion JP. PLoS One. 2012;7:e33060. doi: 10.1371/journal.pone.0033060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.