Abstract
Membrane-protein interaction plays key roles in a wide variety of biological processes. Although various methods have been employed to measure membrane binding of soluble proteins, a robust high-throughput assay that is universally applicable to all proteins is lacking at present. Here we report a new fluorescence quenching assay utilizing enhanced green fluorescence protein (EGFP)-fusion proteins and a lipid containing a dark quencher, N-dimethylaminoazobenzenesulfonyl-phosphatidylethanolamine (dabsyl-PE). The EGFP fluorescence emission intensity showed a large decrease (i.e., >50%) when EGFP-fusion proteins bound the vesicles containing 5 mol% dabsyl-PE. This simple assay, which can be performed using either a cuvette-based spectrofluorometer or a fluorescence plate reader, allowed rapid, sensitive, and accurate determination of lipid specificity and affinity for various lipid binding domains, including two pleckstrin homology domains, an epsin N-terminal homology domain, and a phox homology domain. The assay can also be applied to high-throughput screening of small molecules that modulate membrane binding of proteins.
Keywords: membrane-protein binding, high-throughput assay, dark quencher, green fluorescence protein
Membrane-protein interactions play key roles in a wide variety of biological processes, including cell signaling, membrane trafficking, blood coagulation, and viral infection, among others (1–3). A large number of soluble proteins, both intracellular and extracellular, are known to interact with cell membranes under physiological conditions. Most of them contain lipid binding domains or motifs that mediate membrane binding either through specific recognition of lipid headgroups or by nonspecific electrostatic and/or hydrophobic interactions with membrane lipids (1–3). More recently, it has been reported that many modular protein interaction domains, such as PDZ domains (4–7), can also directly interact with membrane lipids. Also, genomic-scale computation predicts that a large number of noncanonical membrane binding proteins will interact with membranes (8, 9). It is therefore expected that the number of membrane binding proteins will continue to grow with further characterization.
Membrane binding of soluble and/or peripheral proteins has been measured by diverse biochemical and biophysical methods (10, 11), including the lipid overlay assay (12), the sedimentation assay using lipid vesicles (13) or lipid-coated beads (14), various fluorescence methods, and surface plasmon resonance (SPR) analysis. The lipid overlay assay has been popular due to its ease of use but it has major drawbacks, including low sensitivity, poor reliability, and an inability to yield quantitative information (12). The vesicle sedimentation assay can provide quantitative information but it suffers from a difficulty in quantifying vesicle-bound and free proteins, which often makes it necessary to label proteins, and variable pelleting efficiency of different vesicles. The SPR analysis allows the most robust quantitative analysis of membrane-protein interactions with appropriate controls and thus has been a mainstay in biophysical characterization of membrane binding proteins (10, 11). The method offers many advantages over others, including high sensitivity, no requirement for protein labeling, and an ability to provide kinetic information. However, it also has limitations, such as a necessity for expensive instrumentation, uncertainty about the physical nature of lipids coated on the sensor chip, and binding measurements under nonequilibrium conditions. Various fluorescence techniques have been employed to monitor membrane-protein interaction. The change in Trp fluorescence emission intensity in terms of an increase (15), quenching (16), and fluorescence resonance energy transfer (17, 18) can be monitored as a protein binds the membrane if it contains a Trp residue(s) on the membrane binding surface. Although rapid, convenient, and potentially sensitive, this method is not generally applicable because there are many membrane binding proteins without Trp on their membrane binding surfaces and because genetic introduction of Trp to the membrane binding surface can dramatically change the membrane binding property of the protein (19). Site-specific incorporation of a single organic fluorophore into a protein allows highly sensitive monitoring of membrane binding, but this method is limited by experimental inconvenience and the relatively low yield of chemical modification (20). Other fluorescence techniques, including anisotropy (21) and fluorescence correlation spectroscopy analyses (22), have also been used to measure membrane-protein interaction, but their general use has been hampered by their respective drawbacks and limitations.
Most important, none of these methods allow sensitive and robust high-throughput analysis that can be universally applied to all membrane binding proteins. A recent effort to develop an immunochemical plate assay for lipid-protein binding had limited success because many membrane binding proteins cannot tightly bind the lipid molecule isolated from the membrane environment (23). Lack of high-throughput assay greatly hampers large-scale characterization of proteins to identify new and novel membrane binding proteins and screening of small molecule libraries to identify those molecules that can effectively modulate membrane binding of pharmacologically important proteins. To overcome these technical limitations and problems, we have developed a new sensitive and robust high-throughput membrane binding assay that is based on fluorescence quenching of enhanced green fluorescence protein (EGFP) fused to proteins by N-dimethylaminoazobenzenesulfonyl-phosphatidylethanolamine (dabsyl-PE) incorporated in vesicles with various lipid compositions. Our results show that this simple and sensitive assay is applicable to rapid and accurate determination of lipid affinity and specificity of diverse proteins as well as to high-throughput screening of small molecules that can modulate membrane binding of proteins.
MATERIALS AND METHODS
Materials
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), and soy phosphatidylinositol (PtdIns) were from Avanti Polar Lipids and a 1,2-dipalmitoyl derivative of phosphatidylinositol-3-phosphate [PtdIns(3)P], phosphatidylinositol-4-phosphate [PtdIns(4)P], phosphatidylinositol-5-phosphate [PtdIns(5)P], phosphatidylinositol-3,4-bisphosphate [PtdIns(3,4)P2], phosphatidylinositol-3,5-bisphosphate [PtdIns(3,5)P2], phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2], and phosphatidylinositol-3,4,5-trisphosphate [PtdIns(3,4,5)P3], and inositol-1,3,4,5-tetrakisphosphate [Ins(1,3,4,5)P4] were from Cayman Chemical. Dabsyl chloride was purchased from TCI (Portland, OR). All solvents were purchased from Sigma Chemical and Fischer Scientific. Triethylamine was dried by distillation from calcium hydride. Column chromatography was performed using silica gel 60 Å (32–63 mesh) purchased from Sorbent Technologies. 1H NMR spectra were recorded with a Bruker AV-500 spectrometer. Mass spectra were obtained with a Finnigan LCQ Deca mass spectrometer under the positive ion mode.
Dabsyl-PE synthesis
POPE (50 mg, 0.07 mmol) in chloroform (2 ml) was added to a solution of dabsyl chloride (22.6 mg, 0.07 mmol) and triethylamine (0.2 ml) in chloroform (5 ml). The solution was stirred for 6 h at room temperature in the dark. The solvent was removed under reduced pressure and the residue was purified by silica column chromatography (gradient elution of dicholoromethane and methanol (100:0 to 85:15) to afford dabsyl-POPE (36 mg, 0.036 mmol) as an orange solid (yield: 51%). The thin layer chromatography shows that its Rf is 0.25 in acetonitrile/methanol (85:15). The structure of dabsyl-PE was confirmed by 1H NMR and mass spectrometry. 1H NMR (500 MHz in CDCl3) spectrum: chemical shift (δ) = 7.94 (d, J = 8.07 Hz, 2H), 7.81–7.89 (m, 4H), 6.69 (d, J = 8.44 Hz, 2H), 5.22–5.35 (m, 3H), 4.39 (d, J = 10.64 Hz, 1H), 3.92–4.22 (m, 5H), 3.05–3.19 (m, 8H), 2.15–2.32 (m, 4H), 1.92–2.00 (m, 4H), 1.45–1.58 (m, 4H), 1.18–1.32 (m, 44H), and 0.84–0.89 (m, 6H). 13C NMR (125 MHz in CDCl3): δ = 14.1, 22.7, 24.8, 24.9, 27.2, 27.3, 29.2, 29.2, 29,3, 29.3, 29.4, 29.5, 29.6, 29.7, 29.7, 29.8, 31.9, 32.0, 34,1, 34.2, 40.3, 43.7, 62.8, 64.1, 65.1, 70.6, 70.6, 111.4, 122.6, 125.7, 128.0, 129.8, 139.9, 143.7, 152.9, 155.3, 173.6, and 173.8. Mass spectrum (ESI): calculated for C53H88N4O10P1S1 [M+1]+1,005.3, found 1,005.4; calculated for C53H88N4O10P1S1Na [M+Na]+1,027.3, found 1,027.3.
Vesicle preparation
Lipid solutions were mixed according to the final lipid composition and the solvent was evaporated under a stream of nitrogen gas. Tris buffer (20 mM, pH 7.4), containing 0.16 M NaCl was added to the lipid film and the mixture was sonicated for 1 min. Large unilamellar vesicles with 100 nm diameter were then prepared with a Liposofast microextruder (Avestin) using a 100 nm polycarbonate filter.
Protein expression and purification
All lipid binding domains were cloned into either the pET28a vector or the pRSETb vector with a C-terminus EGFP tag. Proteins were expressed in BL21 RIL cells. For expression of the proteins, 500 ml of Luria broth containing 50 μg/ml kanamycin or 100 μg/ml ampicillin was inoculated with BL21 RIL colonies expressing each protein construct. Cells were allowed to grow in the medium at 37°C until an absorbance at 600 nm reached 0.6 AU. Protein expression was induced with the addition of 100 μM isopropyl 1-thio-β-d-galactopyranoside (Research Products, Mount Prospect, IL), at which point cells were moved to a 25°C shaker for 14 h incubation. Cells were harvested by centrifugation (2,500 g for 10 min at 4°C), and the pellet was resuspended in 20 ml of the lysis buffer (50 mM Tris, 300 mM NaCl, 10 mM imidazole, and 10% (v/v) glycerol, pH 7.9). The solution was sonicated for 5 min with 30 s intervals and centrifuged for 30 min (39,000 g at 4°C). The supernatant was transferred to a 50 ml Falcon tube and 1 ml of Ni-NTA agarose (Qiagen) was added. The supernatant was allowed to incubate with the resin for ∼30 min at 4°C with gentle mixing. The supernatant was then poured onto a column, and the resin was washed with 50 ml of 50 mM Tris buffer, pH 7.4, containing 20 mM imidazole and another 50 ml of 50 mM Tris buffer, pH 7.4, containing 40 mM imidazole. Protein was then eluted using 50 mM Tris buffer, pH 7.4, with 300 mM imidazole. Purity of eluted protein was checked by sodium-dodecylsulfate gel electrophoresis using 18% polyacrylamide gel, and the concentration was determined using the Bradford reagents. Protein was frozen in liquid nitrogen and stored at −80°C.
Fluorescence quenching assay
All fluorescence quenching assays were performed using FluoroLog3 spectrofluorometer (Horiba Scientific) at 25°C. Two milliliters of 100 nM protein solution in 20 mM Tris buffer, pH 7.4, containing 0.16 M NaCl was transferred to a quartz cuvette (Hellma Analytics), and the EGFP fluorescence was excited at 460 nm and its emission spectrum was obtained. Typically, the change in emission intensity at 509 nm was monitored after incremental addition of lipid vesicle solution and 1 min incubation with gentle mixing.
Fluorescence plate reader assay
The plate reader assay was performed using the FluoroLog3 spectrofluorometer with the Micromax384 attachment (Horiba Scientific). Nontreated black polystyrene 96-well plates (Corning Incorporated) were used. Protein (100 nm) with the increasing concentration of vesicles with a given lipid composition in 20 mM Tris buffer, pH 7.4, containing 0.16 M NaCl was added to each row of wells and the EGFP fluorescence emission at 509 nm was measured with excitation set at 460 nm.
Ins(1,3,4,5)P4 inhibition assay
C-terminal EGFP-tagged PDK1 plekstrin homology domain (PDK1-PH-EGFP) (100 nm) was incubated with different concentrations of Ins(1,3,4,5)P4 (0, 5, 25, 50, 100, and 200 μM) for 5 min. Then the fluorescence emission of EGFP (excitation at 460 nm and emission at 509 nm) was recorded after incremental addition of POPC/POPS/PtdIns(3,4,5)P3/dabsyl-PE (72:20:3:5) vesicles in both cuvette-based and plate reader assays.
Data analysis
Membrane binding isotherms of proteins were analyzed assuming that each protein binds independently to a site on the vesicle surface composed of n lipids with dissociation constant Kd (10). Values of n and Kd were determined by nonlinear least-squares analysis of the [P]b/[P]o versus [L]o plot using equation 1 (10):
| (Eq. 1) |
where [L]o, [P]o, and [P]b are total lipid, total protein, and bound protein concentrations, respectively and ΔF and ΔFmax indicate the fluorescence intensity decrease and the maximal intensity decrease, respectively. Inhibition of membrane binding of PDK1-PH-EGFP by Ins(1,3,4,5)P4 was analyzed by equation 2 (24):
| (Eq. 2) |
ΔF and ΔFo indicate the fluorescence intensity decrease of EGFP by dabsyl-PE-containing vesicles in the presence and the absence of a given concentration of Ins(1,3,4,5)P4, respectively.
SPR analysis
All SPR measurements were performed at 23°C in 20 mM Tris-HCl, pH 7.4, containing 0.16 M NaCl using a lipid-coated L1 chip in the BIACORE X system as described previously (25, 26). POPC/POPS/phosphoinositide (PtdInsP) (77:20:3) vesicles and POPC vesicles were coated onto the active surface and the control surface, respectively. Equilibrium SPR measurements were done at the flow rate of 5 μl/min to allow sufficient time for the response in resonance units (RUs) of the association phase to reach near-equilibrium values (Req). Assuming a Langmuir-type binding between the protein (P) and protein binding sites (M) on vesicles (i.e., P + M ↔ PM), Req values were then plotted versus the total protein concentration ([P]o), and the Kd value was determined by a nonlinear least-squares analysis of the binding isotherm using an equation, Req = Rmax/(1 + Kd/[P]o). Each data set was repeated three or more times to calculate average and standard deviation values. For kinetic SPR measurements, the flow rate was maintained at 15 μl/min for both association and dissociation phases (27, 28).
RESULTS AND DISCUSSION
Assay strategy
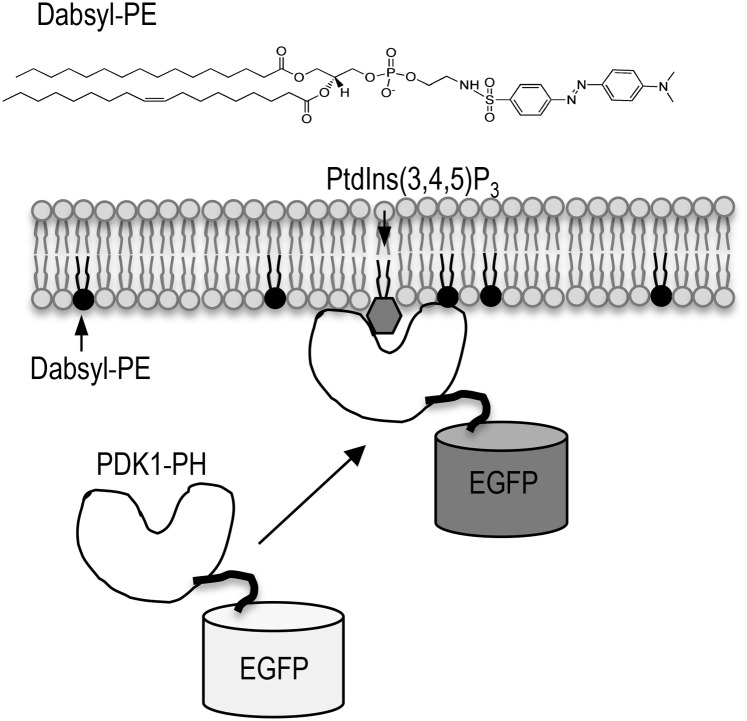
Our basic assay strategy for a universal high-throughput membrane binding assay is two-fold. First, we render comparable fluorescence properties to all proteins by expressing them as EGFP-fusion proteins. We then quantify their membrane binding by monitoring the decrease in EGFP fluorescence as the proteins bind the lipid vesicles incorporating a nonfluorescent quenching dye (dark quencher) (see Fig. 1).
Fig. 1.
The strategy of our fluorescence quenching-based membrane binding assay. To the solution of an EGFP-fusion protein (e.g., PDK1-PH-EGFP) is added vesicles containing their favorite lipids (e.g., PtdIns(3,4,5)P3 shown as a shaded hexagon) and a dark quencher, dabsyl-PE (shown in black). As the protein binds the membrane surface, the fluorescence emission intensity of EGFP is greatly reduced due to quenching by dabsyl-PE, allowing quantitative analysis of membrane-protein interaction.
Fluorescence proteins, including EGFP, have been extensively used to monitor the subcellular localization, dynamics, and interactions of numerous cellular proteins (29, 30). Although some fluorescence proteins, most notably red derivatives, have been reported to induce protein oligomerization, EGFP is known to have much less effect on the structure, function, and dynamics of the parent protein in most cases when attached to either the N or C terminus of the fusion partner with an appropriate linker (29, 30). As is the case with other fusion proteins, EGFP-fusion proteins can be expressed in bacteria in high yield, as stable EGFP generally improves the expression yield of fusion partners, isolated protein domains and truncated proteins in particular. Furthermore, EGFP has high fluorescence quantum yield and does not have detectable affinity for cell membranes.
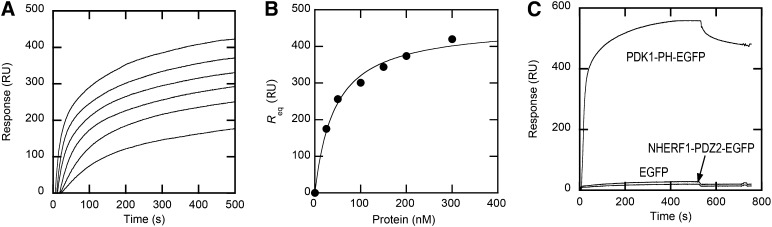
To test the feasibility of using EGFP-tagged proteins for our assay, we first expressed and characterized various lipid binding domains as EGFP-fusion proteins. The EGFP tag was attached to either the N terminus or the C terminus, depending on the protein, as the location of the tag can affect the property of the protein in some cases. C-terminal EGFP-tagged epsin1 N-terminal homology domain (ENTH-EGFP), C-terminal EGFP-tagged Akt1 PH domain (Akt1-PH-EGFP), PDK1-PH-EGFP, and C-terminal EGFP-tagged sorting nexin 27 phox homology domain (SNX27-PX-EGFP) were all expressed in Escherichia coli in high yield. When binding of PDK1-PH-EGFP to POPC/POPS/PtdIns(3,4,5)P3 (77:20:3) vesicles was measured by SPR analysis, its Kd value (44 ± 15 nM) (Fig. 2A, B) was comparable to that of untagged PDK1-PH for the same vesicles (31). A similar trend was seen with all other EGFP-tagged proteins (data not shown). Also, EGFP itself showed negligible binding to the vesicles when compared with PDK1-PH-EGFP (Fig. 2C). This verified the notion that EGFP fusion proteins can be used in place of untagged proteins for membrane binding analysis.
Fig. 2.
Determination of Kd for PDK1-PH-EGFP membrane binding by SPR analysis. A: Equilibrium sensorgrams for PDK1-PH-EGFP interacting with POPC/POPS/PtdIns(3,4,5)P3 (77:20:3) vesicles. The protein concentration was 25, 50, 100, 150, 200, and 300 nM from bottom to top. B: Binding isotherms of PDK1-PH-EGFP with POPC/POPS/PtdIns(3,4,5)P3 (77:20:3) vesicles. The binding isotherm was fit by nonlinear least-squares analysis using the equation: Req = Rmax/(1 + Kd/[P]o) where [P]o, Req, and Rmax are the total protein concentration, the near-equilibrium resonance unit (RU) value for each P0, and the maximal Req value, respectively. Rmax (460 ± 20) RU and Kd (44 ± 15 nM) values were used to construct the theoretical curve (solid line). C: Kinetic sensorgrams for PDK1-PH-EGFP, NHERF1-PDZ2-EGFP, and EGFP (500 nM each) interacting with POPC/POPS/PtdIns(3,4,5)P3 (77:20:3) vesicles coated onto the L1 chip.
We then explored the possibility of using a synthetic lipid that can greatly change the fluorescence emission intensity of EGFP as an EGFP-fusion protein approaches the membrane. Although one would ideally monitor the signal increase for better sensitivity, it is practically impossible to design a simple assay in which the EGFP fluorescence intensity increases as a result of membrane binding. Taking advantage of the high molecular brightness of EGFP, we thus designed an assay that monitors the decrease in EGFP fluorescence intensity by lipid vesicles incorporating a synthetic lipid, dabsyl-PE, which contains a dark quencher dabsyl group (32) in the headgroup (see Fig. 1).
Conditions and efficiency of the EGFP quenching assay
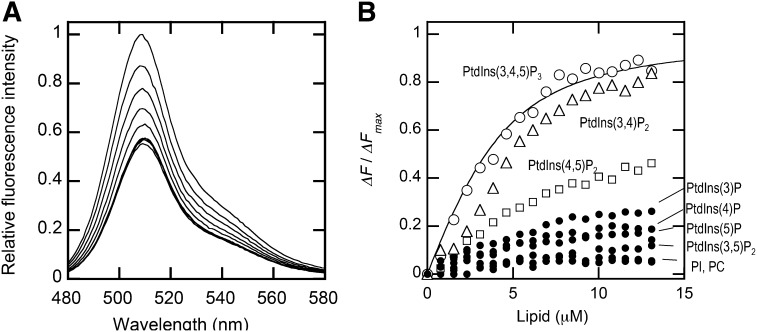
We first measured the binding of PDK1-PH-EGFP to vesicles made of POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (77-x:20:x:3) to find an optimal condition for the assay. The concentration of dabsyl-PE was varied from 3 to 10 mol% with the fixed concentrations of POPS and PtdIns(3,4,5)P3. Both phosphatidylserine (PS) and PtdIns(3,4,5)P3 are necessary for membrane binding of PDK1-PH (31). The maximal quenching of the EGFP fluorescence intensity by dabsyl-PE-containing vesicles gradually increased with the increase in its concentration in the vesicles. When dabsyl-PE = 5 mol%, the maximal quenching reached 50% of the initial intensity (see Fig. 3A). Although a higher concentration of dabsyl-PE gave a larger degree of quenching, we decided to include 5 mol% dabsyl-PE for our assay because dabsyl-PE is an anionic lipid that may promote nonspecific membrane binding of cationic proteins at higher concentrations. The membrane binding isotherm (Fig. 3B) for PDK1-PH-EGFP was constructed from the EGFP fluorescence quenching data and the binding parameters were calculated by the nonlinear least-squares curve fitting using the general membrane binding equation (equation 1) (10). The binding isotherm can also be analyzed using other membrane binding models, including the membrane partitioning model (33). The Kd of PDK1-PH-EGFP for POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (72:20:5:3) vesicles (see Table 1) was comparable to that for POPC/POPS/PtdIns(3,4,5)P3 (77:30:3) vesicles determined by SPR analysis (see above), showing the accuracy of the fluorescence quenching assay. Under the same conditions, POPC/dabsyl-PE (95:5) vesicles did not cause any appreciable fluorescence quenching above the buffer control, confirming that 5 mol% dabsyl-PE does not induce a significant degree of nonspecific vesicle binding, at least for PDK1-PH-EGFP. Also, POPC/POPS/dabsyl-PE/PtdIns(3,4)P2 (72:20:5:3) vesicles gave similar quenching to POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (72:20:5:3) vesicles whereas vesicles containing any other phosphoinositides (PtdInsPs) or phosphatidylinositol (PtdIns) caused much less quenching than did POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (72:20:5:3) vesicles, which is consistent with the reported lipid specificity of PDK1-PH-EGFP (31). This demonstrates that lipid specificity and affinity of a membrane binding protein can be accurately determined by our assay.
Fig. 3.
Fluorescence quenching of the C-terminal EGFP-tagged PDK1 PH domain (PDK1-PH-EGFP) by dabsyl-PE-containing vesicles. A: Fluorescence emission spectra of PDK1-PH-EGFP (100 nM) in the presence of increasing concentrations of POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (72:20:5:3) vesicles were measured in the cuvette-based assay. The total lipid concentrations used were 0, 1.6, 3.9, 5.4, 6.9, 8.5, 9.2, 10.0, and 11.6 μM from top to bottom. Fluorescence emission intensity was normalized against the EGFP intensity value at 509 nm without lipid vesicles. B: Binding isotherms of PDK1-PH-EGFP with vesicles containing various PtdInsPs, i.e., POPC/POPS/dabsyl-PE/PtdInsP (72:20:5:3) vesicles. PtdInsP includes PtdIns(3,4,5)P3, PtdIns(3,4)P2, PtdIns(4,5)P2, PtdIns(3,5)P2, PtdIns(3)P, PtdIns(4)P, PtdIns(5)P, and PtdIns. POPC/dabsyl-PE (95:5) vesicles were also tested as a negative control. The fluorescence decrease upon lipid addition was background corrected by the fluorescence decrease upon addition of the same volume of the buffer solution. ΔF/ΔFmax was calculated as fluorescence intensity decrease at a given lipid concentration divided by the maximal fluorescence decrease. The binding isotherm for POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (72:20:5:3) vesicles was fit by nonlinear least-squares analysis using equation 1. Values of n (40 ± 10) and Kd (35 ± 10 nM) are summarized in Table 1.
TABLE 1.
Determination of dissociation constant for membrane binding for various lipid binding domains by the EGFP quenching assay
| Proteins | Lipids | Kd (nM)a (n in parenthesis) | |
| Cuvette Assay | Plate Reader Assay | ||
| PDK1-PH-EGFP | POPC/POPS/dabsylPE/PtdIns(3,4,5)P3 (72:20:5:3) | 35 ± 10 (n = 40 ± 10) | 44 ± 15 (n = 50 ± 20) |
| Akt1-PH-EGFP | POPC/POPS/dabsylPE/PtdIns(3,4,5)P3 (72:20:5:3) | 25 ± 15 (n = 40 ± 12) | 38 ± 10 (n = 50 ± 15) |
| ENTH-EGFP | POPC/POPS/dabsylPE/PtdIns(4,5)P2 (72:20:5:3) | 45 ± 10 (n = 35 ± 15) | 30 ± 10 (n = 40 ± 10) |
| SNX27-PX-EGFP | POPC/POPS/dabsylPE/PtdIns(3)P (72:20:5:3) | 130 ± 20 (n = 40 ± 20) | 110 ± 25 (n = 30 ± 15) |
Mean ± SD values from triplicate measurements.
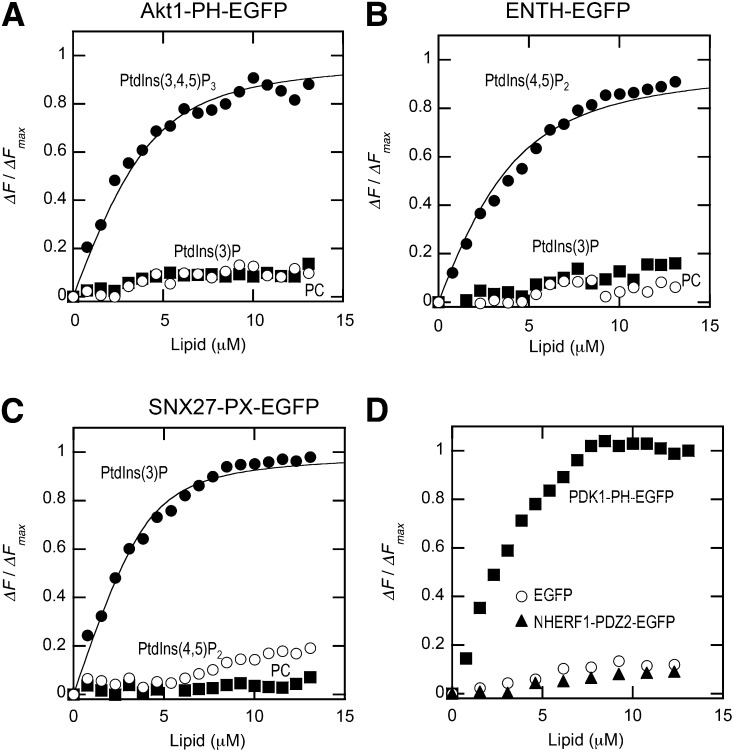
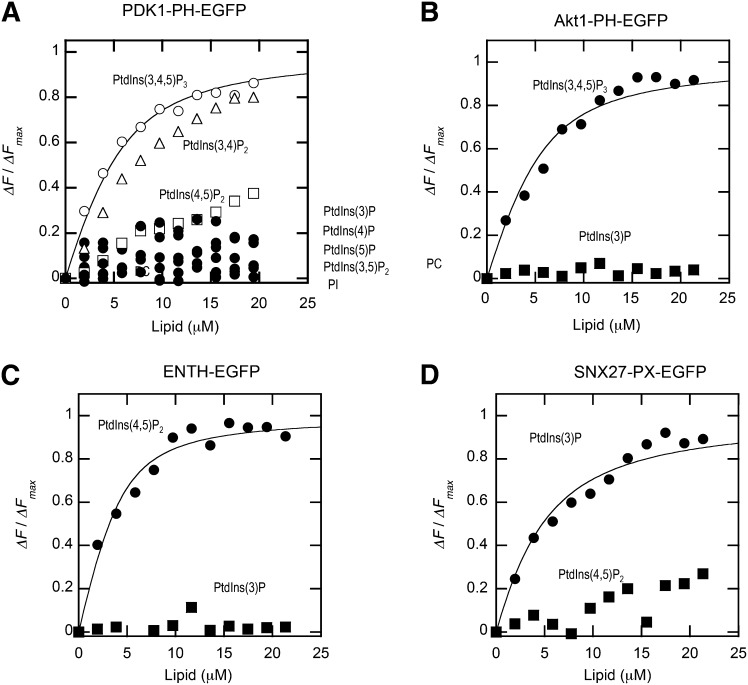
To test the general applicability of our EGFP quenching assay, we measured the binding of three other lipid binding domains, ENTH-EGFP, Akt1-PH-EGFP, and SNX27-PX-EGFP, by the same method. As shown in Fig. 4, all three proteins exhibited the behaviors consistent with their reported membrane binding properties. For instance, ENTH-EGFP tightly and specifically bound POPC/POPS/dabsyl-PE/PtdIns(4,5)P2 (72:20:5:3) vesicles with the Kd value comparable to the reported one (34). Similar results were obtained for Akt1-PH-EGFP and SNX27-PX-EGFP, that are known to have specificity for PtdIns(3,4,5)P3/PtdIns(3,4)P2 (26) and PtdIns(3)P (35), respectively. For all these proteins, >50% quenching of the EGFP signal was achieved by 5 mol% dabsyl-PE-containing vesicles and the background quenching by POPC/dabsyl-PE (95:5) vesicles was negligible above the buffer control. As a negative control, we measured the binding of the C-terminal EGFP-tagged second PDZ domain of NHERF1 (NHERF1-PDZ2-EGFP) (6, 7) and EGFP, which showed no membrane binding by the SPR analysis (Fig. 2C) to POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (72:20:5:3) vesicles. As shown in Fig. 4D, NHERF1-PDZ2-EGFP and EGFP showed negligible binding signals when compared with PDK1-PH-EGFP binding to POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (72:20:5:3) vesicles under the same conditions. Collectively, these results show that our EGFP quenching assay can be used universally for all membrane binding proteins.
Fig. 4.
Binding isotherms of Akt1-PH-EGFP (A), ENTH-EGFP (B), SNX27-PX-EGFP (C), and EGFP and SNX5-PX-EGFP (D) (all 100 nM) with vesicles with various compositions measured in the cuvette-based assay. The binding isotherms for vesicles containing the most preferred lipid (e.g., POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (72:20:5:3) for Akt1-PH-EGFP) were fit by nonlinear least-squares analysis using equation 1. Values of n and Kd are summarized in Table 1. ΔF/ΔFmax was calculated as described for Fig. 3B. For binding of EGFP and NHERF1-PDZ2-EGFP to POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 vesicles (D), binding of PDK1-PH-EGFP (100 nM) to the same vesicles was also shown to demonstrate the negligible membrane binding of these negative controls.
High-throughput EGFP quenching assay
High sensitivity and signal-to-noise ratio of the EGFP quenching assay indicated that it could be used in a plate reader-based high-throughput assay. To quantitatively determine the lipid specificity and affinity simultaneously, we placed in each row of a 96-well plate varying concentrations of vesicles containing different lipids (i.e., POPC/POPS/dabsyl-PE/X (72:20:5:3); X = each of seven PtdInsPs and PtdIns) and allowed them to interact with a fixed concentration of an EGFP-fusion protein. Two control rows contained the buffer solution and POPC/dabsyl-PE (95:5), respectively. As shown in Fig. 5, lipid specificity and affinity for all four EGFP-fusion proteins, PDK1-PH-EGFP, ENTH-EGFP, Akt1-PH-EGFP, and SNX27-PX-EGFP, were determined accurately and rapidly by this 96-well plate reader assay. Kd values for their binding to the preferred lipid vesicles were statistically comparable to those determined by the cuvette-based assay (Table 1). One complete assay for each protein can be completed within minutes and the total amounts of protein and lipid samples for each assay are much less than those required for other assays, including SPR analysis. Because EGFP-tagged proteins and lipid samples can be readily prepared, our assay allows universal high-throughput quantification of lipid specificity and affinity of proteins.
Fig. 5.
Determination of lipid specificity and affinity for PDK1-PH-EGFP (A), Akt1-PH-EGFP (B), ENTH-EGFP (C), and SNX27-PX-EGFP (D) by the fluorescence plate reader assay. For each protein (100 nM), each row of a 96-well plate contained an increasing concentration of vesicles containing each of seven different PtdInsPs [POPC/POPS/dabsyl-PE/PtdInsP (72:20:5:3)]. Two control rows contained the buffer solution and POPC/dabsyl-PE (95:5), respectively. ΔF/ΔFmax was calculated as described for Fig. 3B. The binding isotherms for vesicles containing the most preferred lipid were fit by the nonlinear least-squares analysis using equation 1. Values of n and Kd are summarized in Table 1.
For the above four proteins, 20 mol% of PS was included in all vesicles because PS is known to enhance their membrane binding (26, 31, 34, 36). However, in a more general nonbiased assay, one can run a two-step assay starting with POPC/dabsyl-PE/L1 (95-x:5:x) (L1 = PS, PtdIns, cholesterol, etc.; x = 0–30 mol%) to first detect any requirement for a bulk lipid and then performing a specificity and affinity test with POPC/dabsyl-PE/L1/L2 (75-x:5:20:x) (L2 = PtdInsP, phosphatidic acid, ceramide, diacylglycerol, etc.; x = 0–10 mol%; 20 mol% is an arbitrary concentration for L1). This two-step assay is preferred over a single-step assay employing, e.g., POPC/dabsyl-PE/L (70:5:25) (L = all lipids) because it is not physiologically meaningful to directly compare bulk lipids, such as PS or cholesterols, with signaling lipids, such as PtdIns(3,4,5)P3, which exist in such different concentrations in cell membranes. Also, many membrane binding proteins interact with both bulk lipids and a signaling lipid coincidently (2).
High-throughput screening of membrane binding inhibitors
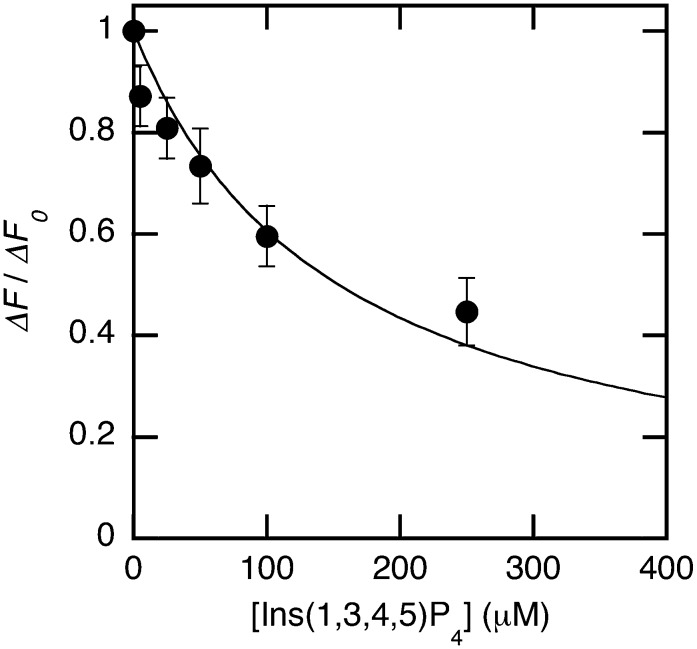
Having established the conditions for the high-throughput membrane binding assay, we then tested if the assay can be used to screen molecules that can modulate the membrane binding of proteins through competition analysis. As a proof of principle, we measured the inhibitory activity of Ins(1,3,4,5)P4 against membrane binding of PDK1-PH. Ins(1,3,4,5)P4 is a soluble molecule that is known to be able to compete with PtdIns(3,4,5)P3 for the lipid binding pocket of proteins (37). Increasing concentrations (0–200 μM) of Ins(1,3,4,5)P4 were added to each row of a 96-well plate and incubated with a fixed concentration (i.e., 100 nM) of PDK1-PH-EGFP. After increasing concentrations of its preferred lipid vesicles [i.e., POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (72:20:5:3)] were added to each row, the final fluorescence intensity readout was recorded and the maximal fluorescence decrease in each row (ΔF) determined after background correction by the buffer solution. As shown in Fig. 6, the analysis of the plot of ΔF/ΔFmax [ΔF without Ins(1,3,4,5)P4] as a function of Ins(1,3,4,5)P4 concentration gave an IC50 value of 150 ± 25 μM. These results demonstrate the feasibility of high-throughput screening for inhibitors of membrane-protein interactions. The same method can be applied to the screening of small molecule libraries against membrane binding of any proteins. Also, the assay can be performed in a more expedited format using a single point lipid determination (e.g., a fixed vesicle concentration giving a half-maximal quenching of EGFP in the absence of an inhibitor) instead of the multiple lipid determination described above.
Fig. 6.
Inhibition assay for membrane binding of PDK1-PH-EGFP. Each row of a 96-well plate contained a fixed concentration of Ins(1,3,4,5)P4, a fixed concentration of PDK1-PH-EGFP (100 nM), and the increasing concentration of POPC/POPS/dabsyl-PE/PtdIns(3,4,5)P3 (72:20:5:3) vesicles (0 to 25 μM). The maximal fluorescence decrease for each concentration of Ins(1,3,4,5)P4 (ΔF) was then determined and converted into ΔF/ΔFo. ΔFo is the maximal fluorescence decrease in the absence of Ins(1,3,4,5)P4. A control row contained the buffer solution. IC50 values were determined by the nonlinear least-squares analysis using equation 2. Each point shows the mean ± SD value from triplicate measurements.
CONCLUSION
We have developed a new fluorescence-based high-throughput assay for membrane-protein binding. Its main advantages are simplicity, sensitivity, speed, and a high degree of flexibility in assay design. It is generally applicable to membrane binding analysis of any proteins (including truncated proteins), modular domains, and small motifs, because most of these proteins can be stably expressed as EGFP-fusion proteins. The assay can be further modified and improved with different fluorescence protein fusion partners and different dark quencher-containing lipids. The assay will facilitate the genomic-scale identification of new and novel membrane binding proteins with unique functionality. It will also accelerate the identification of small molecules that can specifically and potently modulate the membrane binding and activation of many pharmacologically important proteins, such as Akt, PDK, and protein kinase C.
Acknowledgments
The authors thank Kate Korzistka and Ren Sheng for the preliminary work and Charles Delisle for assistance in inhibitor screening.
Footnotes
Abbreviations:
- Akt1-PH-EGFP
- C-terminal enhanced green fluorescence protein-tagged Akt1 plekstrin homology domain
- dabsyl-PE
- N-dimethylaminoazobenzenesulfonyl-phosphatidylethanolamine
- EGFP
- enhanced green fluorescence protein
- ENTH
- epsin N-terminal homology
- ENTH-EGFP
- C-terminal enhanced green fluorescence protein-tagged epsin1 N-terminal homology domain
- Ins(1,3,4,5)P4
- inositol-1,3,4,5-tetrakisphosphate
- NHERF1-PDZ2-EGFP
- C-terminal enhanced green fluorescence protein-tagged second PDZ domain of NHERF1
- PDK1-PH-EGFP
- C-terminal enhanced green fluorescence protein-tagged PDK1 plekstrin homology domain
- PH
- plekstrin homology
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine
- PS
- phosphatidylserine
- PtdIns
- phosphatidylinositol
- PtdIns(3)P
- phosphatidylinositol-3-phosphate
- PtdIns(4)P
- phosphatidylinositol-4-phosphate
- PtdIns(5)P
- phosphatidylinositol-5-phosphate
- PtdIns(3,4)P2
- phosphatidylinositol-3,4-bisphosphate
- PtdIns(3,5)P2
- phosphatidylinositol-3,5-bisphosphate
- PtdIns(4,5)P2
- phosphatidylinositol-4,5-bisphosphate
- PtdIns(3,4,5)P3
- phosphatidylinositol-3,4,5-trisphosphate
- PtdInsP
- phosphoinositide
- PX
- phox homology
- SNX27-PX-EGFP
- C-terminal enhanced green fluorescent protein-tagged sorting nexin 27 phox homology domain
- SPR
- surface plasmon resonance
This work was supported by National Institutes of Health Grants GM-68849 and GM-52598.
REFERENCES
- 1.DiNitto J. P., Cronin T. C., Lambright D. G. 2003. Membrane recognition and targeting by lipid-binding domains. Sci. STKE. 2003: re16. [DOI] [PubMed] [Google Scholar]
- 2.Cho W., Stahelin R. V. 2005. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 34: 119–151 [DOI] [PubMed] [Google Scholar]
- 3.Lemmon M. A. 2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9: 99–111 [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann P. 2006. The prevalence and significance of PDZ domain-phosphoinositide interactions. Biochim. Biophys. Acta. 1761: 947–956 [DOI] [PubMed] [Google Scholar]
- 5.Feng W., Zhang M. 2009. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat. Rev. Neurosci. 10: 87–99 [DOI] [PubMed] [Google Scholar]
- 6.Chen Y., Sheng R., Kallberg M., Silkov A., Tun M. P., Bhardwaj N., Kurilova S., Hall R. A., Honig B., Lu H., et al. 2012. Genome-wide functional annotation of dual-specificity protein- and lipid-binding modules that regulate protein interactions. Mol. Cell. 46: 226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng R., Chen Y., Yung Gee H., Stec E., Melowic H. R., Blatner N. R., Tun M. P., Kim Y., Kallberg M., Fujiwara T. K., et al. 2012. Cholesterol modulates cell signaling and protein networking by specifically interacting with PDZ domain-containing scaffold proteins. Nat. Commun. 3: 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhardwaj N., Stahelin R. V., Langlois R. E., Cho W., Lu H. 2006. Structural bioinformatics prediction of membrane-binding proteins. J. Mol. Biol. 359: 486–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silkov A., Yoon Y., Lee H., Gokhale N., Adu-Gyamfi E., Stahelin R. V., Cho W., Murray D. 2011. Genome-wide structural analysis reveals novel membrane binding properties of AP180 N-terminal homology (ANTH) domains. J. Biol. Chem. 286: 34155–34163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho W., Bittova L., Stahelin R. V. 2001. Membrane binding assays for peripheral proteins. Anal. Biochem. 296: 153–161 [DOI] [PubMed] [Google Scholar]
- 11.Narayan K., Lemmon M. A. 2006. Determining selectivity of phosphoinositide-binding domains. Methods. 39: 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowler S., Kular G., Alessi D. R. 2002. Protein lipid overlay assay. Sci. STKE. 2002: pl6. [DOI] [PubMed] [Google Scholar]
- 13.Rebecchi M., Peterson A., McLaughlin S. 1992. Phosphoinositide-specific phospholipase C-delta 1 binds with high affinity to phospholipid vesicles containing phosphatidylinositol 4,5-bisphosphate. Biochemistry. 31: 12742–12747 [DOI] [PubMed] [Google Scholar]
- 14.Kim Y., Lichtenbergova L., Snitko Y., Cho W. 1997. A phospholipase A2 kinetic and binding assay using phospholipid-coated hydrophobic beads. Anal. Biochem. 250: 109–116 [DOI] [PubMed] [Google Scholar]
- 15.Kraft C. A., Garrido J. L., Leiva-Vega L., Romero G. 2009. Quantitative analysis of protein-lipid interactions using tryptophan fluorescence. Sci. Signal. 2: pl4. [DOI] [PubMed] [Google Scholar]
- 16.Dua R., Wu S. K., Cho W. 1995. A structure-function study of bovine pancreatic phospholipase A2 using polymerized mixed liposomes. J. Biol. Chem. 270: 263–268 [DOI] [PubMed] [Google Scholar]
- 17.Bazzi M. D., Nelsestuen G. L. 1987. Association of protein kinase C with phospholipid vesicles. Biochemistry. 26: 115–122 [DOI] [PubMed] [Google Scholar]
- 18.Nalefski E. A., Slazas M. M., Falke J. J. 1997. Ca2+-signaling cycle of a membrane-docking C2 domain. Biochemistry. 36: 12011–12018 [DOI] [PubMed] [Google Scholar]
- 19.Sumandea M., Das S., Sumandea C., Cho W. 1999. Roles of aromatic residues in high interfacial activity of Naja naja atra phospholipase A2. Biochemistry. 38: 16290–16297 [DOI] [PubMed] [Google Scholar]
- 20.Yoon Y., Lee P. J., Kurilova S., Cho W. 2011. In situ quantitative imaging of cellular lipids using molecular sensors. Nat. Chem. 3: 868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao B., Skidan I., Yang J., Lugovskoy A., Reibarkh M., Long K., Brazell T., Durugkar K. A., Maki J., Ramana C. V., et al. 2010. Small molecule inhibition of phosphatidylinositol-3,4,5-triphosphate (PIP3) binding to pleckstrin homology domains. Proc. Natl. Acad. Sci. USA. 107: 20126–20131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rusu L., Gambhir A., McLaughlin S., Radler J. 2004. Fluorescence correlation spectroscopy studies of Peptide and protein binding to phospholipid vesicles. Biophys. J. 87: 1044–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong D., Smith M. D., Manna D., Bostic H. E., Cho W., Best M. D. 2009. Microplate-based characterization of protein-phosphoinositide binding interactions using a synthetic biotinylated headgroup analogue. Bioconjug. Chem. 20: 310–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dua R., Cho W. 1994. Inhibition of human secretory class II phospholipase A2 by heparin. Eur. J. Biochem. 221: 481–490 [DOI] [PubMed] [Google Scholar]
- 25.Ananthanarayanan B., Stahelin R. V., Digman M. A., Cho W. 2003. Activation mechanisms of conventional protein kinase C isoforms are determined by the ligand affinity and conformational flexibility of their C1 domains. J. Biol. Chem. 278: 46886–46894 [DOI] [PubMed] [Google Scholar]
- 26.Manna D., Albanese A., Park W. S., Cho W. 2007. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J. Biol. Chem. 282: 32093–32105 [DOI] [PubMed] [Google Scholar]
- 27.Stahelin R. V., Cho W. 2001. Differential roles of ionic, aliphatic, and aromatic residues in membrane-protein interactions: a surface plasmon resonance study on phospholipases A2. Biochemistry. 40: 4672–4678 [DOI] [PubMed] [Google Scholar]
- 28.Stahelin R. V., Cho W. 2001. Roles of calcium ions in the membrane binding of C2 domains. Biochem. J. 359: 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaner N. C., Steinbach P. A., Tsien R. Y. 2005. A guide to choosing fluorescent proteins. Nat. Methods. 2: 905–909 [DOI] [PubMed] [Google Scholar]
- 30.Miyawaki A. 2011. Proteins on the move: insights gained from fluorescent protein technologies. Nat. Rev. Mol. Cell Biol. 12: 656–668 [DOI] [PubMed] [Google Scholar]
- 31.Lucas N., Cho W. 2011. Phosphatidylserine binding is essential for plasma membrane recruitment and signaling function of 3-phosphoinositide-dependent kinase-1. J. Biol. Chem. 286: 41265–41272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyagi S., Kramer F. R. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14: 303–308 [DOI] [PubMed] [Google Scholar]
- 33.Peitzsch R. M., McLaughlin S. 1993. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry. 32: 10436–10443 [DOI] [PubMed] [Google Scholar]
- 34.Stahelin R. V., Long F., Peter B. J., Murray D., De Camilli P., McMahon H. T., Cho W. 2003. Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J. Biol. Chem. 278: 28993–28999 [DOI] [PubMed] [Google Scholar]
- 35.Lunn M. L., Nassirpour R., Arrabit C., Tan J., McLeod I., Arias C. M., Sawchenko P. E., Yates J. R., 3rd, Slesinger P. A. 2007. A unique sorting nexin regulates trafficking of potassium channels via a PDZ domain interaction. Nat. Neurosci. 10: 1249–1259 [DOI] [PubMed] [Google Scholar]
- 36.Manna D., Bhardwaj N., Vora M. S., Stahelin R. V., Lu H., Cho W. 2008. Differential roles of phosphatidylserine, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 in plasma membrane targeting of C2 domains. Molecular dynamics simulation, membrane binding, and cell translocation studies of the PKCalpha C2 domain. J. Biol. Chem. 283: 26047–26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komander D., Fairservice A., Deak M., Kular G. S., Prescott A. R., Peter Downes C., Safrany S. T., Alessi D. R., van Aalten D. M. 2004. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 23: 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]