Abstract
In this paper, we report a simple, rapid, and robust method to synthesize surface-enhanced Raman-scattered gold nanoparticles (GNPs) based on green chemistry. Vitis vinifera L. extract was used to synthesize noncytotoxic Raman-active GNPs. These GNPs were characterized by ultraviolet-visible spectroscopy, dynamic light-scattering, Fourier-transform infrared (FTIR), transmission electron microscopy (TEM), X-ray diffraction (XRD), and Raman spectroscopy. The characteristic surface plasmon-resonance band at ~ 528 nm is indicative of spherical particles, and this was confirmed by TEM. The N–H and C–O stretches in FTIR spectroscopy indicated the presence of protein molecules. The predominant XRD plane at (111) and (200) indicated the crystalline nature and purity of GNPs. GNPs were stable in the buffers used for biological studies, and exhibited no cytotoxicity in noncancerous MIO-M1 (Müller glial) and MDA-MB-453 (breast cancer) cell lines. The GNPs exhibited Raman spectral peaks at 570, 788, and 1,102 cm−1. These new GNPs have potential applications in cancer diagnosis, therapy, and ultrasensitive biomarker detection.
Keywords: GNPs, SERS, SPR, Vitis vinifera L., stability
Video abstract
Introduction
Gold nanoparticles (GNPs) have been used for more than 2–3 decades for imaging and targeted therapy. GNPs are of interest due to their unique intrinsic size-dependent properties, such as surface energy and light absorption or scattering, which are attributed to surface plasmon resonance (SPR) and surface-enhanced Raman scattering (SERS). The synthesis of GNPs with a SERS signature involves multiple steps. Plasmonic nanoparticles coated with reporter molecules and covered with protective layer of molecules reduce the toxicity of plasmonic nanoparticles in living cells.1 The application of nanomaterials in therapy and diagnosis of cancerous tissue is of scientific interest due to their unique size-dependent properties. Among all nanomaterials, metals are advantageous due to their SPR and SERS properties.2,3 In addition to the plasmonic property of metal nanoparticles, GNPs have added advantages in cancer therapy and diagnosis due to their biocompatibility and easily controllable shape and size.4,5 Two intravenous preparations – Aurimune™ and AuroLase® – have already been used for clinical applications.6,7 GNPs have been used for more than 50 years for the treatment of rheumatoid arthritis, and are also approved by the FDA for biomedical applications.8 Uni-Gold Recombigen® has been approved for the detection of HIV-1 in plasma, serum, and whole blood.9 Recently, insulin-coated ultrathin GNPs were approved for clinical trials in humans.10
The synthesis of GNPs using green nanotechnology has an advantage over chemical methods in terms of reducing the toxicity of chemicals used in the synthesis process.11,12 Therefore, natural plant products like polysaccharides, phenolics and phytochemicals are commonly used for the synthesis of nanoparticles.13 GNPs have been synthesized using phytochemicals from Terminalia chebula, Breynia rhamnoides, Memecylon edule, Cinnamomum verum, Macrotyloma uniflorum, Rosa hybrid, ginseng soybean and Aloe vera.14–18 Furthermore, Vitis vinifera L. was used for the synthesis of silver nanoparticles.19 These authors showed that phytochemicals in the plant show unique kinetic property to reduce gold salts to form GNPs. Polyphenols such as flavonoids and catechins from tea have been used as reducing agents to synthesize GNPs.20 The subcellular organelle chloroplast reduced the Au salts to produce the GNPs.21
GNPs with SERS signatures are used extensively in biomedical applications due to their inert biocompatible properties and high sensitivity in imaging application.22 Localized SPR and SERS are due to the resonance of free electrons present in the GNPs.23 The introduction of noble metal substrates led to Raman intensities enhanced by as much as 1014–1015 times compared to the weak Raman signal exhibited inherently by the molecules.24 Since its discovery, SERS-based technologies have been used for various biomedical applications, such as live-cell probing and diagnosis of diseased tissue.25,26 The broad-range application of SERS includes spectral specificity, long-term stability compared to fluorescence, and tagging of multiple molecular markers for multiplexing capabilities.27,28 The promising properties of GNPs and an emerging nanotechnology enabled the target-specific and efficient use of this nanocarrier in biomedical applications.29 The general strategy for designing SERS-encoded nanoparticles or SERS nanotags involves the attachment of one or multiple organic dyes as signature reporters onto a metal enhancer in the form of a gold or silver nanoparticle, which is further encapsulated by a polymer biomolecule or a glass shell for protection against aggregation and biocompatibility.30,31 Since the fabrication process reported earlier involves multiple complicated steps, there is a need for rapid synthesis of GNPs with SERS signature.30 The present rapid and simple method uses phytochemicals from fruits of V. vinifera L. to reduce sodium tetrachloroaurate and provide Raman reporter molecules instead of using toxic chemicals for the synthesis and stabilization of metallic nano-particles. The facile synthesis route further demonstrates the great potential of GNPs as a new class of SERS nanoparticles in biomedical applications.
Materials and methods
Chemicals
Sodium tetrachloroaurate (III) dehydrate (99%), L-histidine (>99% purity), L-cysteine (97% purity), cell-culture media Roswell Park Memorial Institute (RPMI) 1640 medium and Dulbecco’s Modified Eagle’s Medium (DMEM), fetal bovine serum (FBS) and poly-L-lysine were procured from Sigma-Aldrich (St Louis, MO, USA). Trypsin–ethylenediaminetet-raacetic acid (EDTA) and antibiotic antimycotic solution were procured from Hi-Media (Mumbai, India). MTT (3[4,5-dimethylthiazol-2-yl] − 2,5-diphenyl tetrazolium bromide) reagent, dimethyl sulfoxide (DMSO), and V. vinifera L. (black grapes) were obtained from (Chennai, India). Gum arabic was purchased from MP Biomedicals (Santa Ana, CA, USA).
Instrumentation
An ultraviolet (UV)-visible spectrophotometer (DU-800; Beckman Coulter, Brea, CA, USA), dynamic light scattering (DLS) analyzer (Zetasizer Nano ZS equipped with 4.0 mW, 633 nm laser, model ZEN3600; Malvern Instruments, Malvern, UK), SpectraMax M4 multimode microplate reader (Molecular Devices, Sunnyvale, CA, USA) at Sankara Nethralaya (Chennai, India), high-resolution transmission electron microscope (HRTEM; Tecnai G14, 140 kV; FEI, Hillsboro, OR, USA), Fourier-transform infrared (FTIR) spectrophotometer (580B; PerkinElmer, Waltham, MA, USA), Raman spectrophotometer (Alpha 300 confocal Raman system equipped with a 532 nm Nd:YAG laser; WITec, Ulm, Germany) at the Indian Institute of Technology (Madras, India), and an X-ray powder diffractometer (PW 1830; Philips, Amsterdam, the Netherlands) at the Indian Institute of Technology (Kanpur, India) were used in the studies.
Cell lines
The breast cancer MDA-MB-453 cell line was procured from the American Type Culture Collection (ATCC), Manassas, VA, USA, and the Müller glial MIO-M1 cell line was a kind gift from Professor Astrid Limb, London, UK.
Preparation of Vitis vinifera L
The V. vinifera L. fruit extract was washed with Milli-Q (MQ; EMD Millipore, Billerica, MA, USA) water and ground in a pestle and mortar to obtain the extract. The extract was centrifuged at 1,000 rpm for 5 minutes. The supernatant containing the extract was filtered through a 0.2 μm syringe filter (EMD Millipore) and used for the synthesis of the GNPs.
Synthesis of GNPs
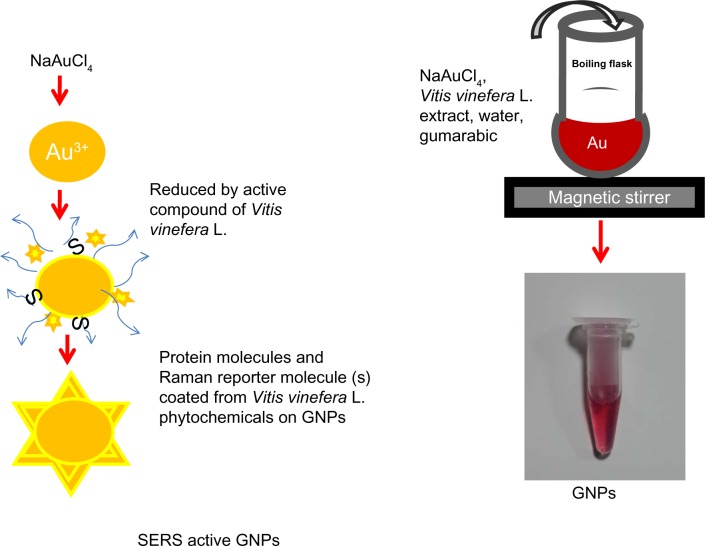
GNPs were synthesized by the reduction of sodium tetrachlo-roaurate by V. vinifera L. fruit extracts. MQ (36 mL of 0.2 μm-filtered) water was heated to 80°C in a boiling flask, followed by the addition of 72 mg of gum arabic and 600 μL of V. vinifera L. fruit extract with constant stirring for 5 minutes. Then, 600 μL (0.1 M NaAuCl4) was added dropwise to the mixture. The color of the reaction mixture changed to red, indicating the synthesis of GNPs. The reaction was stopped by immediate cooling on ice. GNPs with different sizes were obtained by changing the reaction parameters.
Characterization of GNPs
The prepared GNPs were characterized by the UV-visible spectrophotometer to observe the plasmon absorption, with water as blank. The spectra, taken in the scanning range of 400–800 nm, were used to characterize the SPR of the GNPs, using water as a reference. The size distribution and zeta potential of the synthesized GNPs in suspension were measured using DLS. The parameters used in the measurements included viscosity of 0.34 cP, reflective index of 1.054, and temperature of 25°C by the Zetasizer Nano ZS.
TEM analysis
GNPs shape and size range was confirmed by TEM. The samples were prepared by drop-coating on copper grids, followed by drying overnight. The TEM analysis was carried out at an accelerating voltage of 140 kV using HRTEM (Tecnai G14).
XRD measurements
X-ray diffraction (XRD) analysis was performed to identify different phases present in the GNPs with the Philips PW 1830. The instrument was operated at 40 KV voltage and 40 MV current using Cu-Kα radiation (λ = 1.5405 Å, scan rate 1°/minute).
FTIR measurements
FTIR spectroscopy was performed in the range of 500–4,000 cm−1 to study the characteristic functional groups present in as-prepared GNPs and V vinifera L. extract. The FTIR analysis was carried out in the transmittance mode using KBr crystal. Protein estimation was done using the standard Bradford method to confirm the presence of proteins on the surface of GNPs.
Stability of GNPs in various buffers
The stability of the GNPs in different buffers was tested using UV-visible spectrometry. GNPs (1 mL) were added to 0.5 mL of RPMI + FBS RPMI, 0.25% histidine, 0.5% cysteine, borate buffer, and 1% NaCl, followed by incubation for 30 minutes at room temperature. Stability and identity were confirmed by UV-visible spectrophotometry. After 15 days of synthesis, GNPs were again tested for monodispersity as well as long-term stability. The plasmon-resonance band was measured in vitro to confirm the stability of the GNPs in all the solutions.
Cell culture
Human breast cancer (MDA-MB-453) and Müller glial (MIO-M1) cell lines were used to assess the cytotoxicity of GNPs. The cryovials received from the ATCC were rapidly thawed and subsequently cultured in DMEM, supplemented with 10% FBS and 1% antibiotic cocktail (10,000 IU penicillin, 10 mg streptomycin, and 25 μg amphotericin/mL) at 37°C in a 5% CO2 humidified atmosphere. The exponentially growing monolayer cells, upon attaining 90% confluence, were trypsinized using 0.25% trypsin–EDTA solution and used for further experiments.
Cell-toxicity determination using MTT assay
Mitochondrial activity was assessed by MTT assay. The assay was performed to determine the cytotoxicity of the GNPs. Cell viability was measured by trypan blue exclusion in a hemocytometer. Briefly, 5 × 103 cells were seeded in a poly-L-lysine-coated 96-well polystyrene plate. The cells were cultured in a CO2 incubator overnight. The growth medium was removed, and a series of varying concentrations of GNPs − 10, 25, 50, and 100 μM – were added to the culture plate. After incubation for 6, 12, and 24 hours, 10 μL MTT (5 mg/mL) was added to each well. The culture plate was incubated until formazan crystals were formed. The MTT solution was removed followed by the addition of 140 μL DMSO to each well to dissolve the crystals. The optical density was then read by enzyme-linked immunosorbent assay at 570 nm. Percentage cell viability was calculated with respect to control.
Raman spectroscopy for SERS analysis
Raman spectra were recorded using an argon-ion laser as excitation source and scanned from the wave-number region of 2,000–500 cm−1 at a laser power of 8 mW. A 532 nm nm-excitation wavelength was used for recording Raman spectra. The GNPs of various sizes were characterized by Raman spectroscopy. The GNPs were drop-coated on the glass slide, and the Raman spectra were recorded. The spectrum from the V. vinifera L. was used as control. The SERS effect of the dye-doped GNPs was measured according to a previously reported protocol.21 Briefly, 1 g of GNPs was dissolved in 500 μL of rhodamine 6G (R6G; 20 μm) solution and incubated overnight. The doped GNPs were centrifuged and washed twice with water. The dye-doped GNPs were resuspended in 50 μL MQ water. Aliquot (10 μL) was used for the SERS measurement. The R6G solution was used as a control. The Raman enhancement (RE) was calculated for R6G-doped GNPs with respect to R6G dye respectively according to the equation:32
where IR6G GNP and NR6G GNP are Raman intensities of R6G dye-doped GNPs at Raman peak 1,360 cm−1 and number of R6G molecules on GNPs, respectively. IR6GN/R6G is the Raman intensity of R6G and number of dye molecules. The number of molecules was calculated using Avogadro’s number for dye-doped GNPs and R6G with known concentration of R6G. The RE for the GNPs without dye doping was calculated by using Raman spectral intensities from the prominent peak (1,102 cm−1) of the Raman spectrum normalized to the intensities from the V. vinifera L. extract alone. RE (1,102 cm−1) = peak height of enhanced 1,102 cm−1/peak height of enhanced R6G dye molecule × RE of R6G × dilution factor.
Statistical analysis for in vitro cytotoxicity assay
The in vitro assay was run in triplicate with three independent measurements, and statistical analysis was performed by SPSS version 17 (IBM Corporation, Armonk, NY, USA) software. Significant differences between samples were analyzed by one way analysis of variance, and Dunnett’s test performed for post hoc analysis. Significance was set at P < 0.05.
Results and discussion
Synthesis of GNPs
A schematic process for GNPs synthesis is presented in Figure 1. In brief, the synthesis involved the addition of gum arabic,V. vinifera L. extract and NaAuCl4 into preheated water at 45°C and 80°C to NaAuCl4, followed by rapid cooling on ice. The V. vinifera L. extract alone could reduce NaAuCl4 salt to produce GNPs, but these GNPs were not stable and formed aggregates on long-term storage. Therefore, gum arabic was used as a stabilizer to avoid aggregation of GNPs. However, gum arabic alone could not reduce NaAuCl4 to produce the GNPs. Gum arabic is a highly branched polysaccharide structure containing arabinogalactan and glycoproteins. The glycoprotein coating is known to reduce the toxicity of metal nanoparticles.33 The potential adverse effects of nanomaterials on human health are expected to reduce to a greater extent with the use of GNPs synthesized from natural plant compounds, such as polyphenols and phytochemicals, that are known to have abundant antioxidant and anticancer properties.34,35 In the current method, the organic acids, phenolic compounds, and polysaccharides might have reduced the NaAuCl4 into the GNPs.33,34 The GNPs of various sizes were synthesized, as different sizes could have significant effect on cellular signaling processes.36 GNPs of various sizes were synthesized by changing the temperature of the reaction, the concentration of NaAuCl4, and the reducing agent (Table 1). GNPs with a diameter of 14 ± 1 nm (Figure 2B) were synthesized at 80°C. Increasing the time of reduction resulted in the synthesis of the 28 ± 2 nm GNPs (Figure 2D). The 61 ± 2 nm GNPs (Figure 2E) were synthesized at 45°C using higher Au salt concentration. The sizes of the GNPs are dependent on two independent processes, ie, growth and nucleation of the Au atoms during particle synthesis.37,38 The longer the duration of growth and nucleation processes, the bigger the particles synthesized. At higher temperatures with less Au salt, the atoms vibrate strongly at their lattice positions and exchange energy with neighboring atoms.39 The lower temperatures resulted in the reduction in the diffusion and the higher strain energy that led to the formation of bigger particles. The effect of annealing temperature was studied previously by Goh et al with ZnO sacrificial template and polymethylsilsesquioxanes.40 The size of GNPs is inversely proportional to the annealing temperature on ZnO sacrificial template. The size of GNPs decreases with increasing annealing temperature. The zeta potential of the 14 ± 1 nm particles was −32 mV. The bigger GNPs had reduced zeta potential of −22 mV, indicating lower stability of the bigger particles compared to the 14 ± 2 nm GNPs (Table 1). However, the negative zeta potential for all the GNPs indicates the relative stability of these particles in the dispersion medium.
Figure 1.
Schematic illustrating the GNPs synthesis procedure and coating of protein and Raman-active molecules on prepared GNPs.
Abbreviations: GNPs, gold nanoparticles; SERS, surface-enhanced Raman scattering; Au, gold; S, sulphur containing proteins.
Table 1.
Important parameters that effect particle size and zeta potential
| Size of GNPs (mean ± SD) | NaAuCl4 (0.1 mM) | PDI | Zeta potential | Temperature | Time (seconds) | Grape extract | Gum arabic concentration | Volume |
|---|---|---|---|---|---|---|---|---|
| 14 ± 1 | 100 μL | 0.422 | −32 | 80°C | 1–2 | 600 μL | 72 mg | 36 mL |
| 28 ± 2 | 600 μL | 0.625 | −24 | 80°C | 2–3 | 600 μL | 72 mg | 36 mL |
| 61 ± 2 | 600 μL | 0.411 | −22 | 45°C | 2–5 | 600 μL | 72 mg | 36 mL |
Abbreviations: GNPs, gold nanoparticles; SD, standard deviation; PDI, polydispersity index.
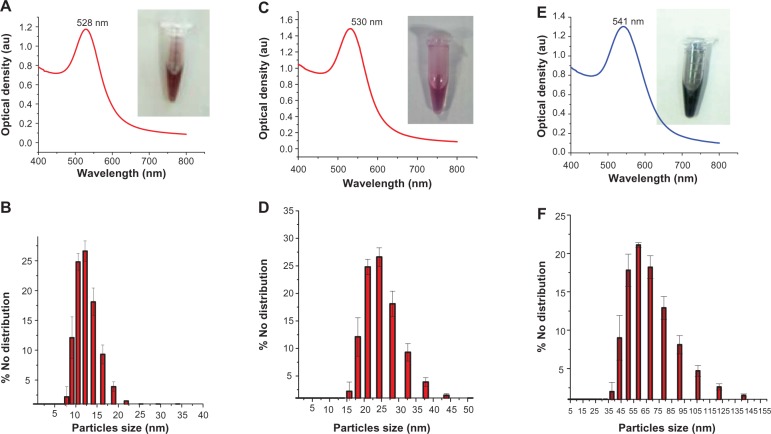
Figure 2.
UV-vis spectra of the different sizes of GNPs: (A) 14 nm; (C) 28 nm; (E) 61 nm. Dynamic light-scattering histograms showing particle distribution of GNPs: (B) 14 ± 1 nm; (D) 28 ± 2 nm; (F) 61 ± 2 nm.
Abbreviation: GNPs, gold nanoparticles; UV-vis, ultraviolet-visible; au, arbitrary unit.
Characterization of GNPs
The maximum absorption at 528 nm for the GNPs with a diameter 14nm indicates that particles are homogeneous isotropic spheres in nature.21,41 Furthermore, the λ maximum of the SPR band can also be used to predict the size of the GNPs.42 The absorption maxima of the SPR band of GNPs depends on the size of the nanoparticles, whereas the absorption intensity depends on the concentration of GNPs of same size. The wavelength maximum at 520–540 nm can be used to indicate the spherical size of the GNPs. The SPR band at 528 nm (Figure 2A) is indicative of spherical particles less than 20 nm in diameter. Consistent with the prediction of particle size from SPR analysis, the average size measured by the DLS number distribution was 14 ± 1 nm (mean ± standard deviation) (Figure 2B). Single clusters of GNPs showed monodispersity of particles. The λ maximum (SPR band) of 28 ± 2 nm and 61 ± 2 nm showed a red shift at 530 nm and 541 nm, respectively (Figure 2C and E) with respect to 14 ± 1 nm (Figure 2A). A similar red shift of the bigger nanoparticles was reported earlier.43 According to Mie theory, the change in the SPR band is due to the adsorption of any material on the surface of GNPs and a change in the dielectric properties of the medium.
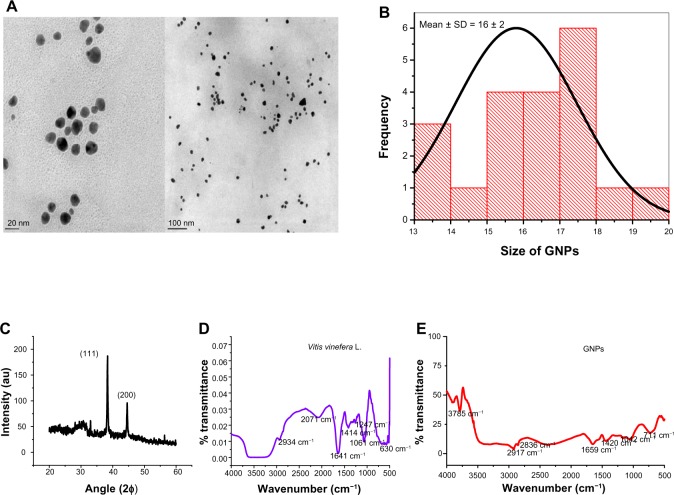
Previous studies found GNPs of less than 20 nm are more suitable for biological application compared to particles of bigger sizes, as their cellular uptake is higher and they can cross the blood–brain barrier in animal models.44,45 Therefore, for the present study, we selected GNPs sized 14 ± 1 nm for characterizations and in vitro study. In accordance with the predicted shape of the particles from absorption measurements, TEM (Figure 3A and B) analysis showed spherical GNPs <20 nm. The GNPs size calculated from the TEM analysis was 16 ± 2 nm (Figure 3B). This value is closer to the size measured by DLS (14 ± 1 nm). The GNPs were characterized by XRD to understand the crystalline nature of the particles. The XRD spectrum exhibited peaks at 38° and 44°, which is indicative of (111) and (200) planes of face-centered cubic gold. The presences of (111) confirm the crystalline nature of the GNPs (Figure 3C). The indexing of the peaks using International Centre for Diffraction Data files confirmed that the characteristic X-ray peaks could be attributed to face-centered cubic Au structure. Earlier, reports of GNPs synthesis exhibited additional diffraction peaks at 64.8° and 78.8°, and these were ascribed to (214) and (311) planes, respectively.21 The diffraction intensity of the (111) plane was higher compared to that of the (200) plane, indicating that (111) planes were predominant in GNPs synthesis. An overwhelmingly strong diffraction peak at the (111) plane compared to other facets was observed in an earlier report on GNPs synthesis.21 The predominance of (111) could be due to lower surface energy compared to other planes, which results in weak bonding ability and chemical reactivity compared to other planes.46,47 The biomolecules from V. vinifera L. fruit extract preferentially adsorbed lower surface energy of the (111) plane and enabled the growth of the Au crystal facets in this plane. The adsorption of V. vinifera L. influenced the surface property and biocompatibility of GNPs. The fibronectin protein nanofibrillar Ag particles synthesized earlier had a predominant (111) plane in XRD indicates the high SERS ability.47
Figure 3.
(A–E) Characterization of GNPs (16 ± 2 nm). (A) TEM showing GNPs shape and size. The analysis was carried out at an accelerating voltage of 200 kV. The particles were measured on two different scales: 20 nm and 100 nm. (B) Histogram showing average size of GNPs measured by TEM. (C) X-ray diffraction spectra from the particles. The particles were recorded at 40 kV voltage and 40 MV current using Cu-Kα radiation (λ = 1.5405 Å, scan rate 1°/minute). (D and E) Fourier-transform infrared spectra of Vitis vinifera L. and GNPs. The spectrum was recorded from 500 to 4,000 cm−1 from the GNPs in the transmittance mode using KBr crystal.
Abbreviations: GNPs, gold nanoparticles; TEM transmission electron microscopy; SD, standard deviation; KBr, potassium bromide; au, arbitrary unit.
V. vinifera L. contains mainly sugar, glucose and fructose, organic acid, tartaric, malic, and citric acid, phenolic compound anthocyanins, tannins, and nitrogenous compound amino acid, peptides, and proteins.48 FTIR analysis was performed to detect the vibration mode of chemical compounds and infer biochemical composition of V. vinifera L. and compound present on GNPs (Figure 3D and E). The sharp peak of GNPs at 3,785 cm−1 is probably an O−H stretch, indicating the presence of phenols from the V. vinifera L. fruit extract. The fruit extract consists of a variety of polyphenols, aldehydes, peptides, and proteins.49,50 The medium peaks of 2,934 cm−1 and 2,071 cm−1 indicate the C−H and C≡H of alkanes and alkynes, respectively (Figure 3D). The GNPs peaks at 2,917 cm−1 and 2,836 cm−1 are indicative of C−H and H−C=O bonds of aldehydes from V. vinifera L. coated on the GNPs. The peaks at 1,641 cm−1 (Figure 3D) and 1,659 cm−1 (Figure 3E) are indicative of N−H bond vibrations from amide groups of the proteins present in V. vinifera L. fruit extract as well as in the GNPs.51,52 The protein content was estimated by the Bradford test found to be 0.056 μg/2 × 107 particles for 61 ± 2 nm- and 0.045 μg/7 × 108 particles for 14 ± 1 nm-sized GNPs. The peak at 1,420 cm−1 is indicative of a C−C stretch from the polyphenols of the V. vinifera L. present on the surface of GNPs. On the other hand, peaks at 1,247 cm−1, 1,061 cm−1, and 1,042 cm−1 might be aliphatic amines with C−N stretches. The peaks at 630 cm−1 and 711 cm−1 could be from alkanes (Figure 3D) present in the V. vinifera L. fruit extract. Furthermore, the 711 cm−1 peak could be from the rocking bond of the alkanes present on the GNPs (Figure 3E). Earlier reports have attributed this peak to the presence of alkanes.53,54 The overlapping peak of the functional group of V. vinifera L. present on the GNPs also corroborates our results in that chemical compounds from the V. vinifera L. fruit extract had been coated on the surface of GNPs and formed chemical bond with GNPs.
Stability and cytotoxicity of GNPs
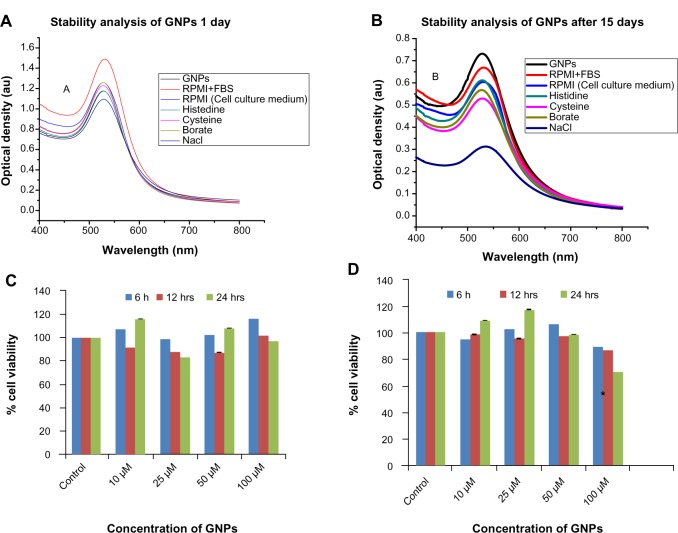
The stability of nanoparticles is an indispensable parameter for biomedical applications. The in vitro stability of the GNPs (Figure 4A and B) was evaluated by monitoring the plasmon wavelength (λ maximum) in borate buffer, RPMI, NaCl, RPMI + FBS, cysteine and histidine, and NaCl at 0 days, and 15 days after the synthesis of GNPs. The plasmon wavelength and bandwidth in the above buffers did not show a shift in SPR band at 0 days, whereas after 15 days the SPR band exhibited a small shift of less than 5 nm. The constant SPR band indicated that GNPs were stable in all buffers and cell-culture media. The data indicated that there was no difference in the SPR band in any of the buffers with respect to GNPs alone (as a control). Our results are consistent with other studies on the stability of GNPs synthesized from plant compounds.14,15
Figure 4.
Stability and cytotoxicity of GNPs (top panel). UV-vis spectra of the GNPs in various buffers at various time periods: (A) 1 day; (B) 15 days. Cytotoxicity measurements of GNPs in different cell lines at various concentrations (bottom panel): (C) MIO-M1 (Müller glial, noncancerous); (D) MDA-MB 453 (breast cancer). The cell viability (% of treated cells with respect to untreated cells) of different cell lines treated with different concentrations: 10, 25, 50, and 100 μM of GNPs at various time periods.
Notes: *Significant difference with respect to control at P< 0.05; error bars represent the standard error of mean.
Abbreviations: GNPs, gold nanoparticles; RPMI+FBS, Roswell Park Memorial Institute medium plus fetal bovine serum; RPMI, Roswell Park Memorial Institute medium; Nacl, sodium chloride; UV-vis, ultraviolet-visible; au, arbitrary unit.
A key parameter in evaluating biocompatibility is centered on the evaluation of potential cytotoxicity of the materials. The in vitro cytotoxicity effect of nanomaterials is most often assessed from mitochondrial enzyme activity using tetrazo-lium salt (MTT assay), with respect to the untreated cell as a control.55,56 Cytotoxicity of the GNPs tested using MTT assay in noncancerous Müller glial (MIO-M1) and breast cancer (MDA-MB-453) cells (Figure 4C and D). The GNPs were nontoxic to the cells up to 100 μM concentration. Cell viability did not decrease after 6 and 12 hours of treatment with increasing concentrations of GNPs from 10 to 100 μM (P < 0.05) in either normal or breast cancer cell lines when compared to the untreated control cells. The cytotoxicity of the GNPs was previously reported to be dependent on GNP concentration, time of treatment, and cell type.16–18 GNPs synthesized using synthetic and toxic chemicals such as LiBH4 penetrate the membranes at high concentrations and decrease cell proliferation via generating reactive oxygen species, in turn decreasing the mitochondrial enzyme activity adenosine triphosphate content of the cell, causing cell damage.57 In contrast, GNPs synthesized using green nanotechnology were nontoxic even at higher concentrations.15 The percentage cell viability was more than 85% until 12 hours of treatment in the Müller glial (MIO-M1) and the breast cancer (MDA-MB-453) cells. However, 100 μM of GNP treatment at 24 hours showed 70.28% cell viability in MDA-MB-453 cells (Figure 4D). These cells showed a significant difference (P < 0.05) when compared to the control at 24 hours treatment with GNPs. On the other hand, Müller glial (MIO-M1) cells showed no significant decrease (P = 0.2) in cell viability after 24 hours of GNP treatment (Figure 4C). According to the ISO-10993-5 guideline,58 a material can be used for biomedical application if the material shows more than 70% cell viability. The GNPs from 10 to 100 μM concentration are safe to use for in vitro studies.
Moreover the toxicity of the metal nanoparticles is reduced by surface coating.59 The GNPs were coated with gum arabic in the present study. This coating might have been responsible for the observed low toxicity in the normal and breast cancer cell lines, as it prevents the direct contact of the metal nanoparticles with the cells and makes these particles more biocompatible than uncoated particles. Gum arabic-coated GNPs have been previously tested in animal models and found to be nontoxic.60
SERS signature on GNPs
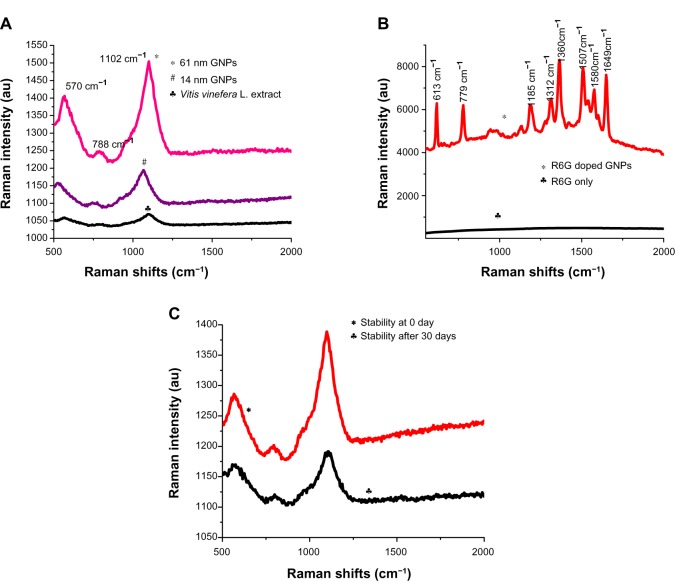
SERS is a unique property of metal nanoparticles that can be used for imaging applications. Therefore, in the current work, GNPs of various sizes (14 ± 1 nm and 61 ± 2 nnm) were tested for the SERS signature without addition of known synthetic Raman-reporter molecules. Synthetic chemicals as Raman reporters have been used before for SERS.29 In our study, Raman intensity was measured from 500 to 2,000 cm−1 (Figure 5A). Raman spectral peak intensities were observed at 570, 788, and 1,102 cm−1. The peak intensity of the different sizes revealed that the intensity of the peak increased with increasing GNPs size. In agreement with the present work, earlier reports indicated that SERS intensity increases with increased particle size.24,61 Raman spectral peak intensities from the V. vinifera L. fruit extract were also observed at 570, 788, and 1,102 cm−1, signifying that the Raman-reporter molecules from the extract were retained on the GNPs during the synthesis process. Interestingly, Raman-signal intensity was higher in GNPs compared to V. vinifera L. fruit extract alone, indicating a SERS effect (Figure 5A). The GNPs were doped with the well-known Raman reporter R6G to study the RE. This enhancement is comparable to the RE attained with Raman reporters from the V. vinifera L. extract. The R6G-doped GNPs showed Raman peak signature for the R6G dye (Figure 5B). A similar signature was reported in an earlier study.21 The RE was calculated as described by Chang et al using the formula given in the Materials and methods section.32
Figure 5.
(A–C) Synthesized GNPs function as SERS nanotag. (A) Raman spectra from GNPs of different sizes: black, grape extract; blue, spectrum from 14 ± 1 nm nanoparticles; pink, spectrum from 61 ± 2 nm particles. (B) Raman spectrum of R6G-doped GNPs: red, R6G-doped particles; black, R6G dye alone. (C) Stability of GNPs after long-term storage: black, 0 days; red, 30 days.
Abbreviations: GNPs, gold nanoparticles; SERS, surface-enhanced Raman scattering; R6G, rhodamine 6G; au, arbitrary unit.
The number of R6G dye molecules in the GNPs used to obtain Raman spectra (Figure 5B) was 120.4 × 1014. The number of dye molecules used as control was 6.23 × 1020. The RE was calculated using the peak heights from the base of the 1,360 cm−1 peak for R6G. The RE for R6G dye-doped GNPs was R6G 3.4 × 107. The RE of 108 has been previously reported for R6G-doped GNPs.21 The peak height from the base of the most prominent peak – 1,102 cm−1 – was used for calculations of RE of GNPs without dye doping. The enhanced Raman intensity for the Raman reporters in GNPs compared to the V. vinifera L. fruit extract was used for calculations (Figure 5A). The RE of R6G-doped GNPs was extrapolated for the reporter molecules from V. vinifera L. fruit extract using Raman peak intensities. The RE for the 61 nm-sized GNPs was 4 × 103. The R6G molecule exhibited higher SERS enhancement compared to the Raman-reporter molecules from V. vinifera L. (Figure 5A and B). SERS activity is known to be dependent on two key factors: (1) resonant surface-plasmon excitation of a metal substrate, and (2) close proximity of analytic molecules to the metal substrate surface.
The surface chemistry of metal substrate was considered as a critical factor for SERS activity because the analytes must be located within 0–4 nm of the substrate surface or the electromagnetic field. Therefore, we proposed that the GNPs had Raman reporter molecule(s) in the close proximity of the Au metal surfaces, resulting in SERS enhancements.62 We hypothesized that the aromatic compounds from the V. vinifera L. extract act as SERS nanotags. The higher SERS activity of R6G may be due to the closer proximity of R6G molecules compared to the Raman reporters from V. vinifera L. fruit extract. The Raman peaks at 570 cm−1 could be due to in-plane bending from carbon and oxygen from the phenolic compounds or aromatic acids, such as p-coumaric acid.63,64 This phenol is present in V. vinifera L. fruit extract and resins. The vibrational peak at 1,102 cm−1 could be from the sugar molecules present in V. vinifera L. extract.65 De Gelder et al reported that the 1,102 cm−1 band is indicative of a trehalose sugar.63 The vibrational peak at 788 cm−1 could be from the histidine amino acid.60 The SERS activity of such biological molecules as histidine, cysteine, and lysine using gold substrate has been reported previously.21,60,63
The long-term stability of the GNPs was assessed for their potential application after storage of the particles. The lack of shift in Raman peaks after 1 month compared to the freshly synthesized particles indicated the stability of the Raman-reporter molecules on GNPs (Figure 5C). On the other hand, stored V. vinifera L. extract was incapable of reducing Au salt to form GNPs with SERS signature. The newly synthesized and stored GNPs can be used for biomedical applications. Earlier studies used SERS active gold nanoparticles for in vivo tumor-cell targeting and sensitive cancer detection.27,29 However, these studies employed GNPs synthesis using NaBH4 as reducing agent and synthetic dyes as SERS-reporter molecules. Our method of synthesis of SERS nanotags is faster and safer than recent syntheses using NaBH4 and ascorbic acid.31
Conclusion
In summary, a single reaction-based, rapid, economical, and environment-friendly method of biosynthesizing SERS nano-particles has been successfully established using V. vinifera L. extract. The synthesized GNPs were spherical in shape and coated with aromatic compounds (proteins/peptides) from V. vinifera L. The phytochemicals from the V. vinifera L. extracts acted both as reducing agents for the synthesis of GNPs and as reporter molecules for SERS. One major advantage of GNPs is SERS ability without the addition of synthetic chemicals or the need for complicated dye-doping procedures, previously reported for the synthesis of SERS nanotags. The identity of the Raman-reporter molecule(s) is presently unknown, and work is being carried out to study the polyphenols, aldehydes, peptides, and protein compounds present in V. vinifera L. extract. As GNPs are nontoxic to cells, the authors believe that the SERS ability of GNPs has great potential in many areas, including cancer diagnosis, therapy, and ultrasensitive biomarker detection.
Acknowledgments
The authors thank the Department of Science and Technology – Nano Mission India (grant no. SR/NM/NS-83/2010) for financial support and Deakin University, Australia for scholarship (Sushma Kalmodia, ID 211823217). The authors also thank Professor Ashutosh Sharma, Indian Institute of Technology, for providing the facility of the DST Centre for Nanotechnology. They also thank Ms Shyama for critical review of manuscript for language proofreading.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Merican Z, Schiller TL, Hawker CJ, Fredericks PM, Blakey I. Self-assembly and encoding of polymer-stabilized gold nanoparticles with surface-enhanced Raman reporter molecules. Langmuir. 2007;23:10539–10545. doi: 10.1021/la702218b. [DOI] [PubMed] [Google Scholar]
- 2.Willets KA, Van Duyne RP. Localized surface plasmon resonance spectroscopy and sensing. Ann Rev Phys Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 3.Lu L, Kobayashi A, Tawa K, Ozaki Y. Silver nanoplates with special shapes: controlled synthesis and their surface plasmon resonance and surface-enhanced Raman scattering properties. Chem Mater. 2006;18:4894–4901. [Google Scholar]
- 4.Steinmetz N. Review of “Cancer Nanotechnology: methods and Protocols (Methods in Molecular Biology)” by Stephen R. Grobmyer (editor), Brij M. Moudgil (editor) Biomed Eng Online. 2010;9:55. [Google Scholar]
- 5.Zhang X, Xing JZ, Chen J, et al. Enhanced radiation sensitivity in prostate cancer by gold-nanoparticles. Clin Invest Med. 2008;31:E160–E167. doi: 10.25011/cim.v31i3.3473. [DOI] [PubMed] [Google Scholar]
- 6.Dobrovolskaia MA, McNeil SE. Immunological properties of engineered nanomaterials. Nat Nanotechnol. 2007;2:469–478. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 7.Chen PC, Mwakwari SC, Oyelere AK. Gold nanoparticles: from nanomedicine to nanosensing. Nanotechnol Sci Appl. 2008;1:45–66. doi: 10.2147/nsa.s3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thakor AS, Jokerst J, Zavaleta C, Massoud TF, Gambhir SS. Gold nanoparticles: a revival in precious metal administration to patients. Nano Lett. 2011;11:4029–4036. doi: 10.1021/nl202559p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention Uni-Gold Recombigen™ HIV: FDA news release 2003Available from: http://www.cdc.gov/hiv/topics/testing/resources/press_releases/FDArelease.htmAccessed August 2, 2013
- 10.Kotok A.Trial of insulin-coated gold nanoparticles approved 2011Available from: http://sciencebusiness.technewslit.com/?p=6957Accessed August 2, 2013
- 11.Pacardo DB, Sethi M, Jones SE, Naik RR, Knecht MR. Biomimetic synthesis of Pd nanocatalysts for the Stille coupling reaction. ACS Nano. 2009;3:1288–1296. doi: 10.1021/nn9002709. [DOI] [PubMed] [Google Scholar]
- 12.Nel A, Xia T, Mädler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 13.Lee KW, Bode AM, Dong Z. Molecular targets of phytochemicals for cancer prevention. Nat Rev Cancer. 2011;11:211–218. doi: 10.1038/nrc3017. [DOI] [PubMed] [Google Scholar]
- 14.Elavazhagan T, Arunachalam KD. Memecylon edule leaf extract mediated green synthesis of silver and gold nanoparticles. Int J Nanomedicine. 2011;6:1265–1278. doi: 10.2147/IJN.S18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aromal SA, Vidhu VK, Philip D. Green synthesis of well-dispersed gold nanoparticles using Macrotyloma uniflorum. Spectrochim Acta A Mol Biomol Spectrosc. 2012;85:99–104. doi: 10.1016/j.saa.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 16.Katti K, Chanda N, Shukla R, et al. Green nanotechnology from cumin phytochemicals: generation of biocompatible gold nanoparticles. Int J Green Nanotechnol Biomed. 2009;1:B39–B52. doi: 10.1080/19430850902931599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla R, Nune SK, Chanda N, et al. Soybeans as a phytochemical reservoir for the production and stabilization of biocompatible gold nanoparticles. Small. 2008;4:1425–1436. doi: 10.1002/smll.200800525. [DOI] [PubMed] [Google Scholar]
- 18.Chandran SP, Chaudhary M, Pasricha R, Ahmad A, Sastry M. Synthesis of gold nanotriangles and silver nanoparticles using Aloe vera plant extract. Biotechnol Prog. 2006;22:577–583. doi: 10.1021/bp0501423. [DOI] [PubMed] [Google Scholar]
- 19.Chaudhary S, Paul S, Sagar S. Biosynthesis of silver nanoparticles using Vitis vinifera extract and evaluation of their antimicrobial activity. Int J Biotechnol Res. 2012;2:1–12. [Google Scholar]
- 20.Nune SK, Chanda N, Shukla R, et al. Green nanotechnology from tea: phytochemicals in tea as building blocks for production of biocompatible gold nanoparticles. J Mater Chem. 2009;19:2912–2920. doi: 10.1039/b822015h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang YX, Zheng J, Gao G, et al. Biosynthesis of gold nanoparticles using chloroplasts. Int J Nanomedicine. 2011;6:2899–2906. doi: 10.2147/IJN.S24785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiti KK, Samanta A, Vendrell M, Soh KS, Olivo M, Chang YT. Multiplex cancer cell detection by SERS nanotags with cyanine and triphenylmethine Raman reporters. Chem Commun (Camb) 2011;28:3514–3516. doi: 10.1039/c0cc05265e. [DOI] [PubMed] [Google Scholar]
- 23.Lee KS, El-Sayed MA. Gold and silver nanoparticles in sensing and imaging: sensitivity of plasmon response to size, shape, and metal composition. J Phys Chem B. 2006;110:19220–19225. doi: 10.1021/jp062536y. [DOI] [PubMed] [Google Scholar]
- 24.Nie S, Emory SR. Probing single molecules and single nanoparticles by surface-enhanced raman scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 25.Kneipp J, Kneipp H, McLaughlin M, Brown D, Kneipp K. In vivo molecular probing of cellular compartments with gold nanoparticles and nanoaggregates. Nano Lett. 2006;6:2225–2231. doi: 10.1021/nl061517x. [DOI] [PubMed] [Google Scholar]
- 26.Kneipp K, Wang Y, Kneipp H, et al. Single molecule detection using surface-enhanced Raman scattering (SERS) Phys Rev Lett. 1997;78:1667–1670. [Google Scholar]
- 27.Lee S, Chon H, Yoon SY, et al. Fabrication of SERS-fluorescence dual modal nanoprobes and application to multiplex cancer cell imaging. Nanoscale. 2012;4:124–129. doi: 10.1039/c1nr11243k. [DOI] [PubMed] [Google Scholar]
- 28.Kho KW, Fu CY, Dinish US, Olivo M. Clinical SERS: are we there yet? J Biophotonics. 2011;4:667–684. doi: 10.1002/jbio.201100047. [DOI] [PubMed] [Google Scholar]
- 29.Jokerst JV, Cole AJ, Van de Sompel D, Gambhir SS. Gold nanorods for ovarian cancer detection with photoacoustic imaging and resection guidance via Raman imaging in living mice. ACS Nano. 2012;6:10366–10377. doi: 10.1021/nn304347g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JF, Huang YF, Ding Y, et al. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nat Nanotechnol. 2010;464:392–395. doi: 10.1038/nature08907. [DOI] [PubMed] [Google Scholar]
- 31.Boca S, Rugina D, Pintea A, Leopold N, Astilean S. Designing gold nanoparticle-ensembles as surface enhanced Raman scattering tags inside human retinal cells. J Nanotechnol. 2012;2012:1–10. [Google Scholar]
- 32.Chang CW, Liao JD, Chang HC, Lin LK, Lin YY, Weng CC. Fabrication of nano-indented cavities on Au for the detection of chemically-adsorbed DTNB molecular probes through SERS effect. J Colloid Interface Sci. 2011;358:384–391. doi: 10.1016/j.jcis.2011.03.032. [DOI] [PubMed] [Google Scholar]
- 33.García I, Marradi M, Penadés S. Glyconanoparticles: multifunctional nanomaterials for biomedical applications. Nanomedicine (Lond) 2010;5:777–792. doi: 10.2217/nnm.10.48. [DOI] [PubMed] [Google Scholar]
- 34.Visioli F, De La Lastra CA, Andres-Lacueva C, et al. Polyphenols and human health: a prospectus. Crit Rev Food Sci Nutr. 2011;51:524–546. doi: 10.1080/10408391003698677. [DOI] [PubMed] [Google Scholar]
- 35.Russo M, Spagnuolo C, Tedesco I, Russo GL. Phytochemicals in cancer prevention and therapy: truth or dare? Toxins (Basel) 2010;2:517–551. doi: 10.3390/toxins2040517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang W, Kim BY, Rutka JT, Chan WC. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 37.Turkevich J, Stevenson PC, Hillier J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss Faraday Soc. 1951;11:55–75. [Google Scholar]
- 38.Kimling J, Maier M, Okenve B, Kotaidis V, Ballot H, Plech A. Turkevich method for gold nanoparticle synthesis revisited. J Phys Chem B. 2006;110:15700–15707. doi: 10.1021/jp061667w. [DOI] [PubMed] [Google Scholar]
- 39.Mittemeijer EJ. Fundamentals of Materials Science: The Microstructure–Property Relationship Using Metals as Model Systems. Heidelberg: Springer; 2011. [Google Scholar]
- 40.Goh LP, Razak KA, Ridhuan NS, Cheong KY, Ooi PC, Aw KC. Direct formation of gold nanoparticles on substrates using a novel ZnO sacrificial templated-growth hydrothermal approach and their properties in organic memory device. Nanoscale Res Lett. 2012;7:563. doi: 10.1186/1556-276X-7-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mie G. Beitrage zer Optik trüber Meiden speziell kolloidaler Metallösungen. Ann Phys. 1908;25:377–445. [Google Scholar]
- 42.Le Ru E, Etchegoin P. Principles of Surface-Enhanced Raman Spectroscopy: And Related Plasmonic Effects. Amsterdam: Elsevier; 2009. [Google Scholar]
- 43.He ST, Yao JN, Jiang P, et al. Formation of silver nanoparticles and self-assembled two-dimensional ordered superlattice. Langmuir. 2001;17:1571–1575. [Google Scholar]
- 44.Semmler-Behnke M, Kreyling WG, Lipka J, et al. Biodistribution of 1.4- and 18-nm gold particles in rats. Small. 2008;4:2108–2111. doi: 10.1002/smll.200800922. [DOI] [PubMed] [Google Scholar]
- 45.Jin H, Heller DA, Sharma R, Strano MS. Size-dependent cellular uptake and expulsion of single-walled carbon nanotubes: single particle tracking and a generic uptake model for nanoparticles. ACS Nano. 2009;3:149–158. doi: 10.1021/nn800532m. [DOI] [PubMed] [Google Scholar]
- 46.Sun YG, Mayers B, Xia YN. Transformation of silver nanospheres into nanobelts and triangular nanoplates through a thermal process. Nano Lett. 2003;3:675–679. [Google Scholar]
- 47.Wang L, Sun YJ, Cui YC, Wang JK, Li Z. Synthesis of silver nanoplates with fibronectin nanofibril template and their SERS applications. Bull Korean Chem Soc. 2013;34:443–446. [Google Scholar]
- 48.Xia EQ, Deng GF, Guo YJ, Li HB. Biological activities of polyphenols from grapes. Int J Mol Sci. 2010;11:622–646. doi: 10.3390/ijms11020622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.López-Vázquez C, Orriols I, Perelló MC, de Revel G. Determination of aldehydes as pentafluorobenzyl derivatives in grape pomace distillates by HS-SPME-GC/MS. Food Chem. 2012;130:1127–1133. [Google Scholar]
- 50.Ali K, Maltese F, Choi YH, Verpoorte R. Metabolic constituents of grapevine and grape-derived products. Phytochem Rev. 2010;9:357–378. doi: 10.1007/s11101-009-9158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajasekharreddy P, Rani PU, Sreedhar B. Qualitative assessment of silver and gold nanoparticle synthesis in various plants: a photobiological approach. J Nanopart Res. 2010;12:1711–1721. [Google Scholar]
- 52.Beattie IR, Haverkamp RG. Silver and gold nanoparticles in plants: sites for the reduction to metal. Metallomics. 2011;3:628–632. doi: 10.1039/c1mt00044f. [DOI] [PubMed] [Google Scholar]
- 53.Vasita R, Katti DS. Structural and functional characterization of proteins adsorbed on hydrophilized polylactide-co-glycolide microfibers. Int J Nanomedicine. 2012;7:61–71. doi: 10.2147/IJN.S26453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dharmadhikari M. Composition of grapes. Newsletter: Vineyard and Vintage View. 1994;9:3–8. [Google Scholar]
- 55.Khlebtsov N, Dykman L. Biodistribution and toxicity of engineered gold nanoparticles: a review of in vitro and in vivo studies. Chem Soc Rev. 2011;40:1647–1671. doi: 10.1039/c0cs00018c. [DOI] [PubMed] [Google Scholar]
- 56.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;16:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 57.AshaRani PV, Low Kah Mun G, Hande MP, Valiyaveettil S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano. 2009;3:279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- 58.Association for the Advancement of Medical Instrumentation, International Organization for Standardization . Biological Evaluation of Medical Devices Part 5: Tests for in Vitro Cytotoxicity. Arlington (VA): AAMI; 2009. [Google Scholar]
- 59.Arnida, Malugin A, Ghandehari H. Cellular uptake and toxicity of gold nanoparticles in prostate cancer cells: a comparative study of rods and spheres. J Appl Toxicol. 2010;30:212–217. doi: 10.1002/jat.1486. [DOI] [PubMed] [Google Scholar]
- 60.Fent GM, Casteel SW, Kim DY, et al. Biodistribution of maltose and gum arabic hybrid gold nanoparticles after intravenous injection in juvenile swine. Nanomedicine. 2009;5:128–135. doi: 10.1016/j.nano.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 61.Schwartzberg AM, Grant CD, Wolcott A, et al. Unique gold nanoparticle aggregates as a highly active SERS substrate. J Phys Chem B. 2004;108:19191–19197. [Google Scholar]
- 62.Claridge SA, Liang HW, Basu SR, Fréchet JM, Alivisatos AP. Isolation of discrete nanoparticle-DNA conjugates for plasmonic applications. Nano Lett. 2008;8:1202–1206. doi: 10.1021/nl0802032. [DOI] [PubMed] [Google Scholar]
- 63.De Gelder J, De Gussem K, Vandenabeele P, Moens L. Reference database of Raman spectra of biological molecules. J Raman Spectrosc. 2007;38:1133–1147. [Google Scholar]
- 64.Zuk M, Dymińska L, Kulma A, et al. IR and Raman studies of oil and seedcake extracts from natural and genetically modified flax seeds. Spectrochim Acta A Mol Biomol Spectrosc. 2011;78:1080–1089. doi: 10.1016/j.saa.2010.12.054. [DOI] [PubMed] [Google Scholar]
- 65.Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P. Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci U S A. 2005;102:11118–11123. doi: 10.1073/pnas.0503410102. [DOI] [PMC free article] [PubMed] [Google Scholar]