Abstract
In the era of functional genomics, the role of transcription factor (TF)–DNA binding affinity is of increasing interest: for example, it has recently been proposed that low-affinity genomic binding events, though frequent, are functionally irrelevant. Here, we investigate the role of binding site affinity in the transcriptional interpretation of Hedgehog (Hh) morphogen gradients. We noted that enhancers of several Hh-responsive Drosophila genes have low predicted affinity for Ci, the Gli family TF that transduces Hh signalling in the fly. Contrary to our initial hypothesis, improving the affinity of Ci/Gli sites in enhancers of dpp, wingless and stripe, by transplanting optimal sites from the patched gene, did not result in ectopic responses to Hh signalling. Instead, we found that these enhancers require low-affinity binding sites for normal activation in regions of relatively low signalling. When Ci/Gli sites in these enhancers were altered to improve their binding affinity, we observed patterning defects in the transcriptional response that are consistent with a switch from Ci-mediated activation to Ci-mediated repression. Synthetic transgenic reporters containing isolated Ci/Gli sites confirmed this finding in imaginal discs. We propose that the requirement for gene activation by Ci in the regions of low-to-moderate Hh signalling results in evolutionary pressure favouring weak binding sites in enhancers of certain Hh target genes.
Keywords: hedgehog signalling, morphogen gradient, enhancers, binding affinity, gene regulation, Ci/Gli
1. Introduction
Enhancers, also known as cis-regulatory elements, are genomic DNA elements in command of the timing, location and levels of gene transcription. These transcriptional regulatory sequences integrate signalling and tissue-specific inputs through binding sites for a myriad of transcription factors (TFs) to specify spatio-temporal patterns of gene expression [1]. Traditionally, enhancers have been identified functionally, in most cases by directly testing the sufficiency of stretches of DNA to drive gene expression in reporter assays. Nowadays, putative enhancers can be mined on a genome-wide basis by biochemical signatures, including histone tail modifications, co-activator binding and DNase accessibility [2,3]. Because hundreds or thousands of chromosomal sites cannot be easily tested for transcriptional activity, some genomic studies accept chromatin signatures associated with enhancer activity as self-validating evidence of enhancer function [4–7]. Another potential biochemical indicator of enhancers is TF or co-activator binding, as assessed on a genome-wide level by ChIP-seq and related techniques [7–12]. These methods have had success in identifying regulatory sequences, although many TF-bound regions do not appear to function as enhancers [13–15]. Other studies use DNA sequence signatures, mainly evolutionary conservation and/or clustering of predicted TF binding motifs, to screen genomes for enhancers [16–19]. These methods have also been successful, although again, they are by no means foolproof: for example, not all functional enhancers show evidence of evolutionary sequence conservation—even if their function is conserved—and conversely, not all highly conserved sequences display regulatory activity [20–23].
Enhancers are increasingly prominent in evolutionary thinking, as they have been shown to be the main agents of morphological diversity during evolution [23–25]. Changes that affect TF binding to enhancers have the potential to modify pleiotropic genes in a tissue-specific manner without compromising the survival of the organism. Sequence alterations such as deletions, insertions and nucleotide substitutions in enhancers have been shown to be responsible for morphological diversity [26]. Because of the complex arrangement of TF binding motifs at enhancers, even tiny changes in regulatory sequences can have significant effects in the transcriptional output by modifying binding affinity, binding site number or changing the spacing between TFs, among many other possible scenarios [1].
Enhancers integrate inputs from different cellular and developmental contexts to produce tissue-specific responses critical during tissue differentiation, proliferation and maintenance. A small number of signalling pathways provide instructive inputs that are used in multiple developmental contexts [27,28]. The highly conserved Hedgehog (Hh) signalling pathway is one of the key regulatory networks mediating cell communication during the development of most animals [29]. The Hh morphogen provides instructive positional information by establishing a signalling gradient that promotes different cell fates at different signal intensities which are interpreted by enhancers that contain binding sites for the effector of the pathway, the TF Cubitus interruptus (Ci). In Drosophila, Hh-receiving cells post-translationally modify Ci, a member of the Gli family of TFs, which activates or represses transcription of key target genes [30]. In the presence of the Hh signal, the activator isoform of Ci (Ci-Act) stimulates transcription of Hh target genes, but in the absence of signalling, a repressor isoform of the same protein (Ci-Rep) inhibits transcription of those genes. Ci recognizes enhancers that contain the same optimal consensus sequence as mammalian Gli factors, GACCACCCA—but, like many other TFs, it can also bind to sequences that deviate from this consensus site [31,32]. Thus, Ci activates or actively represses the transcription of Hh responsive genes depending on the state of signalling.
The Hh signalling gradient has been extensively characterized in the context of the developing wing of Drosophila melanogaster (figure 1a). In the third-instar larval wing imaginal disc, which gives rise to the adult wing, cells in the posterior compartment secrete the Hh morphogen: this signal is received and interpreted by cells of the anterior compartment that express Ci. The short-range Hh signal generates opposing reciprocal gradients of Ci-Act and Ci-Rep (figure 1a) [33,34]. Anterior-compartment cells near the anterior–posterior (A/P) compartment boundary receive maximal levels of Hh signalling and thus form Ci-Act exclusively, hence Hh/Ci regulated enhancers are active: these cells form what we will call the ‘activator zone’. Cells far from the source of Hh do not encounter the ligand and form Ci-Rep only, which represses target enhancers. These cells can be classified into the ‘repressor zone’, which comprises most of the anterior compartment of the wing. Between the activator and repressor zones, there exists a region that receives moderate levels of Hh and produces both Ci-Act and Ci-Rep. We will refer to this region as the ‘mixed zone’. Here, the morphogen response becomes more complex, as Ci binding sites in Hh-responsive enhancers integrate competing inputs with opposing transcriptional functions. How cis-regulatory elements ‘decide’ whether to be active or repressed by Ci in this zone is not well understood, but recent findings [33,35], as well as the results presented here, show that the decision relies in part on the number and sequence of their Ci binding motifs. Bicoid and Dorsal, two morphogens that form signalling gradients during embryogenesis, also regulate key target genes in response to differences in binding site number and affinity [36,37]. However, because of the reciprocal gradients of Ci-Act and Ci-Rep, Hh/Ci-regulated enhancers interpret these differences unconventionally, and drive gene expression in unexpected domains across the gradient [33,35]. A classic response is displayed by the Dorsal target gene twist, which has a proximal enhancer with two low-affinity binding sites that drive limited gene expression in cells with high levels of the morphogen [37]. Improving the affinity of those sites resulted in higher levels of gene expression in a broader domain of the Drosophila embryo [37]. In the case of several Hh/Ci-regulated enhancers, the transcriptional response to changes in affinity is opposite to what is expected from the morphogen gradient model (these observations will be described in more detail below) [38].
Figure 1.
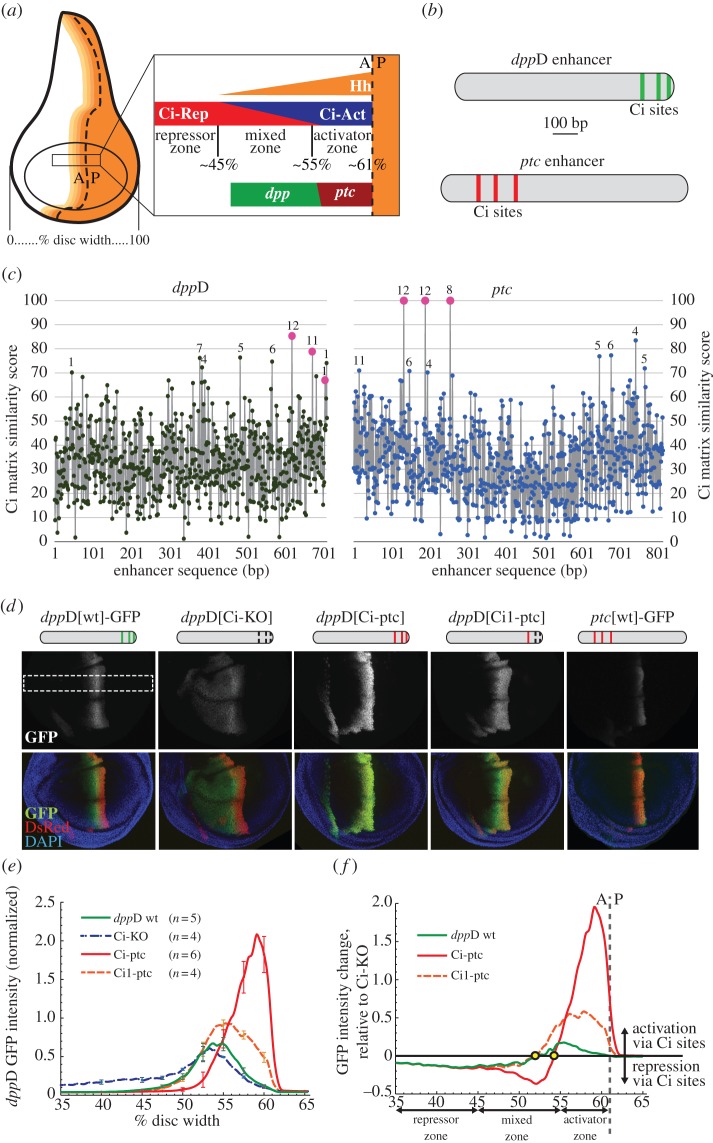
The dppD enhancer requires conserved low-affinity Ci/Gli sites to respond optimally to Hh and Ci in the developing wing. (a) Diagram of the Drosophila third-instar wing imaginal disc, showing the distribution of the Hedgehog signalling gradient across the anterior compartment. The dashed line indicates the anterior–posterior (A/P) boundary separating posterior cells, which secrete Hh, from anterior cells, which express the Ci transcription factor. Magnification of a segment of the wing pouch across the compartment boundary shows distinct zones (repressor, mixed and activator) based on their distance from the source of Hh morphogen. The Hh target genes dpp and ptc respond differently to the gradient; dpp is expressed maximally in the mixed zone, whereas ptc expression is restricted to the activator zone. (b) Ci binding motifs in the dppD and ptc enhancers. (c) Estimated Ci binding affinity and evolutionary conservation across the dppD and ptc enhancers of D. melanogaster. Ci matrix similarity score (see Methods) was plotted for every 9-mer. Known or proposed Ci sites (table 1) are shown as larger dots. For each 9-mer with a score ≥70, numerals indicate the number of Drosophila species (out of 12) in which that sequence is present at or near the orthologous position. (d) Top: diagrams of dppD enhancer constructs, with defined Ci binding sites as vertical bars (broken bars indicate mutated sites). Middle and bottom, confocal images of third-instar larval wing imaginal discs, showing GFP expression driven by dppD-GFP or ptc-GFP reporter transgenes. Red fluorescence is driven by a dppD[Ci-ptc]-DsRed transgene used for GFP fluorescence normalization and positional reference [33]. In dppD[Ci-KO]-GFP, three Ci sites were destroyed by targeted mutation; in dppD[Ci-ptc]-GFP, three Ci sites were converted to optimal motifs taken from ptc; in dppD[Ci1-ptc]-GFP, the 5′ Ci site was optimized, whereas site 2 and 3 were destroyed. The white dashed rectangle indicates the section of the dorsal wing pouch that is measured in the following panels. (e) Normalized GFP fluorescence data collected from the dorsal section of the wing pouch. Error bars indicate 1 s.d. (f) Net effect of wild-type or high-affinity Ci sites on dppD expression (calculated as the normalized transgene expression of dppD[wt] or dppD[Ci-ptc] minus normalized dppD[Ci-KO] expression). Circles indicate the positions on the A/P axis at which Ci input switches from net activation to net repression for each enhancer. (Online version in colour.)
A limited number of direct Hh/Ci target enhancers have been identified in Drosophila over the past two decades (table 1). More recently, new elements have been characterized in vertebrates [50–55]. The highest standard for identification of a direct Ci/Gli target enhancer consists of the following pieces of evidence: (i) the enhancer and parent gene are activated in a pattern consistent with Hh/Gli regulation; (ii) the enhancer contains sites that are biochemically demonstrated to be bound by Gli proteins in vitro or in vivo; and (iii) destruction of Gli sites diminishes the response of the enhancer and/or gene to Hh/Gli in vivo. Most, but not all, of the targets cited above meet that standard of evidence and can be regarded as confirmed direct Hh/Gli targets. Regardless of the species of origin, these enhancers respond to Hh signalling through variations on the same optimal Ci/Gli binding consensus [32,41,43–45,50–52].
Table 1.
Functionally characterized Hh/Ci-regulated enhancers in the Drosophila genome. CiBS: Ci binding site(s). Bases deviating from the Ci/Gli consensus motif GACCACCCA [32,39] are in bold and lower case. CiBS rank: rank of each 9-mer in order of predicted binding affinity for Ci [32].
| enhancer | target gene | sequence of CiBS | CiBS rank | expression | references |
|---|---|---|---|---|---|
| ptc | patched | GACCACCCAa | 1 | wing disc | [33,40], this study |
| GACCACCCAa | 1 | ||||
| GACCACCCAa | 1 | ||||
| dppD | decapentaplegic | GgCCACCtAa | 37 | wing, leg and antennal disc | [33,41], this study |
| GACCgCCCga | 172 | ||||
| tACCtCCCca,b | 1512c | ||||
| dppHO | decapentaplegic | GACCACCag | 240 | wing and haltere disc | [42] |
| cgCCACCCA | 103 | ||||
| wg1.0 | wingless | GAgCAgCCAa | 410 | embryonic ectoderm | [31,35,43], this study |
| GtCCACgCta | 1144d | ||||
| GttCACgCAa | 835 | ||||
| GACCtCCCAa | 4 | ||||
| r1.9 | stripe | ccaCACaCA | 4303e | embryonic ectoderm | [44], this study |
| GACCACCag | 240 | ||||
| knot | knot | GcCCACCCA | 3f,g | wing disc | [45] |
| GaCCACCgc | 248g | ||||
| GgCCACaCA | 43 | ||||
| D-h | hairy | GACCtCCCA | 4 | leg disc | [46] |
| GACCACCat | 235 | ||||
| oc7 | orthodenticle | GcCCtCCCA | 21 | vertex primordium | [47] |
| pCB1.8 | blistered | GcCCACCacb | 816 | wing disc | [48] |
| agCCACCCAb | 101 | ||||
| GACCACagcb | 1259 | ||||
| SRF-A | blistered | GgCCAtCtAb | 735 | wing disc | [49] |
bPotential binding site, not functionally and biochemically validated.
cThis sequence, proposed as a possible Ci binding site by Müller et al. but not tested [41], overlaps another 9-mer with a better (but still poor) Ci site ranking (426). Neither site showed detectable binding to Ci in vitro [33] and neither is well-conserved evolutionarily (see figure 3a and electronic supplementary figure S1b).
dThis sequence, proposed as a Ci binding site by Von Ohlen et al. [31], overlaps another 9-mer with a better (but still poor) Ci site ranking (992). A probe containing both overlapping sequences showed very weak binding to Ci in vitro [35], and the motif identified by Von Ohlen et al. [43] is not evolutionarily well conserved (see figure 3b and supplementary figure S1c).
eThis sequence, proposed as a Ci binding site by Piepenburg et al. [44], overlaps another 9-mer with a better (but still poor) Ci site ranking of 681 (figure 3c).
fThis site was reported as TGCCACCCA, a worse-ranking motif (rank 93), by Hersh et al. [45], but reference genomic sequence (BDGP R5/dm3) gives it as GCCCACCCA, and strong evolutionary sequence conservation supports the latter sequence.
gThese two predicted sites were reported to be functionally irrelevant in vivo by Hersh et al. [45]—but see the previous footnote.
Enhancers of the Drosophila genes patched (ptc) and decapentaplegic (dpp) were two of the earliest-identified direct Hh target sequences [40,41]. The ptc enhancer is directly activated by Hh/Ci in larval imaginal discs via high-affinity Ci sites that perfectly match the optimal Gli binding consensus (figure 1b,c and table 1) [33,40]. By contrast, dpp is activated in the same tissues by an enhancer (designated here as dppD) containing Ci sites of significantly lower affinity, with multiple mismatches to the optimal consensus (figure 1b,c and table 1) [33,41]. In the wing imaginal disc, ptc is expressed in a narrow strip of cells in the activator zone receiving maximal levels of Hh signalling, whereas dpp is expressed in a broader stripe in the mixed zone, farther from the source of morphogen (figure 1a) [56]. These observations present a puzzle: why is a low-affinity Ci target gene such as dpp activated more broadly across the Hh morphogen gradient than a high-affinity target gene like ptc? These results contrast with previous observations of the responsiveness of Bicoid and Dorsal target enhancers with low- and high-affinity sites [36,37]. Wolpert's French flag model of positional information, which has been invoked (in modernized forms) to explain transcriptional responses to Hh signalling [34,38,57,58], would seem to predict that high-affinity targets should be more sensitive to signalling and as a result be expressed in a relatively broad domain across the gradient; by comparison, low-affinity target genes might be expected to have a higher response threshold and thus a more restricted expression domain. Such a model has been recently proposed to explain transcriptional responses to Hh/Gli in the vertebrate neural tube [50]. Yet the expression patterns of ptc and dpp in the wing suggest that different mechanisms may be at work. Furthermore, the effects of opposing activator/repressor TF gradients, acting through the same cis-regulatory sites, have not been satisfactorily explained in any system.
We set out to explore how Ci binding site affinity affects the interpretation of Hh gradients in the developing Drosophila wing and embryonic ectoderm. Here, we present new data that corroborate our recent findings [33,35] that some Hh-responsive enhancers require low-affinity binding sites for normal activation in the regions of relatively low signalling. Not only are these sites important, but their low affinity is equally important: when these non-consensus sites were upgraded to optimal Ci binding motifs, the result is gene expression patterning defects that are consistent with a switch from Ci-mediated activation to Ci-mediated repression [33]. We present evidence consistent with a model in which selective pressure maintains non-consensus, low-affinity Ci binding sites in Hh-responsive enhancers, and propose that this is an evolutionary mechanism for maximizing Hh/Ci-mediated transcriptional activation in the regions of Hh morphogen gradients where Ci-Act and Ci-Rep compete for enhancer binding.
2. Material and methods
(a). Ci binding site prediction, scoring and ranking
A mononucleotide distribution matrix for Ci binding sites, derived from competitive DNA binding assays [32], was downloaded from the Genomatix software suite (www.genomatix.de; Genomatix, Germany). Matrix similarity scores [59] were calculated using data from the first nine nucleotide positions of the Ci matrix, which contain the majority of the information content. The matrix similarity score plots in figures 1c and 2c were generated with Apple Numbers and modified with Adobe Illustrator. Ci site rankings are determined by sorting all possible 9-mers in order of matrix similarity score, such that the optimal motif (GACCACCCA), with a score of 100, has a rank of 1; 9-mers with a lower matrix score than their reverse-complement sequences, such as TGGGTGGTC, are removed from the ranking, so that each high-scoring site is included only once.
Figure 2.
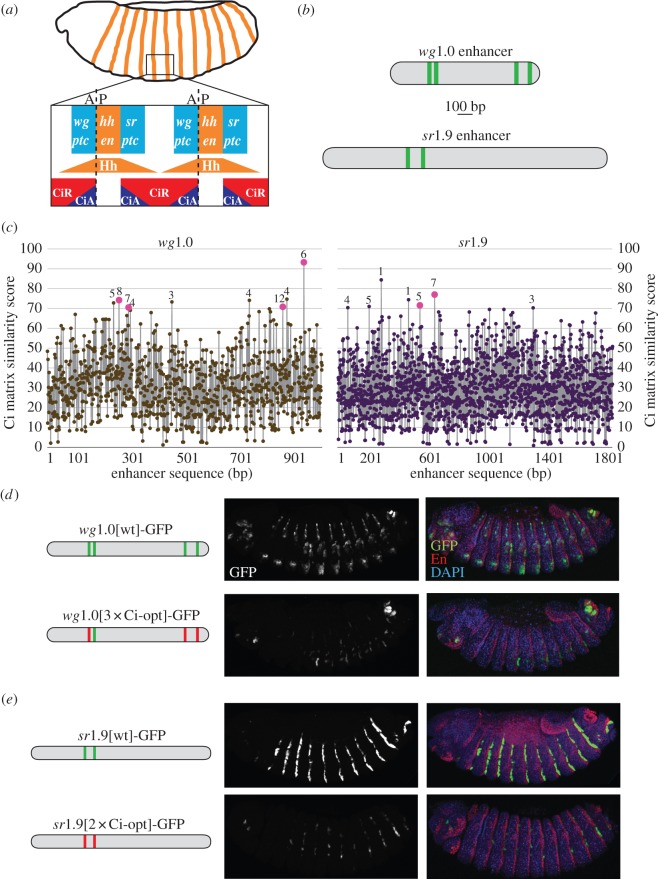
Optimizing the Ci binding affinity in the wingless and stripe embryonic enhancers results in reduced levels of gene expression. (a) Diagram of a stage 14 Drosophila embryo. A closer view of parasegments 5 and 6 shows bidirectional Hh signalling gradients that modulate the transcriptional activity of Ci. Known Hh/Ci target genes respond symmetrically (patched [71,72]) or asymmetrically (e.g. wingless (wg) [73] and stripe (sr) [44]) to the Hh signal. (b) Diagrams of the wg1.0[wt] and sr1.9[wt] enhancers showing Ci binding motifs as vertical lines. (c) Estimated Ci binding affinity across the wg1.0 and sr1.9 enhancers of D. melanogaster. Ci matrix similarity scores and conservation data are indicated as in figure 1c. In (d,e), diagrams on the left show wg1.0 and sr1.9D enhancer constructs; images of transgenic embryos show GFP alone (middle) and merged GFP, En (which marks Hh-producing cells), and DAPI nuclear stain (right). (d) Confocal images of stage 14 transgenic embryos carrying wg1.0[wt] and wg1.0[3xCi-opt], in which three Ci sites have been converted to optimal Ci binding motifs, driving GFP. (e) Confocal images of stage 14 transgenic embryos carrying sr1.9[wt] and sr1.9[2xCi-opt], in which two Ci sites have been converted into optimal Ci binding motifs, driving GFP. A, anterior; P, posterior. (Online version in colour.)
(b). DNA cloning and mutagenesis
Wild-type ptc, dppD, sr1.9 and wg1.0 enhancers were amplified by standard PCR from w1118 genomic DNA. Enhancer constructs were subcloned into the pENTR/D-TOPO plasmid (Invitrogen) by TOPO cloning. Enhancers were subsequently cloned into the pHPdesteGFP transgenesis vector [60] by LR Cloning (Invitrogen), or into the pEAB transgenesis vector (N. C. Evans & S. Barolo 2012, unpublished data) by traditional cloning methods. Targeted binding site mutations were created by overlap extension PCR [61]. Synthetic Hh-responsive enhancers were generated by assembly PCR [61]. See electronic supplementary material, figure S4 for full sequences of wild-type and mutated enhancers investigated in this study.
(c). Transgenesis
Site-directed transformation by embryo injection was performed as previously described [62]. Reporter transgenes were integrated into a phiC31 landing site at genomic position 86FB.
(d). Immunohistochemistry and microscopy
Embryos were fixed and stained using standard methods as previously described [35]. Third-instar wing imaginal discs were dissected and fixed as described [33]. Confocal images were captured on an Olympus FluoView 500 laser scanning confocal microscope mounted on an Olympus IX-71 inverted microscope. Samples to be directly compared were fixed, prepared and imaged under identical confocal microscopy conditions and settings. The primary antibodies used included rabbit anti-EGFP (Invitrogen), diluted 1 : 100, and mouse anti-En (Developmental Studies Hybridoma Bank), diluted 1 : 50. Embryos were staged as described [63].
(e). Quantitation of transgenic reporter expression data
Embryo and wing confocal images were collected and quantified as previously described [33,35]. The MATLAB program Icarus [64] was used to process and plot wing imaginal disc fluorescence data.
(f). Evolutionary sequence alignments
Alignments of enhancer-orthologous sequences from 12 sequenced Drosophila genomes were obtained from the UCSC Genome Browser (www.genome.ucsc.edu), except for the dppD enhancer, for which the UCSC alignment was incomplete; this alignment was performed with Clustal Omega (www.ebi.ac.uk/tools/msa/clustalo), using sequences identified with the EvoPrinter HD online tool [65]. Predicted binding motifs were identified with the GenePalette program [66]; alignment graphics were then modified with Adobe Illustrator.
3. Results and discussion
(a). Many Hh/Ci-regulated enhancers are regulated by non-consensus Ci binding sites
Most of the functionally characterized Hh/Ci-regulated enhancers in Drosophila respond to Hh signalling through non-consensus Ci binding sites (table 1), some of which have been shown to exhibit relatively poor Ci binding affinity in vitro [33,35]. The only known exception is ptc, which encodes the Hh receptor [67,68]. ptc is unique among the known direct Hh/Ci target genes in two ways. First, ptc is regulated by a cluster of highly conserved consensus Ci binding sites of optimal binding affinity (figure 1b,c and see electronic supplementary material, figure S1a) [33,40]. Second, unlike all other known Hh targets in the fly, which respond to Hh in a tissue-restricted pattern, ptc is transcriptionally activated by Hh signalling universally (i.e. in all tissues where Hh signalling occurs), as part of a negative feedback mechanism that regulates the range of signalling [69].
Among the enhancers listed in table 1 is dppD, which is both activated and repressed by Ci in imaginal discs [33,40,41]. The dppD enhancer is regulated by a cluster of Ci binding sites which, though they deviate considerably from the optimal consensus and have low Ci binding affinity in vitro, are required for proper spatial patterning by Hh/Ci in the developing wing (figure 1b,c and see electronic supplementary material, figure S1b) [33,41,69]. This enhancer drives wing and leg expression of the long-range morphogen decapentaplegic (dpp), which encodes a bone morphogenetic protein (BMP) family member that controls wing growth and patterning [70]. Two other Hh-regulated enhancers, wg1.0 and sr1.9, use non-consensus Ci binding sites to drive precise expression patterns in the embryonic ectoderm (table 1 and figure 2). The wg1.0 enhancer responds to Hh via four non-consensus, low-affinity Ci binding sites (table 1 and figure 2b,c and see electronic supplementary material, figure S1c) [35,43] to control the expression of the wingless (wg) gene, which encodes a Wnt-family morphogen [74]. The sr1.9 enhancer relies on two non-consensus Ci binding sites (table 1 and figure 2b,c) to regulate the expression of stripe (sr), a gene required for muscle-pattern formation during embryogenesis [75].
Many of these functionally significant non-consensus Ci binding sites are conserved throughout the evolution of the genus Drosophila (figure 1b and see electronic supplementary material, figure S1). This suggests the possibility of evolutionary pressures maintaining functional low-affinity Ci interactions with enhancers that interpret developmental Hh signalling gradients.
(b). Improving the binding affinity of Ci in the dppD enhancer restricts expression to the activator zone
We noted that the ptc and dppD enhancers, which are regulated by Ci binding sites of very different affinity, drive gene expression in distinct Hh signalling zones of the developing wing (figure 1a and see electronic supplementary material, figure S2) [56]. The ptc enhancer, which contains optimal sites, responds to Hh only in the activator zone, whereas dppD, with its non-consensus, low-affinity sites, responds to Hh in the mixed zone, farther from the source of morphogen (see electronic supplementary material, figure S2) [76]. To determine whether the low affinity of the Ci binding motifs in dppD (which is evolutionarily conserved: figure 3 and see electronic supplementary material, figure S1b) is important for responding to Hh/Ci in the mixed zone, we converted the three low-affinity sites into high-affinity sites taken from the ptc enhancer [33]. We observed that this ‘upgraded’ enhancer, dppD[Ci-ptc], which differs from the wild-type enhancer by only seven nucleotide positions, drives maximal gene expression in the activator zone instead of the mixed zone, similar to ptc (figure 1d). To more precisely determine the transcriptional effect of changes in Ci binding affinity, we used a quantitative reporter gene assay [33] to measure GFP fluorescence across the dorsal portion of the wing pouch and normalized it to a dppD[Ci-ptc]-DsRed reference transgene as an internal control for potential variations in age, fixation and wing shape. We compared normalized GFP transgene expression levels driven by three versions of dppD: wild-type (wt); Ci-KO, in which the Ci sites were destroyed; and Ci-ptc, in which the binding affinity of the sites was improved by targeted base substitutions [33].
Figure 3.
Evolutionary conservation of predicted binding affinity, but not of sequence identity, at many low-to-moderate-affinity Ci binding sites. Twelve-species Drosophila sequence alignments are shown for selected regions of the (a) dppD, (b) wg1.0 and (c) sr1.9 enhancers. Selected 9-mers are shaded, and Ci matrix similarity scores for those motifs are shown to the right. Sequences are from the following Drosophila species, from top to bottom: D. melanogaster, D. simulans, D. sechellia, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. virilis, D. mojavensis and D. grimshawi. Dashes indicate gaps; double-dashes indicate a lack of alignable sequence. Bracketed numbers indicate the number of bases deleted at that position to conserve space. Conserved homeodomain binding motifs are in grey boxes. (Online version in colour.)
In accordance with previous work [41], we found that dppD[Ci-KO] drove a broad expression pattern in the wing that differs from that of the wild-type enhancer in two respects: de-repression in anterior cells, and partial loss of activation in the mixed zone (figure 1e) [33]. We used the expression of dppD[Ci-KO] as a baseline, and measured the difference in fluorescence intensity between it and dppD[wt] or dppD[Ci-ptc] to determine the direct effect mediated by those three Ci binding sites at each position along the A/P axis of the wing disc (figure 1f) [33]. Although the dppD[Ci-KO] expression pattern clearly shows reduced sensitivity to Ci activation and repression, its expression still suggests some regulation by Hh signalling: this is likely due to indirect regulation via a non-Ci input that is itself regulated by Hh/Ci [41], but it could also reflect input from uncharacterized Ci binding sites (figure 3 and see electronic supplementary material, figure S1b). Increased Ci binding affinity provided stronger activation in the activator zone, as well as stronger repression in the repression zone, as expected—but unexpectedly, it also caused a switch from activation to repression in the mixed zone, where dpp (but not ptc) is normally activated (figure 1f) [33].
(c). Finding a happy medium: low-affinity Ci binding sites diversify the Hh response
As we proposed previously, the ectopic repression of dppD[Ci-ptc] in the activator zone may be explained by two biophysical mechanisms [33]. First, it is possible that Ci-Act and Ci-Rep have different binding preferences for distinct Ci motifs, such that Ci-Act prefers certain non-consensus sites while Ci-Rep prefers consensus sites. This scenario may seem unlikely, because Ci-Act and Ci-Rep share the same DNA-binding domain [77,78], but it has not been directly ruled out. An alternative possibility is that strong cooperative interactions occur between Ci-Rep (but not Ci-Act) that result in lower-threshold levels for Ci-Rep (schematic of these models can be found elsewhere [33,79]). Cooperative interactions are pervasive in gene regulation [80] and have been shown to lower threshold responses to other morphogens [37,81]. Fortuitously, these two models predict remarkably different transcriptional outputs for a modified dppD enhancer with a single high-affinity site (dppD[Ci1-ptc]; figure 1d). If the sequence motif itself dictates binding of Ci-Rep versus Ci-Act, then the transcriptional profile of dppD[Ci1-ptc] will be similar to dppD[Ci-ptc], as both enhancers contain only optimal consensus sites of identical sequence. On the other hand, if cooperative interactions between Ci-Rep are responsible for the restricted expression pattern of dppD[Ci-ptc], then dppD[Ci1-ptc] will behave more like dppD[wt], because a single Ci site cannot mediate homomeric cooperative interactions.
We found that dppD[Ci1-ptc] generates a broad stripe that is active in both the activator zone and the mixed zone (figure 1f), which is consistent with the repressor-cooperativity model and inconsistent with the binding-preferences model [33]. These results, and the deep evolutionary conservation of some of the low-affinity Ci sites in dppD[wt], suggest the presence of selective evolutionary pressure maintaining low Ci occupancy at the dppD enhancer. We speculate that dpp requires low-affinity Ci sites, which allow for activation by Hh/Ci but avoid invoking strong cooperative Ci repression in the mixed zone, in order to establish an organizing centre in the middle of the wing for symmetric growth [70].
(d). wg and sr require low-affinity Ci binding sites to respond optimally to Hh/Ci
To determine whether our observations regarding the effects of Ci binding site affinity are unique to dppD or to the developing wing, we examined two other Hh/Ci-regulated enhancers, both of which respond to Hh signalling in the embryonic ectoderm but not the wing. We first tested a 1.0 kb enhancer of the wingless (wg) gene which drives Hh-responsive embryonic stripes anterior to segmental stripes of Hh expression (figure 2a,d) [35,43]. Four Ci binding sites in the wg1.0 enhancer (table 1) have been reported to contribute to activation in Hh-responsive cells [54]. We improved the affinity of the three best-conserved Ci sites (wg1.0[3xCi-opt]; figure 2b,c and see electronic supplementary material, figure S1c) [35]. We observed that, rather than enhancing the transcriptional response to Hh, wg1.0[3xCi-opt] drives reduced expression levels in the embryonic ectoderm (figure 2d) [35].
We also examined the sr1.9 enhancer, which is expressed in Hh-responsive embryonic stripes to the posterior of Hh-positive cells. This element has three non-consensus Ci binding motifs showing significant sequence conservation (figure 2b,c), two of which had been previously identified (table 1) [44]. Destroying two of the predicted Ci sites has been reported to abolish the activity of this element [44], but we found that improving the affinity of these sites, rather than augmenting gene expression, greatly reduced it (figure 2e).
Taken together, these observations are consistent with the idea that wg and sr, like dpp, have Hh-responsive enhancers whose Ci occupancy is tuned at submaximal levels for optimal transcriptional activation in the proper zone of expression. We propose that this regulatory strategy stems from the dual nature of Ci as both an activator and a repressor, and the fact that these opposing activities are exerted through shared binding sites.
(e). Increasing the binding affinity of Ci does not induce significant ectopic expression
We hypothesized that the relatively low binding affinity of these Ci-regulated enhancers might be important, not just for shaping responses to Hh morphogen gradients, but also for maintaining tissue specificity of the Hh response. If this were the case, improving Ci binding affinity in these enhancers might be expected to sensitize them to Hh signalling, and thus might induce ectopic transcriptional responses to Hh/Ci outside of each gene's normal expression pattern. To address this point, we examined our high-affinity versions of the dppD, wg1.0 and sr1.9 enhancers in tissues and at developmental stages where active Hh signalling occurs, but where the gene and enhancer do not normally respond to that signal.
The dppD enhancer normally responds to Hh/Ci in the wing, leg and antennal discs, but not in the embryonic ectoderm (where other genes such as wg, sr and ptc respond to Hh signalling), and not in the morphogenetic furrow of the developing retina (where dpp expression is induced by Hh/Ci, but not by the dppD enhancer; see electronic supplementary material, figure S3a) [82,83]. We did not observe significant ectopic activity of dppD[Ci-ptc] in Hh-responding cells of the embryonic ectoderm, nor in the morphogenetic furrow of the eye (see electronic supplementary material, figure S3a). The only ectopic expression we observed was in part of the dorsal margin of the retina (see electronic supplementary material, figure S3a), which might receive signalling from nearby Hh-positive photoreceptors [84], although this is not part of the normal dpp expression pattern [85].
We next examined the expression of wg1.0[3xCi-opt] and sr1.9[2xCi-opt] in wing imaginal discs, where wg and sr do not normally respond to Hh/Ci, and found that improving Ci affinity did not activate ectopic transcriptional responses to Hh (see electronic supplementary material, figure S3b,c). Consistent with these results, it was previously shown that adding consensus Ci sites to the wing-specific enhancer of vestigial (not a Hh/Ci target gene) fails to induce ectopic Hh responses [49,86]. Our results demonstrate that the tissue-specific Hh responses of enhancers of dpp, wg and sr cannot be explained by low binding affinity for Ci.
(f). Functionally significant non-consensus Ci sites display conservation of motif quality, even in the absence of strict sequence conservation
Evolutionary sequence alignments of functional non-consensus Ci sites reveal multiple possible mechanisms by which the strength of Ci regulatory input into Hh-regulated enhancers may be maintained over evolutionary time, despite significant sequence turnover. Ci site 1 in the dppD enhancer is perfectly preserved across 12 Drosophila species, but this is an exception: most non-consensus Ci motifs, even those for which regulatory function has been demonstrated, are not so strongly conserved, and many have undergone rapid and extensive sequence changes (figure 3). For example, the sequence that comprises Ci site 2 in dppD is conserved and aligned only in the three species most closely related to D. melanogaster; yet examination of nearby sequences reveals that the same motif (CGGGCGGTC) is found nearby in six additional Drosophila species, though it is not aligned with the D. melanogaster motif (figure 3a). In most cases, these motifs share no recognizable flanking sequence with the D. melanogaster site, so it cannot be determined whether this motif is an island of high conservation amid rapidly changing and expanding/contracting flanking sequence, or (probably less likely) the same motif has been independently acquired multiple times during Drosophila evolution.
Ci site 2 in the wg1.0 enhancer has a different evolutionary history: a predicted Ci site is present in all Drosophila species at this position, but the sequence itself is not highly conserved. Three different motifs, with similar predicted affinities, occur at this site (figure 3b), suggesting that although the sequence of the site is evolving rapidly, the quality or predicted affinity of the site is constrained. A similar case of apparent quality constraint coupled with sequence flux occurs at Ci site 2 of the sr1.9 enhancer, where, for example, sequence changes in the Drosophila pseudoobscura lineage diminish the quality of the site, while at the same time creating a new overlapping motif of very similar quality to the D. melanogaster motif (figure 3c).
Ci site 1 in the wg1.0 enhancer seems to have undergone a triplet repeat expansion/contraction in the middle of the site, along with other changes (figure 3b), with the result that some species, such as D. melanogaster, have a single moderate-affinity site, whereas other species have a weaker motif at that position but have gained additional nearby sites. These may be examples of compensatory changes that maintain levels of local Ci occupancy within a region of the enhancer. Another possible case of compensation occurs in the vicinity of Ci site 1 of sr1.9, which is poorly conserved—eight distinct sequence variants occur at this position across 12 species—yet in most cases, overall site quality appears to be well preserved, especially if a neighbouring motif and its variations are taken into account. For example, Drosophila pseudoobscura and Droshophila persimilis have a motif in the position of site 1 that is considerably farther removed from the consensus than that in D. melanogaster (scoring 52.5 compared with 71.8), but have simultaneously acquired changes in a neighbouring sequence that significantly improves its quality as a Ci motif (scoring 83.6 compared with 61.0 in D. melanogaster).
These are anecdotal cases, and the functional significance of these motifs in species other than D. melanogaster has not yet been tested. Nevertheless, careful sequence analysis appears to provide support for our speculation that the poor overall sequence conservation of many low-to-moderate-affinity Ci binding motifs may be deceptive: these local genomic regions may be under selective pressure to maintain Ci occupancies within a certain range, while at the same time allowing a great deal of change at the level of DNA sequence.
(g). Ci is insufficient to activate Hh-responsive gene expression in vivo
To determine whether Ci binding sites, isolated from normal enhancer contexts, are capable of producing a transcriptional response to normal Hh signalling in vivo, we created a transgenic synthetic reporter in which three optimal Ci binding sites lie upstream of a minimal promoter driving GFP expression. This cluster of high-affinity sites (designated HHH) was not sufficient to activate expression in regions of active Hh signalling in imaginal wing discs or in embryos (figure 4). A similar construct bearing four high-affinity sites was previously shown to fail to respond to Hh in leg discs [42]. Our results exemplify a conserved transcriptional strategy known as ‘activator insufficiency’, which is shared by multiple signalling pathways and is thought to be an evolutionary mechanism for preventing ectopic responses to highly pleiotropic signals such as Hh [28].
Figure 4.
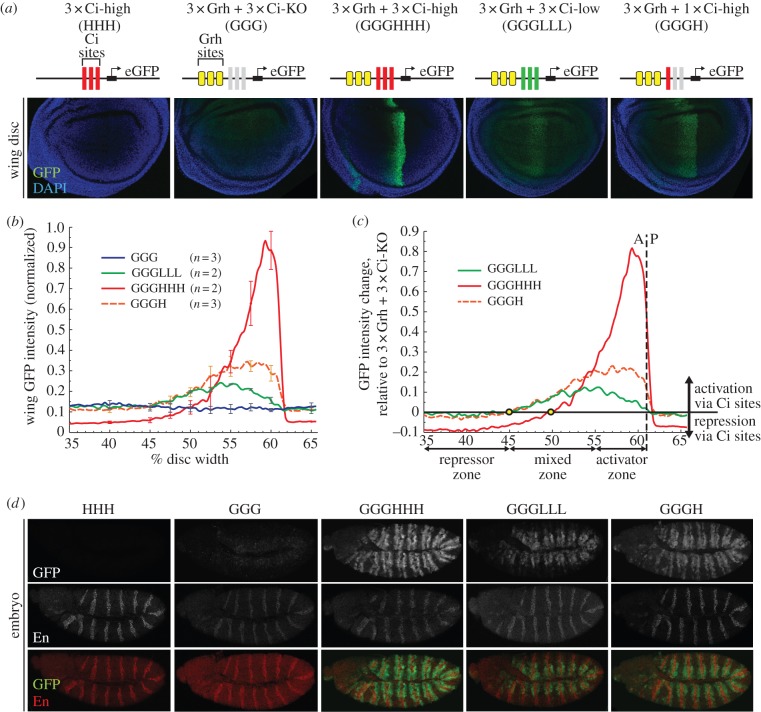
Hedgehog gradient responses of synthetic enhancers in the wing and embryo. (a) Diagrams of synthetic transgenic reporters, with Grainyhead (Grh) sites and low-affinity, high-affinity, and mutant Ci sites indicated. Confocal images of the pouch regions of wing imaginal discs from transgenic third-instar larvae are shown. GFP fluorescence data are internally normalized to a dppD[Ci-ptc]-DsRed reference transgene (not shown). (b) Normalized GFP fluorescence wing data across the dorsal wing pouch. Error bars indicate one standard deviation. (c) Net effect of Ci sites on synthetic transgene expression (normalized GFP expression minus normalized GGG-GFP expression). Circles indicate the positions at which Ci input switches from net activation to net repression. (d) Confocal images of stage 11 transgenic embryos carrying the same synthetic GFP reporters. Top, GFP expression; middle, Engrailed (En) antibody, which marks Hh-producing cells; bottom, merge. (Online version in colour.)
(h). Synthetic Hh/Ci-regulated enhancers recapitulate endogenous expression patterns in the wing
In order to study the functional properties of Ci binding sites outside the context of a complex enhancer sequence, we required to circumvent the insufficiency of Ci sites alone (figure 4a) to activate gene transcription in vivo. We borrowed a clever strategy [87] that combines binding sites for the broadly expressed transcriptional activator Grainyhead (Grh) [88,89] with binding sites for Ci. Grh binding sites have been shown to be sufficient to activate gene transcription in the wing [90]. Using this approach, we were able to create a baseline level of transcription that allowed us to detect activating and repressive inputs from Ci sites, which can then be measured as changes in gene expression in Grh + Ci reporters, relative to a Grh-alone reporter. We generated four versions of synthetic enhancers with three Grh binding sites (GGG) upstream of three high-affinity sites (HHH), three low-affinity sites (LLL), one high-affinity site (H) and three mutant Ci sites (KO) to preserve the spacing between the promoter and Grh (GGG; figure 4a). All of these transgenic constructs drove Hh/Ci-regulated stripes of different width and strength, with the exception of the 3xGrh-only construct (GGG), which, as expected, drove basal levels of expression throughout the wing disc (figure 4a) [89]. We quantitated, normalized and compared GFP fluorescence data from these synthetic reporters as described for figure 1, and observed that GGGHHH is expressed at high levels in the activator zone, GGGLLL is weakly expressed in the mixed zone and GGGH is expressed at moderate levels in the activator and mixed zones (figure 4b).
Next, we subtracted the Grh-only (GGG) expression levels from that of the Grh + Ci reporters to measure Ci-mediated activation and repression across the Hh gradient [33,87]. We found that GGGHHH, the synthetic counterpart of dppD[Ci-ptc], is strongly activated by Ci in the activator zone but is repressed by Ci in the mixed zone, whereas the activity of GGGLLL peaks in the mixed zone and is weaker in the activator zone (figure 4c). GGGH (analogous to dppD[Ci1-ptc]) is activated by Ci in both the activator and mixed zones (figure 4c) [33]. The fact that these synthetic results are strikingly consistent with our observations with dppD (compare figure 4c and figure 1e) indicates that the observed effects of Ci affinity on Hh responses in the wing are not dependent on a particular enhancer context, and demonstrates the utility of synthetic reporters for the quantitative analysis of Ci-regulated transcription in a simple and well-controlled sequence context.
The weak response of GGGLLL in the activator zone, compared with its expression in the mixed zone, is noteworthy (figure 4c). In the case of the native dppD enhancer, diminished expression in the activator zone has been attributed to repression by the homeodomain (HD) TF Engrailed (En), which is expressed in a narrow strip of anterior cells abutting the A/P boundary during late larval stages [91,92]. We analysed the sequences of the synthetics to determine whether we had unknowingly introduced En binding sites (see electronic supplementary material, figure S4e–h), and found a single predicted En site that overlaps with the first Ci site in GGGLLL and GGG (the site is destroyed in GGGHHH and GGGH). This En motif might be responsible for repressing GGGLLL in the activator zone. However, because we did not observe repression of GGG, which has the same En motif, in En-positive cells of the activator zone and the posterior compartment, and because GGGLLL was not repressed in the En-positive posterior compartment (figure 4b), we conclude that these reporters are not directly repressed by En. The restricted activity of the low-affinity Ci binding sites in the mixed zone therefore seems to be encoded in the sequence of the Ci sites themselves. If true, this implies an as-yet-unknown mechanism for interpretation of the Hh gradient in the wing via Ci binding sites, but further research is required.
(i). Synthetic Hh/Ci-regulated enhancers drive ptc-like expression in embryos
To investigate whether the ability of these synthetic enhancers to respond to Hh/Ci is limited to imaginal tissues, we examined embryos at stages when Hh signalling occurs (figure 4d). The Grh activator is expressed in the epidermis of mid- to late-stage embryos [93]. At stage 11, our GGG synthetic construct, containing three Grh binding sites, reported low levels of Grh input in the dorsal ectoderm (figure 4d). At that same stage, our synthetic Grh + Ci reporters (but not GGG) were activated in stripes to the anterior and posterior of each stripe of Hh-expressing cells (figure 4d). This pattern differs from those of the natural Hh/Ci-activated enhancers of wg and sr, whose response is restricted to one side (the anterior and posterior, respectively) of each Hh-positive stripe [43,44]; instead, it more closely resembles that of the ptc gene, which responds symmetrically to stripes of Hh signalling in embryos [71,72]. GGGHHH drove high levels of expression in stripes that span both the dorsal and ventral sides of the embryo, whereas GGGLLL drove moderate levels of expression in dorsal stripes in cells that have the strongest Grh input (figure 4d). GGGH drove activity in a similar pattern to that of GGGHHH, but at lower levels (figure 4d). Contrary to what we observed by improving the affinity of wg1.0 and sr1.9, the high-affinity reporter GGGHHH was not more restricted in its expression than the low-affinity reporter GGGLLL, but instead was more strongly activated (figure 2c,d compared with figure 4d). Therefore, the strongly negative effect of high-affinity Ci sites on expression of the wg and sr enhancers may depend on the sequence context of those regulatory elements.
(j). Deep evolutionary conservation of putative homeodomain binding sites in dppD
The dppD enhancer integrates inputs from other unknown factors besides Ci: this is demonstrated by the dppD[Ci-KO] construct, which is active throughout the anterior compartment of the wing (figure 1a) [33,41]. To investigate the other inputs controlling dppD, we examined the sequence conservation of this element across 12 Drosophila species (see electronic supplementary material, figure S1b). Conserved TF binding motifs are considered likely to be functionally significant [94,95], although there are significant exceptions [22,23,61]. The dppD enhancer contains seven core HD binding motifs (TAAT), of which six are perfectly conserved throughout the genus (see electronic supplementary material, figure S1b). Overrepresentation of conserved HD binding sites has been also shown in some Hh-regulated enhancers in vertebrates [51]. All of the largest blocks of sequence conservation in dppD include at least one HD core motif (figure 3a). Among these conserved potential HD binding sites is a previously identified site (designated as HE in electronic supplementary material, figure S1b) which was shown to repress dppD in posterior cells and has been proposed to mediate repression by En [41].
(k). dppD integrates inputs from conserved putative homeodomain binding sites
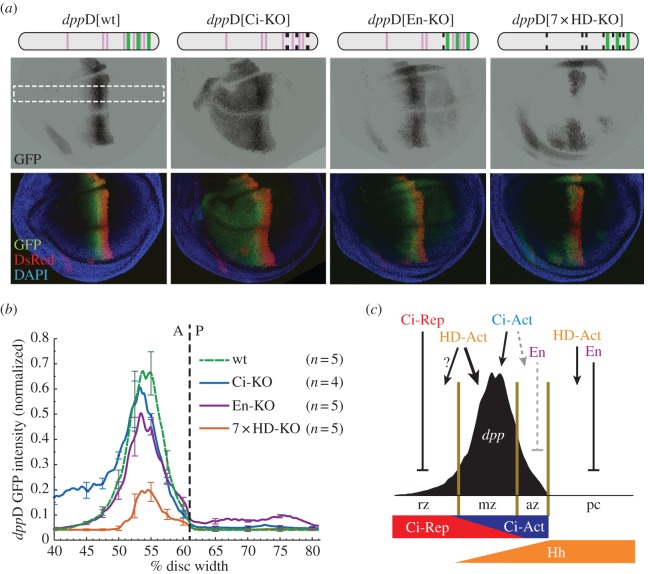
To determine whether these potential HD binding sites contribute to the regulation of dppD, we first tested the contribution of the previously identified En binding site with a targeted mutation (dppD[En-KO]). Consistent with prior findings [41], this mutation resulted in mild de-repression in the posterior compartment, where En is expressed (figure 5a). We next mutated all seven core HD motifs in dppD (7xHD-KO). This mutant enhancer drove a weak, incomplete wing stripe (figure 5a). We quantitated the GFP fluorescence activated by these constructs across the wing to determine the regulatory contribution of putative HD binding sites. By comparing our measurements with wild-type dppD (figure 5b), we found that the predicted En site, in addition to its known role in repression of dppD in posterior cells [41], also contributed to dppD activation in the anterior compartment, in cells lacking En (figure 5b). We also found that at least one of the HD motifs is responsible for activating dppD[En-KO] in posterior cells, as dppD[7xHD-KO] was not active in that compartment. In anterior-compartment cells where dpp is normally expressed, we observed that the loss in activity in dppD[7xHD-KO] was more severe than that caused by mutating three Ci binding sites (dppD[Ci-KO]; figure 5b). The role of HD in activating dppD contrasts with the repressive role of some HD binding proteins in Hh-regulated enhancers in the mouse neural tube [51]. However, we noted that dppD[7xHD-KO] was de-repressed in the retina (data not shown), where Hh signalling is active but dppD[wt] is normally not expressed (see electronic supplementary material, figure 3a).
Figure 5.
Conserved homeodomain (HD) binding motifs integrate regulatory inputs into dppD. (a) Top: diagrams of dppD enhancer constructs, with defined Ci and HD binding sites as vertical bars (wide bars, wild-type low-affinity Ci binding sites; narrow bars, HD motifs; broken bars, mutated sites). Middle and bottom: confocal images of wing imaginal discs from transgenic third-instar larvae are shown. Middle, GFP alone; bottom, GFP merged with dppD[Ci-ptc]-DsRed reference transgene and DAPI nuclear stain. (b) Normalized GFP fluorescence data from transgenic dorsal wing pouches. Error bars indicate 1 s.d. Dashed line shows A/P compartment boundary. (c) Proposed model of the dppD regulatory network across the wing A/P axis. A, anterior; P, posterior; pc, posterior compartment. (Online version in colour.)
Although the identities of the additional dppD inputs remain a mystery, we speculate that the HD TFs Aristaless and Disatalless might be among these factors, based on their expression patterns in the wing and their known genetic relationship with dpp [96,97]. Our results are consistent with a model in which complex regulatory inputs from HD proteins [98] act through highly conserved sites in the dppD enhancer. They also highlight the critical role of low-affinity Ci binding sites, which cooperate with these positive and negative inputs to specify dpp expression in the proper segment of the Hh morphogen gradient (figure 5c). Such a view contrasts sharply with characterizations of low-affinity TF–DNA interactions as functionally inconsequential [99]; to the contrary, certain types of regulatory circuits—especially those regulated by signalling pathways that use activator/repressor switch mechanisms, such as Hh, Wnt, Notch and others—may acquire and maintain low-affinity interactions to extract the maximum amount of information from developmental signalling events [22,28,100].
4. Conclusion
In this study, we have presented in vivo evidence corroborating previous findings [33,35] that multiple tissue-specific enhancers require low-affinity Ci binding sites for optimal activation by Hh/Ci. Most of the Hh target enhancers identified up to this point in Drosophila and mouse are regulated by degenerate Ci/Gli binding sites of low predicted affinity (table 1) [32]. The prevalence of these non-consensus sites in Hh target enhancers across species demonstrates their importance in regulating the Hh response. The transcriptional relevance of low-affinity TF binding is not limited to Hh/Ci regulated enhancers [22]. For instance, two phylogenetically conserved low-affinity binding sites in the mouse Pax6 lens enhancer have been shown to be critical to promote gene expression at the right stage of development [101].
We also provide a mechanistic explanation as to why these Hh/Ci-regulated elements require low-affinity sites to activate transcription in cells with moderate signalling levels. We showed that clusters of high-affinity sites mediate a restricted response in cells with high levels of Hh signalling, most likely as a result of cooperative interactions among Ci-Rep molecules in highly occupied Ci binding sites, whereas clusters of low-affinity sites mediate a broader response by having lower occupancy by Ci [33,35]. Using synthetic enhancer reporters with high- or low-affinity Ci binding sites, we confirmed this effect in the wing, but not in embryos. This tissue-specific discrepancy may imply a context-dependent function for some non-consensus Ci binding sites. As in the Pax6 lens enhancer [101], it is possible that some low-affinity binding sites are required specifically during earlier stages of development to interpret overall lower levels of Hh signalling [102,103].
Finally, we provided clues as to additional regulatory inputs into dppD by showing a requirement for conserved consensus HD binding sites. Cooperation between Glis and HD proteins has been recently shown in the mouse neural tube [51]. In this case, HD proteins are critical to repress Hh-regulated neural tube enhancers, whereas in dppD they are critical to activate gene expression.
The limited number of known, experimentally confirmed, direct Hh/Gli target enhancers may reflect the widespread, practical tendency to search for consensus or near-consensus motifs, and to focus on the highest peaks of TF–DNA binding, when hunting for cis-regulatory sequences. From a biochemical standpoint—for example, when mining ChIP-seq data—low-affinity DNA–binding interactions are troublesome because they are much more common, by definition, than the top 1% of peaks. It is important to note that we do not mean to strictly equate ChIP peak height with TF binding affinity, nor to equate in vitro binding or in silico ‘motif quality’ with in vivo TF occupancy, though these properties may often be roughly correlated. Separating the weak but functional binding events from weak and non-functional binding events is extremely challenging, and some have proposed that low-affinity genome-binding interactions can be categorically ignored [2,99]. This certainly simplifies the problem from a computational perspective, but the findings discussed here and elsewhere [101,104] suggest a risk of discarding functional sequences. Similar challenges confront in silico genomic screens to identify clusters of predicted TF binding sites: these necessarily filter out binding events of low predicted affinity, because there are many more predicted low-affinity binding motifs than consensus high-affinity motifs in any given sequence [105]. Binding site predictions have been supported by taking evolutionary sequence conservation into account [9,32], but this risks filtering out true positives: as shown in our Ci motif alignments, lower-affinity binding sites seem to be less constrained with respect to sequence variation, even in cases when the presence of the site itself is highly conserved. This is presumably because, for each non-consensus binding motif, there are multiple alternative sequences with similar affinity and thus equivalent functionality. Importantly, this type of degenerate motif conservation is easily missed: for example, some of the well-conserved Ci motifs described here are not properly aligned in the UCSC Genome Browser, because they do not constitute contiguous blocks of perfect sequence identity. To avoid these pitfalls, it is important to use phylofootprinting approaches that account for these alignment flaws, such as the one described in [94]. In contrast to most of the low-affinity binding sites discussed here, optimal-affinity Ci motifs in the ptc enhancer have been preserved throughout the evolution of the genus Drosophila, and perhaps much farther: GACCACCCA motifs occur in promoter-proximal regions of multiple vertebrate orthologues of ptc [9,53] (additional data not shown).
Evolutionary enhancer sequence alignments, along with limited experimental data, also suggest that, although many predicted low-affinity sites are poorly conserved, overall TF occupancy on an enhancer may be maintained despite significant sequence turnover. This may occur either through the rapid gain and loss of individual sites, or through the maintenance of relatively weak binding affinity at a site that is unstable at the level of DNA sequence [22,106]. While this last idea requires further direct testing, it is consistent with the fact that Gli sites of moderate predicted affinity have many sequence variants of similar quality, whereas the highest-affinity motifs have far fewer alternatives of similar quality. In other words, there are many more ways to be a weak binding site than a strong site. For example, among all possible 9-mer sequences, there are 654 motifs with Ci matrix similarity scores between 70 and 75 (inclusive), but only 12 motifs with scores between 90 and 95, and one motif with a score above 95. Therefore, weaker binding sites, and the enhancers containing them, have a far greater volume of sequence space in which to roam without strongly impacting transcriptional output [22]. A thermodynamics-based simulation of enhancer evolution has shown that there is a greater number of fit solutions using weak TF sites than using high-affinity sites for a given gene expression problem [107].
Equally consistent with our view of TF binding site evolution is the fact that it is much easier (that is, more likely) to create a low-affinity, non-consensus binding motif with a single mutation than a high-affinity consensus motif. An enhancer-sized DNA sequence can acquire a weak Gli motif with single-nucleotide substitutions at any of a large number of positions, as demonstrated by our simulations (see electronic supplementary material, figure S5). These arguments may help to explain why sequence conservation is not a foolproof test of the functional relevance of non-consensus TF binding sites.
While we cannot offer a simple answer to the technical challenges facing those who hunt enhancers, the findings described in this report lead us to conclude that low-affinity TF–DNA interactions, mediated by non-consensus and often poorly conserved sequence motifs, play important and widespread roles in developmental patterning and cis-regulatory evolution, and therefore cannot be safely ignored.
Acknowledgements
We are particularly grateful to Elliott Ortiz-Soto, Katherine Gurdziel, Dave Parker, Michael White and Barak Cohen for their exceptional contributions to this project. We thank Ingrid Lohmann for generously providing the pHPdesteGFP destination vector, and Niki Evans for making the pEAB transgenic vector. We thank Rachna Pannu for her assistance with the wg enhancer project, and David Lorberbaum and Lisa Johnson for their comments on the manuscript. Confocal imaging was performed at the Microscopy and Image Analysis Laboratory (MIL) at the University of Michigan Medical School; we thank the MIL staff for their assistance.
Funding statement
This work was supported by the Cellular and Molecular Biology Training Grant (NIH T32-GM007315) and a Center for Organogenesis predoctoral fellowship (NIH T32-HD007505) to A.I.R. and by NIH grant GM076509 and NSF grant MCB-1157800 to S.B.
References
- 1.Levine M. 2010. Transcriptional enhancers in animal development and evolution. Curr. Biol. 20, R754–R763 (doi:10.1016/j.cub.2010.06.070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stamatoyannopoulos JA. 2012. What does our genome encode? Genome Res. 22, 1602–1611 (doi:10.1101/gr.146506.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ong C-T, Corces VG. 2011. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12, 283–293 (doi:10.1038/nrg2957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Göke J, Jung M, Behrens S, Chavez L, O'Keeffe S, Timmermann B, Lehrach H, Adjaye J, Vingron M. 2011. Combinatorial binding in human and mouse embryonic stem cells identifies conserved enhancers active in early embryonic development. PLoS Comput. Biol. 7, e1002304 (doi:10.1371/journal.pcbi.1002304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen Y, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (doi:10.1038/nature11243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng J-H, et al. 2013. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev. Cell 24, 324–333 (doi:10.1016/j.devcel.2012.12.011) [DOI] [PubMed] [Google Scholar]
- 7.Yu Y, et al. 2013. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152, 248–261 (doi:10.1016/j.cell.2012.12.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. 2007. Whole-genome ChIP-chip analysis of dorsal, twist, and snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21, 385–390 (doi:10.1101/gad.1509607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vokes SA, et al. 2007. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development (Camb. Engl.) 134, 1977–1989 (doi:10.1242/dev.001966) [DOI] [PubMed] [Google Scholar]
- 10.Visel A, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (doi:10.1038/nature07730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EEM. 2012. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148, 473–486 (doi:10.1016/j.cell.2012.01.030) [DOI] [PubMed] [Google Scholar]
- 12.Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077 (doi:10.1126/science.1232542) [DOI] [PubMed] [Google Scholar]
- 13.Li X-Y, Thomas S, Sabo PJ, Eisen MB, Stamatoyannopoulos JA, Biggin MD. 2011. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol. 12, R34 (doi:10.1186/gb-2011-12-4-r34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biggin MD. 2011. Animal transcription networks as highly connected, quantitative continua. Dev. Cell 21, 611–626 (doi:10.1016/j.devcel.2011.09.008) [DOI] [PubMed] [Google Scholar]
- 15.Maston GA, Landt SG, Snyder M, Green MR. 2012. Characterization of enhancer function from genome-wide analyses. Annu. Rev. Genomics Hum. Genet. 13, 29–57 (doi:10.1146/annurev-genom-090711-163723) [DOI] [PubMed] [Google Scholar]
- 16.Rebeiz M. 2002. SCORE: a computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Proc. Natl Acad. Sci. USA 99, 9888–9893 (doi:10.1073/pnas.152320899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman BP, Pfeiffer BD, Laverty TR, Salzberg SL, Rubin GM, Eisen MB, Celniker SE. 2004. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5, R61 (doi:10.1186/gb-2004-5-9-r61) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goode D, Snell P, Smith D, Cooke J, Elgar G. 2005. Highly conserved regulatory elements around the gene may contribute to the maintenance of conserved synteny across human chromosome 7q36.3. Genomics 86, 172–181 (doi:10.1016/j.ygeno.2005.04.006) [DOI] [PubMed] [Google Scholar]
- 19.Pennacchio LA, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444, 499–502 (doi:10.1038/nature05295) [DOI] [PubMed] [Google Scholar]
- 20.Vavouri T, Lehner B. 2009. Conserved noncoding elements and the evolution of animal body plans. Bioessays 31, 727–735 (doi:10.1002/bies.200900014) [DOI] [PubMed] [Google Scholar]
- 21.Blow MJ, et al. 2010. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 42, 806–810 (doi:10.1038/ng.650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swanson CI, Schwimmer DB, Barolo S. 2011. Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr. Biol. 21, 1186–1196 (doi:10.1016/j.cub.2011.05.056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittkopp PJ, Haerum BK, Clark AG. 2008. Regulatory changes underlying expression differences within and between Drosophila species. Nat. Genet. 40, 346–350 (doi:10.1038/ng.77) [DOI] [PubMed] [Google Scholar]
- 24.Peter IS, Davidson EH. 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144, 970–985 (doi:10.1016/j.cell.2011.02.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haag ES, Lenski RE. 2011. L'enfant terrible at 30: the maturation of evolutionary developmental biology. Development (Camb. Engl.) 138, 2633–2637 (doi:10.1242/dev.066928) [DOI] [PubMed] [Google Scholar]
- 26.Wittkopp PJ, Kalay G. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69 (doi:10.1038/nrg3095) [DOI] [PubMed] [Google Scholar]
- 27.Pires-daSilva A, Sommer RJ. 2003. The evolution of signalling pathways in animal development. Nat. Rev. Genet. 4, 39–49 (doi:10.1038/nrg977) [DOI] [PubMed] [Google Scholar]
- 28.Barolo S, Posakony JW. 2002. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167–1181 (doi:10.1101/gad.976502) [DOI] [PubMed] [Google Scholar]
- 29.Ingham PW, Nakano Y, Seger C. 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406 (doi:10.1038/nrg2984) [DOI] [PubMed] [Google Scholar]
- 30.Hui C-C, Angers S. 2011. Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513–537 (doi:10.1146/annurev-cellbio-092910-154048) [DOI] [PubMed] [Google Scholar]
- 31.Ohlen Von T, Lessing D, Nusse R, Hooper JE. 1997. Hedgehog signaling regulates transcription through Cubitus interruptus, a sequence-specific DNA binding protein. Proc. Natl Acad. Sci. USA 94, 2404–2409 (doi:10.1073/pnas.94.6.2404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. 2006. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124, 47–59 (doi:10.1016/j.cell.2005.10.042) [DOI] [PubMed] [Google Scholar]
- 33.Parker DS, White MA, Ramos AI, Cohen BA, Barolo S. 2011. The cis-regulatory logic of hedgehog gradient responses: key roles for Gli binding affinity, competition, and cooperativity. Science Signal. 4, ra38 (doi:10.1126/scisignal.2002077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashe HL, Briscoe J. 2006. The interpretation of morphogen gradients. Development (Camb. Engl.) 133, 385–394 (doi:10.1242/dev.02238) [DOI] [PubMed] [Google Scholar]
- 35.White MA, Parker DS, Barolo S, Cohen BA. 2012. A model of spatially restricted transcription in opposing gradients of activators and repressors. Mol. Syst. Biol. 8, 614 (doi:10.1038/msb.2012.48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Driever W, Thoma G, Nüsslein-Volhard C. 1989. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature 340, 363–367 (doi:10.1038/340363a0) [DOI] [PubMed] [Google Scholar]
- 37.Jiang J, Levine M. 1993. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72, 741–752 (doi:10.1016/0092-8674(93)90402-C) [DOI] [PubMed] [Google Scholar]
- 38.Wolpert L. 1969. Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 25, 1–47 (doi:10.1016/S0022-5193(69)80016-0) [DOI] [PubMed] [Google Scholar]
- 39.Kinzler KW, Vogelstein B. 1990. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 10, 634–642 (doi:10.1128/MCB.10.2.634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexandre C, Jacinto A, Ingham PW. 1996. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 10, 2003–2013 (doi:10.1101/gad.10.16.2003) [DOI] [PubMed] [Google Scholar]
- 41.Muller B, Basler K. 2000. The repressor and activator forms of Cubitus interruptus control Hedgehog target genes through common generic Gli-binding sites. Development (Camb. Engl.) 127, 2999–3007 [DOI] [PubMed] [Google Scholar]
- 42.Hepker J, Blackman RK, Holmgren R. 1999. Cubitus interruptus is necessary but not sufficient for direct activation of a wing-specific decapentaplegic enhancer. Development (Camb. Engl.) 126, 3669–3677 [DOI] [PubMed] [Google Scholar]
- 43.Ohlen Von T, Hooper JE. 1997. Hedgehog signaling regulates transcription through Gli/Ci binding sites in the wingless enhancer. Mech. Dev. 68, 149–156 (doi:10.1016/S0925-4773(97)00150-0) [DOI] [PubMed] [Google Scholar]
- 44.Piepenburg O, Vorbrüggen G, Jäckle H. 2000. Drosophila segment borders result from unilateral repression of hedgehog activity by wingless signaling. Mol. Cell 6, 203–209 [PubMed] [Google Scholar]
- 45.Hersh BM, Carroll SB. 2005. Direct regulation of knot gene expression by ultrabithorax and the evolution of cis-regulatory elements in Drosophila. Development (Camb. Engl.) 132, 1567–1577 (doi:10.1242/dev.01737) [DOI] [PubMed] [Google Scholar]
- 46.Kwon C, Hays R, Fetting J, Orenic TV. 2004. Opposing inputs by Hedgehog and Brinker define a stripe of hairy expression in the Drosophila leg imaginal disc. Development (Camb. Engl.) 131, 2681–2692 (doi:10.1242/dev.01127) [DOI] [PubMed] [Google Scholar]
- 47.Blanco J, Seimiya M, Pauli T, Reichert H, Gehring WJ. 2009. Wingless and Hedgehog signaling pathways regulate orthodenticle and eyes absent during ocelli development in Drosophila. Dev. Biol. 329, 104–115 (doi:10.1016/j.ydbio.2009.02.027) [DOI] [PubMed] [Google Scholar]
- 48.Nussbaumer U, Halder G, Groppe J, Affolter M. 2000. Expression of the blistered/DSRF gene is controlled by different morphogens during Drosophila trachea and wing development. Mech. Dev. 96, 27–36 (doi:10.1016/S0925-4773(00)00373-7) [DOI] [PubMed] [Google Scholar]
- 49.Halder G, Polaczyk P, Kraus ME, Hudson A, Kim J, Laughon A, Carroll S. 1998. The vestigial and scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev. 13, 3900–3909 (doi:10.1101/gad.12.24.3900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterson KA, et al. 2012. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 26, 2802–2816 (doi:10.1101/gad.207142.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oosterveen T, et al. 2012. Mechanistic differences in the transcriptional interpretation of local and long-range Shh morphogen signaling. Dev. Cell 23, 1006–1019 (doi:10.1016/j.devcel.2012.09.015) [DOI] [PubMed] [Google Scholar]
- 52.Sasaki H, Hui C, Nakafuku M, Kondoh H. 1997. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development (Camb. Engl.) 124, 1313–1322 [DOI] [PubMed] [Google Scholar]
- 53.Agren M, Kogerman P, Kleman MI, Wessling M, Toftgård R. 2004. Expression of the PTCH1 tumor suppressor gene is regulated by alternative promoters and a single functional Gli-binding site. Gene 330, 101–114 (doi:10.1016/j.gene.2004.01.010) [DOI] [PubMed] [Google Scholar]
- 54.Eichberger T, Kaser A, Pixner C, Schmid C, Klingler S, Winklmayr M, Hauser-Kronberger C, Aberger F, Frischauf A-M. 2008. GLI2-specific transcriptional activation of the bone morphogenetic protein/activin antagonist follistatin in human epidermal cells. J. Biol. Chem. 283, 12 426–12 437 (doi:10.1074/jbc.M707117200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasper M, et al. 2006. Selective modulation of hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol. Cell. Biol. 26, 6283–6298 (doi:10.1128/MCB.02317-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Capdevila J, Estrada MP, Sánchez-Herrero E, Guerrero I. 1994. The Drosophila segment polarity gene patched interacts with decapentaplegic in wing development. EMBO J. 13, 71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolpert L. 2011. Positional information and patterning revisited. J. Theor. Biol. 269, 359–365 (doi:10.1016/j.jtbi.2010.10.034) [DOI] [PubMed] [Google Scholar]
- 58.Dessaud E, McMahon AP, Briscoe J. 2008. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development (Camb. Engl.) 135, 2489–2503 (doi:10.1242/dev.009324) [DOI] [PubMed] [Google Scholar]
- 59.Quandt K, Frech K, Karas H, Wingender E, Werner T. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23, 4878–4884 (doi:10.1093/nar/23.23.4878) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boy AL, Zhai Z, Habring-Müller A, Kussler-Schneider Y, Kaspar P, Lohmann I. 2010. Vectors for efficient and high-throughput construction of fluorescent Drosophila reporters using the PhiC31 site-specific integration system. Genesis 48, 452–456 (doi:10.1002/dvg.20637) [DOI] [PubMed] [Google Scholar]
- 61.Swanson CI, Evans NC, Barolo S. 2010. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev. Cell 18, 359–370 (doi:10.1016/j.devcel.2009.12.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl Acad. Sci. USA 104, 3312–3317 (doi:10.1073/pnas.0611511104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Campos-Ortega JA, Hartenstein V. 1985. The embryonic development of Drosophila melanogaster. Berlin, Germany: Springer [Google Scholar]
- 64.Ortiz-Soto E, Ramos AI, Barolo S.Icarus: a simple, customizable tool for sophisticated analysis and normalization of in vivo gene expression patterns. In preparation.
- 65.Yavatkar AS, Lin Y, Ross J, Fann Y, Brody T, Odenwald WF. 2008. Rapid detection and curation of conserved DNA via enhanced-BLAT and EvoPrinterHD analysis. BMC Genomics 9, 106 (doi:10.1186/1471-2164-9-106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rebeiz M, Posakony JW. 2004. GenePalette: a universal software tool for genome sequence visualization and analysis. Dev. Biol. 271, 431–438 (doi:10.1016/j.ydbio.2004.04.011) [DOI] [PubMed] [Google Scholar]
- 67.Chen Y, Struhl G. 1996. Dual roles for patched in sequestering and transducing Hedgehog. Cell 87, 553–563 (doi:10.1016/S0092-8674(00)81374-4) [DOI] [PubMed] [Google Scholar]
- 68.Ingham PW, Taylor AM, Nakano Y. 1991. Role of the Drosophila patched gene in positional signalling. Nature 353, 184–187 (doi:10.1038/353184a0) [DOI] [PubMed] [Google Scholar]
- 69.Freeman M. 2000. Feedback control of intercellular signalling in development. Nature 408, 313–319 (doi:10.1038/35042500) [DOI] [PubMed] [Google Scholar]
- 70.Wartlick O, Mumcu P, Jülicher F, González-Gaitán M. 2011. Understanding morphogenetic growth control: lessons from flies. Nat. Rev. Mol. Cell Biol. 12, 594–604 (doi:10.1038/nrm3169) [DOI] [PubMed] [Google Scholar]
- 71.Hooper JE, Scott MP. 1989. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell 59, 751–765 (doi:10.1016/0092-8674(89)90021-4) [DOI] [PubMed] [Google Scholar]
- 72.Nakano Y, Guerrero I, Hidalgo A, Taylor A, Whittle JRS, Ingham PW. 1989. A protein with several possible membrane-spanning domains encoded by the Drosophila segment polarity gene patched. Nature 341, 508–513 (doi:10.1038/341508a0) [DOI] [PubMed] [Google Scholar]
- 73.Baker NE. 1988. Localization of transcripts from the wingless gene in whole Drosophila embryos. Development (Camb. Engl.) 103, 289–298 [DOI] [PubMed] [Google Scholar]
- 74.Swarup S, Verheyen EM. 2012. Wnt/wingless signaling in Drosophila. Cold Spring Harbor Perspect. Biol. 4, a007930 (doi:10.1101/cshperspect.a007930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frommer G, Vorbrüggen G, Pasca G, Jäckle H, Volk T. 1996. Epidermal egr-like zinc finger protein of Drosophila participates in myotube guidance. EMBO J. 15, 1642. [PMC free article] [PubMed] [Google Scholar]
- 76.Gallet A, Staccini-Lavenant L, Thérond PP. 2008. Cellular trafficking of the glypican dally-like is required for full-strength Hedgehog signaling and Wingless transcytosis. Dev. Cell 14, 712–725 (doi:10.1016/j.devcel.2008.03.001) [DOI] [PubMed] [Google Scholar]
- 77.Pavletich N, Pabo C. 1993. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science 261, 1701–1707 (doi:10.1126/science.8378770) [DOI] [PubMed] [Google Scholar]
- 78.Orenic TV, Slusarski DC, Kroll KL, Holmgren RA. 1990. Cloning and characterization of the segment polarity gene cubitus interruptus Dominant of Drosophila. Genes Dev. 4, 1053–1067 (doi:10.1101/gad.4.6.1053) [DOI] [PubMed] [Google Scholar]
- 79.Whitington T, Jolma A, Taipale J. 2011. Beyond the balance of activator and repressor. 4, pe29 (doi:10.1126/scisignal.2002183) [DOI] [PubMed] [Google Scholar]
- 80.Ptashne M, Gann A. 2002. Genes and signals. Cold Spring Harbor, NY: CSHL Press [Google Scholar]
- 81.Burz DS, Rivera-Pomar R, Jäckle H, Hanes SD. 1998. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 17, 5998–6009 (doi:10.1093/emboj/17.20.5998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heberlein U, Wolff T, Rubin GM. 1993. The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell 75, 913–926 (doi:10.1016/0092-8674(93)90535-X) [DOI] [PubMed] [Google Scholar]
- 83.Heberlein U, Singh CM, Luk AY, Donohoe TJ. 1995. Growth and differentiation in the Drosophila eye coordinated by hedgehog. Nature 373, 709–711 (doi:10.1038/373709a0) [DOI] [PubMed] [Google Scholar]
- 84.Lee JJ, Kessler von DP, Parks S, Beachy PA. 1992. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell 71, 33–50 (doi:10.1016/0092-8674(92)90264-D) [DOI] [PubMed] [Google Scholar]
- 85.Blackman RK, Sanicola M, Raftery LA, Gillevet T, Gelbart WM. 1991. An extensive 3’ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development (Camb. Engl.) 111, 657–666 [DOI] [PubMed] [Google Scholar]
- 86.Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. 1996. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature 382, 133–138 (doi:10.1038/382133a0) [DOI] [PubMed] [Google Scholar]
- 87.Jennings BH, Tyler DM, Bray SJ. 1999. Target specificities of Drosophila enhancer of split basic helix-loop-helix proteins. Mol. Cell. Biol. 19, 4600–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uv AE, Thompson CR, Bray SJ. 1994. The Drosophila tissue-specific factor Grainyhead contains novel DNA-binding and dimerization domains which are conserved in the human protein CP2. Mol. Cell. Biol. 14, 4020–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Uv AE, Harrison EJ, Bray SJ. 1997. Tissue-specific splicing and functions of the Drosophila transcription factor Grainyhead. Mol. Cell. Biol. 17, 6727–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Furriols M, Bray S. 2001. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 11, 60–64 (doi:10.1016/S0960-9822(00)00044-0) [DOI] [PubMed] [Google Scholar]
- 91.Blair SS. 1992. Engrailed expression in the anterior lineage compartment of the developing wing blade of Drosophila. Development (Camb. Engl.) 115, 21–33 [DOI] [PubMed] [Google Scholar]
- 92.Raftery LA, Sanicola M, Blackman RK, Gelbart WM. 1991. The relationship of decapentaplegic and engrailed expression in Drosophila imaginal disks: do these genes mark the anterior–posterior compartment boundary? Development (Camb. Engl.) 113, 27–33 [DOI] [PubMed] [Google Scholar]
- 93.Bray SJ, Kafatos FC. 1991. Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev. 5, 1672–1683 (doi:10.1101/gad.5.9.1672) [DOI] [PubMed] [Google Scholar]
- 94.Kheradpour P, Stark A, Roy S, Kellis M. 2007. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 17, 1919–1931 (doi:10.1101/gr.7090407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meireles-Filho ACA, Stark A. 2009. Comparative genomics of gene regulation-conservation and divergence of cis-regulatory information. Curr. Opin. Genet. Dev. 19, 565–570 (doi:10.1016/j.gde.2009.10.006) [DOI] [PubMed] [Google Scholar]
- 96.Campbell G, Weaver T, Tomlinson A. 1993. Axis specification in the developing Drosophila appendage: the role of wingless, decapentaplegic, and the homeobox gene aristaless. Cell 74, 1113–1123 (doi:10.1016/0092-8674(93)90732-6) [DOI] [PubMed] [Google Scholar]
- 97.Campbell G, Tomlinson A. 1998. The roles of the homeobox genes aristaless and Distal-less in patterning the legs and wings of Drosophila. Development (Camb. Engl.) 125, 4483–4493 [DOI] [PubMed] [Google Scholar]
- 98.Han K, Levine MS, Manley JL. 1989. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell 56, 573–583 (doi:10.1016/0092-8674(89)90580-1) [DOI] [PubMed] [Google Scholar]
- 99.Fisher WW, et al. 2012. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc. Natl Acad. Sci. USA 109, 21 330–21 335 (doi:10.1073/pnas.1209589110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tanay A. 2006. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 16, 962–972 (doi:10.1101/gr.5113606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rowan S, Siggers T, Lachke SA, Yue Y, Bulyk ML, Maas RL. 2010. Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity. Genes Dev. 24, 980–985 (doi:10.1101/gad.1890410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. 2012. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell 148, 273–284 (doi:10.1016/j.cell.2011.10.047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nahmad M, Lander AD. 2011. Spatiotemporal mechanisms of morphogen gradient interpretation. Curr. Opin. Genet. Dev. 21, 726–731 (doi:10.1016/j.gde.2011.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jaeger SA, Chan ET, Berger MF, Stottmann R, Hughes TR, Bulyk ML. 2010. Conservation and regulatory associations of a wide affinity range of mouse transcription factor binding sites. Genomics 95, 185–195 (doi:10.1016/j.ygeno.2010.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Michelson AM. 2002. Deciphering genetic regulatory codes: a challenge for functional genomics. Proc. Natl Acad. Sci. USA 99, 546–548 (doi:10.1073/pnas.032685999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tuch BB, Li H, Johnson AD. 2008. Evolution of eukaryotic transcription circuits. Science 319, 1797–1799 (doi:10.1126/science.1152398) [DOI] [PubMed] [Google Scholar]
- 107.He X, Duque TSPC, Sinha S. 2012. Evolutionary origins of transcription factor binding site clusters. Mol. Biol. Evol. 29, 1059–1070 (doi:10.1093/molbev/msr277) [DOI] [PMC free article] [PubMed] [Google Scholar]