Abstract
Objective
CD14 is a glycosylphosphotidylinositol-(GPI)-anchored membrane glycoprotein expressed on neutrophils and monocytes/macrophages that also circulates as a soluble form (sCD14). Despite the well-recognized role of CD14 in inflammation, relatively little is known about the genetic determinants of sCD14 or the relationship of sCD14 to vascular- and aging-related phenotypes.
Methods and Results
We measured baseline levels of sCD14 in over 5,000 European-American (EA) and African-American (AA) adults 65 years and older from the Cardiovascular Health Study, who were well-characterized at baseline for atherosclerotic risk factors and subclinical cardiovascular disease (CVD), and who have been followed for clinical CVD and mortality outcomes up to 20 years. At baseline, sCD14 generally showed strong positive correlations with traditional cardio-metabolic risk factors and with subclinical measures of vascular disease such as carotid wall thickness and ankle-brachial index (independently of traditional CVD risk factors), and was also inversely correlated with body mass index. In genome-wide association analyses of sCD14, we (a) confirmed the importance of the CD14 locus on chromosome 5q21 in EA; (b) identified a novel African ancestry-specific allele of CD14 associated with lower sCD14 in AA; (c) identified a putative novel association in EA of a non-synonymous variant of PIGC, which encodes an enzyme required for the first step in GPI anchor biosynthesis. Finally, we show that, like other acute phase inflammatory biomarkers, sCD14 predicts incident CVD, and strongly and independently predicts all-cause mortality in older adults.
Conclusion
CD14 independently predicts risk mortality in older adults.
Keywords: CD14, glycosylphosphotidylinositol, coronary heart disease, mortality
INTRODUCTION
CD14 is a glycosylphosphotidylinositol-(GPI)-anchored membrane glycoprotein constitutively expressed on monocytes/macrophages and neutrophils [1]. Cell-surface or membrane (m)CD14 binds to lipopolysaccharide (LPS) and other microbial and non-microbial ligands in the presence of another accessory protein, lipopolysaccharide binding protein (LBP), resulting in activation of various toll-like receptors and downstream pro-inflammatory pathway signaling [2,3]. CD14 also exists as a soluble form (sCD14) [4]. In addition to secretion from liver, sCD14 may be derived by enzymatic cleavage of GPI-anchored cell membrane-bound CD14 [4-9].
In response to inflammation and infection, interleukin (IL)-6 induces hepatic sCD14 expression; therefore circulating sCD14 is regarded as an acute phase protein [10]. Like other acute phase proteins, sCD14 levels increase in the setting of acute and chronic infectious and inflammatory diseases, including atherosclerosis. Similar to membrane-bound CD14, sCD14 is able to mediate LPS-activation of mCD14-negative cells such as endothelial and smooth muscle cells [11,12], events that are important for the development of atherosclerosis and its complications. At higher concentrations, LBP and sCD14 down-regulate LPS-induced responses [13] by transfer of LPS to lipoproteins for subsequent removal.
The genetic and environmental determinants of sCD14 are largely unknown, though CD14 expression appears to be at least partially regulated at the genetic level in a cell-type specific manner. The human CD14 gene on chromosome 5q23-31 contains a common proximal promoter polymorphism encoding a C/T transition at bp -159 from the major transcription start site (TSS) that has been associated with circulating sCD14 levels [14,15]. A regulatory element located further upstream of the CD14 gene is critical for sCD14 expression in hepatocytes, but not in monocytes [9].
Despite its potential importance for atherosclerosis and other aging-related inflammatory diseases, sCD14 has not been well-characterized in prospective, population-based studies of older adults. We therefore measured and analyzed sCD14 in the Cardiovascular Health Study (CHS), a large cohort of older adults with follow-up for incident CVD and mortality for up to 20 years. We first characterized the cross-sectional relationship of sCD14 to other traditional CVD risk factors and subclinical vascular disease, as well as other inflammatory CVD biomarkers (fibrinogen, CRP and IL-6). Second, we conducted genome-wide and CVD candidate gene-centric association scans of sCD14 in the CHS, separately in European Americans (EA) and African Americans (AA). Finally, we assessed the ability of sCD14 and its main genetic determinants to predict incident CVD and mortality during follow-up.
METHODS
CHS Cohort
The CHS is a prospective population-based cohort study of older adult men and women recruited from 4 US field centers: Forsyth County, North Carolina; Sacramento County, California; Washington County, Maryland; and Pittsburgh, Pennsylvania. 5201 participants (original cohort) were recruited in 1988-1989 [16]. An additional 687 primarily black participants were recruited in 1992–1993 (minority cohort) for a total cohort of 5888. The CHS baseline evaluation included demographic, lifestyle and medical histories, physical examination, fasting blood collection, and assessment of subclinical vascular disease by carotid ultrasound and ankle-brachial index (ABI) [17]. Informed consent was obtained using methods approved by institutional review committees at each study site. Genome-wide genotyping was performed on 2,952 EA and 528 AA unrelated CHS participants with sCD14 measurements using the Illumina 370CNV platform. Details of the methods for measurement of plasma biomarkers, genotyping, fine-mapping, ascertainment of clinical CVD events during follow-up adjudication are described in the online-only Data Supplement.
Statistical Analysis
Cross-sectional associations between sCD14 and quantitative traits [body mass index (BMI), lipid levels, fasting glucose, insulin, CRP, IL-6, ABI, and carotid intima media thickness (IMT)] or binary traits (diabetes, hypertension) were assessed using multiple linear regression or logistic regression, respectively. Hypertension is defined as systolic BP>140, diastolic BP>90, or current use of anti-hypertensive medication. CRP and IL-6 were log-transformed to reduce skewness. To test for genetic associations with sCD14, linear regression was performed, assuming an additive genetic model, using PLINK [18] for the genotyped data and Mach2qtl for the imputed data, adjusting for covariates of age, sex, and the first 10 eigenvectors derived from principal components analysis. For AAs, sCD14 was analyzed using log-transformed values in the genetic association analyses to assess statistical significance; means and standard errors were reported from the corresponding untransformed model for comparisons with EAs. Haplotype frequency estimation and association analyses were performed using the haplo.stats package for R [19]. Estimation of sCD14 phenotypic variance explained by genome-wide SNPs was performed using the Genome-wide Complex Trait Analysis (GCTA) method, as described in the online-only Data Supplement.
We performed Cox proportional hazards models to test for association between sCD14 (or other inflammation biomarkers) and risk of incident CVD events or mortality. sCD14 and other biomarkers were analyzed continuously to estimate the effects of a one standard deviation (SD) difference in value, as well as categorically using quintiles. The first quintile represented the reference group. For the continuous analysis, IL-6 and CRP were log transformed. Persons with adjudicated baseline prevalent disease relevant for each outcome were excluded prior to analysis. Four Cox-proportional hazard models containing progressive levels of covariate adjustment were used to assess risk of incident events associated with baseline sCD14 levels. The first model was minimally adjusted for demographic characteristics (age, sex, and race). The second model was additionally adjusted for traditional CVD risk factors (hypertension, systolic blood pressure diabetes, smoking, LDL cholesterol). For analysis of mortality, the second model also included cystatin C, and baseline peripheral artery disease, stroke, coronary heart disease. The third model included additional adjustment for subclinical vascular disease as assessed by ankle brachial index (ABI) and internal carotid wall thickness. The fourth model additionally included adjustment for plasma levels of other inflammation biomarkers, CRP, IL-6 and fibrinogen. Hazard ratios and 95% confidence intervals (CI) (HRs, 95% CI) for biomarkers were compared qualitatively with those of other inflammation biomarkers.
Assessment of association between CD14 genotype and CVD in Women’s Health Initiative
To assess the association between CD14 SNP genotype and CVD, we utilized data from a larger nested case-control sample of incident coronary heart disease and stroke from the Women’s Health Initiative [20]. The sample included 3,042 MI and 2,222 stroke EA cases and 296 MI and 251 stroke AA cases. There were 2,943 EA and 295 AA controls without CHD or stroke. Genotyping and QC of CD14 SNPs was performed using the IBC array at TGen (Phoenix, Arizona). Logistic regression models were adjusted for traditional risk factors (age, BMI, smoking, diabetes, hypertension, cholesterol) and principal components to control for population stratification.
RESULTS
Relationship of sCD14 with baseline CVD risk factors and other inflammation biomarkers
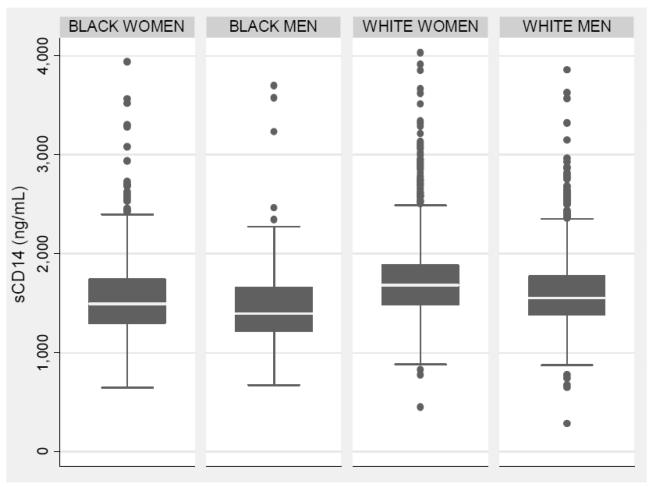
CHS participant characteristics are summarized in Table I in the online-only Data Supplement, by race. sCD14 levels were approximately normally distributed with a mean ± S.D. of 1641 ± 348 (range 284 to 4030) ng/mL. sCD14 levels increased with age and differed significantly by sex (P<0.001) and by race (P<0.001). The mean values in EA women, EA men, AA women, and AA men were 1714 ± 354, 1596 ± 329, 1551 ± 410, and 1454 ± 392 ng/mL, respectively (Figure 1). The Spearman rank correlation coefficient between sCD14 and CRP, fibrinogen, and IL-6 were 0.22, 0.21, and 0.17, respectively (all P<0.001). In age, race, and sex adjusted models, sCD14 was positively correlated with baseline smoking, diabetes, fasting glucose, hypertension and negatively correlated with BMI (all P<0.001) (Table 1). The differences in sCD14 by race and sex persisted upon adjustment for CVD risk factors (Table 1). When adjusted for baseline CHD and CVD risk factors, sCD14 was strongly and independently associated with other inflammatory and acute phase proteins, including CRP, IL-6, and fibrinogen, as well as with increased internal carotid artery wall thickness and decreased ABI (all P<0.001).
Figure 1. Distribution of soluble CD14 according to ethnicity and sex.
Median and inter-quartile range are indicated by the middle band and box edges, respectively. The whiskers indicate upper and lower adjacent values. Dots indicate values outlying 1.5 times the IQR above the 75th percentile and 1.5 times the IQR below the 25th percentile.
TABLE 1.
Association between soluble CD14 and other cardiovascular risk factors and measures of subclinical cardiovascular disease at the CHS baseline exam
| Baseline Characteristic | Model A Beta ± SE |
Model B Beta ± SE |
Model C Beta ± SE |
|---|---|---|---|
| Age (years) | 7.13 ± 0.86* | 6.60 ± 0.88* | 4.34 ± 0.90* |
| Female sex | 119 ± 9.7* | 123 ± 9.6* | 118 ± 10.3* |
| African American race | −155 ± 13.3* | −164 ± 13.5* | −185 ± 13.3* |
| Current smoking | 96 ± 15* | 84 ± 15* | 41 ± 15** |
| Diabetes | 59 ± 10* | 66 ± 11* | 35 ± 11** |
| Hypertension | 74 ± 10* | 78 ± 10* | 54 ± 10* |
| Systolic blood pressure (mm Hg) | 0.49 ± 0 .22 | 0.26 ± 0.22 | 0.18 ± 0.22 |
| LDL cholesterol (mg/dL) | 0.34 ± 0.12 | 0.44 ± 0.13** | 0.43 ± 0.13** |
| HDL cholesterol (mg/dL) | −1.02 ± 0.32** | −0.96 ± 0.34** | 0.38 ± 0.34 |
| Triglycerides (mg/dL) | 17.1 ± 11.1 | 4.62 ± 11.54 | −30 ± 13.4 |
| Glucose (mg/dL) | 1.07 ± 0.13* | 1.00 ± 0.16* | 0.73 ± 0.16* |
| Insulin (mg/dL) | 0.84 ± 0.18* | 0.70 ± 0.18* | 0.40 ± 0.18 |
| BMI (kg/m2) | −4.87 ± 1.03* | −4.87 ± 1.03* | −13.0 ± 1.12* |
| Waist circumference (cm) | −1.14 ± 0.38** | 0.316 ± 0.674 | −1.15 ± 0.658 |
| C-reactive protein (g/L) | 95 ± 4.5* | 101 ± 4.7* | 62 ± 6.0* |
| Interleukin-6 (pg/dL) | 117 ± 7.8* | 117 ± 8.1* | 51 ± 9.3* |
| Fibrinogen (mg/dL) | 1.28 ± 0.069* | 1.26 ± 0.070* | 0.55 ± 0.085* |
| Internal carotid wall thickness (mm) | 75.6 ± 8.80* | 55.6 ± 8.98* | 36.7 ± 8.96* |
| Ankle-brachial index | −273 ± 27.8 * | −198 ± 28.9* | −113 ± 29.1* |
Triglycerides, C-reactive protein, and interleukin-6 were natural log-transformed.
Model A: adjusted for age, race, and sex.
Model B: adjusted for age, race, sex, smoking, diabetes, hypertension, systolic blood pressure, BMI.
Model C: adjusted for age, race, sex, smoking, diabetes, hypertension, systolic blood pressure, BMI, LDL cholesterol, HDL cholesterol, C-reactive protein, interleukin-6, and fibrinogen.
In all models, the estimates for age, race, and sex were adjusted only for the two remaining variables.
P<0.0001
P<0.01
Estimation of proportion of sCD14 phenotypic variation explained by genome-wide SNPs
To our knowledge, family-based estimates of sCD14 heritability have not been previously reported. Therefore, we estimated the amount of phenotypic variance of sCD14 explained by “distant” genetic relationships between individuals with the CHS EA population by applying the GCTA method using all genotyped autosomal SNPs. The estimate of sCD14 phenotypic variance explained by genome-wide SNPs was 33 ± 17% (P = 0.03).
Genome-wide association scan of sCD14 in EA and AA
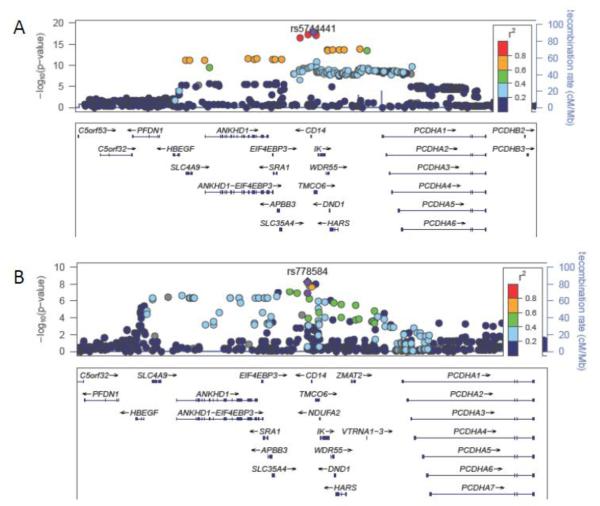
In the genome-wide association scan of 2,952 EA and 528 AA, two regions, 1q24 and 5q31, reached genome-wide significance in EA (Figure I in the online-only Data Supplement); the chromosome 5q31 peak was also genome-wide significant in AA (Figure II in the online-only Data Supplement). Results for the lead (index) SNPs associated with sCD14 in each genomic region are summarized in Table 2. In EA, the strongest genome-wide association signal was located on chromosome 5q31, centered around the CD14 structural gene. The 5q31 region of association spanned ~500 kb, encompassed 16 additional genes and included an extended block of LD containing 165 SNPs with P<5 × 10−8 (Figure IIIA in the online-only Data Supplement). The minor allele at the EA index SNP rs5744441 (MAF=0.22; imputation quality score = 0.99) was associated with 101 ± 10 ng/mL lower sCD14 (P= 3.0 × 10−23). The rs5744441 SNP is located about 3 kb upstream of CD14.
Table 2.
Top genome-wide association results for soluble CD14 in European Americans and African Americans
| Population | Chromosome | Number SNPS with P<5×10−8 |
Top SNP in region |
Position Hg18 |
Candidate genes | Minor allele | Minor allele frequency |
Effect size (Standard Error) [ng/mL sCD14] |
P-value |
|---|---|---|---|---|---|---|---|---|---|
| European- Americans |
5q31 | 144 | rs5744441 | 139997031 |
HBEGF, SLC4A9,
ANKHD1, EIF4EBP3, SRA1, APBB3, SLC35A4, CD14, IK, WDR55, TMCO6, DND1, HARS, HARS2, NDUFA2,ZMAT2, VTRNA1 |
G | 0.22 | −101 (10) | 2.98 × 10−23 |
| African- Americans |
5q31 | 1 | rs778584 | 139985396 | CD14 | T | 0.31 | 128 (24) | 5.5 × 10−9 |
| European- Americans |
1q24 | 19 | rs1063412 | 170677590 | PIGC | A | 0.43 | −48 (8.5) | 1.40 × 10−8 |
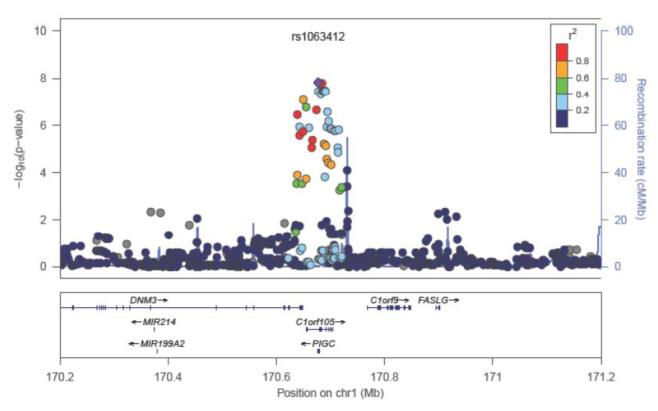
In the EA GWAS, a second region on chromosome 1q24.3 achieved genome-wide significance (Table 2 and Figure IV in the online-only Data Supplement). The index SNP rs1063412 at 1q24.3 is a C/T polymorphism that encodes a Pro266Ser amino acid substitution within the second exon of PIGC (MAF=0.43). According to the HapMap, the C allele (Pro266) is the minor allele in European and Asian populations, but is the major allele in African populations. The C allele (MAF=0.43 in CHS EA) was associated with 48 ± 8 ng/mL lower sCD14 (P=1.4 × 10−8).
In AA, there was also a genome-wide significant association at the CD14 locus, but the specific SNPs associated with sCD14 were distinct from those in EA (Table 2 and Figure IIIB in the online-only Data Supplement). The index SNP in AA rs778584 (MAF=0.31) was located in the 3′ flanking region and was associated with 128 ng/mL ± 24 higher sCD14 (P= 5.5 × 10−9). The EA index SNP at the CD14 locus rs5744441 was less common (MAF=0.08) in AA and associated more weakly with sCD14 (Beta=-108 ± 38 ng/mL; P=0.003). In AA, the PIGC rs1063412 C allele (Pro266) was considerably more common (MAF=0.83) than in EA. Nonetheless, the PIGC rs1063412 Pro266 variant was not significantly associated with sCD14 in AA (P=0.95) (Figure V in the online-only Data Supplement).
Haplotype analysis and fine-mapping of CD14 gene region
To further explore the CD14 gene variants associated with sCD14 in EA and AA, we utilized denser CD14 genotype data from the IBC array which contains 15 SNPs covering the CD14 coding, intronic, untranslated, and flanking regions. In EA, the most strongly associated SNP on the IBC array was rs5744455 (Beta=-101 ± 9.5 ng/mL; P=3.5 × 10−26), located about 50 bp upstream of the transcription start site. On the basis of 1000 Genomes CEU data, this SNP is in strong LD with the index GWAS SNP, rs5744441 (LD r2 = 1.0). After conditioning on rs5744455, P-values for 3 additional SNPs in the CD14 region were significant at the Bonferroni-adjusted threshold of P<0.003: rs2569191 and rs778584 (which are in strong LD; r2=0.97) and rs4914, suggesting the possibility of allelic heterogeneity at the CD14 locus. Together with rs5744455, rs4914 (a synonymous coding variant) and rs778584 define four common CD14 EA haplotypes (Table 3): Compared to referent haplotype 1, haplotype 4 (tagged by the minor allele of rs5744455) was associated with lower sCD14, while haplotype 3 (tagged by the minor allele of rs4914) and haplotype 2 (tagged by the minor allele of rs778584) were each associated with higher sCD14 levels.
Table 3.
Association of CD14 haplotypes with soluble CD14 in European Americans and African Americans
| CD14 SNP | European Americans | African Americans | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | rs778584 | rs4914 | rs5744455 | rs5744451 | Freq | Beta (SE) | P-value | Freq | Beta (SE) | P-value |
| H1 | C | C | G | A | 0.17 | Referent | --- | 0.44 | Referent | -- |
| H2 | T | C | G | A | 0.49 | 67 (11) | 6.15e-08 | 0.31 | 51 (25) | 0.04 |
| H3 | C | G | G | A | 0.12 | 87 (15) | 3.23e-08 | 0.07 | 26 (44) | 0.55 |
| H4 | C | C | A | A | 0.21 | −46 (12) | 0.0002 | 0.08 | −93 (40) | 0.02 |
| H5 | C | C | G | C | <.01 | NA | NA | 0.09 | −262 (37) | 3.2e-013 |
SE = standard error
The global P-value for CD14 haplotype association with sCD14 in EA = 6.65e-31
The global P-value for CD14 haplotype association with sCD14 in AA = 1.28e-12
In AA, several other SNPs on the IBC array had more significant P-values than the index SNP from the AA GWAS rs778584. The lead SNP from the IBC analysis was rs5744451 (MAF=0.09), which was associated with 279 ± 36 ng/mL lower sCD14 (P= 1.3 × 10−14). rs5744451 is located in the 5′ flanking region of CD14. This SNP was not successfully imputed and hence not included in the GWAS analysis. After conditioning on rs5744451, three additional CD14 SNPs (rs5744455, rs2569191, and rs778584) had P<0.003. Haplotype analysis confirmed that rs5744451 tags a second CD14-lowering haplotype that is common among AA but rare among EA (Table 3).
Relationship of sCD14 to incident CVD and mortality
Among the 5,462 EA and AA participants with sCD14 measured at the CHS baseline exam, there were 820 incident MI, 1,547 incident CHD, 945 incident strokes and 3,820 deaths during the 20-year follow-up period. Of the 3,820 deaths, 1519 were due to cardiovascular causes and 2,290 were due to non-cardiovascular causes. The relationships between sCD14 and these outcomes are shown in Table 4. When minimally-adjusted for age, sex, race, baseline sCD14 was associated with increased risk of MI, CHD, stroke, and mortality. Additional adjustment for traditional risk factors and measures of subclinical vascular disease attenuated these associations, though the associations with CHD, stroke, and mortality remained significant. Moreover, the association of sCD14 with mortality and stroke (but not MI or CHD) was independent of the other studied biomarkers (CRP, IL-6, fibrinogen). In the fully-adjusted model 4, sCD14 was associated similarly with death due to cardiovascular causes (HR= 1.13; 95%CI = 1.05 to 1.22; p= 0.001) and death due non-cardiovascular causes (HR= 1.11; 95%CI = 1.04 to 1.18; p=0.001). Table II in the online-only Data Supplement shows the association of each inflammation biomarker with mortality in multivariable models including all the inflammation biomarkers. In these models, sCD14 and IL-6 were each independently associated with mortality, while CRP and fibrinogen were not. When analyzed categorically, there was a graded, linear increase in mortality risk with each successive sCD14 quintile. Compared to the bottom quintile, the hazard ratios (95% confidence intervals) for the 2nd, 3rd, 4th, and 5th quintile were 1.00 (0.87 - 1.14), 1.02 (0.89 - 1.16), 1.12 (0.98 - 1.27), and 1.27 (1.11 - 1.45), respectively. The HRs comparing top to bottom quintiles for cardiovascular death and non-cardiovascular were 1.26 (95%CI 1.01 -1.57; p=0.04) and 1.32 (95%CI 1.11 - 1.57; p=0.002), respectively.
Table 4.
Estimated hazard ratios for association between 1 standard deviation unit increase in soluble CD14 and incident CVD events and mortality
| Mortality (n=3,820 ) | MI (n=870) | CHD (n=1,547) | Stroke (n=945) | |||||
|---|---|---|---|---|---|---|---|---|
| Model | HR (95%CI) | P | HR (95%CI) | P | HR (95%CI) | P | HR (95%CI) | P |
| Minimal (1) | 1.25 (1.21, 1.29) |
1.2 × 10−42 | 1.10 (1.03, 1.19) |
0.006 | 1.14 (1.09, 1.20) |
2.4 × 10−7 | 1.15 (1.07, 1.23) |
5.2 × 10−5 |
| Multivariable (2) | 1.11 (1.07, 1.15) |
5.5 × 10−9 | 1.07 (0.99, 1.15) |
0.075 | 1.11 (1.06, 1.18) |
7.5 × 10−5 | 1.12 (1.04, 1.20) |
0.002 |
| Subclinical (3) | 1.09 (1.06, 1.13) |
1.3 × 10−6 | 1.03 (0.96, 1.11) |
0.38 | 1.07 (1.01, 1.13) |
0.017 | 1.11 (1.03, 1.19) |
0.004 |
| Biomarkers (4) | 1.09 (1.05, 1.13) |
2.9 × 10−5 | 0.96 (0.89, 1.05) |
0.39 | 1.01 (0.95, 1.07) |
0.73 | 1.08 (1.00, 1.17) |
0.041 |
Model Definitions: 1) Age, sex, race
2) Model 1 + smoking, diabetes, hypertension, SBP, LDL (baseline CVD, cystatin C)
3) Model 2 + carotid IMT and ankle-brachial index
4) Model 3 + C-reactive protein, interleukin-6, and fibrinogen
Among the sCD14-associated SNPs identified in our GWAS, there was no association with risk of mortality in either ethnicity of CHS (Table III in the online-only Data Supplement). In CHS, the minor alleles of CD14 rs4914 and rs778584 were each nominally associated with 1.3 to 1.6 fold increased risk of stroke in AA, while rs778584 was associated with decreased risk of CHD in EA (P=0.025). However, these results were not significant when corrected for multiple hypothesis testing. Nor was there significant evidence for association between CD14 genotype and risk of CHD or stroke in a second, large case-control sample from WHI (Table IV in the online-only Data Supplement).
DISCUSSION
In a bi-racial population of older adults, we have characterized the epidemiologic and genetic associations of sCD14, as well as relationships of this inflammatory biomarker to incident CVD and mortality. Baseline sCD14 was correlated positively with EA ethnicity, female sex, traditional CVD risk factors (smoking, hypertension, diabetes), and other inflammatory biomarkers (IL-6, CRP, fibrinogen), and negatively correlated with BMI. We identified two genome-wide significant sCD14-associated loci, the CD14 structural locus on chromosome 5q21 and a novel missense variant of PIGC, which encodes an enzyme required for the first step in GPI anchor biosynthesis. We also showed that sCD14 predicts incident CVD and mortality in older adults.
Using a combined GWAS and fine-mapping approach, we identified intra- and inter-population allelic heterogeneity at the CD14 locus, including two new CD14 variants associated with lower sCD14 levels. The significant, non-redundant variants at the CD14 locus explain 3.7% of the trait variability in EA versus 9.0% in AA. One of these variants (haplotype 4 tagged by rs5744455 in Table 3) was associated with lower sCD14 in both EA and AA. rs5744455 may represent a functional variant, as this SNP is located about 50 bp upstream of the TSS within an ENCODE DNaseI hypersensitivity site in CD14+ monocytes and promyelocytic cells. DNaseI hypersensitivity sites are characterized by open chromatin and tend to be transcriptionally active. The other newly discovered sCD14-lowering variant was present only among AA (haplotype 5 tagged by rs5744451 in Table 3. Of the SNPs associated with haplotype 5, rs5744430 lies within a 700 bp hepatocyte-specific enhancer region located ~6 kb 5′ of the TSS [9,21]. rs5744430 is also located within several putatively functional genomic elements indicative of chromatin accessibility, including an ENCODE DNaseI hypersensitivity site, histone methylation and acylation enrichment sites, and hepatocyte-specific transcription factor binding elements for HNF4A- and FOXA1, as determined by genome-wide ChIP-Seq [22].
Two other CD14 variants (haplotypes 2 and 3 in Table 3) were associated with higher sCD14 levels. Haplotype 2 (tagged by rs778584 in our data set) contains the previously characterized -159 T allele of rs2569190, which is located in the proximal promoter [23]. The CD14 -159 T allele polymorphism increases transcription of CD14 in monocytes by decreasing binding affinity to the inhibitory Sp3 subunit of an Sp transcription factor site [15] and has been associated with higher sCD14 concentration [14] as well as greater density of mCD14 in some studies [24] but not in others [25,26].
Despite the greater burden of CVD risk factors and higher levels of other commonly measured inflammation biomarkers among AA, we found that levels of sCD14 were lower in AA than EA. Whether the ethnic differences in sCD14 are due to environmental and/or genetic factors is uncertain. The CD14 gene locus shows evidence of balancing selection, particularly among European populations [27], which could be due to its impact on the magnitude of host immune response to local pathogens. It is also noteworthy that the two CD14 alleles tagged by rs778584 and rs4914 associated with higher sCD14 are both more common in EA than AA, while the sCD14-lowering haplotype tagged by rs5744451 is common only among AA. Assuming independence of SNPs, and an additive genetic model, differences in allele frequencies between the identified SNPs accounts for approximately 23% of the inter-population differences in sCD14 levels. Therefore, allele frequency differences at the CD14 locus provide some potential explanation for the higher sCD14 levels observed in EA.
CD14 is one of more than 100 cell-surface proteins anchored to the plasma membrane via GPI. Assembly of GPI-linked proteins occurs in the endoplasmic reticulum and involves more than 30 different genes [28]. The first step of the biosynthesis, the transfer of N-acetylglucosamine from UDP-N-acetylglucosamine to phosphatidylinositol, is mediated by at least three genes (PIGA, PIGH and PIGC). Somatic mutations in the X-linked gene phosphatidylinositol glycan class A (PIGA) in hematopoietic stem cells cause paroxysmal nocturnal hemoglobinuria (PNH), which manifests as bone-marrow failure, hemolytic anemia, and thrombosis. As a result of the GPI anchoring defect, PNH patients do not express mCD14 on their monocytes, yet have normal plasma levels of sCD14 [6]. Notably, the Pro266Ser amino acid substitution encoded by PIGC rs1063412 is predicted to be functionally deleterious by Condel [29]. The association of higher sCD14 with a functional coding variant of PIGC suggests that defective GPI anchor synthesis may be a mechanism that results in increased release of intracellular CD14 into the circulation from myeloid cells. While the association reached genome-wide significance in EA and is certainly biologically plausible, verification of the association between PIGC rs1063412 and sCD14 in an independent EA population is necessary to authenticate the finding.
Germline mutations in GPI anchor genes have been identified in congenital autosomal recessive disorders that characterized by a diverse range of phenotypes. To our knowledge, this is the first report of a common, inherited variant of a gene involved in GPI anchor biosynthesis associated with a complex trait. A recent GWAS identified a SNP located about 70 kb downstream of PIGC associated with waist-hip ratio (WHR) [30]. However, there is no LD between the WHR-associated rs1011731 variant and the sCD14-associated PIGC coding variant. In the current study, we were unable to detect an association between the PIGC locus and sCD14 in AA. Even though the variant is more common in AA, our AA sample size was considerably smaller than our EA sample; therefore, power was only 43% to detect an effect of similar magnitude in AA. Other possible explanations for the lack of replication in AA include (1) a false-positive result (type I error) or overestimation of the effect (“winner’s curse”) in EA; (2) effect modification by another genetic or environmental factor that is differentially expressed between EA and AA; (3) rs1063412 may not be true causal variant and the frequency of the true causal variant differs between EA and AA and/or is not well-tagged in AA.
Our results show that sCD14 levels are associated with both subclinical vascular disease (carotid IMT and ABI) and with risk of future clinical CVD in older adults, independently of other risk factors. The reason for the negative association between sCD14 and BMI is unclear, though the inter-relationships between the CD14 system and obesity appear to be complex. sCD14 can both potentiate and down-regulate responses to LPS. For example, adipose tissue CD14 gene expression is increased in obese humans [31], while mice harboring CD14 mutations have decreased body fat [31,32]. In contrast, administration of exogenous recombinant human sCD14 is associated with decreased visceral fat and adipocyte inflammatory gene expression in high-fat-fed mice. Another possible explanation for the paradoxical association with sCD14 observed in CHS is that lower BMI could constitute a marker for frailty and chronic disease in older adults. Therefore, additional studies of sCD14 and BMI involving younger populations and longitudinal data on BMI may be required.
Prior studies of sCD14 and subclinical atherosclerosis have been inconsistent, though most have included primarily younger and middle-aged subjects [33-36]. In a smaller prospective European study of healthy middle-aged men, sCD14 levels did not differ between CHD cases and controls [37]. Despite the association between sCD14 levels and CVD risk in CHS, we found little evidence of association between CD14 genotype and risk of CHD or stroke, in samples totaling over 4,000 CHD cases and 3,000 stroke cases. These results are consistent with those of a recent meta-analysis of previously published studies (11,813 CHD cases and 6,196 controls) which showed no evidence of association between the CD14 C-159T promoter polymorphism and CHD in Europeans or subjects from India, though a possible association between -159 T and CHD was observed among East Asians [38].
Several limitations of the current study should be noted. Replication in an independent sample of EA, and also studies including larger numbers of AA, will be required to validate the PIGC association and to determine whether the finding is ethnicity-specific. The remaining phenotypic variability in sCD14 not ascribed to heritable factors or loci identified in our analyses suggests a contribution of combined effects of rare variants not tagged by current genotyping arrays, non-additive genetic factors, as well environmental factors. Therefore, genotyping arrays of even greater density and/or targeted sequencing of the CD14 locus may ultimately be required to identify the full spectrum of trait-associated variants. Finally, further study of the inverse relationship of sCD14 with body mass, as well as generalization of these association findings to younger adults is needed.
In population studies of older adults, serum levels of CRP and IL-6 predict adverse health outcomes and mortality. Though sCD14 is an acute phase protein, and correlates with CRP and IL-6, we found that sCD14 and IL-6 were strong and independent risk factors for mortality in older adults. Accounting for IL-6 and sCD14, there was no association of CRP or fibrinogen with mortality. sCD14 levels are increased in a number of acute and chronic inflammatory conditions and higher levels have been associated with disease severity [39], including mortality in Gram-negative sepsis [40]. Conversely, in mouse models, CD14 deficiency has been associated with resistance to LPS-induced shock [41], while mice harboring TLR4 and CD14 mutations have increased bone mineral density, decreased body fat, greater insulin sensitivity, live long lives and show no evidence of infection [32]. Increasing evidence suggests that pro-inflammatory mediators play a direct role in aging-related chronic diseases such as atherosclerosis, diabetes and cancer. Whether higher levels of sCD14 or other inflammatory mediators are directly causal, or indicative of chronic, low-level infection and other immune alterations in older adults, or simply reflect worsening subclinical and clinical disease, is difficult to distinguish. Nonetheless, sCD14 may have potential clinical utility as a marker of aging, disease risk, or disease progression in older adults.
Supplementary Material
Supplemental Table I. Characteristics of CHS participants with soluble CD14 measured
Supplemental Table II: Risk of CVD outcomes and mortality associated with soluble CD14 and other inflammation biomarkers
SUPPLEMENTAL TABLE III: Association of sCD14-associated SNPs with incident CVD outcomes and mortality in EA and AA
SUPPLEMENTAL TABLE IV: Association of sCD14-associated SNPs with incident CHD and stroke in EA and AA from the Women’s Health Initiative
Supplemental Figure I. Manhattan Plot of soluble CD14 GWAS in European Americans
Supplemental Figure II. Manhattan Plot of soluble CD14 GWAS in African Americans
Supplemental Figure III: Regional association plots for chromosome 5q31.
Shown are soluble CD14 regional association plots generated using LocusZoom for the chromosome 5q31 locus in (A) European Americans; (B) African Americans. The color of each single nucleotide polymorphism (SNP) indicates the level of pairwise linkage disequilibrium (LD) based on r-squared relative to the lead SNP in the region. r-squared values were calculated from HapMap CEU for European Americans and YRI for African Americans. SNPs with missing LD information are shown in gray.
Supplemental Figure IV: Regional association plot for chromosome 1q24 in European Americans.
Shown is the soluble CD14 regional association plots generated using LocusZoom the chromosome 1q24 locus in European Americans. The color of each single nucleotide polymorphism (SNP) indicates the level of pairwise linkage disequilibrium (LD) based on r-squared relative to the lead SNP in the region. r-squared values were calculated from HapMap CEU. SNPs with missing LD information are shown in gray.
Supplemental Figure V. Regional association plot of 1q24 region in African Americans
Figure 2.
Figure 3.
ACKNOWLEDGEMENTS
Sources of Funding: The research reported in this article was supported by contracts HHSN268201200036C, N01-HC-85239, N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, and grant HL080295R01 and HL71862 from the National Heart, Lung, and Blood Institute (NHLBI), with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the National Institute on Aging (NIA).
Footnotes
Disclosures: None.
A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm..
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Alex P. Reiner, Departments of Medicine and Epidemiology and Genome Sciences, University of Washington, Seattle WA, 98195, USA
Ethan M. Lange, Department of Genetics, University of North Carolina, Chapel Hill, NC, 27599, USA
Nancy S. Jenny, Department of Medicine and Pathology, University of Vermont, Burlington, Vermont, USA
Paulo H.M. Chaves, Department of Medicine, Family Center for Geriatric Research and Education, Herbert Wertheim College of Medicine, Florida International University, Miami, FL 33199
Jaclyn Ellis, Department of Genetics, University of North Carolina, Chapel Hill, NC, 27599, USA.
Jin Li, Department of Genetics, University of North Carolina, Chapel Hill, NC, 27599, USA.
Jeremy Walston, Department of Medicine, Johns Hopkins School of Medicine, Baltimore, MD, USA.
Leslie A. Lange, Department of Genetics, University of North Carolina, Chapel Hill, NC, 27599, USA
Mary Cushman, Department of Medicine and Pathology, University of Vermont, Burlington, Vermont, USA.
Russell P. Tracy, Department of Medicine and Pathology, University of Vermont, Burlington, Vermont, USA
REFERENCES
- 1.Haziot A, Chen S, Ferrero E, Low MG, Silber R, Goyert SM. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J Immunol. 1988;141:547–52. [PubMed] [Google Scholar]
- 2.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–3. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 3.Tobias PS, Ulevitch RJ. Lipopolysaccharide binding protein and CD14 in LPS dependent macrophage activation. Immunobiology. 1993;187:227–32. doi: 10.1016/S0171-2985(11)80341-4. [DOI] [PubMed] [Google Scholar]
- 4.Bazil V, Baudys M, Hilgert I, Stefanová I, Low MG, Zbrozek J, Horejsí V. Structural relationship between the soluble and membrane-bound forms of human monocyte surface glycoprotein CD14. Mol Immunol. 1989;26:657–62. doi: 10.1016/0161-5890(89)90048-5. [DOI] [PubMed] [Google Scholar]
- 5.Bazil V, Strominger JL. Shedding as a mechanism of down-modulation of CD14 on stimulated human monocytes. J Immunol. 1991;147:1567–1574. [PubMed] [Google Scholar]
- 6.Durieux JJ, Vita N, Popescu O, Guette F, Calzada-Wack J, Munker R, Schmidt RE, Lupker J, Ferrara P, Ziegler-Heitbrock HW, et al. The two soluble forms of the lipopolysaccharide receptor, CD14: characterization and release by normal human monocytes. Eur J Immunol. 1994;24:2006–12. doi: 10.1002/eji.1830240911. [DOI] [PubMed] [Google Scholar]
- 7.Bufler P, Stiegler G, Schuchmann M, Hess S, Krüger C, Stelter F, Eckerskorn C, Schütt C, Engelmann H. Soluble lipopolysaccharide receptor (CD14) is released via two different mechanisms from human monocytes and CD14 transfectants. Eur J Immunol. 1995;25:604–10. doi: 10.1002/eji.1830250244. [DOI] [PubMed] [Google Scholar]
- 8.Su GL, Dorko K, Strom SC, Nüssler AK, Wang SC. CD14 expression and production by human hepatocytes. J Hepatol. 1999;31:435–42. doi: 10.1016/s0168-8278(99)80034-8. [DOI] [PubMed] [Google Scholar]
- 9.Pan Z, Zhou L, Hetherington CJ, Zhang DE. Hepatocytes contribute to soluble CD14 production, and CD14 expression is differentially regulated in hepatocytes and monocytes. J Biol Chem. 2000;275:36430–5. doi: 10.1074/jbc.M003192200. [DOI] [PubMed] [Google Scholar]
- 10.Bas S, Gauthier BR, Spenato U, Stingelin S, Gabay C. CD14 is an acute-phase protein. J Immunol. 2004;172:4470–9. doi: 10.4049/jimmunol.172.7.4470. [DOI] [PubMed] [Google Scholar]
- 11.Pugin J, Schürer-Maly CC, Leturcq D, Moriarty A, Ulevitch RJ, Tobias PS. Lipopolysaccharide activation of human endothelial and epithelial cells is mediated by lipopolysaccharide-binding protein and soluble CD14. Proc Natl Acad Sci U S A. 1993;90:2744–8. doi: 10.1073/pnas.90.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juan TS, Kelley MJ, Johnson DA, Busse LA, Hailman E, Wright SD, Lichenstein HS. Soluble CD14 truncated at amino acid 152 binds lipopolysaccharide (LPS) and enables cellular response to LPS. J Biol Chem. 1995;270:1382–7. doi: 10.1074/jbc.270.3.1382. [DOI] [PubMed] [Google Scholar]
- 13.Kitchens RL, Thompson PA. Modulatory effects of sCD14 and LBP on LPS-host cell interactions. J Endotoxin Res. 2005;11:225–9. doi: 10.1179/096805105X46565. [DOI] [PubMed] [Google Scholar]
- 14.Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FD. A Polymorphism* in the 5′ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol. 1999;20:976–83. doi: 10.1165/ajrcmb.20.5.3494. [DOI] [PubMed] [Google Scholar]
- 15.LeVan TD, Bloom JW, Bailey TJ, Karp CL, Halonen M, Martinez FD, Vercelli D. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J Immunol. 2001;167:5838–44. doi: 10.4049/jimmunol.167.10.5838. [DOI] [PubMed] [Google Scholar]
- 16.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–76. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 17.O’Leary DH, Polak JF, Wolfson SK, Jr, Bond MG, Bommer W, Sheth S, Psaty BM, Sharrett AR, Manolio TA. Use of sonography to evaluate carotid atherosclerosis in the elderly. The Cardiovascular Health Study. CHS Collaborative Research Group. Stroke. 1991;22:1155–63. doi: 10.1161/01.str.22.9.1155. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, Schaid DJ. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Hered. 2003;55:56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
- 20.Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control Clin Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 21.Hetherington CJ, Kingsley PD, Crocicchio F, Zhang P, Rabin MS, Palis J, Zhang DE. Characterization of human endotoxin lipopolysaccharide receptor CD14 expression in transgenic mice. J Immunol. 1999;162:503–9. [PubMed] [Google Scholar]
- 22.Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, Suh BB, Hinrichs AS, Clawson H, Zweig AS, Kirkup V, Fujita PA, Rhead B, Smith KE, Pohl A, Kuhn RM, Karolchik D, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–5. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang DE, Hetherington CJ, Tan S, Dziennis SE, Gonzalez DA, Chen HM, Tenen DG. Sp1 is a critical factor for the monocytic specific expression of human CD14. J Biol Chem. 1994;269:11425–34. [PubMed] [Google Scholar]
- 24.Hubacek JA, Rothe G, Pit’ha J, Skodova Z, Stanek V, Poledne R, Schmitz G. C(-260)->T polymorphism in the promoter of the CD14 monocyte receptor gene as a risk factor for myocardial infarction. Circulation. 1999;99:3218–3220. doi: 10.1161/01.cir.99.25.3218. [DOI] [PubMed] [Google Scholar]
- 25.Heesen M, Blomeke B, Schluter B, Heussen N, Rossaint R, Kunz D. Lack of association between the −260 C->T promoter polymorphism of the endotoxin receptor CD14 gene and the CD14 density of unstimulated human monocytes and soluble CD14 plasma levels. Intens. Care Med. 2001;27:1770–1775. doi: 10.1007/s001340101106. [DOI] [PubMed] [Google Scholar]
- 26.Ito D, Murata M, Tanahashi N, Sato H, Sonoda A, Saito I, Watanabe K, Fukuuchi Y. Polymorphism in the promoter of lipopolysaccharide receptor CD14 and ischemic cerebrovascular disease. Stroke. 2000;31:2661–2664. doi: 10.1161/01.str.31.11.2661. [DOI] [PubMed] [Google Scholar]
- 27.Ferrer-Admetlla A, Bosch E, Sikora M, Marquès-Bonet T, Ramírez-Soriano A, Muntasell A, Navarro A, Lazarus R, Calafell F, Bertranpetit J, Casals F. Balancing selection is the main force shaping the evolution of innate immunity genes. J Immunol. 2008;181:1315–22. doi: 10.4049/jimmunol.181.2.1315. [DOI] [PubMed] [Google Scholar]
- 28.Kinoshita T, Fujita M, Maeda Y. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J Biochem. 2008;144:287–94. doi: 10.1093/jb/mvn090. [DOI] [PubMed] [Google Scholar]
- 29.González-Pérez A, López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–9. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, Thorleifsson G, Zillikens MC, Speliotes EK, Mägi R, Workalemahu T, White CC, Bouatia-Naji N, Harris TB, Berndt SI, Ingelsson E, Willer CJ, Weedon MN, Luan J, Vedantam S, Esko T, Kilpeläinen TO, Kutalik Z, Li S, Monda KL, Dixon AL, Holmes CC, Kaplan LM, Liang L, Min JL, Moffatt MF, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernández-Real JM, Pérez del Pulgar S, Luche E, Moreno-Navarrete JM, Waget A, Serino M, Sorianello E, Sánchez-Pla A, Pontaque FC, Vendrell J, Chacón MR, Ricart W, Burcelin R, Zorzano A. CD14 modulates inflammation-driven insulin resistance. Diabetes. 2011;60:2179–86. doi: 10.2337/db10-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson GB, Riggs BL, Platt JL. A genetic basis for the “Adonis” phenotype of low adiposity and strong bones. FASEB J. 2004;18:1282–1284. doi: 10.1096/fj.04-1572fje. [DOI] [PubMed] [Google Scholar]
- 33.Risley P, Jerrard-Dunne P, Sitzer M, Buehler A, von Kegler S, Markus HS, Carotid Atherosclerosis Progression Study Promoter polymorphism in the endotoxin receptor (CD14) is associated with increased carotid atherosclerosis only in smokers: the Carotid Atherosclerosis Progression Study (CAPS) Stroke. 2003 Mar;34:600–4. doi: 10.1161/01.STR.0000055941.61801.5A. [DOI] [PubMed] [Google Scholar]
- 34.Hung J, McQuillan BM, Chapman CM, Thompson PL, Beilby JP. Promoter polymorphism of the gene for CD14 receptor is not associated with sub-clinical carotid atherosclerosis in a community population. Eur J Cardiovasc Prev Rehabil. 2004;11:344–9. doi: 10.1097/01.hjr.0000129741.07723.6c. [DOI] [PubMed] [Google Scholar]
- 35.Amar J, Ruidavets JB, Bal Dit Sollier C, Bongard V, Boccalon H, Chamontin B, Drouet L, Ferrières J. Soluble CD14 and aortic stiffness in a population-based study. J Hypertens. 2003;21:1869–77. doi: 10.1097/00004872-200310000-00014. [DOI] [PubMed] [Google Scholar]
- 36.Amar J, Ruidavets JB, Bal dit Sollier C, Bongard V, Boccalon H, Chamontin B, Drouet L, Ferrières J. CD14 C(-260)T gene polymorphism, circulating soluble CD14 levels and arteriosclerosis. J Hypertens. 2004 Aug;22(8):1523–8. doi: 10.1097/01.hjh.0000133724.16947.a3. [DOI] [PubMed] [Google Scholar]
- 37.Morange PE, Tiret L, Saut N, Luc G, Arveiler D, Ferrieres J, Amouyel P, Evans A, Ducimetiere P, Cambien F, Juhan-Vague I, PRIME Study Group TLR4/Asp299Gly, CD14/C-260T, plasma levels of the soluble receptor CD14 and the risk of coronary heart disease: The PRIME Study. Eur J Hum Genet. 2004;12:1041–9. doi: 10.1038/sj.ejhg.5201277. [DOI] [PubMed] [Google Scholar]
- 38.Zhang HF, Zhong BL, Zhu WL, Xie SL, Qiu LX, Zhu LG, Wang Y, Lei L. CD14 C-260T gene polymorphism and ischemic heart disease susceptibility: a HuGE review and meta-analysis. Genet Med. 2009;11:403–8. doi: 10.1097/GIM.0b013e3181a16cb0. [DOI] [PubMed] [Google Scholar]
- 39.Mikuls TR, LeVan TD, Sayles H, Yu F, Caplan L, Cannon GW, Kerr GS, Reimold AM, Johnson DS, Thiele GM. Soluble CD14 and CD14 polymorphisms in rheumatoid arthritis. J Rheumatol. 2011;38:2509–16. doi: 10.3899/jrheum.110378. [DOI] [PubMed] [Google Scholar]
- 40.Landmann R, Zimmerli W, Sansano S, Link S, Hahn A, Glauser MP, Calandra T. Increased circulating soluble CD14 is associated with high mortality in gram-negative septic shock. J Infect Dis. 1995;171:639–44. doi: 10.1093/infdis/171.3.639. [DOI] [PubMed] [Google Scholar]
- 41.Haziot A, Ferrero E, Kontgen F, Hijiya N, Yamamoto S, Silver J, Stewart CL, Goyert SM. Resistance to endotoxin shock and reduced dissemination of gram-negative bacteria in CD14-deficient mice. Immunity. 1996;4:407. doi: 10.1016/s1074-7613(00)80254-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table I. Characteristics of CHS participants with soluble CD14 measured
Supplemental Table II: Risk of CVD outcomes and mortality associated with soluble CD14 and other inflammation biomarkers
SUPPLEMENTAL TABLE III: Association of sCD14-associated SNPs with incident CVD outcomes and mortality in EA and AA
SUPPLEMENTAL TABLE IV: Association of sCD14-associated SNPs with incident CHD and stroke in EA and AA from the Women’s Health Initiative
Supplemental Figure I. Manhattan Plot of soluble CD14 GWAS in European Americans
Supplemental Figure II. Manhattan Plot of soluble CD14 GWAS in African Americans
Supplemental Figure III: Regional association plots for chromosome 5q31.
Shown are soluble CD14 regional association plots generated using LocusZoom for the chromosome 5q31 locus in (A) European Americans; (B) African Americans. The color of each single nucleotide polymorphism (SNP) indicates the level of pairwise linkage disequilibrium (LD) based on r-squared relative to the lead SNP in the region. r-squared values were calculated from HapMap CEU for European Americans and YRI for African Americans. SNPs with missing LD information are shown in gray.
Supplemental Figure IV: Regional association plot for chromosome 1q24 in European Americans.
Shown is the soluble CD14 regional association plots generated using LocusZoom the chromosome 1q24 locus in European Americans. The color of each single nucleotide polymorphism (SNP) indicates the level of pairwise linkage disequilibrium (LD) based on r-squared relative to the lead SNP in the region. r-squared values were calculated from HapMap CEU. SNPs with missing LD information are shown in gray.
Supplemental Figure V. Regional association plot of 1q24 region in African Americans