Abstract
Background
Acidithiobacillus caldus is a sulfur oxidizing extreme acidophile and the only known mesothermophile within the Acidithiobacillales. As such, it is one of the preferred microbes for mineral bioprocessing at moderately high temperatures. In this study, we explore the genomic diversity of A. caldus strains using a combination of bioinformatic and experimental techniques, thus contributing first insights into the elucidation of the species pangenome.
Principal Findings
Comparative sequence analysis of A. caldus ATCC 51756 and SM-1 indicate that, despite sharing a conserved and highly syntenic genomic core, both strains have unique gene complements encompassing nearly 20% of their respective genomes. The differential gene complement of each strain is distributed between the chromosomal compartment, one megaplasmid and a variable number of smaller plasmids, and is directly associated to a diverse pool of mobile genetic elements (MGE). These include integrative conjugative and mobilizable elements, genomic islands and insertion sequences. Some of the accessory functions associated to these MGEs have been linked previously to the flexible gene pool in microorganisms inhabiting completely different econiches. Yet, others had not been unambiguously mapped to the flexible gene pool prior to this report and clearly reflect strain-specific adaption to local environmental conditions.
Significance
For many years, and because of DNA instability at low pH and recurrent failure to genetically transform acidophilic bacteria, gene transfer in acidic environments was considered negligible. Findings presented herein imply that a more or less conserved pool of actively excising MGEs occurs in the A. caldus population and point to a greater frequency of gene exchange in this econiche than previously recognized. Also, the data suggest that these elements endow the species with capacities to withstand the diverse abiotic and biotic stresses of natural environments, in particular those associated with its extreme econiche.
Introduction
The genus Acidithiobacillus consists of a group of obligatory acidophilic, Gram-negative, rod shaped bacteria that derive energy from the oxidation of reduced sulfur compounds to support autotrophic growth [1]. Acidithiobacillus species are involved in the bioleaching of metal sulfides, the desulfurization of coal and natural gas and the decontamination of industrial wastes, and for all these reasons are considered a biotechnologically relevant group of bacteria [2]. Significant intrinsic diversity, judged in terms of both genetic and physiological heterogeneity, has been recognized within the Acidithiobacillus genus. Several molecular typing studies have classified available strains into lineages [3] and specific assignment of some of these linages has recently been revised [4]–[6].
Due to its ability to oxidize reduced sulfur compounds at moderately high temperatures, Acidithiobacillus caldus is the primary sulfur oxidizer in coal piles and spoils and in mineral concentrate reactors operating at temperatures above 40°C [7], [8]. Several aspects of its physiology have been studied in representative strains including sulfur oxidation [9]–[13], central carbon metabolism [14], resistance to arsenic, copper, iron and other heavy metals [15]–[20] and attachment and growth on minerals [21]. Also, a number of broad host range plasmids and a Tn21-like transposon have been characterized for the species [15], [22]–[24]. According to these and other studies strain specific properties are apparent and further support the existence of divergent strain lineages within the A. caldus species. Yet, very little is known about the underlying genomic diversity of A. caldus and its influence in niche adaptation and strain differentiation.
In the absence of publicly available metagenomic datasets that support extensive populational comparative analyses for these biotechnologically relevant bacteria, lineage specific adaptations need to be addressed using comparative genomic approaches. Here, we report the whole genome sequence comparison of the A. caldus type strain (ATCC 51756) with that of A. caldus strain SM-1 [14]. The type strain was originally isolated from a coal spoil at the Kingsbury mine in UK after enrichment culture at pH 2.8 [25] and the SM-1 strain was obtained from a reactor used in low grade gold-bearing concentrate bioleaching operating at 40–50°C and pH 1.0–1.5 in China [26].
This work presents the first contribution to the elucidation of the species pangenome and the first comprehensive study of the flexible genome of an Acidithiobacillus species. Furthermore, it sheds light into the repertoire of mobile genetic elements (MGEs) found in this econiche. Using molecular approaches, we show the occurrence, distribution and excision capacity of these MGEs in the type strain and other cultivated strains of the species from diverse geographical origins, demonstrating that isolates from different parts of the world are consistently variable at the whole genome level. In addition, we predict the potential function of some of the accessory gene products carried by these MGEs and provide inferences on the ecological significance of these functions in strain linages adaptation.
Results and Discussion
Global Strain Comparison
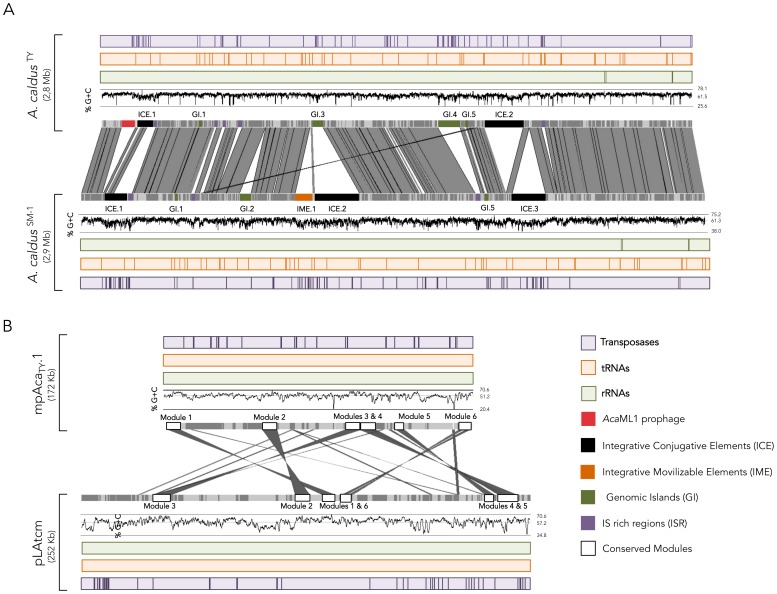
A. caldus ATCC 51756 (A. caldus TY) and A. caldus SM-1 (A. caldus SM-1) are 100% identical at the 16S rRNA gene level and the average nucleotide identity (ANI) of their genomes is 97.9%. The genomes of both strains are compartmentalized in one chromosome, one megaplasmid and a number of smaller plasmids differing in G+C content from the mean value of either chromosome ( Table 1 ). The A. caldus TY chromosome is syntenic with that of A. caldus SM-1, with similar overall architecture and global structural properties ( Figure 1a ). However, the whole genome comparison shows evidence of several integration events with elements of exogenous origin ( Table 2 ). Pairwise BLAST comparisons indicated that the type strain has 633 unique genes while A. caldus SM-1 has 872 genes with no homologs in A.caldus TY. The majority of the strain-specific genes fall within the chromosomal foreign regions and the additional genome compartments (Table S1). One of the foreign elements identified in the type strain's genome is a 72 Kb inducible temperate bacteriophage integrated within srrA tmRNA encoding gene whose genomic features have recently been described [27]. The remainder of the foreign elements are described in the sections below.
Table 1. Replicons in ATCC 51756 and SM-1 A. caldus strains.
| A. caldus ATCC 51756 | A. caldus SM-1 | ||||||||
| General Features | Chromosome | mpAcaTY1 | pAcaTY1 | pAcaTY2 | Chromosome | pLAtcm | pLAtc3 | pLAtc2 | pLAtc1 |
| Accession number | ACVD00000000 | ACVD00000000 | ACVD00000000 | ACVD00000000 | NC_015850 | NC_015851 | NC_015852 | NC_015853 | NC_015854 |
| Size (Kb) | 2,778 | 171.8 | 27.5 | 9.8 | 2,932 | 251.8 | 29.7 | 14.1 | 9.8 |
| Total CDS | 2,669 | 215 | 33 | 21 | 2,848 | 255 | 10 | 13 | 28 |
| Non redundant CDS | 2,522 | 144 | 33 | 21 | 2,635 | 216 | 10 | 13 | 28 |
| G+C content (%) | 61.7 | 54.4 | 59.3 | 50.3 | 61.0 | 57.2 | 59.8 | 57.7 | 58.8 |
| rRNA genes | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| tRNA genes | 47 | 0 | 0 | 0 | 47 | 0 | 0 | 0 | 0 |
| Prophage regions | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Integrated MGEs | 6 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 |
| Transposases | 90 | 15 | 2 | 2 | 140 | 50 | 1 | 0 | 1 |
| IS fragments | 44 | 16 | 1 | 1 | 32 | 7 | 0 | 1 | 1 |
| Mega/Plasmid Functional Modules | |||||||||
| Replication/Integration | - | repA-like, kfrA | repA | - | repA, repC | repA, res | res1, res2 | ||
| Maintenance/Partition | xerD | pasAB, parAB | toxinX, copG, xerD | xerD | copG | parA, parG | vapC, mazE, parA | ||
| Conjugation/Mobilization | trbN | traD, mobA | - | - | mobABCDE | mobA | trbJ, trbL |
Abbreviations: Chr Chromosome; Mpl Megaplasmid; P Plasmid; CDS Protein Coding Sequence; MGEs Mobile genetic elements; IS Insertion sequence. SM-1 plasmid designation is that of You et al., 2011 [14].
Figure 1. Genomic comparison of A. caldus strains ATCC 51756 and SM-1.
Similarities and differences between sequenced A. caldus strains revealed by in silico analysis of (A) the chromosome and (B) the megaplasmids. Chromosomes are represented using ACT. Features are color-coded as follows, grey, CDS or coding sequences; light purple, transposases; light orange tRNAs; light green, rRNA; red, prophage AcaML1; black, ICE or integrative conjugative element; orange, IME or integrative mobilizable element; green, GI or genomic island; purple, ISR or IS rich regions; white, conserved gene modules numbered according to Table S3a.
Table 2. Types of Integrative MGEs in A. caldus strains ATCC 51756 and SM-1.
| General features | Att sites | MGE Functional Modules | ||||||||
| MGE_ID | Type of MGE | Size (Kb) | # ORFs | G+C % | attB | attL/R | Phage genes | T4SS | Toxin-Antitoxin System | Partition system |
| AcaML1 | Prophage | 59,363 | 72 | 65.48 | srrA | DR (10, 10) | Int, Xis | - | - | |
| ICEAcaTY.1 | ICE-1* | 71,96 | 65 | 56.99 | Met-CAT | DR (77, 46) | Int | - | HigB/higA | ParAB |
| GIAcaTY.1 | GI-1¢ | 13,719 | 13 | 58.59 | Arg-CCG | DR (77,77) | Int | - | VapC/Phd MazF/MazE | - |
| GIAcaTY.3 | GI-3 | 45,079 | 35 | 58.15 | Lys-CTT | DR (76, 45) | Int(t) | - | HipAB VapC/Phd | - |
| GIAcaTY.4 | GI-4 | 104,967 | 94 | 57.00 | Ser-GGA, TspO | DR (738, 738) | Int | TraA, TraU | - | |
| GIAcaTY.5 | GI- 5& | 9,952 | 11 | 58,82 | Arg-TCT | DR (77, 45) | Int | - | - | |
| ICEAcaTY.2 | ICE-2# | 183,318 | 183 | 57.44 | Asn-GTT | DR (77, 47) | Int, Xis, cI, cII, Cro | Trb | VapC/YefM HicA/HicB | ParAB |
| ICEAcaSM.1 | ICE-1* | 104,341 | 98 | 57.02 | Met-CAT | DR (77,46) | Int | Trb | MazF/MazE | ParAB |
| GIAcaSM.1 | GI-1¢ | 13,644 | 15 | 58.53 | Arg-CCG | DR (77,77) | Int | - | VapC/Phd ORF/MazE | - |
| GIAcaSM.2 | GI- 2 | 50,579 | 45 | 56.55 | Ser-GCT | DR (77, 46) | Int | - | HipAB (2) | - |
| IMEAcaSM.1 | IME-1 | 80,923 | 58 | 56.75 | Ser-CGA | DR (89, 18) | Int | VirD2, VirD4, TraY | - | |
| ICEAcaSM.2 | ICE- 2# | 208,271 | 202 | 58.47 | Lys-CTT | DR (76,50) | Int, Xis, cI, cII, Cro | Trb | HicA/HicB | ParAB |
| GIAcaSM.5 | GI- 5& | 12,358 | 12 | 58.42 | Arg-TCT | DR (77,45) | Int | - | - | |
| ICEAcaSM.3 | ICE-3 | 157,371 | 154 | 56.86 | Arg-ACG | DR (77,47) | Int | Trb | PRTRC |
ICE_Type 1 elements are >98% similar in about 56 Kb of their sequences;
ICE_Type 2 elements are >90% similar in about 100 Kb of their sequences,
GI_Type 1 elements are almost identical,
GI_Type 5 elements are almost identical.
Abbreviations: ICE, Integrative Conjugative Element; GI, Genomic Island; IME, Integrative Movilizable Element; Int, Integrase; Int(t), Truncated Integrase; Xis, Excisionase; cI, CI-family phage regulator, cII, CII-family phage regulator; Cro, CRO-family phage regulator; T4SS, Type IV secretion system.
Origin and Function of Horizontally Transferred Genes in Each Strain
Based on sequence similarity analysis to data in current databases, nearly one third of predicted foreign genes in each strain appears to have originated within the gammaproteobacteria, mostly within the Acidithiobacilli (Table S2). Putative horizontally transferred genes with possible origins among members of the betaproteobacteria (49 in strain A. caldus TY and 47 in strain A. caldus SM-1) and the alphaproteobacteria (26 and 18 in each strain respectively) were also identified.
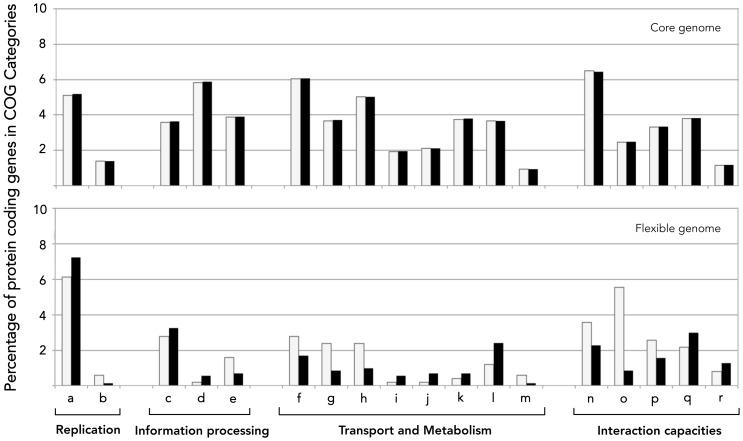
Excluding genes with no predicted functional assignment, which were greatly enriched in A. caldus TY and A. caldus SM-1 flexible gene pools, COG gene categories represented in both flexible genomes were enriched in DNA metabolism, cell wall biogenesis, motility, secretion and defense mechanisms ( Figure 2 ). In turn, functions related to energy metabolism and inorganic ion transport were differentially represented in the flexible portion of these two genomes ( Figure 2 ). Specific functions associated with the flexible gene pool of each strain are described in greater detail bellow.
Figure 2. Functional categorization of A. caldus strain-specific genes.
Bars represent the percentage of gene functions falling into the COG categories indicated. (A) Core genome, (B) Flexible genome of A. caldus ATCC 51756 (light grey) and A. caldus SM-1 (black). Genes with unknown functions were excluded from both graphs. These represent 35% of the core genome of the species and 64–71% of the flexible genome of A. caldus type strain and A. caldus SM-1, respectively. (a) Replication, recombination and repair; (b) Cell cycle control, cell division, chromosome partitioning; (c) Transcription; (d) Translation, ribosomal structure and biogenesis; (e) Posttranslational modification, protein turnover, chaperones; (f) Energy production and conversion; (g) Carbohydrate transport and metabolism; (h) Amino acid transport and metabolism; (i) Lipid transport and metabolism; (j) Nucleotide transport and metabolism; (k) Coenzyme transport and metabolism; (l) Inorganic ion transport and metabolism; (m) Secondary metabolites biosynthesis, transport and catabolism; (n) Cell wall/membrane/envelope biogenesis; (o) Cell motility; (p) Signal transduction mechanisms; (q) Intracellular trafficking, secretion, and vesicular transport; (r) Defense mechanisms.
Plasmids
The A. caldusTY plasmid content is distinct form that of A. caldus SM-1 both in terms of the number of elements and their gene content ( Table 1 ). Unlike all other Acidithiobacillus species sequenced so far, both A. caldus strains carry a large megaplasmid (>150 Mb). This additional genome compartment has a common backbone made up of 67 highly conserved protein coding genes (97% similar at the predicted amino acid level on average) organized in 6 discrete modules ( Figure 1b ). Within the backbones are genes encoding proteins possibly required for megaplasmid partitioning and replication (ParAB, XerD, GyrAB, ssBP, ssDNA exonuclease, DNA helicase) and several others of unknown function (Table S3a). Between 22 and 36% of the megaplasmid-encoded genes have well conserved orthologs in the A. caldus chromosome, including nitrogen assimilation and regulation functions, a Kdp potassium transporting ATPase and several transposases (Table S3a). This suggests these were recently mobilized from the megaplasmid to the chromosome or vice versa. The larger A. caldus SM-1 megaplasmid also carries accessory genes predicted to be involved in iron uptake, heavy metal tolerance, nucleotide metabolism and several pseudogenes.
Typical plasmid replication, maintenance and/or mobilization gene modules were identified on the smaller plasmids ( Table 1 , Table S3b). Plasmid pAcaTY.1 resembles pLAtc2 and pLAtc3 and previously characterized members of the pTcM1 plasmid family [22]. These four plasmids share an 11 Kb region containing genes for replication, partitioning, a Pin-like invertase and an IncQ-like mobilization relaxase of the MobA-type, as well as small highly conserved proteins of unknown function. Plasmid pAcaTY.2 and pLAtc1 are different from all known Acidithiobacilli plasmids and from each other. Yet, it is likely that pAcaTY.2 corresponds to pTK1 plasmid reported in the original description of the type strain [28].
Transposable Elements
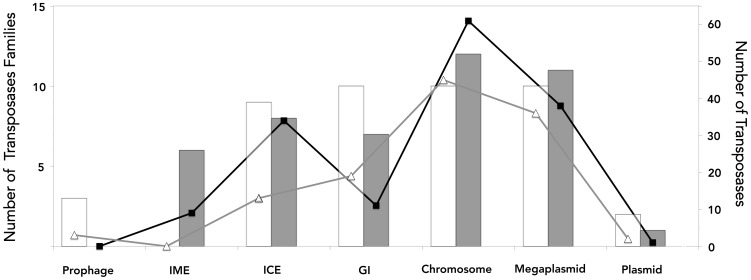
Prediction and classification of transposases using TnpPred [29] indicated that nearly 5% of the predicted genes of both strains encode transposases belonging to 20 different IS families (Table S4a). Although the diversity of IS types is similar between strains, strain SM-1 has more transposases (total 154) than the type strain (total 97) ( Figure 3 ). Four additional IS families were found in the current analysis of strain SM-1 with respect to its genome annotation [14]. The IS family distribution and relative abundance varied between replicons in both strains ( Figure 3 ). The most abundant IS families in the chromosome and the megaplasmid were IS5 and IS256 for the type strain and ISL3, IS5 and IS21 in A. caldus SM-1.
Figure 3. Diversity and abundance of IS families in A. caldus sequenced genomes.
Bars represent the number of insertion sequence families predicted in the A. caldus type strain (white) and SM-1 strain (grey) genomes associated to the core and flexible gene complement. Lines represent the total number of sequences in each category in the A. caldus type strain (triangle) and SM-1 strain (square). Abbreviations: ICE, integrative conjugative element; IME, integrative mobilizable element; GI, genomic islands.
In both strains, a large number of these IS elements were found within large integrative elements in the chromosome and in the non-shared gene blocks of megaplasmids mpAcaTY and pLAtcm. Several of these ISs were found to cluster in flexible genomic regions that do not fit the criteria of genomic islands (GI), integrative conjugative elements (ICE) or even composite transposons. These IS rich regions (ISR) also included accessory functions related mostly to O-antigen biosynthesis, sugar modification and transport (Table S4b).
Integrative Mobile Genetic Elements
Strains A. caldus TY and A. caldus SM-1 have 6 and 7 integrated MGEs respectively, 3 of which are fully or partially conserved in both strains ( Figure 1a ). The unique MGEs that differ in genomic integration site, size, G+C, and gene content, have been named according to their position in the core genome of the species. In turn, conserved elements have been similarly named. Table 2 provides general information on the integrated MGEs detected.
Briefly, all these elements are flanked by pairs of direct repeats (DR, attL and attR), generally involving tRNA genes and encode a repeat-adjacent integrase (int, COG4974), predicted to mediate unidirectional site-specific recombination between the attachment site of the element (attI) and the bacterial attachment site (attB). Five of these elements encode components of type IV secretion systems (T4SS) of IncP plasmids producing P-type pili (trb-type) for conjugative plasmid transfer that define them as ICE (Table S5). ICE-2 and ICE-3 type elements have all the essential components of a functional T4SS (VirB1 to VirB11), including the VirD2 relaxase, the VirD4 coupling protein that links the secretion system and relaxosome at the cytoplasmic membrane. One of the MGEs present in the A. caldus SM-1 genome encodes only the relaxase and the coupling protein typically present in non-conjugative mobilizable plasmids, and is thus classified as an Integrative Mobilizable Element (IME). The remaining elements flanked by direct repeats identified herein, that are larger than 5 Kb and encode an integrase gene or fragments of it and yet lack clear signatures for mobilization or conjugation, are designated as GIs.
All A. caldus ICE elements encode ParAB or ParB/ThiF partitioning protein systems, presumably involved in segregation of the excised forms during host cell division ( Table 2 ). Both these system have been demonstrated to play a role in ICE partitioning in other microbes [30], [31]. Also, all predicted ICE elements and some of the GIs harbor addictional modules consisting of a pair of genes encoding a type II toxin-antitoxin system consisting of a stable toxin (VapC, MazF, HigB, HicA, RelE) and a labile antitoxin (YefM, MazE, HigA, HicB, RelB) [32]. Resembling their roles in plasmid maintenance, toxin-antitoxin modules have been suggested to function as ICE post-segregational killing systems [33], [34], yet other roles can not be excluded.
The two largest ICE elements (ICE.2 type) are highly similar to each other over about 50% of their respective sequences (∼100,000 Kb), with at least 90% nucleotide sequence identity. In addition to a well conserved integrase (COG4974), both elements contain an excisionase (COG1257). Also, three ORFs encoding orthologs of the classical cI (COG2932), cII (pfam05269) and Cro (COG4197) phage regulators are conserved between the ICE.2 type elements. The relative levels of these regulators are known to control the lysogenic/lytic switch of phages such as Lambda [35] and ICE such as ICESt1 [36]–[38], suggesting that they may participate in the control of the integration/excision events. However, the integration site and accessory gene complement of both elements is distinct, suggesting that they are members of a family of elements that frequently occur in Acidithiobacilli genomes and confer different functional properties depending on their specific cargo and/or niche ( Table 3 ).
Table 3. Accessory genes present in A. caldus strains ATCC 51756 and SM-1 flexible genome.
| Genes/Operon | Description | Function | A. caldus TY MGEs | A. caldus SM-1 MGEs |
| Ion uptake, homeostasis and/or tolerance | ||||
| phoO/P | Phosphate selective porin | Phosphorous assimitation | pLAtc2 | |
| amtB | Ammonia uptake permease | Nitrogen assimilation | mpAcaTY1 | |
| gnlA | Glutamine synthetase I | Nitrogen assimilation | mpAcaTY1 | |
| omr | TonB-dependent outer membrane receptor | Iron homeostasis | pLAtcm | |
| mntH | Mn2+/Fe2+ transporter | Iron homeostasis | pLAtcm | |
| kdpABCE | Potassium transporting ATPase | Potassium homeostasis | ICEAcaTY.1 | pLAtcm, ICEAcaSM.1 |
| merTPAB | Mercury resistance | Ion detoxification | IMEAcaSM.1 | |
| arsADR | Arsenic resistance | Ion detoxification | pLAtc3 | |
| czcD | Heavy metals trasport, CDF family protein | Ion detoxification | pLAtcm | |
| czcCBA | Heavy metals trasport, RND family efflux pump | Ion detoxification | pLAtcm | |
| TISS, TolC | Type I secretion system | Ion detoxification | ISRAcaTY.6 | IMEAcaSM.1 |
| Energy metabolism | ||||
| cyoABCD/ctaAB | bo3-like terminal oxidase | Growth in low O2 conditions | pAcaTY1 | ICEAcaSM.1 |
| ndhDF | NADH dehydrogenase, subunits 4 & 5 | Alternative electron transport | IMEAcaSM.1 | |
| hupSL-hypFCDEAB | Group 2 Ni-Fe hydrogenase | Hydrogen metabolism | GIAcaTY.4 | |
| hyfBCEFG | Group 4 hydrogenase | Hydrogen metabolism | GIAcaTY.4 | |
| car | Beta-CA- type Carbonic anhydrase | CO2/pH homeostasis | GIAcaTY.4 | |
| Phage/MGEs resistance mechanisms | ||||
| rms | Restriction modification system | Defense against MGEs | GIAcaTY.1, ISRAcaTY.4 | GIAcaSM.1, ICEAcaSM.3 |
| CRISPR/cas | CRISPR/Cas system | Defense against MGEs | ICEAcaTY.2 | ICEAcaSM.2 |
| Substrate colonization | ||||
| fleQ, flhA-FLEN, flgEFG | Flagella biosyntehsis | Motility/Adhesion | ICEAcaTY.2 | |
| cheYZA, mcp | Chemotaxis system | Motility/Adhesion | ICEAcaTY.2 | ICEAcaSM.1 |
| pilL/NOPQRSUMV | Type IV adhesion pili | Motility/Adhesion | ICEAcaTY.2 | ICEAcaSM.2 |
| wba/wcb | O-polysaccharide biosynthesis | LPS biosynthesis | GIAcaTY.3, ISRAcaTY.1 | |
| rfb | Unbranched O-polysaccharide biosynthesis | LPS biosynthesis | GIAcaSM.2 | |
| gtf | Gycosyltransferases | LPS/EPS biosynthesis | ISRAcaTY.2. ISRAcaTY.3 | ISRAcaSM.2 |
Abbreviations: ps = pseudogene; CDF = cation diffusion facilitator; RND = resistance-nodulation-cell division.
Occurrence of Integrated MGEs in A. caldus Strains
In order to gain insight into the distribution of the identified ICEs, IME and GIs, their occurrence was analyzed in five strains of A. caldus from diverse geographical origins. The presence of the predicted elements was assessed by PCR of the borders (attL/attR) and/or the integration site in the genome (attB) and the nature of the recovered amplicons was confirmed by sequencing ( Table 4 ).
Table 4. Concurrence and MGEs distribution of A. caldus strains.
| Strain | AcaML1 | ICEAcaTY.1 | ICEAcaTY.2 | ICEAcaSM.2 | ICEAcaSM.3 | IMEAcaSM.1 | GIAcaTY.1 | GIAcaSM.2 | GIAcaTY.3 | GIAcaTY.4 | GIAcaTY.5 |
| TY | + | + | + | − | −, s | −, s | + | −, s | + | + | + |
| BC13 | + | + | + | −, s | + | −, s | + | −, s | + | + | + |
| F | + | + | + | + | −, s | + | + | −, s | + | + | + |
| CSH12 | + | + | + | −, s | + | −, s | + | −, s | + | −, s | + |
| MNG | − | −, s | −, s | + | + | −, s | + | −, s | −, s | −, s | −, s |
| #6 | − | −, s | −, s | + | + | + | + | −, s | −, s | −, s | −, s |
(+) attR and attL present; (−) attR and attL absent; (s) attB present.
Patterns of occurrence of the integrated MGEs do not follow geographic distribution of the strains, but rather their relatedness, as assessed by a resolutive phylogenetic gene marker era (unpublished data). Elements that are present in the type strain genome also tend to occur in its closely related strain BC13, and also often in the Australian strain CSH-12 and strain F from South Africa. This is also true in the case of the prophage. In contrast, elements that occur in the Chinese strain SM-1 are recurrently present in strain #6 from South Africa and absent from the rest of the genomes analyzed. The IME occurs more stochastically, being found in the SM-1 strain and two South African strains (F and # 6). MGEs distribution profiles could reflect patterns of occurrence of cognate receptors for temperate bacteriophages such as AcaML1, or mating pair formation determinants in each linage. Also, occurrence could reflect phage related selection patterns. One exception seems to be the genomic island GI-1 which is invariably present in all the strains evaluated. In this case, the most conservative explanation is that its integration probably occurred in a common ancestor and that after gene loss and selective decay, it was subsequently fixed.
Excision/Integration Capacities of Acidithiobacillus ICE/IMEs
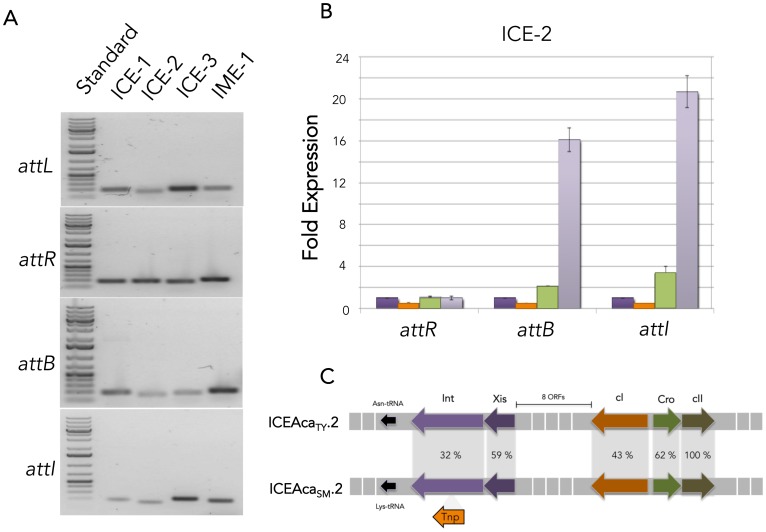
To evaluate the capacity of the detected ICEs and IMEs to excise out of their host chromosomes, end point PCR analysis of genomic DNA obtained from stationary phase A. caldus strains was performed. These analyses revealed the co-existence of the integrated (attL, attR) and excised (attB, attI) forms of all elements in the strains tested ( Figure 4a ). Sequencing of the resulting bands confirmed the nature of the amplicons as that expected from the excision of the cognate elements from their target tRNAs. These results indicate that the elements are capable of excision and formation of a circular intermediate suitable for further transfer to recipient hosts.
Figure 4. Excision capacities of Acidithiobacillus ICE and IME.
A Agarose gel electrophoresis of end point PCR products generated with primers for attL, attR, attB and attI showing excisive recombination of ICE-1, ICE-2, ICE-3 and IME-1 in cognate A. caldus strains ATCC51756, F, CSH12 and F respectively (A). DNA damaging agents effect on ICE-2 excision frequency (B). Relative fold induction of the integrated form (attR) and the excised form (attB, attI) of ICE-2 in the type strain assessed by real time PCR upon treatment with Mit C or mitomycin C (1 µg/ml, 16 hs), light purple; UV-C radiation (200 J/m2, 5 min), light green; and Fe3+ or ferric iron (250 mM, 45 min), orange, with respect to control conditions (no DNA damaging treatment), dark purple. Abbreviations: ICE, integrative conjugative element; IME, integrative mobilizable element.
Quantitative PCR analysis of the relative ratios of the integrated (attR) and excised (attB) forms of the ICE-2 type elements (which carry a predicted phage-type regulatory gene module) under control conditions and upon infliction of DNA damage revealed changes in the observed proportions of attB/attI ( Figure 4b ). This suggests that a phage-type of mechanism underlies ICEAcaTY.2 and ICEAcaSM.2 site-specific excision. cI orthologs, like the one encoded in ICE-2 type elements have been found in other ICE, including ICESt1-3 from Streptococcus thermophilus [36], ICEBs1 from Bacillus subtilis [37], SXT from V. cholerae [38] and ICEAfe1 from A. ferrooxidans [33]. Furthermore, conditions that elicit the SOS (DNA damage) response have been found to derepress the excision of these ICE [33], [36]–[38] and to promote the transfer of ICEBs1 and SXT-related elements from enterobacteria [36]–[39]. Such regulation is reminiscent of DNA damage induced derepression of the site-specific excision of numerous prophages during lytic cycle [40] and suggests that this mechanism also controls the dynamics of integration/excision of ICE-2 type elements in A. caldus and possibly also their transfer to a healthy host.
Functional Clues Derived from A. caldus Flexible Genome Analysis
Although each of the A. caldus sequenced strains carries a distinct repertoire of MGEs, and despite differences in their origin and culturing histories, accessory genes present in their flexible genomes shared common themes. These genes partitioned in the five mayor functional categories described in detail below. Several of these functional categories have previously been shown to be over-represented in GIs and metagenomic GIs in other model systems and econiches and are related to surface exposed proteins or cellular structures known to be bacteriophage recognition targets [41]. As such many of these functions are probably relevant to population genomics control mediated by bacteriophages prevalent in acidic econiches [42]. In turn, several others reflect differences in niche explotation. MGE-specific distribution of the accessory genes is provided in Table 3 and gene functions partition between strains is schematized in Figure 5 .
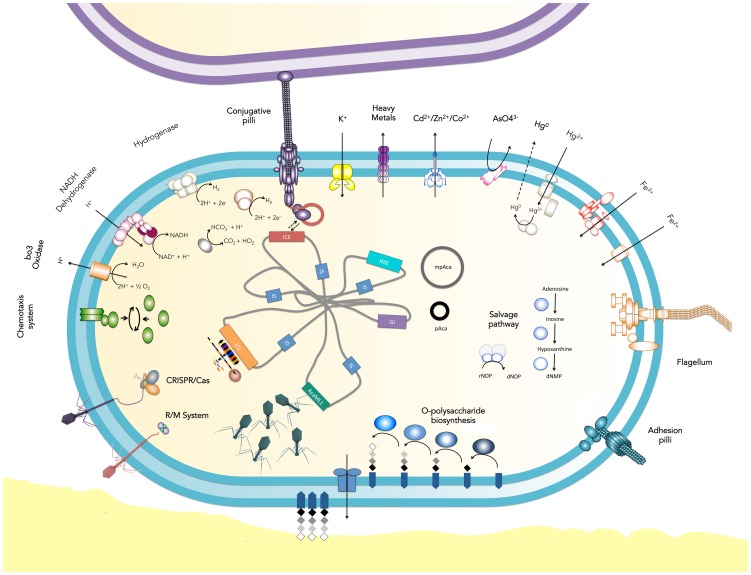
Figure 5. Functional repertoire of the A. caldus mobilome.
Schematic representation of the predicted shared (full) and exclusive (empty) accessory gene products present in the A. caldus TY and A. caldus SM-1 mobilome and the diversity of MGEs identified in this study.
1. Ion uptake, homeostasis and/or tolerance
Several uptake systems for ions such as ammonium, iron and potassium were found to form part of the A. caldus flexible gene pool. Ammonia is a growth-limiting factor in biomining environments. Several copies of the AmtB transporter encoding gene are frequently found in the genomes of bioleaching acidophiles [43], [44] and other free-living bacteria [45]. Although nitrogen fixation genes are commonly associated with MGEs [46], ammonia transport functions are typically chromosomal. The pLAtcm megaplasmid encoded amtB may contribute to increased ammonia uptake in acidic conditions.
Conversely, iron loads in acidic econiches can be 1018 times the concentration found in neutral pH environments and thus acidophiles like A. caldus have to cope with issues related to potential iron toxicity [18]. Encoding additional transporters for ferrous/ferric iron uptake, which tightly regulate metal entrance, may contribute to iron homeostasis. Also, TonB-dependent outer membrane receptors may serve as receptors for colicins and bacteriophages [47], [48] afecting survival and competition in the environment and ultimately genomic stability.
In addition to the potassium transporting ATPase encoding operon kdpABCF present in the A. caldus core genome, three copies also occur in the flexible genome within ICE.1 type elements and the pLAtcm megaplasmid. The KdpABCF complex is involved in high affinity ATP driven potassium uptake against steep concentration gradients and plays several physiological roles [49]. Increased operon copy number in the Acidithiobacilli may have a primary role in pH homeostasis where an inside positive membrane potential is suggested to be generated by potassium ions [50].
The A. caldus SM-1 flexible gene pool was enriched in metal detoxification systems for mercury, arsenic and other less well defined heavy metals. This is probably related to the fact that the SM-1 strain was isolated from gold bearing mineral concentrates [26], which often contain arsenopyrite and other heavy metal entrapped minerals. A truncated version of the mercuric reductase encoding operon merTPAB (lacking the regulatory genes merR and merD) is present in the IMEAcaSM.1. As shown in other bacteria, unregulated expression of this operon could confer A. caldus with increased mercurial detoxification capacities [51]. It has been well established in other microorganisms that horizontal dissemination of genes responsible for resistance to inorganic mercury can occur via MGEs such as transposons, plasmids and GIs in neutrophiles [52]–[54] but no evidence has been reported on their occurrence on IME or ICE type elements nor other MGEs in acidophiles. Conversely, arsenic resistance has been previously described in A. caldus and specifically linked to transposon TnAtcArs [15] and plasmid pLAtc3 [14]. In addition, two type I secretion systems (TolC-type) for efflux of antibiotics, dyes or heavy metals, a CzcD cation diffusion facilitator family member and a CzcCBA resistance-nodulation-cell division family efflux pump of Cd2+, Zn2+ and Co2+, also form part of the A. caldus flexible genome ( Table 3 ). Similar metal detoxification pumps have also been found to form part of the flexible genomes of other free-living bacteria [54], [55]. These findings are in agreement with previous evidence generated for closely related acidophiles like A. ferrooxidans, showing that changes in genome structure and gene copy number develop in response to environmental effects and/or toxic metals exposure [56], [57]. In the case of biomining acidophiles, transfer and spread of these functions could contribute to rapid adaptation and survival of the species in highly acidic and metal rich environments containing millimolar quantities of metals [58].
2. Energy metabolism
The flexible gene pool of both strains also contained components of several alternative electron-transporting pathways ( Table 3 ). Protein subunits of a terminal oxidase complex encoded in the type strain plasmid pAcaTY1 are 99% identical to those present in the SM-1 strain ICEAcaSM.1 and 51–63% identical to the bo3 quinol oxidase encoded within the A. caldus core genome. In addition to the oxidase subunits, the flexible genome gene clusters also encode two heme maturation proteins which convert heme B (protoheme IX) to heme O (CtaA) and heme O to heme A (CtaB) [59], suggesting that this could be a mixed oxidase of the ba3 type. A ba3 type oxidase has been identified in the extremely thermophilic bacterium Thermus thermophilus HB8 [60] and shown to be expressed under limited O2 supply [61]. Since oxygen solubility in water diminishes with temperature (∼25% less at 40°C than at 25°C), this alternative oxidase may have been selected for by A. caldus more than once to provide a growth advantage at its higher optimum growth temperature. Alternatively, heme A could replace heme O in the active site of the oxidase depending on growth conditions, as documented in other microorganisms [62], providing additional metabolic flexibility.
The ndhFD gene pair encoding essential membrane components of respiratory complex I (NDH-1) occurred in three locations of the A. caldus core genome as well as the SM-1 flexible genome. Multiple and divergent copies of ndhD and ndhF have been reported in other genomes and are suggested to make up many distinct NDH-1 complexes that may feed electrons into the respiratory chain or other electron transport systems conferring further metabolic flexibility [63].
Accessory genes in this functional category also include a group 2 cytoplasmic uptake NiFe-hydrogenase (hup operon) and a group 4 hydrogenase (hyf operon). Orthodox respiratory hupSL-encoded hydrogenase activity is presumed to be responsible for recycling endogenous hydrogen (H2) produced during nitrogen (N2) fixation in aerobic diazotrophes [64]. Co-occurrence of the hup operon with nifA and two orthologs of the regulator PII, as well as sequence similarity, suggests that the gene cluster found in GIAcaTY.4 could have been acquired from a diazotroph. However, lack of N2 fixation genes in A. caldus type strain [43] suggests that the presumed role of the Ni-Fe hydrogenase is that of a respiratory hydrogenase adapted for use of exogenous H2 [65]. The role of the distinct HyfBCEFG hydrogenase present in GIAcaTY.4 is less obvious. In Escherichia coli, the group-4 hydrogenase (hydrogenase-4), is coupled to the fermentative formate dehydrogenase, oxidizing formate to CO2 and reducing 2H+ to H2 under fermentative conditions [66]. This is very likely the case of the hydrogenase-4 HyfBCEFG present in the core genome of both A. caldus strains, which co-occur with genes for carbon fixation and presumably interact with the formate dehydrogenase encoded a few genes upstream. However, the flexible genome hyf gene cluster is found immediately adjacent to a beta-CA-type carbonic anhydrase encoding gene, the product of which catalyzes the reversible hydration of CO2 to bicarbonate (CO2+H2O = HCO3 −+H+) [67]. This suggests that this H2-evolving hydrogenase could reduce H+ -ions to yield H2 (gas) and contribute to pH and CO2 homeostasis. Occurrence of alternative terminal oxidases, NDH1 subunits and hydrogenases on MGEs has previously been reported in other microorganisms [54], [68], [69], further indicating that certain adaptive strategies are shared between acidic and neutral environments.
3. Phage/MGEs resistance mechanisms
The two A. caldus strains examined carry CRISPR/Cas systems and restriction modification complexes in their flexible genomes. Both features help to build a defense against invading mobile genetic elements and potentially undesirable genetic cargo.
CRISPR loci present in A. caldus TY and A. caldus SM-1 ICE.2 type elements have a similar organization, consisting of two repeat–arrays of variable size and spacer content and five genes encoding highly similar Cas-like proteins. In spite of the inter-strain conservation, these proteins are highly divergent from other known and characterized CRISPR associated proteins, including those from its close relative A. ferrooxidans [33]. The total number of repeats (23 and 27 respectively) and the diversification of spacer sequences suggests that these loci may be active. None of the spacer sequences is shared between the two loci, yet two spacers from the ICEAcaTY.2 CRISPR and two others from the ICEAcaSM.2 CRISPR share sequence similarity to A. caldus pTcM-1 family plasmids and one spacer of each locus shares sequence similarity to Acidiphilium spp. plasmids with whom the Acidithiobacilli share the niche.
Diverse lines of evidence suggest that CRISPR/Cas cassettes are actively propagated between genomes by horizontal gene transfer. Anomalous nucleotide frequencies of CRISPR/Cas systems compared to cognate chromosomes, conservation of certain cas gene arrays in phylogenetically distant genomes and the occurrence of diversified CRISPR/Cas systems in closely related strains or species, are evidence that these cassettes disseminate via horizontal gene transfer as discrete units [70], [71]. Plasmid [72] and GI mediated mobilization [54], [73]–[75] and chromosomal conjugation [75] have been proposed to underlie the dissemination of these cassettes. However, the extent to which these mechanisms operate and actually contribute to intra-species and inter-species CRISPR/Cas propagation remains obscure. The presence of CRISPR/Cas systems within ICE, as described herein for A. caldus type strain and strain SM-1 and shown recently also in the case of A. ferrooxidans [33], suggests that additional pathways for propagation of chromosomally encoded CRISPR/Cas cassettes exists.
Accessory genes in the A. caldus flexible genome (ICEAca SM.3 and shared GI.1) also include a type II restriction modification (RM) system, most likely selected for as protection against invading foreign DNA after sequence-specific methylation of defined target sequences of the host chromosome [76]. Type II RM systems occur frequently on mobile genetic elements [77]. The presence and functionality of RM systems such as these may have implications in phage infection and restriction, DNA uptake and natural competence.
4. Substrate colonization
Several cellular functions that play relevant roles in chemo-sensing, surface attachment, autoaggregation and biofilm formation form part of the A. caldus flexible genome ( Table 3 ) and many other free-living and pathogenic bacteria [78].
In addition to gene clusters in the conserved core genome encoding flagella and chemotaxis genes [14], [43], the A. caldus type strain flexible genome carries a 38-gene cluster encoding all necessary genes for basal body, hook and flagella filament biosynthesis [79] and all essential chemotaxis genes that mediate signal specific induced motility [80]. The major structural component of the flagellum, the FlaB flagelin, is less than 70% similar to the core encoded flagelin, suggesting that variable extracellular structures are determined by the presence of this ICE. This probably influences surface attachment [81] or phage recognition [82]. In addition, a 12 ORFs locus encoding type IV adhesion pili, promoting bacterial attachment to, and colonization of a wide variety of surfaces [83] is exclusively present within the ICE.2 type elements.
Three different loci encoding regulation, synthesis and export functions related to the biosynthesis of O-polysaccharide, the outermost domain of lipopolysaccharide (LPS), also form part of the A. caldus flexible genome (GIAcaSM.2, GIAcaTY.3, ISRAcaTY.1). In most Gram-negative bacteria, LPS is one of the major constituents of the outer leaflet of the outer membrane. Several bacteria specifically use LPS to adhere to animal and plant host tissues and various abiotic surfaces and recent reports indicate that it has a role in autoaggregation and biofilm formation [84]–[86]. It also serves as a bacteriophage receptor in certain bacteria [87].
Occurrence of all these traits, contributing to different developmental stages of the formation of biofilms, enhance bacterial survival in the environment and provide resistance against a variety of abiotic and biotic stress factors (e.g. bacteriophages). Also, close proximity of the bacterial cells within biofilms has been shown to increase plasmid dispersal by conjugation, as well as DNA release and transformation [88], [89]. No information is yet available on the effect of biofilms in ICEs and IMEs dispersal but it can be expected to be similar to that of conjugative and mobilizable plasmids. In addition, these capacities may improve substrate utilization and thereafter growth and proliferation of A. caldus in biomining environments.
5. Nucleotide metabolism
The A. caldus SM-1 flexible genome also encodes several functions related to nucleotide salvage to recover bases and nucleosides that result from RNA and DNA degradation.
Specifically, pLAtcm megaplasmid encodes a guanine/adenosine deaminase required for irreversible deamination of adenosine to the nucleoside inosine, a purine nucleoside phosphorylase for deribosylation of inosine to hypoxanthine and a phosphorybosiltransferase which adds activated ribose-5-phosphate to bases creating nucleotide monophosphates. Accessory genes present in ICEAcaSM.3 also encode the beta subunit of the class Ia aerobic ribonucleotide reductase (nrdB). The latter is the small subunit of a functionally critical enzyme complex in nucleotide metabolism that catalyzes the rate limiting step in the synthesis of deoxyribonucleotides used for DNA replication and repair [90]. No reports on the occurrence of ribonucleotide reductases on MGEs other than bacteriophages could be found in the literature [91]. It is possible that an ICE-encoded NrdB subunit (which is well conserved with respect to the chromosomal copy of nrdB form A. caldus type strain) could activate the chromosomally encoded catalytic subunit NrdA and thereafter supply dNTPs during ICE replication or DNA damage.
Conclusions
Comparative genomic analysis of A. caldus sequenced strains revealed that they differ in about 20% of their respective genomes, which is close to that observed for highly flexible species like E. coli [92]. Both strains vary not only in the number and types of independent replicons that they host, but also in the MGE content and accessory gene distribution of their flexible genomes.
In this work we uncovered a considerable diverse repertoire of MGEs, including at least three families of ICE elements, a family of IME elements and several GIs, complementing previous efforts to characterize plasmids [28], insertion sequences [93], and bacteriophages [22] from biomining Acidithiobacilli. Most ICE and the IME are widespread between strains of the species obtained from diverse geographical origins, indicating that conserved or variant versions of them still occur in the A. caldus population as active excising elements that may be transferred horizontally when needed. In addition, A. caldus SM-1 and type strain are the first Acidithiobailli to be characterized carrying a megaplasmid.
Deep bioinformatic analyses of strain-specific segments allowed identification and functional categorization of the accessory gene pool. Several of the features found (e.g. restriction modification systems, metal resistance systems, hydrogenases, etc.) have been linked to the flexible gene pool in other microorganisms inhabiting completely different econiches, yet facing similar challenges (e.g. bacteriophage infection, limiting nutrients or energy supply) [54], [94], [95]. Yet others, (e.g. mercurial detoxification system, CRISPR/Cas systems) had not been unambiguously mapped to integrative MGEs prior to this report.
Despite differences in the isolation and culturing histories of the two strains under comparison, the nature of several relevant physiological features identified along their flexible genome segments were highly similar (e.g. defense mechanism against MGEs, chemotaxis gene cluster, adhesion pilli gene cluster). However, genes that were common to both flexible genomes were distributed in different types of elements in each strain, indicating that certain features have been selected for more than once in the evolution of the species. Conversely, other functions in the flexible genome clearly reflect the local environmental conditions in which each strain thrives (e.g. several heavy metal homeostasis strategies in strain SM-1).
Collectively, the differences we report here suggest that significant, yet uncharacterized, diversity exists within this particular group of acidophiles and that gain and/or loss of mobile genetic elements and their cargo also plays an important role in strain differentiation and adaptation to acidic econiches.
Materials and Methods
Nucleotide sequences
The Acidithiobacillus caldus ATCC 51756 draft genome sequence (ACVD00000000, [43]) was assembled using the Phred/Phrap/Consed software package [96]. A total of 41,813 high-quality reads with an average length of 745 bp were assembled into 57 contigs with at least twofold coverage. Remaining gaps were closed by further sequencing gap-spanning PCR products. Final assembly, consisting of one chromosome, one megaplasmid and two smaller plasmids, was deposited at GenBank under the accession number PRJNA36585. Sequences for A. caldus SM-1 (NC_015850-54) were obtained directly from the GenBank database.
Sequence annotation
Gene prediction was performed using CRITICA [97] and Glimmer [98]. Non-coding genes were predicted using tRNAscan-SE [99]. Predicted proteins were functionally annotated using protein alignments to NCBI nr [100] and categorized against the COG database [101] and the GO database [102]. Coding sequences were further characterized using the suite of protein comparison and classification programs available in InterproScan [103] and the Prosite database [104]. Insertion sequence family assignments were made using TnpPred [29]. CRISPRs were predicted using CRISPI [105], CRISPR Finder [106] and CRT1 CRISPR Recognition Tool version 1.1 [107]. Comparative genomic tools such as Microbesonline [108] and RAST [109] were also employed.
Sequence comparisons
Sequence comparisons were conducted using MUMmer [110], MAUVE [111] and the Artemis Comparison Tool with a minimum score cut-off of 121 and a minimum percentage identity cut-off of 100% [112]. To retrieve paralogs and/or redundant proteins, the predicted proteome of each strain was analyzed against itself using BLASTp and a 95% identity (E<1e-10) as cutoff. To retrieve orthologs predicted proteomes of each strain were cross-compared using bidirectional BLASTp, a reciprocal 95% identity (E<1e-10) cutoff and in-house bioperl scripts. Chromosomal maps were made using the CGview server [113].
MGE identification
Mobile genetic elements present in the genome of A. caldus genomes were identified using an in-house designed pipeline. First, all integrases in the genome of each strain were mapped using BLASTp and a collection of query sequences retrieved from the NCBI database. Second, regions adjacent to each predicted integrase were recovered and their occurrence elsewhere in the genome (as direct repeats) assessed using BLASTn. Regions flanked by the identified repeats were recovered and analyzed for their GC content and skew [114] and occurrence on the other sequenced strain of the species using sequence comparison tools. Finally, gene content of the predicted elements was analyzed to search for hallmarks of horizontal gene transfer.
Bacterial strains and culture conditions
The bacterial strains used in this study are listed in Table S6. A. caldus strains were grown in mineral salts medium (MSM) with trace elements [8] and 5 g/L S0 or 5 mM tetrathionate as energy substrate. Cultures were grown at 40°C and pH 2.5 in aerobic conditions (150 rpm). Stock solutions of tetrathionate were filter-sterilized and added to the autoclaved (121°C for 15 min) MSM, whereas ethanol-sterilized powdered sulfur was added to MSM prior to autoclaving at 105°C for 30 min.
General DNA techniques
A. caldus stationary phase cultures to be used for nucleic acid purification, were centrifuged at 6,000 g to remove solid sulfur precipitates prior to cell harvest. The cell pellet was resuspended in 9K salt solution for further washing. Washed cells were collected by centrifugation at 10,000 g for 15 min. DNA isolation and routine manipulations were carried out following standard protocols [115]. DNA quantifications were performed in the NanoDrop 2000 Spectrophotometer (Thermo Scientific).
End point PCR
Oligonucleotide primers used in this study are listed in Table S7. Polymerase chain reaction (PCR) products were amplified with DNA polymerase Dreamtaq (ThermoScientific) and were purified from agarose gels with the Mini Elute Gel Extraction Kit (Qiagen). Each PCR reaction contained 10 ng of template DNA, 0.5 µM of required primers, and 0.2 mM of each deoxyribonucleotide in a volume of 25 µl of 1× PCR buffer containing 1.5 mM MgCl2. PCR conditions were as follows: initial denaturing step at 95°C for 2 min followed by 28–30 amplification cycles (denaturation at 95°C for 20 sec, annealing at the appropriate temperature depending on the specific primers pairs for 20 sec and elongation at 72°C) and a final elongation step at 72°C for 10 min. DNA sequencing was carried out by Macrogen Inc. (Seoul, Korea).
Real-time PCR
Real-time PCR reactions were performed in the RotorGene Q PCR System (Qiagen) using the KAPA SYBR FAST qPCR Kit (Kapa Biosystems). The 20 µl PCR reactions contained 2 µl of a 1∶100 diluted cDNA sample; 200 nM of each primer and 1× KAPA Master Mix. The cycling protocol was as follows: initial denaturation for 10 min at 95°C followed by 40 cycles of 3 s at 95°C, 20 s at 60°C; 1 s at 72°C. Fluorescence was measured after the extension phase at 72°C. The PCR products were subjected to a melting curve analysis, that commenced at 52°C and increased at 0.5°C s-1 up to 95°C, with a continuous fluorescent measurement. Specific amplification was confirmed by a single peak in the melting curve. For each experimental condition stationary phase genomic DNA was extracted from two independent cultures. The reactions for each target gene were performed in triplicate and in the same PCR run. DNA 10-fold dilutions (ranging from 10 ng to 1 pg) of corresponding PCR amplicons were used to generate a 5-point standard curve for every gene by using the Cycle Threshold (Ct) value versus the logarithm of each dilution factor. Reaction efficiency (E = (10(−1/slope))−1) for every gene was derived from the slope of the corresponding standard curves. Amplicon quantities were calculated from the standard curve by the software Rotor Gene Q Series Software 2.0.2 (Qiagen) set with default parameters. Each experiment included a no template control.
Mitomycin C and UV treatment
To produce DNA damage, 1 L mid exponential phase cells (3 days old) were concentrated 100 fold and treated with 1 µg/mL mitomycin C (CalBiochem) or exposed to UV-C (3 min at 200 J/m2, 254 nm) for 5 min and then allowed to recover for two generation times (16 h) in fresh media at 40°C and 200 rpm. Total genomic DNA was extracted as described above. The attL and attR recombination sites flanking the ICE and the chromosomal attachment site (attB) were PCR amplified using 10 ng of total genomic DNA and the primers listed in Table S7. All experiments were performed in triplicate.
Supporting Information
A. caldus strain-specifc gene lists.
(XLSX)
Taxonomic origin of A. caldus ATCC 51756 and A. caldus SM-1 exclusive genes.
(XLSX)
Megaplasmid mpAcaTY.1 and plasmids pAcaTY.1 and pÀcaTY.2 annotation and gene module information.
(XLSX)
Predicted IS families and features associated to IS-rich regions in the genome of A. caldus ATCC 51756 and A. caldus SM-1.
(XLSX)
ICE and IME Trb gene cluster synteny analysis.
(XLSX)
A. caldus strains used in this study.
(XLSX)
Oligonucleotide primers used in this study.
(XLSX)
Acknowledgments
We thank Mónica Gonzalez for her excellent technical assistance. Also, we gratefully acknowledge Shelly Dean for providing bacterial strains used in this study.
Funding Statement
This work was supported by the Fondo Nacional de Desarrollo Científico y Tecnológico Fondecyt (http://www.conicyt.cl/fondecyt/) [grant numbers 1100887, 1090451], and the Programa de Financiamiento Basal de la Comisión Nacional de Investigación Científica y Tecnológica (http://www.conicyt.cl/pia/category/lineas-del-programa/creacion-consolidacion-centros/ [grant number PFB16]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kelly DP, Wood AP (2005) Genus I. Acidithiobacillus Kelly and Wood 2000. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergey's Manual of Systematic Bacteriology, New York: Springer. 2nd edn, vol 2, part B, pp. 60–62. [Google Scholar]
- 2. Rawlings DE, Johnson DB (2007) The microbiology of biomining: development and optimization of mineral-oxidizing microbial consortia. Microbiology 153: 315–324. [DOI] [PubMed] [Google Scholar]
- 3. Ni Y-Q, He K-Y, Bao J-T, Yang Y, Wan D-S, et al. (2008) Genomic and phenotypic heterogeneity of Acidithiobacillus spp. strains isolated from diverse habitats in China. FEMS Microbiol Ecol 64: 248–259. [DOI] [PubMed] [Google Scholar]
- 4. Hallberg KB, Amouric A, Brochier-Armanet C, Bonnefoy V, Johnson DB (2010) Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 71–73: 167–70. [DOI] [PubMed] [Google Scholar]
- 5. Hallberg KB, González-Toril E, Johnson DB (2010) Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 14: 9–19. [DOI] [PubMed] [Google Scholar]
- 6. Hedrich S, Johnson DB (2013) Acidithiobacillus ferridurans, sp. nov.; an acidophilic iron-, sulfur- and hydrogen-metabolizing chemolithotrophic Gammaproteobacterium. Int J Syst Evol Microbiol In press. [DOI] [PubMed] [Google Scholar]
- 7. Hallberg KB, Lindström EB (1994) Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology 140: 3451–3456. [DOI] [PubMed] [Google Scholar]
- 8. Dopson M, Lindström EB (1999) Potential role of Thiobacillus caldus in arsenopyrite bioleaching. Appl Environ Microbiol 65: 36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen L, Ren Y, Lin J, Liu X, Pang X, et al. (2012) Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS One 7: e39470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mangold S, Valdés J, Holmes DS, Dopson M (2011) Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus . Front Microbiol 2: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rzhepishevska OI, Valdés J, Marcinkeviciene L, Gallardo CA, Meskys R, et al. (2007) Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Appl Environ Microbiol 73: 7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bugaytsova Z, Lindström EB (2004) Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus . Eur J Biochem 271: 272–280. [DOI] [PubMed] [Google Scholar]
- 13. Dopson M, Lindström EB, Hallberg KB (2002) ATP generation during reduced inorganic sulfur compound oxidation by Acidithiobacillus caldus is exclusively due to electron transport phosphorylation. Extremophiles 6: 123–129. [DOI] [PubMed] [Google Scholar]
- 14. You XY, Guo X, Zheng HJ, Zhang MJ, Liu LJ, et al. (2011) Unraveling the Acidithiobacillus caldus complete genome and its central metabolisms for carbon assimilation. J Genet Genomics 38: 243–252. [DOI] [PubMed] [Google Scholar]
- 15. Kotze AA, Tuffin IM, Deane SM, Rawlings DE (2006) Cloning and characterization of the chromosomal arsenic resistance genes from Acidithiobacillus caldus and enhanced arsenic resistance on conjugal transfer of ars genes located on transposon TnAtcArs. Microbiology 152: 3551–3560. [DOI] [PubMed] [Google Scholar]
- 16. Dopson M, Lindström EB, Hallberg KB (2001) Chromosomally encoded arsenical resistance of the moderately thermophilic acidophile Acidithiobacillus caldus . Extremophiles 5: 247–255. [DOI] [PubMed] [Google Scholar]
- 17. Xia L, Yin C, Cai L, Qiu G, Qin W, et al. (2010) Metabolic changes of Acidithiobacillus caldus under Cu(2+) stress. J Basic Microbiol 50: 591–598. [DOI] [PubMed] [Google Scholar]
- 18. Osorio H, Martínez V, Nieto PA, Holmes DS, Quatrini R (2008) Microbial iron management mechanisms in extremely acidic environments: comparative genomics evidence for diversity and versatility. BMC Microbiol 8: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aston JE, Peyton BM, Lee BD, Apel WA (2010) Effects of ferrous sulfate, inoculum history, and anionic form on lead, zinc, and copper toxicity to Acidithiobacillus caldus strain BC13. Environ Toxicol Chem 29: 2669–2675. [DOI] [PubMed] [Google Scholar]
- 20. Mangold S, Potrykus J, Björn E, Lövgren L, Dopson M (2013) Extreme zinc tolerance in acidophilic microorganisms from the bacterial and archaeal domains. Extremophiles 17: 75–85. [DOI] [PubMed] [Google Scholar]
- 21. Edwards KJ, Bond PL, Banfield JF (2000) Characteristics of attachment and growth of Thiobacillus caldus on sulphide minerals: a chemotactic response to sulphur minerals? Environ Microbiol 2: 324–232. [DOI] [PubMed] [Google Scholar]
- 22. van Zyl LJ, Deane SM, Louw LA, Rawlings DE (2008) Presence of a family of plasmids (29 to 65 kilobases) with a 26-kilobase common region in different strains of the sulfur-oxidizing bacterium Acidithiobacillus caldus . Appl Environ Microbiol 74: 4300–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rawlings DE (2005) The evolution of pTF-FC2 and pTC-F14, two related plasmids of the IncQ-family. Plasmid 53: 137–147. [DOI] [PubMed] [Google Scholar]
- 24. Tuffin IM, de Groot P, Deane SM, Rawlings DE (2005) An unusual Tn21-like transposon containing an ars operon is present in highly arsenic-resistant strains of the biomining bacterium Acidithiobacillus caldus . Microbiology 151: 3027–3039. [DOI] [PubMed] [Google Scholar]
- 25. Marsh RM, Norris PR (1983) The isolation of some thermophilic, autotrophic, iron- and sulfur-oxidizing bacteria. FEMS Microbiol Lett 17: 311–315. [Google Scholar]
- 26. Liu X, Lin J, Zhang Z, Bian J, Zhao Q, et al. (2007) Construction of conjugative gene transfer system between E. coli and moderately thermophilic, extremely acidophilic Acidithiobacillus caldus MTH-04. J Microbiol Biotechnol 17: 162–167. [PubMed] [Google Scholar]
- 27. Tapia P, Flores FM, Covarrubias PC, Acuña LG, Holmes DS, et al. (2012) Complete genome sequence of temperate bacteriophage AcaML1 from the extreme acidophile Acidithiobacillus caldus ATCC 51756. J Virol 86: 12452–12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leibniz Institute DSMZ- German Collection of Microorganisms and Cell Cultures web site. Available: https://www.dsmz.de/catalogues/details/culture/DSM-8584.html. Accessed 2013 May 10.
- 29. Riadi G, Medina-Moenne C, Holmes DS (2012) TnpPred: A web service for the robust prediction of prokaryotic transposases. Comp Funct Genomics 2012: 678761–678766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qiu X, Gurkar AU, Lory S (2006) Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa . Proc Natl Acad Sci U S A 103: 19830–19835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hickey WJ, Chen S, Zhao J (2012) The phn Island: A new genomic island encoding catabolism of polynuclear aromatic hydrocarbons. Front Microbiol 3: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerdes K, Christensen SK, Lobner-Olesen A (2005) Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol 3: 371–382. [DOI] [PubMed] [Google Scholar]
- 33. Bustamante P, Covarrubias PC, Levicán G, Katz A, Tapia P, et al. (2012) ICEAfe1, an actively excising genetic element from the biomining bacterium Acidithiobacillus ferrooxidans . J Mol Microbiol Biotechnol 22: 399–407. [DOI] [PubMed] [Google Scholar]
- 34. Wozniak RA, Waldor MK (2009) A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet 5: e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Court DL, Oppenheim AB, Adhya SL (2007) A new look at bacteriophage lambda genetic networks. J Bacteriol 189: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bellanger X, Morel C, Decaris B, Guédon G (2007) Derepression of excision of integrative and potentially conjugative elements from Streptococcus thermophilus by DNA damage response: implication of a cI-related repressor. J Bacteriol 189: 1478–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD (2005) Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A 102: 12554–12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beaber JW, Hochhut B, Waldor MK (2004) SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427: 72–74. [DOI] [PubMed] [Google Scholar]
- 39. Rodriguez-Valera F, Ussery DW (2012) Is the pan-genome also a pan-selectome? F1000 Research 1: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, et al. (2009) Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7: 828–836. [DOI] [PubMed] [Google Scholar]
- 41. McGrath BM, O'Halloran JA, Pembroke JT (2005) Pre-exposure to UV irradiation increases the transfer frequency of the IncJ conjugative transposon-like elements R391, R392, R705, R706, R997 and pMERPH and is recA-dependent. FEMS Microbiol Lett 243: 461–465. [DOI] [PubMed] [Google Scholar]
- 42. Court DL, Oppenheim AB, Adhya SL (2007) A new look at bacteriophage lambda genetic networks. J Bacteriol 189: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Valdés J, Quatrini R, Hallberg K, Dopson M, Valenzuela PDT, et al. (2009) Draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus caldus ATCC 51756 reveals metabolic versatility in the Acidithiobacillus genus. J Bacteriol 191: 5877–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Valdés J, Pedroso I, Quatrini R, Tettelin H, Eisen JA, et al. (2008) Acidithiobacillus ferrooxidans metabolism: From genome sequence to industrial applications. BMC Genomics 9: 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soupene E, Chu T, Corbin RW, Hunt DF, Kustu S (2002) Gas channels for NH(3): proteins from hyperthermophiles complement an Escherichia coli mutant. J Bacteriol 184: 3396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hsiao WW, Ung K, Aeschliman D, Bryan J, Finlay BB, et al. (2005) Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet 1: e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jakes KS, Finkelstein A (2010) The colicin Ia receptor, Cir, is also the translocator for colicin Ia. Mol Microbiol 75: 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rabsch W, Ma L, Wiley G, Najar FZ, Kaserer W, et al. (2007) FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. J Bacteriol 189: 5658–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Greie JC (2011) The KdpFABC complex from Escherichia coli: a chimeric K+ transporter merging ion pumps with ion channels. Eur J Cell Biol 90: 705–710. [DOI] [PubMed] [Google Scholar]
- 50. Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA (2009) Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol 55: 1–79. [DOI] [PubMed] [Google Scholar]
- 51. Horn JM, Brunke M, Deckwer WD, Timmis KN (1994) Pseudomonas putida strains which constitutively overexpress mercury resistance for biodetoxification of organomercurial pollutants. Appl Environ Microbiol 60: 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bogdanova ES, Minakhin L, Bass I, Volodin A, Hobman J, et al. (2001) Class II broad-spectrum mercury resistance transposons in Gram-positive bacteria from natural environments. Res Microbiol 152: 503–514. [DOI] [PubMed] [Google Scholar]
- 53. Osborn AM, Bruce KD, Strike P, Ritchie DA (1997) Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev 19: 239–262. [DOI] [PubMed] [Google Scholar]
- 54. Ivars-Martinez E, Martin-Cuadrado AB, D'Auria G, Mira A, Ferriera S, et al. (2008) Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J 2: 1194–1212. [DOI] [PubMed] [Google Scholar]
- 55. Yagi JM, Sims D, Brettin T, Bruce D, Madsen EL (2009) The genome of Polaromonas naphthalenivorans strain CJ2, isolated from coal tar-contaminated sediment, reveals physiological and metabolic versatility and evolution through extensive horizontal gene transfer. Environ Microbiol 11: 2253–2270. [DOI] [PubMed] [Google Scholar]
- 56. Kondratyeva TF, Muntyan LN, Karavaiko GI (1995) Zinc-and arsenic-resistant strains of Thiobacillus ferrooxidans have increased copy numbers of chromosomal resistance genes. Microbiology 141: 1157–1162. [DOI] [PubMed] [Google Scholar]
- 57.Holmes DS, Haq RU (1989) Adaptation of Thiobacilus ferrooxidans for industrial applications. In: Salley J, McCready RGL, Wichlacz PL, editors. Biohydrometallurgy. Ottawa: Canadian Centre for Mineral and Energy Technology. pp. 116–127. [Google Scholar]
- 58. Dopson M, Baker-Austin C, Koppineedi PR, Bond PL (2003) Growth in sulfidic mineral environments: metal resistance mechanisms in acidophilic microorganisms. Microbiology 149: 1959–1970. [DOI] [PubMed] [Google Scholar]
- 59. Svensson B, Lübben M, Hederstedt L (1993) Bacillus subtilis CtaA and CtaB function in haem A biosynthesis. Mol Microbiol 10: 193–201. [DOI] [PubMed] [Google Scholar]
- 60. Zimmermann BH, Nitsche CI, Fee JA, Rusnak F, Münck E (1988) Properties of a copper-containing cytochrome ba3: a second terminal oxidase from the extreme thermophile Thermus thermophiles . Proc Natl Acad Sci U S A 85: 5779–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Keightley JA, Zimmermann BH, Mather MW, Springer P, Pastuszyn A, et al. (1995) Molecular genetic and protein chemical characterization of the cytochrome ba3 from Thermus thermophilus HB8. J Biol Chem 270: 20345–20358. [DOI] [PubMed] [Google Scholar]
- 62. Sone N, Fujiwara Y (1991) Haem O2 can replace haem A in the active site of cytochrome c oxidase from thermophilic bacterium PS3. FEBS Lett 288: 154–158. [DOI] [PubMed] [Google Scholar]
- 63. Klughammer B, Sültemeyer D, Badger MR, Price GD (1999) The involvement of NAD(P)H dehydrogenase subunits, NdhD3 and NdhF3, in high-affinity CO2 uptake in Synechococcus sp. PCC7002 gives evidence for multiple NDH-1 complexes with specific roles in cyanobacteria. Mol Microbiol 32: 1305–1315. [DOI] [PubMed] [Google Scholar]
- 64. Vignais PM, Billoud B (2007) Occurrence, classification, and biological function of hydrogenases: an overview. Chem Rev 107: 4206–4272. [DOI] [PubMed] [Google Scholar]
- 65. Ng G, Tom CG, Park AS, Zenad L, Ludwig RA (2009) A novel endo-hydrogenase activity recycles hydrogen produced by nitrogen fixation. PLoS One 4: e4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Andrews SC, Berks BC, McClay J, Ambler A, Quail MA, et al. (1997) A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenlyase system. Microbiology 143: 3633–3647. [DOI] [PubMed] [Google Scholar]
- 67. Supuran CT (2008) Carbonic anhydrases–an overview. Curr Pharm Des 14: 603–614. [DOI] [PubMed] [Google Scholar]
- 68. Colbeau A, Magnin JP, Cauvin B, Champion T, Vignais PM (1990) Genetic and physical mapping of an hydrogenase gene cluster from Rhodobacter capsulatus . Mol Gen Genet 220: 393–399. [Google Scholar]
- 69. Cava F, Zafra O, Magalon A, Blasco F, Berenguer J (2004) A new type of NADH dehydrogenase specific for nitrate respiration in the extreme thermophile Thermus thermophilus . J Biol Chem 279: 45369–45378. [DOI] [PubMed] [Google Scholar]
- 70. Horvath P, Coûté-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, et al. (2009) Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol 131: 62–70. [DOI] [PubMed] [Google Scholar]
- 71. Portillo MC, Gonzalez JM (2009) CRISPR elements in the Thermococcales: evidence for associated horizontal gene transfer in Pyrococcus furiosus . J Appl Genet 50: 421–430. [DOI] [PubMed] [Google Scholar]
- 72. Godde JS, Bickerton A (2006) The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol 62: 718–729. [DOI] [PubMed] [Google Scholar]
- 73. Brüggemann H, Lomholt HB, Tettelin H, Kilian M (2012) CRISPR/cas loci of type II Propionibacterium acnes confer immunity against acquisition of mobile elements present in type I P. acnes . PLoS One 7: e34171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ho Sui SJ, Fedynak A, Hsiao WW, Langille MG, Brinkman FS (2009) The association of virulence factors with genomic islands. PLoS One 4: e8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shah SA, Garrett RA (2010) CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol 162: 27–38. [DOI] [PubMed] [Google Scholar]
- 76. Pingoud A, Fuxreiter M, Pingoud V, Wende W (2005) Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci 62: 685–707. [DOI] [PubMed] [Google Scholar]
- 77. Furuta Y, Abe K, Kobayashi I (2010) Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res 38: 2428–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li YH, Tian X (2012) Quorum sensing and bacterial social interactions in biofilms. Sensors 12: 2519–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Aldridge P, Hughes KT (2002) Regulation of flagellar assembly. Curr Opin Microbiol 5: 160–165. [DOI] [PubMed] [Google Scholar]
- 80. Hamer R, Chen PY, Armitage JP, Reinert G, Deane CM (2010) Deciphering chemotaxis pathways using cross species comparisons. BMC Syst Biol 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sauer K, Camper AK (2001) Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol 183: 6579–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhilenkov EL, Popova VM, Popov DV, Zavalsky LY, Svetoch EA, et al. (2006) The ability of flagellum-specific Proteus vulgaris bacteriophage PV22 to interact with Campylobacter jejuni flagella in culture. Virol J 3: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sakai D, Komano T (2002) Genes required for plasmid R64 thin-pilus biogenesis: identification and localization of products of the pilK, pilM, pilO, pilP, pilR, and pilT genes. J Bacteriol 184: 444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yan Q, Hu X, Wang N (2012) The novel virulence-related gene nlxA in the lipopolysaccharide cluster of Xanthomonas citri ssp. citri is involved in the production of lipopolysaccharide and extracellular polysaccharide, motility, biofilm formation and stress resistance. Mol Plant Pathol 12: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nesper J, Lauriano CM, Klose KE, Kapfhammer D, Kraiss A, et al. (2001) Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect Immun 69: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nakao R, Senpuku H, Watanabe H (2006) Porphyromonas gingivalis galE is involved in lipopolysaccharide O-antigen synthesis and biofilm formation. Infect Immun 74: 6145–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Seed KD, Faruque SM, Mekalanos JJ, Calderwood SB, Qadri F, et al. (2012) Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLoS Pathog 8: e1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nguyen KT, Piastro K, Gray TA, Derbyshire KM (2010) Mycobacterial biofilms facilitate horizontal DNA transfer between strains of Mycobacterium smegmatis . J Bacteriol 192: 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Maeda S, Ito M, Ando T, Ishimoto Y, Fujisawa Y, et al. (2006) Horizontal transfer of nonconjugative plasmids in a colony biofilm of Escherichia coli . FEMS Microbiol Lett 255: 115–120. [DOI] [PubMed] [Google Scholar]
- 90. Nordlund P, Reichard P (2006) Ribonucleotide reductases. Annu Rev Biochem 75: 681–706. [DOI] [PubMed] [Google Scholar]
- 91. Dwivedi B, Xue B, Lundin D, Edwards RA, Breitbart M (2013) A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC Evol Biol 13: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, et al. (2008) The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190: 6881–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cádiz R, Gaete L, Jedlicki E, Yates J, Holmes DS, et al. (1994) Transposition of IST2 in Thiobacillus ferrooxidans . Mol Microbiol 12: 165–170. [DOI] [PubMed] [Google Scholar]
- 94. Fernández-Gómez B, Fernández-Guerra A, Casamayor EO, González JM, Pedrós-Alió C, et al. (2012) Patterns and architecture of genomic islands in marine bacteria. BMC Genomics 13: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. López-Pérez M, Gonzaga A, Martin-Cuadrado AB, Onyshchenko O, Ghavidel A, et al. (2012) Genomes of surface isolates of Alteromonas macleodii: the life of a widespread marine opportunistic copiotroph. Sci Rep 2: 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gordon D (2003) Viewing and editing assembled sequences using Consed. Curr Protoc Bioinformatics 11: 11.2. [DOI] [PubMed] [Google Scholar]
- 97. Badger JH, Olsen GJ (1999) CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol 16: 512–524. [DOI] [PubMed] [Google Scholar]
- 98. Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23: 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mulder N, Apweiler R (2007) InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol 396: 59–70. [DOI] [PubMed] [Google Scholar]
- 104. Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, et al. (2006) The PROSITE database. Nucleic Acids Res 34: D227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Rousseau C, Gonnet M, Le Romancer M, Nicolas J (2009) CRISPI: a CRISPR interactive database. Bioinformatics 25: 3317–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Grissa I, Vergnaud G, Pourcel C (2007) CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35: W52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, et al. (2007) CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, et al. (2009) MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 38: D396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. (2008) The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Delcher AL, Salzberg SL, Phillippy AM (2003) Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics 10: 10.3. [DOI] [PubMed] [Google Scholar]
- 111. Darling AC, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, et al. (2005) ACT: the Artemis Comparison Tool. Bioinformatics 21: 3422–3423. [DOI] [PubMed] [Google Scholar]
- 113. Grant JR, Stothard P (2008) The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36: W181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhang R, Zhang CT (2004) A systematic method to identify genomic islands and its applications in analyzing the genomes of Corynebacterium glutamicum and Vibrio vulnificus CMCP6 chromosome I. Bioinformatics 20: 612–622. [DOI] [PubMed] [Google Scholar]
- 115. Nieto PA, Covarrubias PC, Jedlicki E, Holmes DS, Quatrini R (2009) Selection and evaluation of reference genes for improved interrogation of microbial transcriptomes: case study with the extremophile Acidithiobacillus ferrooxidans . BMC Mol Biol 10: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. caldus strain-specifc gene lists.
(XLSX)
Taxonomic origin of A. caldus ATCC 51756 and A. caldus SM-1 exclusive genes.
(XLSX)
Megaplasmid mpAcaTY.1 and plasmids pAcaTY.1 and pÀcaTY.2 annotation and gene module information.
(XLSX)
Predicted IS families and features associated to IS-rich regions in the genome of A. caldus ATCC 51756 and A. caldus SM-1.
(XLSX)
ICE and IME Trb gene cluster synteny analysis.
(XLSX)
A. caldus strains used in this study.
(XLSX)
Oligonucleotide primers used in this study.
(XLSX)