Abstract
HDL removes cell cholesterol and protects against atherosclerosis. ApoA-I provides a flexible structural scaffold and an important functional ligand on the HDL surface. We propose structural models for apoA-IMilano (R173C) and apoA-IParis (R151C) mutants that show high cardioprotection despite low HDL levels. Previous studies established that two apoA-I molecules encircle HDL in an antiparallel, helical double-belt conformation. Recently, we solved the atomic structure of lipid-free Δ(185–243)apoA-I and proposed a conformational ensemble for apoA-IWT on HDL. Here we modify this ensemble to understand how intermolecular disulfides involving C173 or C151 influence protein conformation. The double-belt conformations are modified by belt rotation, main-chain unhinging around Gly, and Pro-induced helical bending, and they are verified by comparison with previous experimental studies and by molecular dynamics simulations of apoA-IMilano homodimer. In our models, the molecular termini repack on various-sized HDL, while packing around helix-5 in apoA-IWT, helix-6 in apoA-IParis, or helix-7 in apoA-IMilano homodimer is largely conserved. We propose how the disulfide-induced constraints alter the protein conformation and facilitate dissociation of the C-terminal segment from HDL to recruit additional lipid. Our models unify previous studies of apoA-IMilano and demonstrate how the mutational effects propagate to the molecular termini, altering their conformations, dynamics, and function.
Keywords: Milano and Paris mutations, double-belt and trefoil/tetrafoil conformations, protein-lipid interactions, protein dynamics and function, atherosclerosis
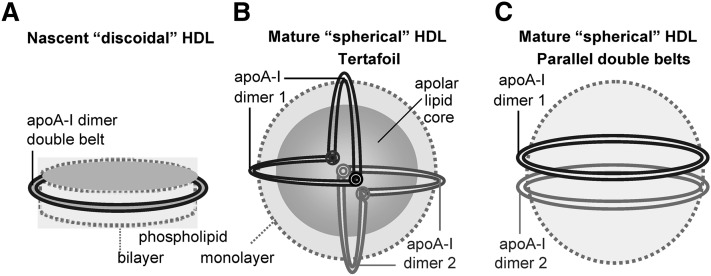
HDLs are water-soluble nanoparticles that remove excess cell cholesterol and protect against cardiovascular disease. Plasma levels of HDLs and their major protein, apoA-I (243 amino acids), are well-established negative risk factors for atherosclerosis, even though genetic factors that raise HDL levels do not necessarily confer additional cardioprotection (Refs. 1, 2 and references therein). Plasma HDL carries from two to five copies of apoA-I depending on the particle size (8–12 nm) and varies in particle shape (nascent “discoidal” or mature “spherical”, Fig. 1), protein conformation, biochemical composition, and functional properties (1). The nascent HDL particle is generally envisioned as a contorted disc composed of a cholesterol-containing phospholipid bilayer that is encircled by two antiparallel apoA-I molecules in a highly α-helical dynamic “double-belt” conformation (3–5). Micelle-like lipid packing has also been proposed in some studies (Ref. 6 and references therein). ApoA-I on HDL activates LCAT that converts nascent HDL into mature spherical particles containing a core of apolar lipids, mainly cholesterol esters, and a small amount of triacylglycerides. ApoA-I on mature HDL is proposed to form a double belt similar to that on nascent HDL but bent in a “trefoil/tetrafoil” fashion to confer 2D curvature to the surface of spherical particles (7, 8) (Fig. 1B).
Fig. 1.
Cartoon representation of apoA-I conformations on various HDL. (A) Nascent “discoidal” HDL contains two copies of apoA-I encircling the phospholipid bilayer in an antiparallel, double-belt conformation (4, 5). This conformation is also shown as a ribbon diagram in supplementary Fig. II. (B) Mature spherical HDL contains a core of apolar lipids (mainly cholesteryl esters and triacylglycerides) surrounded by the protein-containing phospholipid monolayer. Large HDL contains two molecular dimers of apoA-I that are proposed to form a tetrafoil on the particle surface (7, 8). We proposed that each double belt in this tetrafoil is bent around two flexible Gly-containing hinges (circles) (9). The two dimers are in different shades of gray. (C) If the double-belt bending around the flexible hinges is impaired, the two apoA-I double belts are expected to form two parallel rings on the HDL surface. We propose that this parallel arrangement, which was first proposed in the cross-linking studies of rHDL(A-IM/A-IM) (10), may be a general property of apoA-IM and, possibly, apoA-IP homodimers (Fig. 8 and supplementary Fig. V). Importantly, these and all other models presented in this work illustrate an overall protein arrangement but not the small local conformational adjustments that are probably involved in the double-belt formation by the mutant proteins.
ApoA-I on the HDL surface forms a flexible structural scaffold and is an important ligand for many functional interactions that direct HDL metabolism (1, 7–11). The amino acid sequence of apoA-I starts with a G-repeat (residues 1–43) followed by ten Pro-punctuated tandem 11/22-mer repeats (residues 44–243) (supplementary Fig. I). These repeats have high propensity to form amphipathic α-helices that are the major lipid surface-binding motif in apolipoproteins (12). Importantly, Pro-induced helical kinks are in registry in the two antiparallel protein molecules of the double belt (3–5), conferring the overall belt curvature that maintains the shape and size of HDL particles.
The cardioprotective effects of apoA-I are due, in part, to its essential role in cholesterol removal from peripheral cells via reverse cholesterol transport (1, 11, 13). At the initial rate-limiting steps of this complex pathway, monomeric lipid-poor or lipid-free apoA-I and small nascent HDL recruit cell cholesterol via the ATP-binding cassette transporters ABCA1 and ABCG1, respectively (14, 15). ABCA1-mediated lipid efflux from cells to apoA-I leads to HDL biogenesis. The nascent HDL particle undergoes LCAT-driven maturation followed by continuous remodeling by LCAT and other plasma factors (1, 11). At the final step of reverse cholesterol transport, lipids from the core of mature HDL are taken up by the HDL receptor SRB1 and are either used for steroid hormone synthesis or excreted via bile, while the protein-containing surface remnants reenter reverse cholesterol transport.
In addition to their central role in reverse cholesterol transport, apoA-I and HDL possess anti-inflammatory, antioxidant, antithrombolytic, and other beneficial properties (Ref. 16 and references therein). Efforts to improve HDL quality and quantity have been aimed at developing novel HDL-based therapies for cardiovascular disease to complement statins and other lipid-lowering drugs (17, 18). These efforts have utilized human wild-type apoA-I (apoA-IWT), its mimetics, and its R173C mutant termed apoA-IMilano (apoA-IM) (18).
OVERVIEW OF FUNCTION AND STRUCTURE OF APOA-IMILANO AND APOA-IPARIS
ApoA-IMilano (R173C) and apoA-IParis (R151C) are natural variants of human apoA-I that cause no deleterious health effects and may even improve cardioprotection despite abnormally low levels of plasma apoA-I and HDL and high levels of HDL triacylglycerides in mutation carriers, who are all heterozygotes (19–21). High cardioprotection by low levels of plasma HDLMilano and HDLParis (HDLM and HDLP) led to a hypothesis that apoA-IP and, particularly, apoA-IM are gain-of-function mutations (Refs. 22–24 and references therein). This prompted the development of HDL-based therapies utilizing apoA-IM (18, 23, 25). Although pilot studies of such injection therapies reported a significant reduction in atherosclerotic lesions in animals and humans (Refs. 18, 23, 26–30 and references therein), the advantages of using apoA-IM over apoA-IWT are subject to debate. Some studies using targeted replacement of apoA-IWT with apoA-IM supported improved cardioprotection by apoA-IM (18, 27, 30), while others reported similar effects of the two proteins (31). Also, some studies reported enhanced ability of apoA-IM and Milano sera to recruit cell cholesterol (32), while others reported similar effects of apoA-IM and apoA-IWT in vivo and in vitro (33–35). Furthermore, enhanced antioxidant activity of apoA-IM was reported in some studies (22) but not in other studies (36). In summary, there is no clear consensus regarding the gain-of-function effects of Milano and Paris mutations. The molecular basis underlying distinct functional properties of these naturally occurring mutants is unclear and is explored in the current work. Our main focus is on apoA-IM that has been better studied experimentally than any other apoA-I variant, which helps validate our structural models of the disulfide-linked apoA-IM homodimer.
Importantly, mutation sites R151C and R173C are located in the middle of the 22-mer helical repeats 6 and 7, respectively (supplementary Fig. I), in the “left” helical faces that are juxtaposed in the double belt (5, 37). This location of a single Cys in apoA-I facilitates formation of an intermolecular disulfide that preserves the helical packing in an antiparallel double belt with Pro kinks in registry (5, 37, 38). Therefore, the intermolecular disulfides formed by C151 or C173 are compatible with the antiparallel double-belt conformation. This suggests that apoA-IM and apoA-IP homodimers (A-IM/A-IM and A-IP/A-IP) on HDL form double belts resembling that of apoA-IWT but differing in helix registry: 5/5′ in apoA-IWT, 6/6′ in apoA-IP, and 7/7′ in apoA-IM (where N indicates Molecule 1 and N′ Molecule 2 of the dimer).
The overall conformational similarity of apoA-IWT, apoA-IP, and apoA-IM on HDL is supported by the normal size of plasma HDL in Milano and Paris carriers.2 However, the size distribution in human HDLM is shifted toward smaller particles (39) and includes an additional subclass of unusually small (∼7.8 nm) protease-sensitive particles containing a disulfide-linked apoAM homodimer. These unusually small HDL, which are a hallmark of the Milano mutation in humans, are potent acceptors of cell cholesterol in vitro, a property that was proposed to contribute to the enhanced cardioprotection by HDLM (40). In addition to apoA-IM homodimer and to apoA-IWT monomer, HDL in Milano carriers also contains comparable amounts of apoA-IM monomer and apoA-IM/apoA-II heterodimer (A-IM/A-II), in which the second-major HDL protein, apoA-II (77 amino acids), is disulfide-linked via its single Cys6 to Cys173 of apoA-IM (41). Similar heterodimers are found in human HDLP (42, 43). Protein conformations on these particles remain unknown and are addressed in the current work.
Low levels of plasma HDL and apoA-I in Milano and Paris carriers probably result from rapid proteolytic degradation of HDLM (43, 44), which may be due to several factors. First, apoA-IM and apoA-IP have impaired ability of to activate LCAT, leading to a low cholesterol ester to triacylglyceride ratio in the core of HDLM and HDLP (42, 45), which is expected to destabilize HDL (46) and promote apoA-I dissociation and clearance. Second, differences in structural stability between apoA-IM and apoA-IWT are also expected to contribute to the enhanced catabolism of apoA-IM. Compared with apoA-IWT, monomeric and disulfide-linked dimeric apoA-IM in solution was reported to have slightly lower α-helical content (36, 47, 48), altered aromatic packing in the N-terminal region (47), reduced structural stability, and reduced unfolding cooperativity (35, 49, 50). Notably, recent hydrogen-deuterium exchange (HX) studies of lipid-free apoA-I by Phillips and colleagues assigned very similar secondary structures to specific segments of apoA-IWT and apoA-IM but revealed that the N-terminal half of the apoA-IM molecule adopts two alternative conformations, one similar to that of apoA-IWT and another more accessible to solvent (48). Together, these and other studies indicate that, despite the overall structural similarity of apoA-IWT and apoA-IM, there are also subtle differences in the conformation and dynamics of their N-terminal domains, with apoA-IM displaying a less stable, more dynamic structure in solution and on the lipid, which is expected to contribute to its enhanced catabolism.
The structural differences resulting from R173C mutation apparently propagate to both molecular termini. In fact, studies of reconstituted HDL (rHDL) by Calabresi and colleagues showed that rHDL(A-IM/A-IM) are more labile to proteolysis than their wild-type counterparts (32, 40, 51), especially in the hydrophobic C-terminal region that forms the primary lipid-binding site in apoA-I (52). These observations prompted the authors to propose that the C-terminal ends of apoA-I can transiently dissociate from the surface of HDL(A-IM/A-IM) and bind to the cell surface, helping recruit additional cellular lipids (32, 51). These studies also inferred different C-terminal conformations for apoA-IWT and apoA-IM on rHDL (10). Such C-terminal dissociation appears at odds with Lys cross-linking/mass spectrometry studies by the group of Sorci-Thomas and Thomas, which reported that the C-terminal ends wrap around the lipid acyl chains in rHDL(A-IM/A-IM) (10). In summary, previous studies suggest that R173C mutation slightly destabilizes apoA-I and alters the conformation and dynamics of its N- and C-terminal regions in solution and/or on HDL. The mechanism via which the effects of R173C mutation propagate to the molecular termini and alter their conformation and dynamics in solution and on the lipid surface is unclear and is proposed, for the first time, in the current work.
NEW INSIGHTS FROM THE ATOMIC STRUCTURE OF LIPID-FREE Δ(185–234)APOA-I
This work builds upon the conformational ensemble that we recently proposed for apoAWT on various HDL (9). As a starting model to derive this ensemble, we used the X-ray crystal structure of the C-terminally truncated lipid-free apoA-I, Δ(185–234)apoA-I, solved by Mei and Atkinson to 2.2 Å resolution (53). This first atomic-resolution structure of apoA-I provided tremendous insights into the structure and function of this important protein in solution and on HDL, including the key role of the 5/5′ helix pair whose central opening creates a unique lipid presentation tunnel for the LCAT reaction (53, 54). Full ramifications of this atomic structure are just beginning to emerge. They include new insights into the structural and functional differences between lipid-free (53) and lipid-poor apoA-I monomer (55), novel insights into the pathogenic pathway of apoA-I destabilization and misfolding in amyloidosis (56), better insights into the structural and functional roles of apoA-II on HDL (57), and the conformational ensemble of apoA-IWT on HDL of various shapes and sizes (9).
Here, we use this ensemble as a basis to derive possible conformations of apoA-IM and apoA-IP homo- and heterodimers on various HDL. The proposed conformations of apoA-IM homodimers are verified by comparison with biophysical studies by several groups (10, 40, 48, 51). These conformations differ from all previously proposed models of apoA-IM or apoA-IP (10, 38, 58, 59) which were based, in part, on the low-resolution structure of the N-terminally truncated Δ(1–43)apoA-I (3) and, hence, lacked the correct conformation of the 1–43 segment. As evident from the crystal structure of Δ(185–234)apoA-I, this segment forms an integral part of the four-segment bundle in apoA-I monomer or dimer (53) (Fig. 1A), which is incorporated for the first time in our proposed models (Fig. 2). These models unify previous experimental studies of various forms of apoA-IM. They reconcile the reports on the lipid-bound (10) versus lipid-dissociated state (32) of the C-terminal regions in rHDL(A-IM/A-IM) and help explain why the unusually small HDL(A-IM/A-IM) provides an efficient acceptor of cell cholesterol (40) at the critical rate-limiting step of reverse cholesterol transport (1, 14, 15).
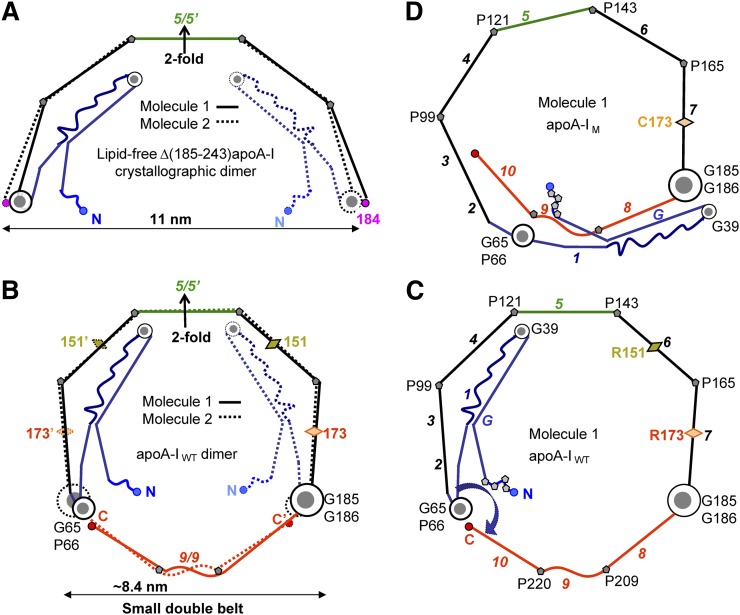
Fig. 2.
Proposed models of the small double belts of apoA-IWT (B, C) and apoA-IM (D) that were derived by using as a starting model the 2.2 Å resolution X-ray crystal structure of Δ(185–243)apoA-I (PDB ID 3R2P) (53). The models in these and other figures are drawn to scale and represent idealized planar double belts viewed down the normal to the plane. The conformational changes, which we propose are involved in the adaptation of the 3D protein structure to lipid-surface binding, including opening of the four-segment bundle, have been described previously (9, 53). (A) Left panels show molecular dimers comprised of Molecule 1 (solid lines) and Molecule 2 (dashed lines), and right panels show only Molecule 1. Sequence repeats are color-coded as follows: blue, N-terminal repeats G-1 (residues 1–65); black, repeats 2–7 (residues 66–185), except for the central repeat 5 (residues 121–143, in green); red, C-terminal repeats 8–10 (residues 186–243). Sequence repeats (shown in Fig. 1) are numbered in italics in (C) and (D). Pro positions are marked, and key Gly-containing flexible hinges are shown in gray-and-white circles: G185-G186 (largest circle), G65-P66 (medium circle), G39 (smallest circle). Locations of Milano and Paris mutations R173C and R151C are shown by diamonds.
APOA-IMILANO HOMODIMER IN A SMALL, DOUBLE-BELT CONFORMATION
Small ∼7.8 nm HDL(A-IM/A-IM) found in plasma of Milano carriers have been proposed to importantly contribute to cardioprotection by providing efficient acceptors for cell cholesterol (40). Furthermore, ∼7.8 nm rHDL containing one disulfide-linked apoA-IM homodimer and ∼76 POPC molecules readily form in vitro and have been extensively studied experimentally (10, 60). These studies provide important structural constraints for our models. To derive the models for apoA-IM homodimer, we used the conformation of apoA-IWT in a small double belt as a starting model (Fig. 2B, C; also shown by a ribbon diagram in supplementary Fig. II). This conformation was obtained as previously described (9) by using the modular protein sequence (supplementary Fig. I) together with the crystallographic dimer of lipid-free Δ(185–243)apoA-I (53) (Fig. 2A) and assuming that the C-terminal segment 185–243 forms a highly helical closed double belt on HDL, similar to that observed in the low-resolution structure of Δ(1–43)apoA-I (3).
Our working assumption is that apoA-I adaptation to changing lipid load occurs via the polypeptide hinge motion around flexible regions observed in the crystal structure, such as those encompassing conserved glycines G39 and G65 in the N-terminal domain or G129 in the central repeat 5 (9, 53). Another working assumption is that, similar to apoA-IWT, apoA-IM homodimer forms a closed antiparallel dynamic double belt with Pro kinks in registry (38), which is supported strongly by the cross-linking/mass spectrometry studies (10). To obtain the molecular packing in this double belt, we first rotated Molecule 2 of the apoA-IM dimer around the center of the belt to align the C173-C173′ pair to form a disulfide. The resulting double belt had two apparent drawbacks: steric clashes between the overlapping N-terminal regions and lack of Pro kink registry in the double-belt parts distant from C173 (supplementary Fig. III). To alleviate the steric clashes, the N-terminal residue segment 1–65 in the two molecules was rotated around the flexible hinge region containing G65-P66 (circular arrow in supplementary Fig. III-B). To improve the Pro kink registry, the double belt was constricted via small changes in the Pro-induced kink angles to bring the C termini closer together until optimal helical packing was achieved. Fig. 2D shows the resulting conformation of Molecule 1, and Fig. 3A shows the corresponding small symmetrical double belt of the disulfide-linked apoA-IM homodimer.
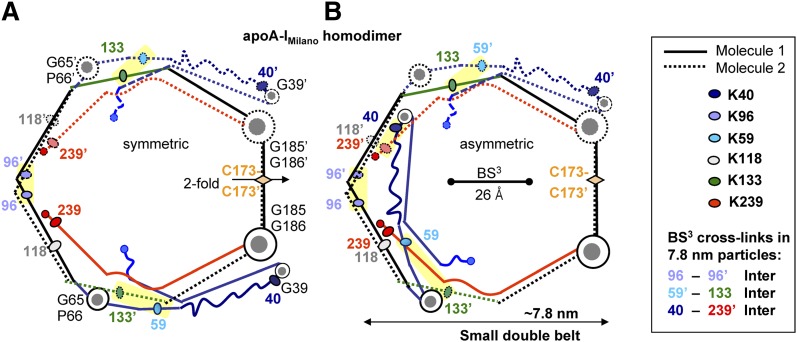
Fig. 3.
Chemical cross-links identified in ∼7.8 nm rHDL(A-IM/A-IM) (10) mapped on the proposed model of the small A-IM/A-IM double belt. (A) Symmetric double-belt contains two apoA-IM molecules in a conformation depicted in Fig. 2D. The two-fold symmetry axis passes through theC173-C173′ disulfide. (B) In Molecule 1 of the asymmetric double belt, segment 1–65 (in blue) is folded back around the G65-P66 hinge. Line and color coding is as in Fig. 2. The major intermolecular cross-links are highlighted. Cross-linked Lys are shown by ovals; residue numbers are color-coded. These cross-links are facilitated by the lysine locations in the “left” faces of the amphipathic α-helices, which are proximal in the double belt (5, 38). Thus, K96 is located in the middle of the “left” helical face that is facing its symmetry mate in the double belt, thereby facilitating the K96-K96′ cross-link. Similarly, K133 is located in the “left” helical face, facilitating its cross-link with L59 that is located in the “left” face of the short helix observed in the crystal structure (9). Also, K239 is located in the “left” helical face, which is expected to facilitate its cross-linking to K40 located in the turn between the helices in the crystal structure (53). Bar size shows the maximal Cα-Cα distance of 26 Å that can be cross-linked by BS3 used by Bhat and colleagues (10).
In this symmetrical double belt, Molecules 1 and 2 are related via the two-fold symmetry axis that passes through the C173-C173′ disulfide (Fig. 3A). Fig. 3B shows an alternative asymmetric molecular arrangement in the double belt of similar size, in which Molecules 1 and 2 differ in the orientation of the residue segment 1–65 (in blue). Several lines of experimental evidence support rotation of this segment around the G65-P66-containing hinge in apoA-IM. First, HX studies inferred two alternative conformations for lipid-free apoA-IM, one similar to that of lipid-free apoA-IWT and another having more solvent-exposed N-terminal half of the molecule (48). We propose that this increased solvent exposure results from rotation of the 1–65 residue segment around the G65-P66-containing hinge (circular arrow in Fig. 2D). Second, this rotation is expected to disrupt the aromatic clusters in the N-terminal half of the molecule, which stabilize the four-segment bundle in apoA-I (53). Such a disruption is indicated strongly by the spectroscopic studies showing largely intact secondary structure but altered aromatic packing and reduced protein stability and unfolding cooperativity of lipid-free apoA-IM as compared with apoA-IWT (35, 36, 47). We hypothesize that the rotation of the 1–65 segment away from the 4-segment bundle reflects a small destabilizing effect of R173C mutation (35, 36, 48) in repeat 7 that is an integral part of the bundle (53). Such destabilization may result from the disruption of the salt bridge network involving R173 and other charged groups from repeats 7, 2, and 3 in the 4-segment bundle (48, 53), as well as from the structural constraints imposed by the intermolecular disulfide formed by C173. In summary, accumulative evidence suggests strongly that in apoA-IM in solution, segment 1–65 can adopt alternative rotamer positions around the flexible hinge containing G65-P66. We hypothesize that these rotamer positions can be kinetically trapped on small HDL, resulting in the two alternative double-belt conformations depicted in Fig. 3.
Figs. 2 and 3 and other stick models in this article illustrate the overall protein arrangement, showing the relative orientation of the N-terminal segment 1–65 with respect to the protein double belt, but not its exact position on the lipid. The four-helix bundle containing this segment undergoes a conformational change upon transfer from solution to HDL to make available the apolar helical faces for lipid binding. This change was proposed to involve rotations of the α-helices around their axes and around their connecting hinges containing G39 and G65 (Ref. 9 and references therein). The resulting all-atom structures that depict the proposed protein conformations on the lipid were explored in molecular dynamics (MD) simulations described below.
The diameter of the idealized planar double belts shown in Fig. 3 is nearly 8 nm. The actual lipoprotein diameter is probably smaller, since Pro and Gly can induce not only kinks but also out-of-plane “wobble” of α-helices. This results in nonplanar double-belt geometry reported in the crystallographic (3, 53) and in MD simulations described below (and in Refs. 10, 38, 58 and references therein). Therefore, the actual double belts depicted in Fig. 3 are expected to have diameters under 8 nm. We hypothesize that these small double belts reflect structural aspects of the ∼7.8 nm particles, such as the small plasma HDLM or small model rHDL(A-IM/A-IM) (39, 40, 47).
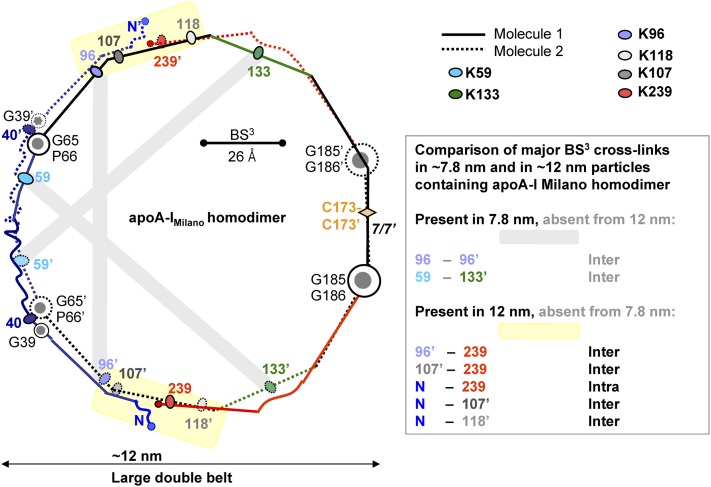
To test this hypothesis, we used the results of the protein cross-linking/mass-spectrometry studies of the 7.8 nm rHDL(A-IM/A-IM) (10). Mapping the observed Lys cross-links onto our models shows an excellent agreement between the data and the models, including K96-K96′, K59-K133′ (Fig. 3A, B, yellow highlight), and all other cross-links reported by Bhat and colleagues (10) (data not shown). This includes K96-K118 cross-link found on rHDL of all sizes (10); our models suggest that on small rHDL this cross-link may be inter- or intramolecular (Fig. 3), whereas on larger particles it is exclusively intramolecular (Figs. 4 and 5). Notably, location of K59, K96, K133, and K239 in the “left” helical face positions them closer to their cross-linking counterparts in the apoA-I dimer and thereby facilitates their chemical cross-links. Importantly, the K40-K239 cross-link reported in Bhat and colleagues is compatible only with the asymmetric orientation of segment 1–65, providing experimental evidence for the small asymmetric double belt (Fig. 3B). In summary, the belt size and conformation of the apoA-IM homodimers depicted in Fig. 3 are in excellent agreement with the cross-linking studies of the ∼7.8 nm rHDL(A-IM/A-IM) (10) and suggest two alternative double-belt conformations differing in the orientation of the N-terminal segment 1–65.
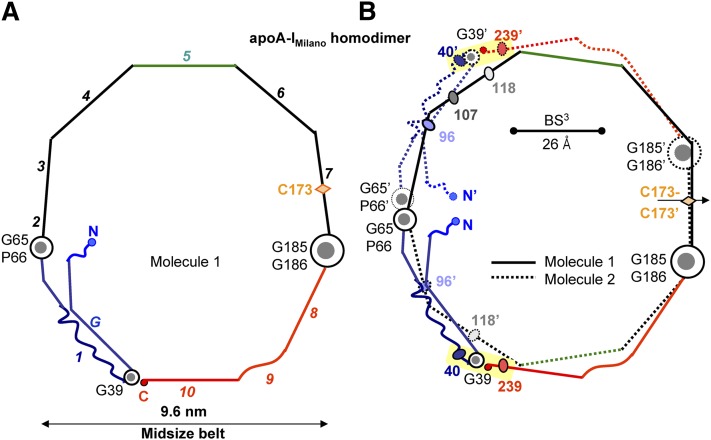
Fig. 4.
Proposed model of the mid-sized A-IM/A-IM double belt is compatible with the K40-K239 cross-link. In the midsized belt, the polypeptide chain in each molecule is shown bent back around G39 (A). The two belt-forming molecules are related via the two-fold (B). Yellow highlight shows chemical cross-link K40-K239 that was found on the large ∼12 nm rHDL(A-IM/A-IM) and is compatible with the midsized belt conformation (10). Bar size shows the upper limit for the Cα-Cα distance (26 Å) that can be cross-linked by BS3 (10).
Fig. 5.
Chemical cross-links identified in large and small rHDL(A-IM/A-IM) (10) mapped on the proposed model of the large A-IM/A-IM double belt. Cross-links that have been found only in large ∼12.5 nm rHDL are highlighted in yellow; gray highlights indicate cross-links that have been found in small but not in large rHDL(A-IM/A-IM). Color coding is as in Fig. 3.
CONFORMATIONS OF APOA-IMILANO HOMODIMER IN MIDSIZE AND LARGE DOUBLE BELTS
Previous studies postulated that the double-belt size in apoA-IWT can be increased by sequential unhinging of the N-terminal segment around two flexible hinges encompassing G65-P66 and G39 (53), accompanied by incremental changes in registry of the C-terminal repeats 8–10 (9). Here, we applied these postulates to derive the conformations of apoA-IM homodimer in the midsized (d ∼ 9.6 nm, Fig. 4) and in the large, fully expanded double belts (d ∼ 12 nm, Fig. 5). The resulting models comprise a closed double belt with maximal helical kink registry and satisfy all other previously described structural constraints (9). Therefore, we hypothesize that our models reflect key aspects of the protein conformations on plasma HDLM that also range in size from about 7.8 to 12 nm, as well as on rHDL(A-IM/A-IM) that form either small (∼7.8 nm) or large (∼12.5 nm) particles (Refs. 10, 39, 40, 47; also see footnote 2).
To test this hypothesis, we mapped the chemical cross-links identified in rHDL(A-IM/A-IM) onto our models. Bhat and colleagues reported that two Lys cross-links, K96-K96′ and K59-K133′, were observed in the small rHDL(A-IM/A-IM) but were absent from the large particles (10). This observation is in excellent agreement with our models (yellow highlights in Fig. 3 and gray highlights in Fig. 5). Furthermore, several cross-links were observed in the large, but not the small, particles; these include K96-K239, K107-K239, and multiple cross-links between the N-terminal α-amino group and K107, K118, or K239 (10). Again, all these lysines are located in the “left” face of the amphipathic α-helices, facilitating their cross-linking. These results are in excellent agreement with our model of the large particles (yellow highlights in Fig. 5). The only exception is the K40-K239 cross-link that was detected in all particles but that was clearly incompatible with the fully expanded double belt (Fig. 5) (10). However, the intramolecular cross-link K40-K239 is compatible with the midsized double belt in which the N-terminal segment is folded back upon itself around G39 (Fig. 4). The intermolecular cross-link K40-K239′ is also compatible with the small asymmetric double belt (Fig. 3B). Importantly, on the basis of this cross-link, Bhat and colleagues proposed that the two midsized double belts of apoA-IM homodimer, such as those shown in Fig. 4B, are arranged as two parallel rings on large rHDL (Fig. 1C) (10). As discussed below, we propose that such a parallel arrangement of the two double belts (four copies of apoA-IM) is possible on plasma HDLM containing two copies of dimeric apoA-IM. In summary, the cross-linking data (10) strongly support our models of apoA-IM homodimer on HDL of various sizes. Importantly, our models differ from those previously proposed, as we incorporate alternative N-terminal conformations suggested by the results of the recent HX/mass spectrometry and high-resolution X-ray crystallographic studies (48, 53), which were not available at the time of the cross-linking studies by Bhat and colleagues.
TESTING THE MODELS OF APOA-IMILANO HOMODIMERS BY MD SIMULATIONS
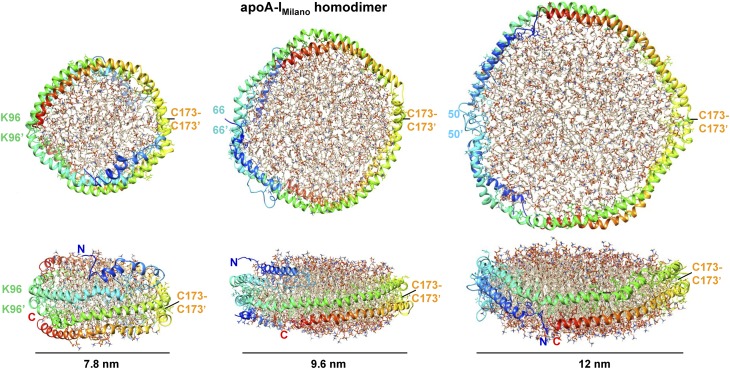
To test the energetic plausibility of the proposed models, we performed all-atom MD simulations using three different systems containing apoA-IM homodimer. The systems contained a small, midsized, or large symmetrical protein double belt, disulfide-linked via the Cys173 pair, and varying amounts of lipid. The overall protein arrangement in the starting models followed the diagrams in , respectively. In the starting model of the large particle, apoA-I residues 1 and 2 were in a random coil conformation; residues 3–55 were in a conformation similar to that in the crystal structure of Δ(185–243)apoA-I but extended around the G39-containing hinge (rather than folded back) to allow for a fully expanded belt (Fig. 5); and residues 56–243 formed an idealized 3/11 helix (11 residues per three turns) bent in a circular arc, such that the 1–55 segment closed the circle without overlap. All helices were oriented with their apolar sides facing the lipid interior. The two protein molecules were arranged in an antiparallel double belt allowing C173-C173′ disulfide bond formation. The helical axes in the double belt were ∼10 Å apart. A circular patch of 232 POPC molecules in a bilayer conformation was centered within the double belt. In the midsized particle, the starting protein model was created similarly, except that the conformation of the residue segment 3–55 was exactly as in the crystal structure (i.e., folded back around the G39-containing hinge as in Fig. 4), and the diameter of the circular arc was reduced accordingly to allow residues 39–243 form a closed belt. A circular bilayer patch of 178 POPC molecules was added. In the small particle, the starting protein model was created similarly as in the midsized particle, except that the helical belt was further constricted to adhere to the overall geometry shown in Fig. 3A. A circular patch of 76 POPC was added.
The MD simulations were performed using NAMD 2.9 software (61) as previously described (62). Briefly, each system was ionized and charge-neutralized with 0.15 M NaCl by using the ‘‘Add Ions’’ plug-in in Visual Molecular Dynamics software (63). The TIP3P water model was used to solvate the system (64). The CHARMM 22 (65, 66) and 27 (67, 68) force fields were used for protein and lipid, respectively. After solvation and ionization, 10,000 steps of conjugant gradient minimization were performed for each model. Next, the protein was frozen while the system was heated to 310 K and equilibrated for 0.5–1.0 ns until the gap between lipid and protein was reduced. Afterwards, the protein was unfrozen, and the complete models were equilibrated at 310 K for 5 ns for the large particles or for 15 ns for the midsized and small particles.
Fig. 6 shows the all-atom models of the small, midsized, and large HDL(apoA-IM/apoA-IM) after MD simulations. The overall arrangement of the protein and lipid in each model was retained, indicating its energetic plausibility. The main changes in the protein conformation upon MD simulations involved formation of a less regular helical structure that was kinked around prolines and partial movement of the N-terminal segment in the small double belt from the edge of the bilayer to its top or bottom. A similar shift in segment 1–65 was proposed to occur during metabolic remodeling of plasma HDL and its maturation from nascent “discoidal” to small “spherical” particles (9). Notably, the distances between the Ca atoms of the cross-linkable lysines shown in Figs. 3–5 were under 26 Å after MD simulations, showing that the models were consistent with the Lys cross-linking data reported by Bhat et al. In sum, our MD simulations show that the proposed models of apoA-IM homodimer on the small, midsized, and large HDL are energetically plausible and agree with the existing cross-linking data.
Fig. 6.
All-atom models of rHDL(A-IM/A-IM) particles of various sizes after MD simulations. Upper panels show top views and lower panels show side views. The two protein molecules in each model are in rainbow colors from the N to the C terminus (blue to red). The position of the C173-C173′ disulfide in each model is indicated. Residue pairs that are located across from the C173 pair and are approximately in registry in the two molecules are indicated. This includes K96-K96′ pair that formed a BS3 cross-link only on the small rHDL(A-IM/A-IM) but not in their larger counterparts (10).
HYPOTHETICAL MODELS OF APOA-I/APOA-II HETERODIMERS
In addition to the disulfide-linked apoA-IM homodimer, we also propose conformations of the disulfide-linked heterodimers, A-IM/A-II and A-IP/A-II (supplementary Fig. IV). Again, our models differ from those previously proposed (59), as we incorporate the N-terminal conformation of apoA-I suggested by the atomic structure of Δ(185–243)apoA-I that only recently became available. Because apoA-II-containing heterodimers are not readily available in amounts sufficient for detailed structural studies (59), these tentative models await their experimental verification.
CONFORMATIONAL ENSEMBLES OF APOA-IM, APOA-IP, AND APOA-IWT HOMODIMERS COMPARED
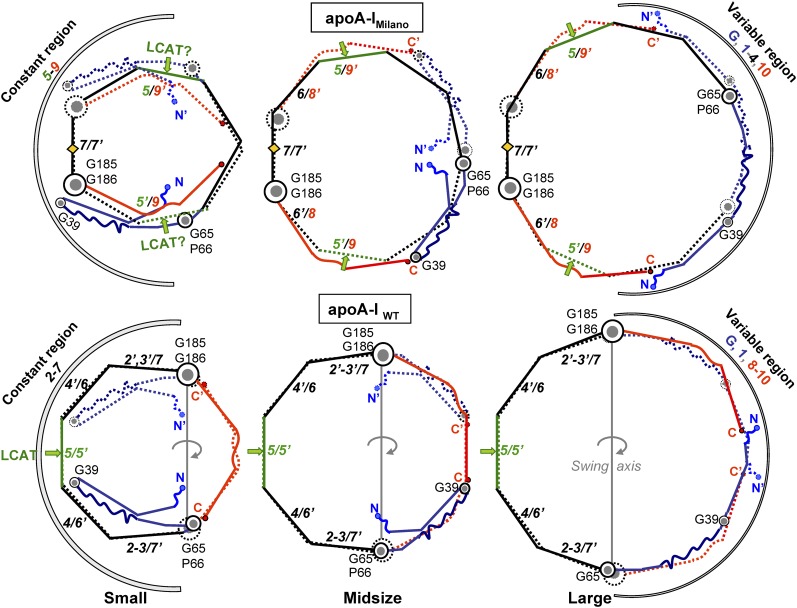
The proposed conformations of the apoA-IM homodimer in the double belts of various sizes are shown in Fig. 7 (top). Hypothetical conformations of similar double belts containing apoA-IP homodimer are reported in supplementary Fig. V. The conformational ensemble previously proposed for apoA-IWT (9) is shown for comparison (Fig. 7, bottom). An important feature of this ensemble is that registry of repeats 2–7 is conserved in double belts of different sizes (9). This “constant” region (residues 66–185 in apoA-IWT) is centered at the 5/5′ helix pair (in green). In contrast, packing of the N- and C-terminal repeats G–1 (residues 1–65, in blue) and 8–10 (residues 186–243, in red) varies on the double belts of different sizes. Importantly, the two flexible hinges encompassing G65-P66 and G185-G186, which delineate the constant and variable regions in apoA-IWT, are aligned in all double belts. This alignment facilitates swing motion of the variable and constant regions around the axis connecting G65-P66 and G185-G186 (Fig. 7, bottom panels). We postulated that this out-of-plane swing motion is essential for maturation and growth of HDLWT (9), as it helps to confer 2D surface curvature to the spherical HDL particle and to accommodate additional copies of apoA-IWT in the trefoil/tetrafoil arrangement first proposed by Davidson's group (7, 8) (Fig. 1B).
Fig. 7.
Conformational ensemble proposed for the small, midsized, and large double belts of apoA-IM (top) and apoA-IWT (bottom). The double belts are rotated by ∼180°C with respect to their orientations in Figs. 2–5.Green arrows point to the center of repeat 5. Repeat pair 5/5′, which was proposed to form the lipid presentation tunnel for the LCAT reaction (53), is replaced with 5/9′ pairing in apoA-IM homodimer (green arrows). Arcs indicate constant and variable regions. Gray line in the bottom panels shows swing axis in the apoA-IWT double belt, which connects G65-P66 and G185-G186 hinge regions. These hinges are in registry in the double belt of apoA-IWT (bottom) but in neither in apoA-IM (top) nor in apoA-IP homodimers (supplementary Fig. V), which is expected to impede tetrafoil formation by these homodimers (Fig. 1).
Our models suggest that, despite the overall similarity of the double belts formed by apoA-IWT, apoA-IM, and apoA-IP homodimers, there are also important structural and dynamic differences resulting from the Cys mutations and the intermolecular disulfides in the mutant proteins (Fig. 7 and supplementary Fig. V). First, all double belts in our models have constant and variable regions, albeit in different locations: the constant region in apoA-IM is centered at the disulfide-linked 7/7′ pair; in apoA-IP at the disulfide-linked 6/6′ pair; and in apoA-IWT at the 5/5′ pair (Fig. 7 and supplementary Fig. V). As revealed by the high-resolution crystal structure, the central opening in the 5/5′ pair is uniquely suited to provide a lipid presentation tunnel for the LCAT reaction on HDL (53, 54). Disruption of this tunnel in the apoA-IM and apoA-IP homodimers results in impaired LCAT activation (53), explaining the low levels of mature HDL in plasma of Milano and Paris carriers. In our models, 5/5′ helix pairing in apoA-IWT is replaced with 5/7′ and 5′/7 pairings in apoA-IP (supplementary Fig. V) or with approximate 5/9′ and 5′/9 pairings in apoA-IM double belts (Fig. 7). This suggests that the single lipid presentation tunnel in the double belt of apoA-IWT is replaced with two inefficient “semitunnels” in apoA-IM or apoA-IP (Fig. 7 and supplementary Fig. V, green arrows).
Second, flexible hinges G65-P66 and G185-G186 are aligned in apoA-IWT double belts of various sizes but not in their apoA-IM or apoA-IP counterparts (Fig. 7 and supplementary Fig. V). As a result, the concerted out-of-plane swing motion of the two dimer-forming molecules around these highly conserved hinges (69), which we proposed to be important for HDL maturation (9), is probably impaired in the apoA-IM and apoA-IP double belts, further contributing to the impaired maturation of HDLM and HDLP. Furthermore, the impaired swing motion of the double belt is expected to prevent its tetrafoil conformation on large spherical HDL that contains four copies of apoA-I (Fig. 1B). We speculate that, instead of the tetrafoil, Milano and Paris homodimers form two parallel double belts encircling the large particle (Fig. 1C), similar to those postulated in the cross-linking studies of large rHDL(A-IM/A-IM) (10).
Third, our models, supported by the cross-linking data (10), suggest that the preferred double-belt conformation of apoA-IM homodimer has the N-terminal segment folded back around the flexible hinge containing G39 but not G65-P66 (Fig. 7, top left and center). This preferred conformation resembles one of the three alternative N-terminal conformations proposed for apoA-IWT (Fig. 7, bottom center). In summary, despite the overall similarity, there are also important structural and dynamic differences between apoA-IM and apoA-IWT homodimers. These differences include a shift in the conformational distribution in the N-terminal part and the impaired swing motion of the A-IM/A-IM double belt. The latter is expected to result in parallel arrangement of the two Milano double belts on large HDL, as opposed to tetrafoil arrangement postulated for apoA-IWT (Fig. 1B, C). These differences in the apoA-I structure and dynamics on HDLWT and HDLM are expected to influence their interactions with functional ligands in reverse cholesterol transport and with many other HDL-associated proteins (16, 70).
EFFECTS OF THE DISULFIDE LINK ON LIPID RECRUITMENT BY APOA-IM
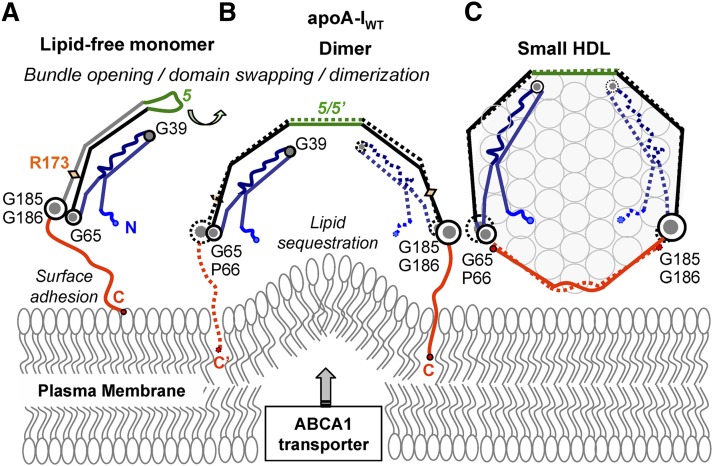
Our models suggest distinct molecular mechanisms for lipid efflux from cells to apoA-IWT and to apoA-IM. Fig. 8 illustrates these events for apoA-IWT, starting with the monomeric lipid-poor/free protein that is proposed to serve as the primary acceptor of cell cholesterol via ABCA1 transporter at the critical rate-limiting step of reverse cholesterol transport (1, 13–15). Lipid-free (53) and lipid-poor (55) apoA-IWT monomer in solution is proposed to adopt a globular helix-bundle structure similar to that observed in the crystallographic dimer, but with the C-terminal half domain-swapped from Molecule 2 to Molecule 1 via the central repeat 5 (Fig. 7A) that is disordered in solution (53). We proposed that binding of this monomer to phospholipid surface, possibly mediated by ABCA1 transporter that can dimerize in the plasma membrane (71), is accompanied by apoA-I dimerization and lipid sequestration via the hydrophobic C-terminal segment 8–10 that forms the primary lipid-binding site (Fig. 8B) (72). This process generates small nascent HDL containing two copies of apoA-IWT (Fig. 8C).
Fig. 8.
Proposed structural changes in the apoA-IWT protein and lipid during the initial rate-limiting steps of reverse cholesterol transport. Lipid-poor/free monomeric apoA-IWT forms a globular four-segment bundle as previously described (53, 55) (A). Upon adhesion to the lipid surface via the flexible hydrophobic C-terminal segment 185–243 (in red), apoA-IWT dimerizes via the domain swapping around repeat 5 (in green) (53), which is followed by lipid sequestration (9) (B). This process generates small nascent HDL containing one copy of A-IWT–A-IWT double belt wrapped around the perimeter of a phospholipid bilayer (C). In C, nascent discoidal HDL are shown face-up, and phospholipid head groups are show in circles. Lipid efflux from the plasma membrane to apoA-I is mediated by the lipid transporter ABCA1 that forms protrusions on the plasma membrane and thereby increases lipid availability to apoA-I (Refs. 13–15 and references therein).
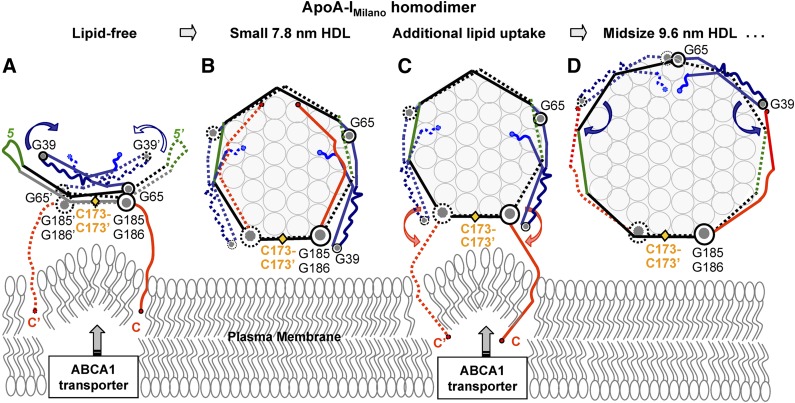
Lipid efflux to the disulfide-linked homodimers is illustrated in Fig. 9 for apoA-IM and in supplementary Fig. VI for apoA-IP. On the basis of the previous structural and stability studies of apoA-IM and apoA-IWT (10, 36, 47, 48, 53), we propose that each molecule in the apoA-IM homodimer adopts a conformation similar to that of the apoA-IWT monomer but with two alternative orientations of the 1–65 segment. One orientation is similar to that of the apoA-IWT and has the N-terminal segment folded back around G39 and G65-P66 to form a 4-segment bundle (Fig. 9A). In the second orientation, the N-terminal segment is folded back around G39 but is expanded around G65-P66 away from the bundle (Fig. 9A, blue arrow). The second orientation is probably preferred on HDL, as suggested by the cross-linking studies of rHDL of various sizes (10), and is depicted in the small symmetric double belt on the 7.8 nm particles (Figs. 3A and 9B). As described below, this orientation of the N-terminal segment, together with the close spacing between the two G185-G186 hinge pairs in the Milano homodimer, is expected to influence the conformation, dynamics, and function of the molecular C termini.
Fig. 9.
Proposed structural changes in apoA-IM homodimer and in lipid during the initial step of reverse cholesterol transport. (A) Solution structure of apoA-IM homodimer is similar to that of apoA-IWT monomer (Fig. 8A) but has a slightly destabilized four-segment bundle in which the N-terminal segment 1–65 can rotate around G65-P66 hinges (blue arrows). C-terminal segment 185–243 (repeats 8–10, in red) sequesters lipid to form unusually small ∼7.8 nm HDL that are a hallmark of the Milano mutation (39, 40, 47) (B). Transient dissociation of the C-terminal segment from these small HDL enables it to recruit additional lipid (C) and form larger particles (D). Nascent discoidal HDL(A-IM/A-IM) are shown face-up, and phospholipid head groups are show in circles.
In apoA-I, the hydrophobic C-terminal segment 8–10 (residues 185–243) is largely unordered in solution but becomes predominantly helical on the lipid (Refs. 52, 53, 72 and references therein). Additional mobility of this segment is conferred by the flexible hinge G185-G186. In the double belt, the spacer between these hinges is composed of sequence repeats 2–7 in apoA-IWT (Figs. 1A and 6B), repeats 5–7 in apoA-IP (supplementary Fig. V), and repeat 7 in apoA-IM (Figs. 2–4 and 8). The latter spacing is ∼33 Å long (that is, the length of a 22-mer α-helix) as compared with 110 Å spacing observed in the crystallographic dimer of Δ(185–243)apoA-I (53) (Fig. 1A) and depicted in the model of apoA-IWT (Fig. 8B). We propose that this reduced spacing between the two G185-G186 hinges in the apoA-IM homodimer restricts the amount of lipid sequestered by its C-terminal segment (Fig. 8A, B). This concept helps explain why the apoA-IM homodimer readily forms unusually small 7.8 nm HDL in vivo and in vitro (32, 39, 40).
Our models also help explain why these small particles form efficient acceptors of additional lipid from cells (32, 40). In our model of the small double belt of apoA-IWT, helices 8-10 pack against each other to close the belt and form its integral part (Fig. 6, bottom left, and Fig. 8, right). Therefore, spontaneous dissociation of these helices has low probability, as it involves a steep energy penalty for transient solvent exposure of lipid acyl chains around the HDL perimeter. In contrast, in the small symmetrical double belt of apoA-IM, helices 8-10 pack against the N-terminal and central helices G-1 and 4-6 to form two two-segment pairs (Fig. 7, top left, and Fig. 9B). As illustrated in Fig. 9C, such packing allows dissociation of the C-terminal segment from the particle edge without breaking the double belt and exposing HDL lipids to solvent. We hypothesize that such dissociation of the C-terminal segment, which is facilitated by its rotation around the G185-G186 hinges, occurs transiently in apoA-IM homodimer, enabling it to recruit additional lipid (Fig. 9C). Our hypothesis is supported strongly by the studies of Calabresi and colleagues showing that ∼7.8 nm plasma HDLM acts as an efficient acceptor of cell cholesterol via the ABCA1 transporter and that the C-terminal segment in this HDLM is highly labile to proteolysis (40). The authors proposed that transient dissociation of the C-terminal regions is specific to HDLM and is important for cell surface interactions (32, 40). Our models are in excellent agreement with this idea and suggest a molecular basis underlying the increased structural lability and functional activity of the C-terminal segment in the apoA-IM homodimer on small HDL (Fig. 9).
SUMMARY AND FUTURE STUDIES
Our models suggest a pathway via which apoA-IM homodimer recruits lipids to form nascent HDL. We propose that this recruitment is facilitated by the increased mobility of the C-terminal segment but is restricted by its close spacing that limits the amount of sequestered lipids (Fig. 9C). This increased mobility and reduced spacing of the C-terminal segment suggests that lipid sequestration by apoA-IM homodimer occurs at a faster pace but in smaller increments as compared with apoA-IWT. Whether the net effect benefits cell cholesterol recruitment by apoA-IM remains unclear. However, it is clear that the differences in the repeat registry in apoA-IM (7/7′), apoA-IP (6/6′), and apoA-IWT (5/5′) alter the protein conformation and dynamics not only in the central LCAT-activating region of the double belt (53) but also in the variable N- and C-terminal regions (Figs. 7–9). Moreover, Milano and Paris disulfides disrupt the registry of the two major Gly-containing flexible hinges in apoA-I double belt (Fig. 7 and supplementary Fig. V), G65-P66, and G185-G186, and thereby impair the hinge motion that, we think, is necessary for the tetrafoil conformation of apoA-I on mature HDL (Fig. 1B). Therefore, on large mature HDL, two pairs of Milano or Paris homodimers are expected to form parallel rings, as opposed to the tetrafoil postulated for apoA-IWT. In addition to these global structural changes in the mutant proteins, local adjustments in their conformations are expected, which are beyond the scope of the current work. Together, these structural and dynamic differences are expected to influence functional interactions of the Milano and Paris mutants with various metabolic partners that drive reverse cholesterol transport, as well as with nearly 100 other HDL-associated proteins (1, 16, 70).
Even though the proposed molecular arrangement of the disulfide-linked apoA-IM homodimer is supported strongly by the cross-linking studies of rHDL(A-IM/A-IM) (10), our models have clear limitations. First, the accuracy of the cross-linking structural studies and the resulting models is limited, in part, by the length of the cross-linker arm that defines the maximal Cα-Cα distance (26 Å for BS3) (10). Second, the exact arrangement of the N-terminal segment 1–65 on HDL remains tentative, especially on the small discoidal particles that mimic nascent HDL. MD simulations suggested that this segment can shift from the edge of the bilayers to its top or bottom (Fig. 6A). Such a shift was previously proposed to facilitate maturation of small discoidal HDL into spherical HDL (9), and this will be further explored in future studies. Third, our proposed conformational ensemble of the disulfide-linked apoAM homodimer may be incomplete. For example, it is consistent with but does not explicitly include the dynamic structure in the central region of apoA-I containing repeat 5. Highly dynamic structure in this region was reported in several studies in solution and on HDL (48, 53, 73, 74), and it was proposed to help mediate the protein conformational switch from solution to HDL (53), as well as help adapt the double-belt length to HDL of various sizes (73, 74). This highly dynamic structure in repeat 5 can be incorporated into our models, providing them with additional plasticity. Another limitation is that, unlike the disulfide-linked apoA-IM homodimer, the proposed models of the disulfide-linked apoAP homodimer and of the Milano and Paris heterodimers have not been tested experimentally.
Despite these limitations, our models help postulate testable hypotheses for experimental studies. For example, the N-terminal opening around the Gly-containing hinges in apoA-I in solution and on HDL is currently being probed by engineering structure-based disulfides that prevent such opening. The proposed pairing of helical repeat 6 with 8 and of helical repeat 5 with 9 in HDL(A-IM/A-IM) or close proximity of the molecular N-termini in the midsized but not the small or large HDL(A-IM/A-IM) (Fig. 7, top panels) can be probed by FRET or EPR studies of the labeled protein. These and other aspects of the proposed models can also be probed by using extensive cross-linking following the protocols developed by Davidson's team (7, 8). These experimental approaches together with other methods, such as HX/mass spectrometry, can be used to probe the validity of the proposed models of apoA-IP homodimers and of the Milano and Paris heterodimers with apoA-II. The ultimate test for the proposed models, which is high-resolution structural analysis of HDL, remains a major challenge in the field.
Supplementary Material
Acknowledgments
The authors thank University of Alabama at Birmingham Information Technology and Department of Mechanical Engineering for use of the commodity cluster Cheaha that they jointly maintain.
Footnotes
Abbreviations:
- A-IM/A-II
- apoA-IMilano/apoA-II heterodimer
- A-IM/A-IM
- apoA-IMilano homodimer
- apoA-IM
- apoA-IMilano (R173C)
- apoA-IP
- apoA-IParis (R151C)
- apoA-IWT
- wild-type apolipoprotein A-I
- HDLM
- HDL from plasma of Milano carriers
- HDLP
- HDL from plasma of Paris carriers
- HX
- hydrogen-deuterium exchange
- MD
- molecular dynamics
- rHDL
- reconstituted HDL
Discoidal rHDL composed of the disulfide-linked Milano or Paris homodimer can contain either two or four apoA-I molecules, whereas similar rHDL composed of apoA-IWT can contain two, three, or four apoA-I molecules. This difference influences the particle size distribution in discoidal rHDL(A-IM/A-IM), which forms either small or large particles (4, 32, 45), but not in plasma HDL. In fact, in addition to the disulfide-linked apoA-I homodimer, plasma HDL of Milano or Paris carriers, who are all heterozygotes, can contain one copy of unpaired apoA-IWT and/or one or more copies of apoA-II homo- or heterodimer, leading to a continuous-sized distribution in these particles.
This work was supported by the National Institutes of Health Grants GM-067260 (to O.G.), HL-026355 (to D.A. and O.G.) and HL-102515 (to J.P.S.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six figures.
REFERENCES
- 1.Rothblat G. H., Phillips M. C. 2010. High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Curr. Opin. Lipidol. 21: 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voight B. F., Peloso G. M., Orho-Melander M., Frikke-Schmidt R., Barbalic M., Jensen M. K., Hindy G., Hólm H., Ding E. L., Johnson T., et al. 2012. Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet. 380: 572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA. 94: 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., Harvey S. C. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274: 31755–31758 [DOI] [PubMed] [Google Scholar]
- 5.Brouillette C. G., Anantharamaiah G. M., Engler J. A., Borhani D. W. 2001. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim. Biophys. Acta. 1531: 4–46 [DOI] [PubMed] [Google Scholar]
- 6.Sorci-Thomas M. G., Owen J. S., Fulp B., Bhat S., Zhu X., Parks J. S., Shah D., Jerome W. G., Gerelus M., Zabalawi M., et al. 2012. Nascent high density lipoproteins formed by ABCA1 resemble lipid rafts and are structurally organized by three apoA-I monomers. J. Lipid Res. 53: 1890–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva R. A., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., Davidson W. S. 2008. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. USA. 105: 12176–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang R., Silva R. A., Jerome W. G., Kontush A., Chapman M. J., Curtiss L. K., Hodges T. J., Davidson W. S. 2011. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol. 18: 416–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gursky O. 2013. Crystal structure of Δ(185–243)apoA-I suggests a mechanistic framework for the protein adaptation to the changing lipid load in good cholesterol: from flatland to sphereland via double belt, belt buckle, double hairpin and trefoil/tetrafoil. J. Mol. Biol. 425: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhat S., Sorci-Thomas M. G., Calabresi L., Samuel M. P., Thomas M. J. 2010. Conformation of dimeric apolipoprotein A-I Milano on recombinant lipoprotein particles. Biochemistry. 49: 5213–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lund-Katz S., Phillips M. C. 2010. High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell. Biochem. 51: 183–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segrest J. P., Garber D. W., Brouillette C. G., Harvey S. C., Anantharamaiah G. M. 1994. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv. Protein Chem. 45: 303–369 [DOI] [PubMed] [Google Scholar]
- 13.Rosenson R. S., Brewer H. B., Jr, Davidson W. S., Fayad Z. A., Fuster V., Goldstein J., Hellerstein M., Jiang X. C., Phillips M. C., Rader D. J., et al. 2012. Cholesterol efflux and atheroprotection: advancing the concept of reverse cholesterol transport. Circulation. 125: 1905–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oram J. F., Heinecke J. W. 2005. ATP-binding cassette transporter A1: a cell cholesterol exporter that protects against cardiovascular disease. Physiol. Rev. 85: 1343–1372 [DOI] [PubMed] [Google Scholar]
- 15.Yvan-Charvet L., Wang N., Tall A. R. 2010. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol. 30: 139–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon S. M., Hofmann S., Askew D. S., Davidson W. S. 2011. High density lipoprotein: it's not just about lipid transport anymore. Trends Endocrinol. Metab. 22: 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao X., Yuan S. 2010. High density lipoproteins-based therapies for cardiovascular disease. J. Cardiovasc. Dis. Res. 1: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tardif J. C. 2010. Emerging high-density lipoprotein infusion therapies: fulfilling the promise of epidemiology? J. Clin. Lipidol. 4: 399–404 [DOI] [PubMed] [Google Scholar]
- 19.Franceschini G., Sirtori C. R., Capurso A., 2nd, Weisgraber K. H., Mahley R. W. 1980. A-IMilano apoprotein. Decreased high density lipoprotein cholesterol levels with significant lipoprotein modifications and without clinical atherosclerosis in an Italian family. J. Clin. Invest. 66: 892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sirtori C. R., Calabresi L., Franceschini G., Baldassarre D., Amato M., Johansson J., Salvetti M., Monteduro C., Zulli R., Muiesan M. L., et al. 2001. Cardiovascular status of carriers of the apolipoprotein A-IMilano mutant: the Limone sul Garda study. Circulation. 103: 1949–1954 [DOI] [PubMed] [Google Scholar]
- 21.Bruckert E., von Eckardstein A., Funke H., Beucler I., Wiebusch H., Turpin G., Assmann G. 1997. The replacement of arginine by cysteine at residue 151 in apolipoprotein A-I produces a phenotype similar to that of apolipoprotein A-I Milano. Atherosclerosis. 128: 121–128 [DOI] [PubMed] [Google Scholar]
- 22.Bielicki J. K., Oda M. N. 2002. Apolipoprotein A-I(Milano) and apolipoprotein A-I(Paris) exhibit an antioxidant activity distinct from that of wild-type apolipoprotein A-I. Biochemistry. 41: 2089–2096 [DOI] [PubMed] [Google Scholar]
- 23.Cimmino G., Ibanez B., Vilahur G., Speidl W. S., Fuster V., Badimon L., Badimon J. J. 2009. Up-regulation of reverse cholesterol transport key players and rescue from global inflammation by ApoA-I(Milano). J. Cell. Mol. Med. 13(9B): 3226–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calabresi L., Sirtori C. R., Paoletti R., Franceschini G. 2006. Recombinant apolipoprotein A-I Milano for the treatment of cardiovascular diseases. Curr. Atheroscler. Rep. 8: 163–167 [DOI] [PubMed] [Google Scholar]
- 25.Kaul S., Shah P. K. 2005. ApoA-IMilano / phospholipid complexes emerging pharmacological strategies and medications for the prevention of atherosclerotic plaque progression. Curr. Drug Targets Cardiovasc. Haematol. Disord. 5: 471–479 [DOI] [PubMed] [Google Scholar]
- 26.Nissen S. E., Tsunoda T., Tuzcu E. M., Schoenhagen P., Cooper C. J., Yasin M., Eaton G. M., Lauer M. A., Sheldon W. S., Grines C. L., et al. 2003. Effect of recombinant ApoA-I Milano on coronary atherosclerosis in patients with acute coronary syndromes: a randomized controlled trial. JAMA. 290: 2292–2300 [DOI] [PubMed] [Google Scholar]
- 27.Kaul S., Coin B., Hedayiti A., Yano J., Cercek B., Chyu K. Y., Shah P. K. 2004. Rapid reversal of endothelial dysfunction in hypercholesterolemic apolipoprotein E-null mice by recombinant apolipoprotein A-I(Milano)-phospholipid complex. J. Am. Coll. Cardiol. 44: 1311–1319 [DOI] [PubMed] [Google Scholar]
- 28.Chiesa G., Parolini C., Sirtori C. R. 2008. Acute effects of high-density lipoproteins: biochemical basis and clinical findings. Curr. Opin. Cardiol. 23: 379–385 [DOI] [PubMed] [Google Scholar]
- 29.Speidl W. S., Cimmino G., Ibanez B., Elmariah S., Hutter R., Garcia M. J., Fuster V., Goldman M. E., Badimon J. J. 2010. Recombinant apolipoprotein A-I Milano rapidly reverses aortic valve stenosis and decreases leaflet inflammation in an experimental rabbit model. Eur. Heart J. 31: 2049–2057 [DOI] [PubMed] [Google Scholar]
- 30.Ibanez B., Giannarelli C., Cimmino G., Santos-Gallego C. G., Alique M., Pinero A., Vilahur G., Fuster V., Badimon L., Badimon J. J. 2012. Recombinant HDL(Milano) exerts greater anti-inflammatory and plaque stabilizing properties than HDL(wild-type). Atherosclerosis. 220: 72–77 [DOI] [PubMed] [Google Scholar]
- 31.Parolini C., Chiesa G., Gong E., Caligari S., Cortese M. M., Koga T., Forte T. M., Rubin E. M. 2005. Apolipoprotein A-I and the molecular variant apoA-I(Milano): evaluation of the antiatherogenic effects in knock-in mouse model. Atherosclerosis. 183: 222–229 [DOI] [PubMed] [Google Scholar]
- 32.Calabresi L., Canavesi M., Bernini F., Franceschini G. 1999. Cell cholesterol efflux to reconstituted high-density lipoproteins containing the apolipoprotein A-IMilano dimer. Biochemistry. 38: 16307–16314 [DOI] [PubMed] [Google Scholar]
- 33.Wang W. Q., Moses A. S., Francis G. A. 2001. Cholesterol mobilization by free and lipid-bound apoAI(Milano) and apoAI(Milano)-apoAII heterodimers. Biochemistry. 40: 3666–3673 [DOI] [PubMed] [Google Scholar]
- 34.Weibel G. L., Alexander E. T., Joshi M. R., Rader D. J., Lund-Katz S., Phillips M. C., Rothblat G. H. 2007. Wild-type ApoA-I and the Milano variant have similar abilities to stimulate cellular lipid mobilization and efflux. Arterioscler. Thromb. Vasc. Biol. 27: 2022–2029 [DOI] [PubMed] [Google Scholar]
- 35.Alexander E. T., Weibel G. L., Joshi M. R., Vedhachalam C., de la Llera-Moya M., Rothblat G. H., Phillips M. C., Rader D. J. 2009. Macrophage reverse cholesterol transport in mice expressing ApoA-I Milano. Arterioscler. Thromb. Vasc. Biol. 29: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu X., Wu G., Zeng W., Xue H., Chen B. 2005. Cysteine mutants of human apolipoprotein A-I: a study of secondary structural and functional properties. J. Lipid Res. 46: 1303–1311 [DOI] [PubMed] [Google Scholar]
- 37.Li L., Li S., Jones M. K., Segrest J. P. 2012. Rotational and hinge dynamics of discoidal high density lipoproteins probed by interchain disulfide bond formation. Biochim. Biophys. Acta. 1821: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klon A. E., Jones M. K., Segrest J. P., Harvey S. C. 2000. Molecular belt models for the apolipoprotein A-I Paris and Milano mutations. Biophys. J. 79: 1679–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franceschini G., Frosi T. G., Manzoni C., Gianfranceschi G., Sirtori C. R. 1982. High density lipoprotein-3 heterogeneity in subjects with the apo-AI Milano variant. J. Biol. Chem. 257: 9926–9930 [PubMed] [Google Scholar]
- 40.Favari E., Gomaraschi M., Zanotti I., Bernini F., Lee-Rueckert M., Kovanen P. T., Sirtori C. R., Franceschini G., Calabresi L. 2007. A unique protease-sensitive high density lipoprotein particle containing the apolipoprotein A-I(Milano) dimer effectively promotes ATP-binding Cassette A1-mediated cell cholesterol efflux. J. Biol. Chem. 282: 5125–5132 [DOI] [PubMed] [Google Scholar]
- 41.Weisgraber K. H., Rall S. C., Jr, Bersot T. P., Mahley R. W., Franceschini G., Sirtori C. R. 1983. Apolipoprotein A-I Milano detection of normal A-I in affected subjects and evidence for a cysteine for arginine substitution in the variant A-I. J. Biol. Chem. 258: 2508–2513 [PubMed] [Google Scholar]
- 42.Daum U., Langer C., Duverger N., Emmanuel F., Benoit P., Denèfle P., Chirazi A., Cullen P., Pritchard P. H., Bruckert E., et al. 1999. Apolipoprotein A-I (R151C) Paris is defective in activation of lecithin: cholesterol acyltransferase but not in initial lipid binding, formation of reconstituted lipoproteins, or promotion of cholesterol efflux. J. Mol. Med. 77: 614–622 [DOI] [PubMed] [Google Scholar]
- 43.Perez-Mendez O., Bruckert E., Franceschini G., Duhal N., Lacroix B., Bonte J. P., Sirtori C., Fruchart J. C., Turpin G., Luc G. 2000. Metabolism of apolipoproteins AI and AII in subjects carrying similar apoAI mutations, apoAI Milano and apoAI Paris. Atherosclerosis. 148: 317–325 [DOI] [PubMed] [Google Scholar]
- 44.Roma P., Gregg R. E., Meng M. S., Ronan R., Zech L. A., Franceschini G., Sirtori C. R., Brewer H. B., Jr 1993. In vivo metabolism of a mutant form of apolipoprotein A-I, apo A-IMilano, associated with familial hypoalphalipoproteinemia. J. Clin. Invest. 91: 1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calabresi L., Franceschini G., Burkybile A., Jonas A. 1997. Activation of lecithin cholesterol acyltransferase by a disulfide-linked apolipoprotein A-I dimer. Biochem. Biophys. Res. Commun. 232: 345–349 [DOI] [PubMed] [Google Scholar]
- 46.Sparks D. L., Davidson W. S., Lund-Katz S., Phillips M. C. 1995. Effects of the neutral lipid content of high density lipoprotein on apolipoprotein A-I structure and particle stability. J. Biol. Chem. 270: 26910–26917 [DOI] [PubMed] [Google Scholar]
- 47.Calabresi L., Vecchio G., Longhi R., Gianazza E., Palm G., Wadensten H., Hammarström A., Olsson A., Karlström A., Sejlitz T., et al. 1994. Molecular characterization of native and recombinant apolipoprotein A-IMilano dimer. The introduction of an interchain disulfide bridge remarkably alters the physicochemical properties of apolipoprotein A-I. J. Biol. Chem. 269: 32168–32174 [PubMed] [Google Scholar]
- 48.Chetty P. S., Ohshiro M., Saito H., Dhanasekaran P., Lund-Katz S., Mayne L., Englander W., Phillips M. C. 2012. Effects of the Iowa and Milano mutations on apolipoprotein A-I structure and dynamics determined by hydrogen exchange and mass spectrometry. Biochemistry. 51: 8993–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gianazza E., Eberini I., Sirtori C. R., Franceschini G., Calabresi L. 2002. Size is a major determinant of dissociation and denaturation behaviour of reconstituted high-density lipoproteins. Biochem. J. 366: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexander E. T., Tanaka M., Kono M., Saito H., Rader D. J., Phillips M. C. 2009. Structural and functional consequences of the Milano mutation (R173C) in human apolipoprotein A-I. J. Lipid Res. 50: 1409–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calabresi L., Tedeschi G., Treu C., Ronchi S., Galbiati D., Airoldi S., Sirtori C. R., Marcel Y., Franceschini G. 2001. Limited proteolysis of a disulfide-linked apoA-I dimer in reconstituted HDL. J. Lipid Res. 42: 935–942 [PubMed] [Google Scholar]
- 52.Saito H., Dhanasekaran P., Nguyen D., Holvoet P., Lund-Katz S., Phillips M. C. 2003. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 278: 23227–23232 [DOI] [PubMed] [Google Scholar]
- 53.Mei X., Atkinson D. 2011. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286: 38570–38582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Segrest J. P., Jones M. K., Catte A., Thirumuruganandham S. P. 2012. Validation of previous computer models and MD simulations of discoidal HDL by a recent crystal structure of apoA-I. J. Lipid Res. 53: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jayaraman S., Cavigiolio G., Gursky O. 2012. Folded functional lipid-poor apolipoprotein A-I obtained by heating of high-density lipoproteins: Relevance to HDL biogenesis. Biochem. J. 442: 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gursky O., Mei X., Atkinson D. 2012. Crystal structure of the C-terminal truncated apolipoprotein A-I sheds new light on the amyloid formation by the N-terminal segment. Biochemistry. 51: 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao X., Yuan S., Jayaraman S., Gursky O. 2012. Effect of apolipoprotein A-II on the structure and stability of human high-density lipoprotein: Implications for the role of apoA-II in HDL metabolism. Biochemistry. 51: 4633–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rocco A. G., Mollica L., Gianazza E., Calabresi L., Franceschini G., Sirtori C. R., Eberini I. 2006. A model structure for the heterodimer apoA-IMilano-apoA-II supports its peculiar susceptibility to proteolysis. Biophys. J. 91: 3043–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rocco A. G., Sensi C., Gianazza E., Calabresi L., Franceschini G., Sirtori C. R., Eberini I. 2010. Structural and dynamic features of apolipoprotein A-I cysteine mutants, Milano and Paris, in synthetic HDL. J. Mol. Graph. Model. 29: 406–414 [DOI] [PubMed] [Google Scholar]
- 60.Calabresi L., Vecchio G., Frigerio F., Vavassori L., Sirtori C. R., Franceschini G. 1997. Reconstituted high-density lipoproteins with a disulfide-linked apolipoprotein A-I dimer: evidence for restricted particle size heterogeneity. Biochemistry. 36: 12428–12433 [DOI] [PubMed] [Google Scholar]
- 61.Kalé L., Skeel R., Bhandarkar M., Brunner R., Gursoy A., Krawetz N., Phillips J., Shinozaki A., Varadarajan K., Schulten K. 1999. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 151: 283–312 [Google Scholar]
- 62.Jones M. K., Catte A., Patterson J. C., Gu F., Chen J., Li L., Segrest J. P. 2009. Thermal stability of apolipoprotein A-I in high-density lipoproteins by molecular dynamics. Biophys. J. 96: 354–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humphrey W., Dalke A., Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14: 33–38 [DOI] [PubMed] [Google Scholar]
- 64.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79: 926–935 [Google Scholar]
- 65.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4: 187–217 [Google Scholar]
- 66.MacKerell A. D., Jr, Bashford D., Dunbrack R. L., Jr, Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102: 3586–3616 [DOI] [PubMed] [Google Scholar]
- 67.Feller S. E., Pastor R. W. 1997. Length scales of lipid dynamics and molecular dynamics. Pac. Symp. Biocomput. 1997: 142–150 [PubMed] [Google Scholar]
- 68.Schlenkrich M., Brickmann J., MacKerell A., Jr, Karplus M. 1996. An empirical potential energy function for phospholipids: criteria for parameter optimization and applications. In Biological Membranes: A Molecular Perspective from Computation and Experiment. K. M. Merz and B. Roux, editoirs. Birkhauser, Boston. 31–81. [Google Scholar]
- 69.Bashtovyy D., Jones M. K., Anantharamaiah G. M., Segrest J. P. 2011. Analyses of sequence conservation of apolipoprotein A-I afford novel insights into HDL structure-function. J. Lipid Res. 52: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaisar T. 2012. Proteomics investigations of HDL: challenges and promise. Curr. Vasc. Pharmacol. 10: 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagata K. O., Nakada C., Kasai R. S., Kusumi A., Ueda K. 2013. ABCA1 dimer-monomer interconversion during HDL generation revealed by single-molecule imaging. Proc. Natl. Acad. Sci. USA. 110: 5034–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mei X., Atkinson D. 2013. Probing the structure and function of apoA-I C-terminal domain during HDL formation. (Abstract 2021 in ASBMB Annual Meeting. Boston MA, April 20–24, 2013.) [Google Scholar]
- 73.Maiorano J. N., Jandacek R. J., Horace E. M., Davidson W. S. 2004. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 43: 11717–11726 [DOI] [PubMed] [Google Scholar]
- 74.Martin D. D., Budamagunta M. S., Ryan R. O., Voss J. C., Oda M. N. 2006. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J. Biol. Chem. 281: 20418–20426 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.