Abstract
AIDS, caused by the retrovirus HIV, remains the largest cause of morbidity in sub-Saharan Africa yet almost all genetic studies have focused on cohorts from Western countries. HIV shows high co-morbidity with tuberculosis (TB), as HIV stimulates the reactivation of latent tuberculosis (TB). Recent clinical trials suggest that an effective anti-HIV response correlates with non-neutralising antibodies. Given that Fcγ receptors are critical in mediating the non-neutralising effects of antibodies, analysis of the extensive variation at Fcγ receptor genes is important. Single nucleotide variation and copy number variation (CNV) of Fcγ receptor genes affects the expression profile, activatory/inhibitory balance, and IgG affinity of the Fcγ receptor repertoire of each individual. In this study we investigated whether CNV of FCGR2C, FCGR3A and FCGR3B as well as the HNA1 allotype of FCGR3B is associated with HIV load, response to highly-active antiretroviral therapy (HAART) and co-infection with TB. We confirmed an effect of TB-co-infection status on HIV load and response to HAART, but no conclusive effect of the genetic variants we tested. We observed a small effect, in Ethiopians, of FCGR3B copy number, where deletion was more frequent in HIV-TB co-infected patients than those infected with HIV alone.
Introduction
AIDS, caused by the T-lymphotropic retrovirus HIV, remains the largest cause of morbidity in sub-Saharan Africa [1]. African countries currently have the highest disease burden of HIV, with 9.2% prevalence in Addis Ababa in Ethiopia and over 10% in Dar-es-Salaam in Tanzania, yet almost all genetic studies have focused on cohorts from Western countries [2]. In Africa, HIV shows high co-morbidity with tuberculosis (TB), as HIV stimulates the reactivation of latent TB, and we and others have shown that TB co-infection is associated with a higher viral load (VL) prior to treatment and a poorer response to treatment [3-5] . This presents challenges to the standard treatment regimens of both HIV and TB [6,7].
The most effective treatment for HIV and TB would be an effective vaccine; several are currently in clinical trials for HIV (e.g., STEP trial, RV144), and for TB, the vaccine Bacillus Calmette-Guérin (BCG) remains ineffective against pulmonary TB in adults. An effective vaccine is likely to stimulate the production of potent broadly neutralising antibodies that are able to neutralise the pathogen. However, in the recent RV144 trial, when looking for correlates of protection from HIV-1 infection, it was found that neutralising antibodies and cytotoxic T lymphocyte (CTL) responses were absent in protected patients [8]. In contrast, HIV -1 infection inversely correlated with gp120 V1V2-specific and antibody-dependent cell cytotoxicity (ADCC)- and antibody-dependent cell-mediated viral inhibition (ADCVI)-mediating non-neutralising antibodies. Given that Fcγ receptors are critical in mediating the non-neutralising effects of antibodies, this suggests an important role for Fcγ receptors in recruiting innate immune cells to sites of HIV infection. The interaction between the Fc region of IgG and Fcγ receptors is critical for mediating the biological effects of the humoral immune response, such as ADCC and ADCVI. The ratio of activatory/inhibitory signals generated by engagement of different Fcγ receptors by IgG determines the threshold for induction of IgG-mediated responses. In addition, it has been shown that Fcγ receptor function is critical in mounting an effective response to HIV infection in experimental animals [9].
Fcγ receptor genetic variation has been associated with infectious and inflammatory disease in both genomewide [10] and candidate gene studies [11,12], and it is known that at least some of this variation affects function, both in terms of subcellular localisation, cell type expression and IgG subtype affinity binding [13,14]. Two studies have suggested that genetic variation of host Fcγ receptors may affect various aspects of HIV infection and progression. The FCGR2A gene encodes an activating receptor expressed on macrophages and neutrophils, and a coding polymorphism (rs1801274 c.497AG [p.His131Arg]) has been associated with susceptibility to perinatal HIV infection [15] and variation in HIV progression to AIDS [16], with His131 homozygotes showing increased perinatal transmission and more rapid progression to AIDS. It is known that the two different alleles differ markedly in their affinity for IgG2 [13,17], and it was shown not only that anti-gp120 IgG2 complexes were present in individuals chronically infected with HIV, but that HIV-1 immune complexes were internalised more efficiently by monocytes from donors who were homozygous for the His131 allele.
Initial studies have focused on two alleles of one gene, but the genetic variation of the FCGR region is extensive and complex. In particular, the FCGR3A and FCGR3B genes are 97% identical and the product of an 80kb duplication that occurred after the divergence between macaque and human-chimpanzee lineages (~25 million years ago) [18]. They encode two different Fcγ receptors, with FCGR3A being expressed on natural killer (NK) cells, monocytes, dendritic cells, and macrophages and FCGR3B being expressed on neutrophils, mast cells, and eosinophils. Both genes exhibit copy number variation, deletion of the FCGR3B gene is associated with both SLE and RA, and is likely to cause ectopic expression of the FCGR2B inhibitory receptor on NK cells [19,20]. The FCGR2C gene is an activating receptor, formed as a fusion gene of FCGR2A and FCGR2B during the duplication of the ancestral FCGR3 gene and is expressed on NK cells. FCGR2C shows copy number variation related to the copy number variation of FCGR3A and FCGR3B, such that deletion or duplication of FCGR3A or FCGR3B results in concomitant deletion or duplication of FCGR2C. There is also the additional complication of a variant (rs10917661; c.169CT) which converts a glutamine to a stop codon, rendering FCGR2C non-functional [21]. FCGR3B also carries two major alleles that produce isoforms differing by 4 amino acids called human neutrophil antigen 1a and 1b (HNA1a and HNA1b ; rs76714703)[22] . These alleles affect binding to IgG1 and IgG3 and phagocytosis of opsonized particles.
In this study we investigated whether copy number variation of FCGR2C, FCGR3A and FCGR3B as well as HNA1 allelic variation of FCGR3B is associated with HIV load, response to HAART and co-infection with TB in two African populations.
Methods
Samples and ethics statement
Patient sample and clinical data collection was as previously described [5,23]. DNA extraction was performed using QIAamp DNA Maxi kit in a single laboratory. The study protocol was approved by the Institutional Review Board at the Faculty of Medicine, Addis Ababa University and Ethiopian Science and Technology Ministry; the regional ethical review board in Stockholm at the Karolinska Institutet and the ethical review committee of Muhimbili University of Health and Allied Sciences. Written informed consent was obtained from each subject before the start of this study. All samples had previously been shown to be homozygous for the CCR5 32bp insertion allele [5], where the deletion allele is known to be protective against HIV progression. The breakdown of samples analysed is shown in Table 1.
Table 1. Summary of samples analysed.
| Ethiopian | Tanzanian | ||
|---|---|---|---|
| Samples analysed | n | 720 | 347 |
| Samples analysed with detailed clinical data | n | 618 | 347 |
| FCGR3 copy number genotypes and detailed clinical data | n | 607 | 344 |
| CD4<200 with baseline VL data and genotypes | n | 517 | 167 |
| males | 201 | 72 | |
| females | 316 | 95 | |
| CD4 (mean +/- sd) | 92.30 +/-52.978 | 93.33 +/- 61.274 | |
| VL x 105 (mean +/- sd) | 3.32 +/- 6.78 | 4.87 +/- 10.66 | |
| CD4<200 with at least one CD4 follow-up datapoint for response analysis | n | 373 | 137 |
| males | 128 | 59 | |
| females | 256 | 78 | |
| CD4 (mean +/- sd) | 97.89 +/-52.835 | 97.05 +/- 60.252 | |
| VL x105 (mean +/- sd) | 2.98 +/-5.43 | 4.58 +/- 8.44 |
Copy number analysis
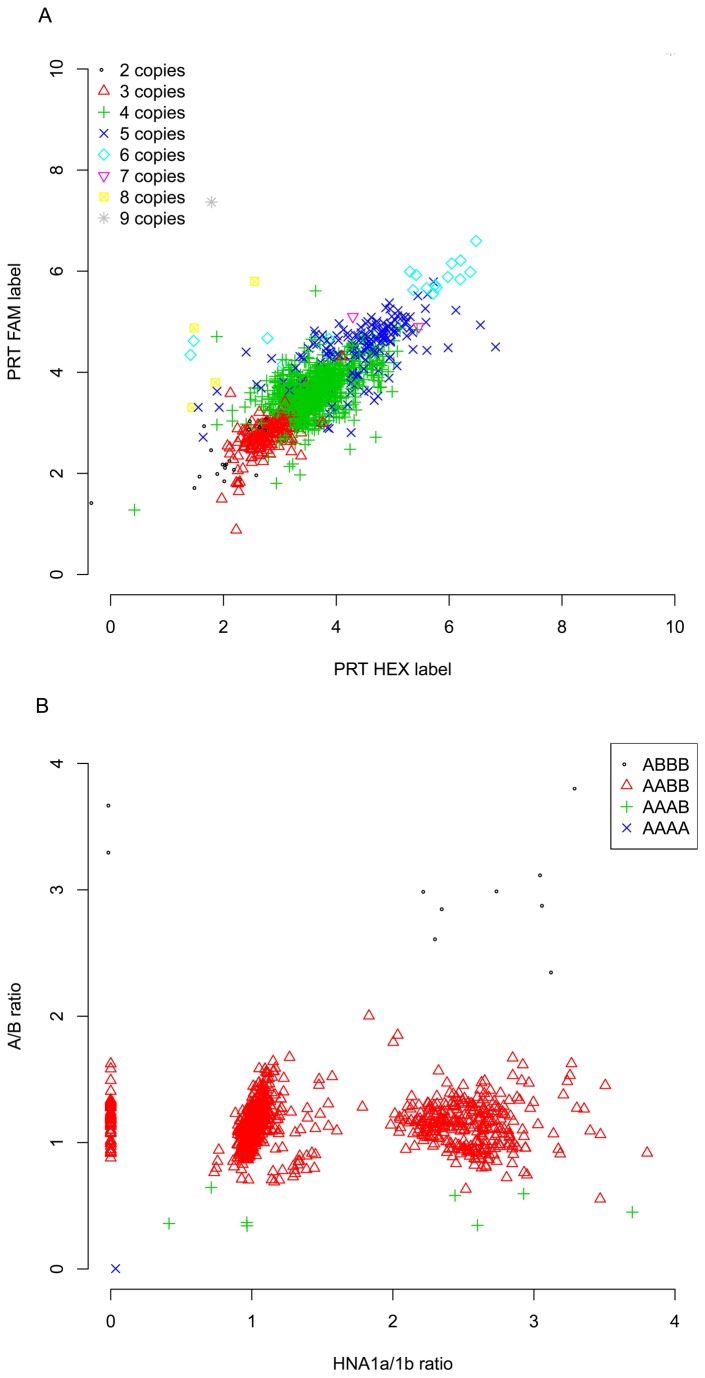
Copy number analysis was performed as described previously [18]. Briefly, duplicate calls from a paralogue ratio test (PRT, [24]) are combined with three independent assays measuring restriction enzyme digestion ratios using a maximum-likelihood framework, which calls the most-likely integer copy number given the results from the five assays. Raw data from the duplicate PRTs show very strong concordance with limited clustering and a small number of outliers (Figure 1a), showing that duplicate PRT by itself is not sufficient to reliably call copy number in all samples, and the extra information provided by restriction enzyme variant ratios (REDVR) is needed. The raw results for two REDVR assays are shown in Figure 1b. The two assays distinguish FCGR3A from FCGR3B using the arginine to stop change (C733T, y axis) and distinguish HNA1a and HNA1b on FCGR3B using the C147T nonsynonymous change [22,25,26]. Clear clustering of samples is observed, and the samples are classified according to the number of FCGR3A and FCGR3B copies observed. Note that, for clarity, only samples with four copies of FCGR3 (FCGR3A and FCGR3B) are shown.
Figure 1. Analysis of raw copy number quantification data.
a) Correlation between individual PRT copy number estimates.
Copy number typing uses duplicate PRTs combined with restriction enzyme digest ratios to infer copy number. Individual raw PRT results from the FAM-labelled experiment (y-axis) and the HEX-labelled experiment are plotted for all samples, colour-coded according to the estimated integer copy number of each sample.
b) Analysis of restriction enzyme digest ratios .
Raw ratios for the A/B assay and the HNA1a/1b assay are plotted for all samples with a total FCGR3 copy number estimate of 4. Each point is coloured according to the FCGR3A:FCGR3B ratio estimate.
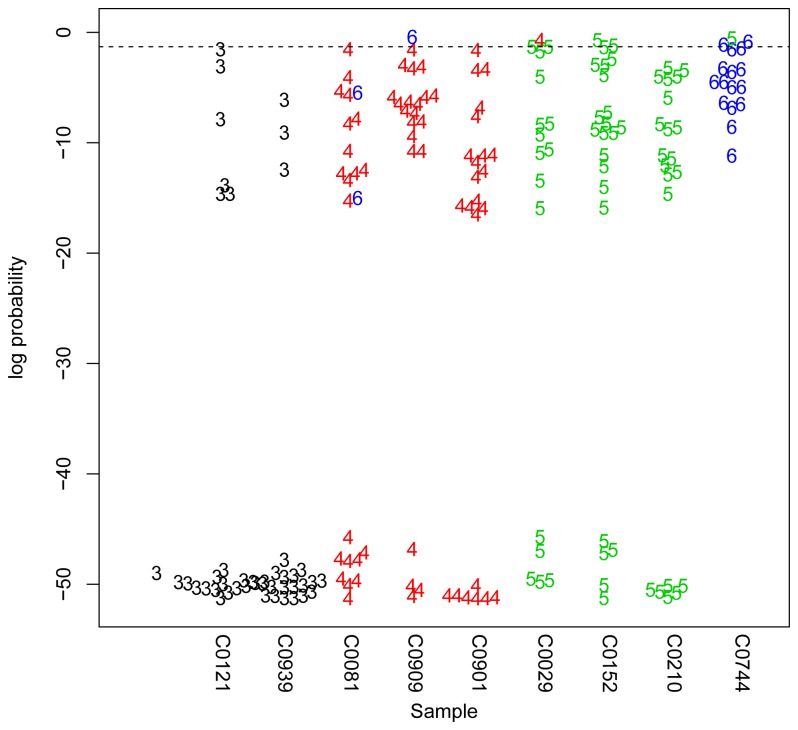
Integer copy numbers were inferred from all assays combined using the maximum-likelihood approach described previously [25], which generates an associated quality score for each copy number call. As previously, copy number estimates with a quality score of p<0.05 (equivalent to an odds ratio of 20:1 of this copy number being correct against any other copy number being correct). Positive control samples from the Human Random Control collection (Health Protection Agency, Salisbury, UK) were run with every experiment and provided a measure of the reproducibility of the assay (Figure 2). Of 190 repeat tests, 183 (96%) passed the quality score threshold. Of these 183, 2 (1%) gave the incorrect copy number score, suggesting an error rate for total copy number of 1-2%. Individual gene copy numbers were inferred using the total copy number score and the observed ratio value of the particular assay. For example, from a 1:1 ratio of the FCGR3A:FCGR3B REDVR with a total copy number of 4, we would infer a copy number of 2 for FCGR3A and 2 for FCGR3B.
Figure 2. Positive control total FCGR3 copy number estimates from repeat tests.
The nine positive control DNA samples shown were repeated in every experiment and total FCGR3 copy number calculated. Each point represents an individual copy number measurement (as indicated by the colour and number), plotted on the y-axis representing the quality score as a log probability of the copy number being an alternative to the copy number shown. The dotted line shows the quality score threshold of 0.05, with samples below this threshold being accepted. The lower the point on the y-axis, the more confident we are of the copy number. 190 tests are shown, with 183 being below the quality threshold. Of these 183, 2 show the incorrect copy number, both in C0081 calling a 6 instead of a 4.
Statistical analysis
To analyse the effect of genotype on HIV load at initiation of HAART, we initially constructed a generalised linear model using SPSS 20.0 (IBM) and a gamma-log link, using Type III sum of squares and Wald estimation. Initial VL was used as the dependent variable, with population and disease status as fixed predictor factors, CD4 count and genotype as scalar predictor variables.
To examine the effect of genotype on CD4+ count following initiation of HAART, we constructed a generalised linear mixed model, using STATA, where the dependent variable (CD4+ count) was modelled as a Gaussian distribution. In this model, we assigned population and disease status as fixed factors, initial CD4 count and time since HAART initiation as scalar covariates and integer copy number as an ordinal covariate. The model was calculated using Type III sum of squares, with a variance correction to allow for multiple CD4+ timepoint readings from a single patient.
Two-tailed t-tests were performed in Microsoft Excel, assuming unequal variances of the two samples.
Results and Discussion
Distribution of copy number alleles
Previous studies have shown that the deletion and duplication alleles for both FCGR3A and FCGR2B are relatively rare in the population [18,25]. Our data support this: in both Tanzanian and Ethiopian populations the frequency of heterozygotes for deletions and duplications is approximately 5%, and the frequency of homozygous deletions or duplications is less than 1% (table 2).
Table 2. FCGR3A copy number frequencies.
| copy number | Ethiopian HIV Count (frequency) | Ethiopian HIV+TB Count (frequency) | Tanzanian HIV Count (frequency) | Tanzanian HIV+TB Count (frequency) |
|---|---|---|---|---|
| 0 | 0 (0) | 1 (<0.01) | 0 (0) | 0 (0) |
| 1 | 13 (0.05) | 24 (0.05) | 4 (0.02) | 20 (0.14) |
| 2 | 249 (0.90) | 390 (0.88) | 186 (0.92) | 121 (0.83) |
| 3 | 15 (0.05) | 23 (0.05) | 12 (0.06) | 3 (0.02) |
| 4 | 1 (<0.01) | 4 (<0.01) | 0 (0) | 1 (<0.01) |
| 5 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| total | 278 | 442 | 202 | 145 |
| mean | 2.01 | 2.01 | 2.04 | 1.91 |
Both the FCGR3B deletion allele and the FCGR2C active allele have a functional effect. For carriers of the FCGR3B deletion allele, not only is there a gene dosage effect resulting in a lower expression of FCGR3B on the surface of neutrophils [27], but a change in the expression pattern of the adjacent FCGR2B gene, which is not copy number variable itself. The FCGR2B gene encodes the only known inhibitory Fc gamma receptor, and deletion of FCGR3B results in ectopic expression of FCGR2B on NK cells, possibly as a result of a NK-specific regulatory element being brought into closer physical proximity to the FCGR2B gene [19,20]. Again, as observed previously, the frequency of deletions and duplications of the FCGR3B gene is higher than for the FCGR3A gene (table 3),
Table 3. FCGR3B copy number frequencies.
| copy number | Ethiopian HIV Count (frequency) | Ethiopian HIV+TB Count (frequency) | Tanzanian HIV Count (frequency) | Tanzanian HIV+TB Count (frequency) |
|---|---|---|---|---|
| 0 | 1 (<0.01) | 9 (0.02) | 0 (0) | 0 (0) |
| 1 | 38 (0.14) | 69 (0.16) | 20 (0.10) | 28 (0.19) |
| 2 | 177 (0.64) | 306 (0.69) | 166 (0.82) | 109 (0.75) |
| 3 | 57 (0.21) | 54 (0.12) | 16 (0.08) | 6 (0.04) |
| 4 | 4 (0.01) | 3 (<0.01) | 0 (0) | 1 (<0.01) |
| 5 | 0 (0) | 1 (<0.01) | 0 (0) | 1 (<0.01) |
| 6 | 1 (<0.01) | 0 (0) | 0 (0) | 0 (0) |
| 7 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| total | 278 | 442 | 202 | 145 |
| mean | 2.10 | 1.95 | 1.98 | 1.88 |
The FCGR2C gene encodes an activating receptor that is normally expressed on NK cells [21]. Not only is FCGR2C copy number variable but there is a common variant (rs10917661; p. Q57*; c.169CT) which results in a pseudogene, so that, in Europeans, most people do not express FCGR2C. In this study, we combine measurement of FCGR2C copy number together with detection of the allelic status of FCGR2C to determine, for each individual, the number of copies of FCGR2C that are predicted to encode a full-length functional gene. We show there is an appreciable frequency of the active FCGR2C allele in both African populations tested (table 4), although we note that this may be an overestimate, as we did not determine the “non-classical” FCGR2C null allele, likely to be caused by a single nucleotide variant in the donor splice site of intron 7, causing skipping of exon 7 and subsequent frameshift causing early termination of the polypeptide chain [19].
Table 4. FCGR3C (active) copy number frequencies.
| copy number | Ethiopian HIV Count (frequency) | Ethiopian HIV+TB Count (frequency) | Tanzanian HIV Count (frequency) | Tanzanian HIV+TB Count (frequency) |
|---|---|---|---|---|
| 0 | 211 (0.76) | 375 (0.86) | 162 (0.81) | 128 (0.88) |
| 1 | 59 (0.21) | 54 (0.12) | 36 (0.18) | 14 (0.10) |
| 2 | 6 (0.02) | 7 (0.02) | 1 (<0.01) | 3 (0.02) |
| total | 276 | 436 | 199 | 145 |
| rs10917661 Q57 allele frequency | 0.13 | 0.08 | 0.10 | 0.07 |
| rs10917661 Q57 allele count | 71 | 68 | 38 | 20 |
| rs10917661 *57 allele count | 481 | 804 | 360 | 270 |
We initially compared the mean copy number between the HIV-only and HIV-TB co-infected cohorts in both Tanzanians and Ethiopians. While there was no significant difference between FCGR3A copy number (table 2), we did find a lower mean copy number for FCGR3B in the HIV-TB co-infected cohort in both populations, which was significant in the Ethiopian cohort (table 3, two-tailed t-test p=0.002). Although not significant in the smaller Tanzanian cohort (two-tailed t-test p=0.078), a striking frequency difference in FCGR3B 1 copy individuals is observed in the Tanzanian HIV only (10%) and HIV-TB (19%) cohorts. We also observe a lower mean active copy number for FCGR2C in the HIV-TB co-infected cohort in both populations (table 4), which was significant again only in the Ethiopian cohort (two-tailed t-test p=0.004), which is expected given the close relationship between FCGR3B and FCGR2C copy number. Taken together, this might suggest greater expression of inhibitory over activatory FCGR2 receptors on NK cells is associated with co-infection of TB with HIV. Indeed, the role of Fc receptors and NK cells in TB is unclear and under-explored, with most work focused on the cellular T-cell-mediated response. In mice NK cells respond to Mycobacterium tuberculosis infection yet are not critical in protection [28], but deletion of the inhibitory Fc receptor FCGR2B expressed on B cells resulted in more effective mycobacterial containment [29]. Although the Ethiopian cohort is larger than the Tanzanian cohort, and therefore has more power to detect a significant effect, our data should be interpreted cautiously, as our observation was only significant in the Ethiopian cohort, and the HIV-TB coinfected and HIV-only cohorts were different arms of the study and quantitative differences may be due to subtle batch effects. The observed effect size is on the edge of the size of effect that the study is powered to predict with the Ethiopian cohort having 80% power to predict a difference in means of 0.14, and the Tanzanian cohort having 80% power to predict a difference in means of 0.15. At the moment, we suggest that this is an intriguing result that awaits further study.
Association of Fcγ receptor copy number and allelic variation with baseline viral load and progression post HAART administration
We then analysed the effect of the allelic variation of copy number of the three genes, and HNA1 allelic status, on HIV load just prior to initiation of HAART in patients whose CD4 count was less than 200. As previously published [5], we saw an effect of population of origin and TB-co-infection status, but no effect of the genetic variants we tested (table 5). We also analysed the effect of allelic variation on immune reconstitution following initiation of HAART, as measured by the response in CD4 count over the course of 48 weeks. Again, whilst observing an effect of time and baseline CD4 count on CD4 count during HAART, we found no effect of FCGR2C, FCGR3A, FCGR3B copy number and HNA1 allelic status on immune reconstitution (table 6).
Table 5. Tests of association of genotype with HIV load pre-HAART n=684.
| Genotype | FCGR3A copy number | FCGR3A copy number | FCGR3B copy number | FCGR3B copy number | HNA1 ratio | HNA1 ratio | FCGR2C copy number | FCGR2C copy number |
|---|---|---|---|---|---|---|---|---|
| Covariate | beta coefficient (95%CI) (copies/mL) | P value | beta coefficient (95%CI) (copies/mL) | P value | beta coefficient (95%CI) (copies/mL) | P value | beta coefficient (95%CI) (copies/mL) | P value |
| Population (Ethiopian=1, Tanzanian=2) | -0.58 (-0.88,-0.28) | <0.001 | -0.58 (-0.88,-0.28) | <0.001 | -0.52 (-0.84,-0.20) | 0.001 | -0.57 (-0.87,-0.27) | <0.001 |
| No TB Co-infection | -0.53 (-0.79,-0.27) | <0.001 | -0.54 (-0.80,-0.27) | <0.001 | -0.54 (-0.81,-0.28) | <0.001 | -0.53 (-0.79,-0.27) | <0.001 |
| CD4 count | -0.003 (-0.006, -0.001) | 0.01 | -0.003 (-0.006, -0.001) | 0.007 | -0.003 (-0.006, -0.001) | 0.007 | -0.003 (-0.006, -0.001) | 0.008 |
| Copy number | 0.084 (-0.27, 0.43) | 0.64 | 0.056 (-0.13, 0.24) | 0.56 | 0.19 (-0.21, 0.58) | 0.35 | 0.142 (-0.20, 0.36) | 0.58 |
Table 6. Tests of association of genotype with CD4 count during HAART n=1823.
| Genotype | FCGR3A copy number | FCGR3A copy number | FCGR3B copy number | FCGR3B copy number | HNA1 ratio | HNA1 ratio | FCGR2C copy number |
FCGR2C copy number
|
|
|---|---|---|---|---|---|---|---|---|---|
| Statistic | beta coefficient (95%CI) (cells/mm3) | P value | beta coefficient (95%CI) (cells/mm3) | P value | beta coefficient (95%CI) (cells/mm3) | P value |
beta coefficient (95%CI) (cells/mm3) |
P value | |
| Time after HAART initiation (weeks) | 2.59 (2.36,2.82) | <0.001 | 2.59 (2.36,2.82) | <0.001 | 2.59 (2.36,2.82) | <0.001 | 2.60 (2.36,2.84) |
<0.001 | |
| Baseline CD4 (cells/mm3) | 0.86 (0.75, 0.97) | <0.001 | 0.86 (0.75,0.97) | <0.001 | 0.86 (0.75,0.97) | <0.001 | 0.86 (0.74,0.97) |
<0.001 | |
| Population (Ethiopian=1, Tanzanian=2) | 16.15 (2.17, 30.1) | 0.024 | 15.80 (1.74,29.85) | 0.028 | 17.97 (3.43,32.5) | 0.02 | 16.16 (1.98,30.3) |
0.03 | |
| No TB Co-infection | 14.98 (2.05, 27.91 | 0.023 | 14.58 (1.59,27.56) | 0.028 | 14.81 (1.89,27.7) | 0.03 | 15.49 (2.37,28.9) |
0.02 | |
| copy number | 6.88 (-9.95,23.71) | 0.423 | -2.47 (-12.25,7.31) | 0.621 | -8.74 (-28.15,10.68) | 0.38 | -2.25 (-15.4,10.91) | 0.74 | |
It is known that the allelic sequence and copy number variation of Fcγ receptor genes determine the expression profile, activatory/inhibitory balance, and IgG affinity of the Fc receptor repertoire of each individual. Given the known importance of Fc genetic variation on antibody mediated immune responses we hypothesised that allelic and copy number variation might be associated with baseline viral load (HIV load prior to HAART administration) and progression (CD4 count during HAART treatment). For the two African populations studied, there was no evidence of association with the variants we have examined. The lack of association of Fc receptor genetic variation with HIV progression and baseline viral load would support studies in primate challenge models suggesting that non-neutralising antibodies provide limited or no protection against HIV [30]. However our data and primate challenge data contrasts with data from trials such as the RV144 trial which indicated a role for non-neutralising antibodies in mediating the protection found in 31% of their patients [8]. Alternatively, it may be that epistatic interactions between different FCGR alleles and IgG allotypes are important in host control of HIV. Indeed, epistasis has been described for KIR/HLA-C mediated control of HIV [31]. Larger epidemiological studies combined with functional approaches are needed to provide the power to test this possibility in a thorough manner.
Acknowledgments
Thanks to Samantha de Werff and Mahnaz Abbasian, Mark Jobling for access to ABI3130xl capillary electrophoresis platform, and the patients for participation in this study.
Funding Statement
This work was supported by the Wellcome Trust [grant number 087663] and a United Kingdom Medical Research Council New Investigator award [grant number GO801123] to E.J.H.; European & Developing Countries Clinical Trials Partnership [grant numbers CT.2005.32030.001, CG_TA.05.40204_005]; and the Swedish International Development Cooperation Agency/ Department for Research Cooperation [grant numbers HIV-2006-031, SWE 2007–270, VR 521-2011-3437]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD et al. (2013) Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380: 2197-2223. [DOI] [PubMed] [Google Scholar]
- 2. An P, Winkler CA (2010) Host genes associated with HIV/AIDS: advances in gene discovery. Trends Genet 26: 119-131. doi: 10.1016/j.tig.2010.01.002. PubMed: 20149939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toossi Z, Mayanja-Kizza H, Hirsch CS, Edmonds KL, Spahlinger T et al. (2001) Impact of tuberculosis (TB) on HIV‐1 activity in dually infected patients. Clin Exp Immunol 123: 233-238. doi: 10.1046/j.1365-2249.2001.01401.x. PubMed: 11207653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Day JH, Grant AD, Fielding KL, Morris L, Moloi V et al. (2004) Does tuberculosis increase HIV load? J Infect Dis 190: 1677-1684. doi: 10.1086/424851. PubMed: 15478075. [DOI] [PubMed] [Google Scholar]
- 5. Hardwick RJ, Amogne W, Mugusi S, Yimer G, Ngaimisi E et al. (2012) β-defensin Genomic Copy Number Is Associated With HIV Load and Immune Reconstitution in Sub-Saharan Africans. J Infect Dis 206: 1012-1019. doi: 10.1093/infdis/jis448. PubMed: 22837491. [DOI] [PubMed] [Google Scholar]
- 6. Kwan CK, Ernst JD (2011) HIV and tuberculosis: a deadly human syndemic. Clin Microbiol Rev 24: 351-376. doi: 10.1128/CMR.00042-10. PubMed: 21482729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pawlowski A, Jansson M, Sköld M, Rottenberg ME, Källenius G (2012) Tuberculosis and HIV co-infection. PLOS Pathog 8: e1002464 PubMed: 22363214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD et al. (2012) Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366: 1275-1286. doi: 10.1056/NEJMoa1113425. PubMed: 22475592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ et al. (2007) Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449: 101-104. doi: 10.1038/nature06106. PubMed: 17805298. [DOI] [PubMed] [Google Scholar]
- 10. Khor CC, Davila S, Breunis WB, Lee YC, Shimizu C et al. (2011) Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat Genet 43: 1241-1246. doi: 10.1038/ng.981. PubMed: 22081228. [DOI] [PubMed] [Google Scholar]
- 11. Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD et al. (2006) Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439: 851-855. doi: 10.1038/nature04489. PubMed: 16482158. [DOI] [PubMed] [Google Scholar]
- 12. Robinson JI, Carr IM, Cooper DL, Rashid LH, Martin SG et al. (2012) Confirmation of association of FCGR3B but not FCGR3A copy number with susceptibility to autoantibody positive rheumatoid arthritis. Hum Mutat 33: 741-749. doi: 10.1002/humu.22031. PubMed: 22290871. [DOI] [PubMed] [Google Scholar]
- 13. Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N et al. (2009) Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood 113: 3716-3725. doi: 10.1182/blood-2008-09-179754. PubMed: 19018092. [DOI] [PubMed] [Google Scholar]
- 14. Niederer HA, Clatworthy MR, Willcocks LC, Smith KGC (2010) FcγRIIB, FcγRIIIB, and systemic lupus erythematosus. Ann N Y Acad Sci 1183: 69-88. doi: 10.1111/j.1749-6632.2009.05132.x. PubMed: 20146709. [DOI] [PubMed] [Google Scholar]
- 15. Brouwer KC, Lal RB, Mirel LB, Yang C, Eijk AM et al. (2004) Polymorphism of Fc receptor IIa for IgG in infants is associated with susceptibility to perinatal HIV-1 infection. AIDS 18: 1187-1194. doi: 10.1097/00002030-200405210-00012. PubMed: 15166534. [DOI] [PubMed] [Google Scholar]
- 16. Forthal DN, Landucci G, Bream J, Jacobson LP, Phan TB et al. (2007) FcγRIIa genotype predicts progression of HIV infection. J Immunol 179: 7916-7923. PubMed: 18025239. [DOI] [PubMed] [Google Scholar]
- 17. Warmerdam PA, Van de Winkel JG, Vlug A, Westerdaal NA, Capel PJ (1991) A single amino acid in the second Ig-like domain of the human Fc gamma receptor II is critical for human IgG2 binding. J Immunol 147: 1338-1343. PubMed: 1831223. [PubMed] [Google Scholar]
- 18. Machado LR, Hardwick RJ, Bowdrey J, Bogle H, Knowles TJ et al. (2012) Evolutionary history of copy-number-variable locus for the low-affinity fcγ receptor: mutation rate, autoimmune disease, and the legacy of Helminth infection. Am J Hum Genet 90: 973-985. doi: 10.1016/j.ajhg.2012.04.018. PubMed: 22608500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van der Heijden J, Breunis WB, Geissler J, de Boer M, van den Berg TK et al. (2012) Phenotypic variation in IgG receptors by nonclassical FCGR2C alleles. J Immunol 188: 1318-1324. doi: 10.4049/jimmunol.1003945. PubMed: 22198951. [DOI] [PubMed] [Google Scholar]
- 20. Mueller M, Barros P, Witherden AS, Roberts AL, Zhang Z et al. (2012) Genomic Pathology of SLE-Associated Copy-Number Variation at the FCGR2C/FCGR3B/FCGR2B Locus. Am J Hum Genet 92: 28-40. PubMed: 23261299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Metes D, Ernst LK, Chambers WH, Sulica A, Herberman RB et al. (1998) Expression of functional CD32 molecules on human NK cells is determined by an allelic polymorphism of the FcγRIIC gene. Blood 91: 2369–2380. PubMed: 9516136. [PubMed] [Google Scholar]
- 22. Bux J (2008) Human neutrophil alloantigens. Vox Sang 94: 277-285. doi: 10.1111/j.1423-0410.2007.01031.x. PubMed: 18208407. [DOI] [PubMed] [Google Scholar]
- 23. Mugusi SF, Ngaimisi E, Janabi MY, Mugusi FM, Minzi OM et al. (2012) Risk factors for mortality among HIV-positive patients with and without active tuberculosis in Dar es Salaam, Tanzania. Antiviral Therapy 17: 265-274. PubMed: 22293579. [DOI] [PubMed] [Google Scholar]
- 24. Armour JAL, Palla R, Zeeuwen PLJM, den Heijer M, Schalkwijk J et al. (2007) Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res 35: e19. doi: 10.1093/nar/gkl1089. PubMed: 17175532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hollox EJ, Detering JC, Dehnugara T (2009) An integrated approach for measuring copy number variation at the FCGR3 (CD16) locus. Hum Mutat 30: 477-484. doi: 10.1002/humu.20911. PubMed: 19143032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ory PA, Clark MR, Kwoh EE, Clarkson SB, Goldstein IM (1989) Sequences of complementary DNAs that encode the NA1 and NA2 forms of Fc receptor III on human neutrophils. J Clin Invest 84: 1688-1691. doi: 10.1172/JCI114350. PubMed: 2478590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Willcocks LC, Lyons PA, Clatworthy MR, Robinson JI, Yang W et al. (2008) Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med 205: 1573-1582. doi: 10.1084/jem.20072413. PubMed: 18559452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Junqueira-Kipnis AP, Kipnis A, Jamieson A, Juarrero MG, Diefenbach A et al. (2003) NK cells respond to pulmonary infection with Mycobacterium tuberculosis, but play a minimal role in protection. J Immunol 171: 6039-6045. PubMed: 14634116. [DOI] [PubMed] [Google Scholar]
- 29. Maglione PJ, Xu J, Casadevall A, Chan J (2008) Fcγ receptors regulate immune activation and susceptibility during Mycobacterium tuberculosis infection. J Immunol 180: 3329-3338. PubMed: 18292558. [DOI] [PubMed] [Google Scholar]
- 30. Burton DR, Hessell AJ, Keele BF, Klasse PJ, Ketas TA et al. (2011) Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc Natl Acad Sci USA 108: 11181-11186. doi: 10.1073/pnas.1103012108. PubMed: 21690411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bashirova AA, Thomas R, Carrington M (2011) HLA/KIR restraint of HIV: surviving the fittest. Annu Rev Immunol 29: 295-317. doi: 10.1146/annurev-immunol-031210-101332. PubMed: 21219175. [DOI] [PMC free article] [PubMed] [Google Scholar]