Abstract
Background
An outbreak of 29 cases of Pneumocystis jirovecii pneumonia (PCP) occurred among renal and liver transplant recipients (RTR and LTR) in the largest Danish transplantation centre between 2007 and 2010, when routine PCP prophylaxis was not used.
Methods
P. jirovecii isolates from 22 transplant-cases, 2 colonized RTRs and 19 Pneumocystis-control samples were genotyped by restriction fragment length polymorphism and multi-locus sequence typing analysis. Contact tracing were used to investigate transmission. Potential risk factors were compared between PCP cases and matched non-PCP transplant patients.
Results
Three unique Pneumocystis genotypes were shared among 19 of the RTRs, LTRs and a colonized RTR in 3 distinct clusters, two of which overlapped temporally. In contrast, Pneumocystis-control samples harbored a wide range of genotypes. Evidence of possible nosocomial transmission was observed. Among several potential risk factors, only CMV viremia was consistently associated with PCP (P = 0.03; P = 0.009). Mycophenolate mofetile was associated with PCP risk only in the RTR population (P = 0.04).
Conclusion
We identified three large groups infected with unique strains of Pneumocystis and provide evidence of an outbreak profile and nosocomial transmission. LTRs may be infected in PCP outbreaks simultaneously with RTRs and by the same strains, most likely by inter-human transmission. Patients are at risk several years after transplantation, but the risk is highest during the first 6 months post-transplantation. Since patients at risk cannot be identified clinically and outbreaks cannot be predicted, six months of PCP chemoprophylaxis should be considered for all renal and liver transplant recipients.
Keywords: Pneumocystis jirovecii, pneumocystis pneumonia, liver transplant, renal transplant, outbreak
Introduction
Pneumocystis jirovecii pneumonia (PCP) is a common opportunistic infection in immunocompromised patients, and is now believed to result from recent acquisition in most cases [1, 2]. Recipients of solid organ transplants are at increased risk of developing PCP, with substantial variation in risk between transplant types as well as between centers [3–6]. As a consequence, the indications for and duration of PCP prophylaxis have been debated. In many centers, the incidence of PCP after renal or liver transplantation has been very low despite the absence of routine PCP prophylaxis [7].
In PCP outbreaks among renal transplant recipients (RTRs), epidemiological studies with genotyping have suggested inter-human transmission of Pneumocystis [3, 8–15]. It remains unclear if such outbreaks are due to the introduction of specific P. jirovecii strains into the hospital environment, to changes in immunosuppressive regimens, or to other factors affecting patient susceptibility. The relative importance of asymptomatic carriers and other reservoirs as the source of transmission in these outbreaks remains unclear as well. In this single center report, we describe an outbreak of PCP among both RTRs and liver transplant recipients (LTRs). To examine possible transmission of Pneumocystis, we genotyped isolates obtained from RTRs and LTRs during the outbreak (cases) and isolates obtained from other patients with PCP diagnosed at the same hospital and during the same period (Pneumocystis-controls). In addition, we compared risk factors for PCP in renal and liver transplanted patients by matching non-infected transplant patients (transplant-controls) 2:1 to PCP cases.
Results
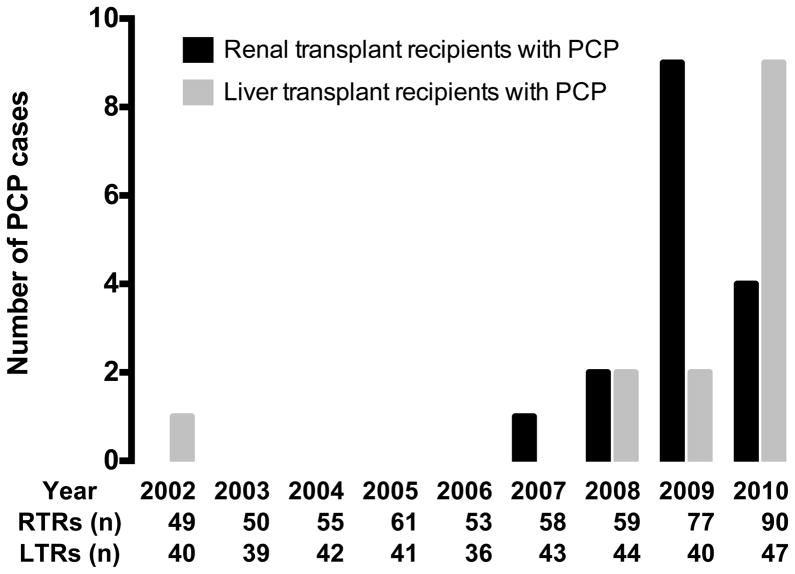
An outbreak of PCP in 29 solid organ transplant patients occurred between 2007 and 2010 (Figure 1), and included 16 renal and 13 liver transplant recipients. The majority were diagnosed by direct immunofluorescence microscopy (DFA), but one RTR (6 %) and four LTRs (29 %) were diagnosed by single-round polymerase chain reaction (PCR). General characteristics of cases are shown in table 1a and 1b. In the preceding period, from 2002 to 2007, only one transplant patient was diagnosed with PCP. This outbreak occurred with no apparent change in the overall use of immunosuppressive regimens. Routine PCP prophylaxis was not utilized between 2002 and 2010. Standard trimethoprim/sulfamethoxazole PCP prophylaxis was initiated at the end of the outbreak, in the fall of 2009 for RTRs and in early 2011 for LTRs.
Figure 1.
Incidence of PCP and number of transplants performed, 2002 to 2010.
Table 1a.
Risk factor analysis for renal transplant recipients compared to transplant-controls.
| Renal transplant recipients | |||
|---|---|---|---|
| Variable | Cases n = 16 |
Transplant-controls n = 32 |
P-value |
| Male | 8 (50 %) | 21 (66 %) | 0,4 |
| Length of admission after RTx, days (mean) | 21 | 19 | 0,6 |
| Age at PCP symptoms/risk date, years (mean) | 46 | 46 | 0,9 |
| PCP admission, days (median) | 25 (10–95) | - | - |
| Death in relation to PCP | 0 (0 %) | - | - |
| PCP prophylaxis | 1 (6 %) | 6 (19 %) | 0,4 |
| Immunosuppression | |||
| Tacrolimus | 5 (31 %) | 9 (28 %) | 1 |
| Cyclosporine | 7 (44 %) | 20 (63%) | 0,2 |
| Everolimus | 2 (13 %) | 3 (9 %) | 1 |
| Sirolimus | 0 | 1 (3%) | 1 |
| MMF | 16 (100 %) | 24 (75 %) | 0,04 |
| Azathioprine | 0 | 7 (22 %) | 0,08 |
| Corticosteroids | 16 (100 %) | 32 (100 %) | 1 |
| Corticosteroid dosage1, mg/day (median) | 10 | 10,5 | 0,8 |
| High dose corticosteroids2 | 0 | 1 (3%) | 1 |
| ≥250 mg prednisolone/day for > 2 days | 3 (19 %) | 4 (13 %) | 0,7 |
| Induction therapy |
Cases n = 153 |
Transplant-controls n = 32 |
|
| Thymoglobulin | 4 (27 %) | 7 (22 %) | 0,7 |
| Simulect | 5 (33 %) | 15 (47 %) | 0,5 |
| Zenapax | 4 (27 %) | 8 (25 %) | 1 |
| Rituximab + IVIG | 1 (7 %) | 0 (0 %) | 0,3 |
| ATGAM | 1 (7 %) | 2 (6 %) | 1 |
| Rejection | |||
| Graft rejection, total | 6 (38 %) | 6 (19 %) | 0,2 |
| Graft rejection <6 months | 4 (25 %) | 4 (13 %) | 0,4 |
| Last graft rejection to PCP/risk date4, days (median) | 149 (62–4868) | 131 (33–4911) | 0,7 |
| CMV | |||
| CMV infection prior to PCP/risk date | 2 (13 %) | 3 (9 %) | 1 |
| Time from last positive CMV-PCR to PCP/risk date, days (median) | 49 (25–73) | 29 (16–2154) | 1 |
| CMV prophylaxis at PCP symptoms/risk date5 | 5 (31 %) | 8 (25 %) | 0,7 |
| CMV infection concurrent/current with PCP/risk date6 | 5 (31 %) | 2 (6%) | 0,03 |
| Other | |||
| Diabetes type 1 | 2 (13 %) | 1 (3 %) | 0,3 |
| Diabetes type 2 | 1 (6 %) | 2 (6 %) | 1 |
| Hepatitis C | 1 (6 %) | 1 (3 %) | 1 |
| Charlson co-morbidity index | 0,8 | ||
| 1 | 5 (31 %) | 14 (44 %) | |
| 2 | 2 (13 %) | 3 (9 %) | |
| 3 | 0 (0 %) | 1 (3 %) | |
| Neutropenia | 1 (6 %) | 1 (3%) | 1 |
| ABO incompatibility | 1 (7 %)3 | 0 | 0,3 |
| Graft cold ischemia, hours (median), n = 10 and 22 | 14 (1–22) | 17,5 (2–29) | 0,1 |
Variables expressed as medians are tested by a Mann-Whitney-Wilcoxon test. RTx = renal transplantation
From 30 days prior to presentation of clinical symptoms/risk date.
Prednisolone or equivalent corticosteroid dosage of ≥ 20 mg/day given for at least 1 month prior to clinical symptoms.
One patient was transplanted in another country and not included.
From the time of biopsy.
Prescription of valganciclovir within 7 days before PCP/risk date.
For transplant-controls, current CMV infection was defined as infection within 2 months before the risk date.
Table 1b.
Risk factor analysis for liver transplant recipients compared to transplant-controls.
| Liver transplants recipients | |||
|---|---|---|---|
| Variable | Cases n = 141 |
Transplant- controls n = 28 |
P-value |
| Male | 10 (71 %) | 18 (64 %) | 0,7 |
| Length of admission after LTx, days (mean) | 27 | 42 | 0,2 |
| Age at PCP symptoms/risk date, years (mean) | 51 | 49 | 0,8 |
| PCP admission, days (median) | 24 (8–120) | - | - |
| Death in relation to PCP | 1 (7 %) | - | - |
| PCP prophylaxis | 0 | 4 (14 %) | 0,3 |
| Immunosuppression | |||
| Tacrolimus | 10 (71 %) | 19 (68 %) | 1 |
| Cyclosporine | 4 (29 %) | 8 (29 %) | 1 |
| Everolimus | 0 (0 %) | 1 (4%) | 1 |
| MMF | 12 (86 %) | 26 (93 %) | 0,6 |
| Corticosteroids | 14 (100 %) | 28 (100 %) | 1 |
| Corticosteroid dosage2, mg/day (median) | 15 | 15,5 | 0,9 |
| High dose corticosteroids3 | 4 (29 %) | 10 (36 %) | 0,7 |
| ≥250 mg prednisolone/day for >2 days | 3 (21 %) | 7 (25 %) | 1 |
| Anti-lymphocyte | 1 (7 %) | 1 (4%) | 1 |
| Rejection | |||
| Graft rejection, total | 4 (29 %) | 7 (25 %) | 1 |
| Graft rejection < 6 months | 3 (21 %) | 7 (25 %) | 1 |
| Last graft rejection to PCP/risk date4, days (median) | 122 (26–226) | 97 (75–135) | 0,3 |
| CMV | |||
| CMV infection prior to PCP/risk date | 3 (21 %) | 2 (7 %) | 0,3 |
| Time from last positive CMV- PCR to PCP/risk date, days (median) | 48 | 76 | 0,6 |
| CMV prophylaxis at PCP symptoms/risk date5 | 9 (64 %) | 16 (57 %) | 0,7 |
| CMV infection concurrent/current with PCP/risk date6 | 4 (29 %) | 0 (0 %) | 0,009 |
| Other | |||
| Diabetes type 1 | 0 | 0 | - |
| Diabetes type 2 | 5 (36 %) | 6 (21 %) | 0,5 |
| Hepatitis C | 1 (7 %) | 1 (4 %) | 1 |
| Charlson co-morbidity index | 0,1 | ||
| 1 | 3 (21 %) | 10 (36 %) | |
| 2 | 6 (43 %) | 3 (11 %) | |
| 6 | 0 (0 %) | 1 (4 %) | |
| Neutropenia | 3 (21 %) | 1 (4%) | 0,1 |
| Blood loss, liters (median), n =14 and 24 | 7,8 | 8,5 | 0,2 |
| Fulminant liver failure | 2 (14 %) | 5 (18%) | 1 |
| Partial organ transplantation | 0 (0 %) | 3 (11 %) | 0,5 |
| ALT (median), n = 13 and n = 25 | 28 (9–172) | 52 (16–287) | 0,2 |
| Alkaline phosphatase (median), n = 13 and 25 | 82 (45–1050) | 163 (34–792) | 0,6 |
Variables expressed as medians are tested by a Mann-Whitney-Wilcoxon test. LTx = liver transplantation
One LTR was diagnosed in 2002 and was not part of the outbreak, but is included in the risk factor analysis.
From 30 days prior to presentation of clinical symptoms/risk date.
Prednisolone or equivalent corticosteroid dosage of ≥ 20 mg/day given for at least 1 month prior to clinical symptoms.
From the time of biopsy.
Prescription of valganciclovir within 7 days before PCP/risk date.
For transplant-controls, current CMV infection was defined as infection within 2 months before the risk date.
Genotype analysis
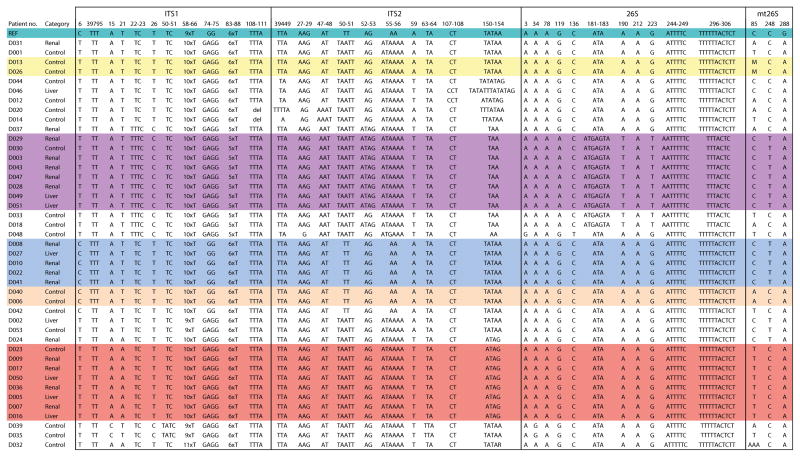
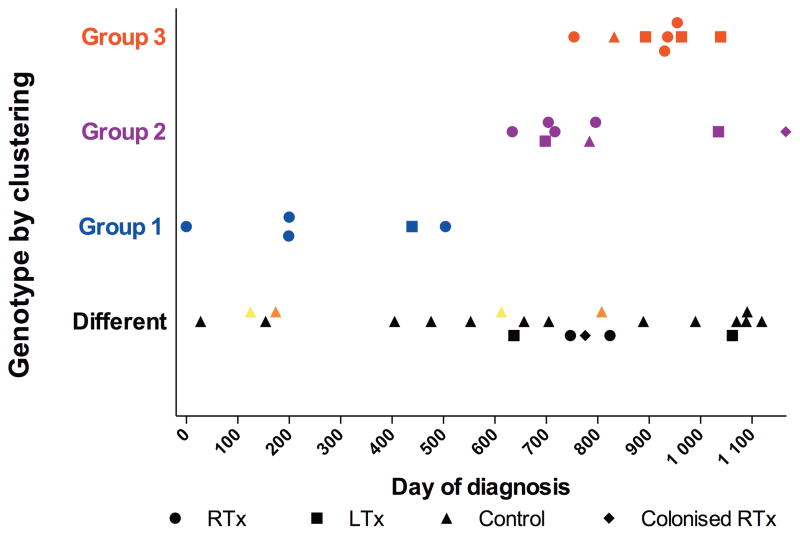
To understand the epidemiology of this outbreak, we undertook genotypic analysis of the 22 available Pneumocystis isolates with sufficient DNA, using restriction fragment length polymorphism (RFLP) and multi-locus sequence typing (MLST). Twenty-one were successfully analyzed by both methods and one by MLST only; genotyping results were identical by both methods. Three genotype patterns (groups 1, 2 and 3) were observed in 18 of 22 samples (Figure 2a and 2b). Each group was temporally clustered, with groups 2 and 3 overlapping in time (figure 3). Four outbreak cases were not part of the clusters, but resulted from infection with different Pneumocystis genotypes. MLST analysis of two additional isolates from Pneumocystis-colonized RTRs identified one as a genotype-group 2 isolate and the other as a unique genotype.
Figure 2.
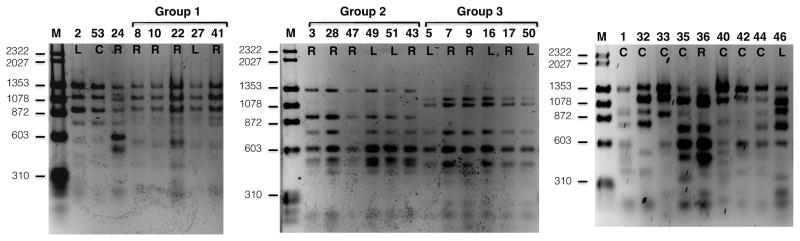
Figure 2a. Restriction fragment length polymorphism (RFLP) analysis of Pneumocystis samples from outbreak patients and PCP controls. Group number refer to the one of the three major genotypes shared among 18 of 22 RTRs and LTRs. PCR products were digested with Dra1 and analyzed by agarose gel electrophoresis. Samples from the same group were run in adjacent lanes to facilitate visualization. Digestion with Hpy188I confirmed the groupings (data not shown). The numbers represent the patient number, and the letters underneath represent the clinical group: R, RTR; L, LTR; C, control. Sample 36 is part of group 3 (confirmed on other gels). Sample 2 is from an LTR and 24 is from an RTR with non-outbreak genotypes. Sample 48 were analyzed by RFLP, but not included due to space limitations. Molecular weight markers (M) are located in the left lane of each gel.
Figure 2b. Multi-locus sequence typing (MLST) analysis of Pneumocystis samples from outbreak patients and PCP controls. A shows results for ITS1 and ITS2, and B for 26S and mt26S sequences. Only those sites that showed a polymorphism in any sequence are included; numbering is based on the reference sequence, which is shown in the top row. Groups 1–3 are indicated on the left and color coded. Category refers to classification of patients as RTR (Renal), LTR (Liver) and controls. D029 and D031 are considered colonized RTRs. Two pairs of control samples that had an identical MLST pattern are also color coded (D013 and D026; D040 and D006). Samples D002 (LTR) and D053 (control) had an identical RFLP pattern by two enzymes but differed at a single locus in the 26S gene. All sequences have been deposited with GenBank, and have been assigned GenBank numbers; ITS1 and ITS2 KC470767-KC470809; 26s, KC470724-KC470766; mt26s, KC470810-KC470852. GenBank numbers for the reference sequences are as follows: ITS1 and ITS2, U07220; 26S, L13615; mt26S, M58605. For ITS2, position 1 in the figure corresponds to nucleotide 313 of U07220.
Figure 3.
Pneumocystis cases and Pneumocystis-controls plotted by genotype-group and date of diagnosis. Day “0”: Diagnosis of first outbreak case, September 21, 2007. The three groups are color coded. Control patients were admitted at the same hospital as the cases. Two pairs of Pneumocystis-controls had the same genotype and were colored accordingly. Two controls were infected with outbreak strains. One, a patient treated with immunosuppressants for glomerulonephritis was admitted to the same ward as the RTRs; the other was a patient with myelodysplastic syndrome admitted to the Department of Hematology in another building at the hospital.
Pneumocystis-control samples with sufficient DNA for analysis were available from 19 PCP patients, the majority with hematological malignancies. Nine were successfully analyzed by both methods, and 10 by MLST only. Unlike the transplant isolates, Pneumocystis-controls generally had diverse genotypes, though 2 control patients who were hospitalized during the outbreak were infected with group 2 or 3 isolates. The Pneumocystis-controls were admitted at the same hospital as the cases, but to different floors, with one exception: one patient with glomerulonephritis was admitted to the same floor as RTRs infected with the same Pneumocystis genotype (group 2) and thus appears to be part of the outbreak.
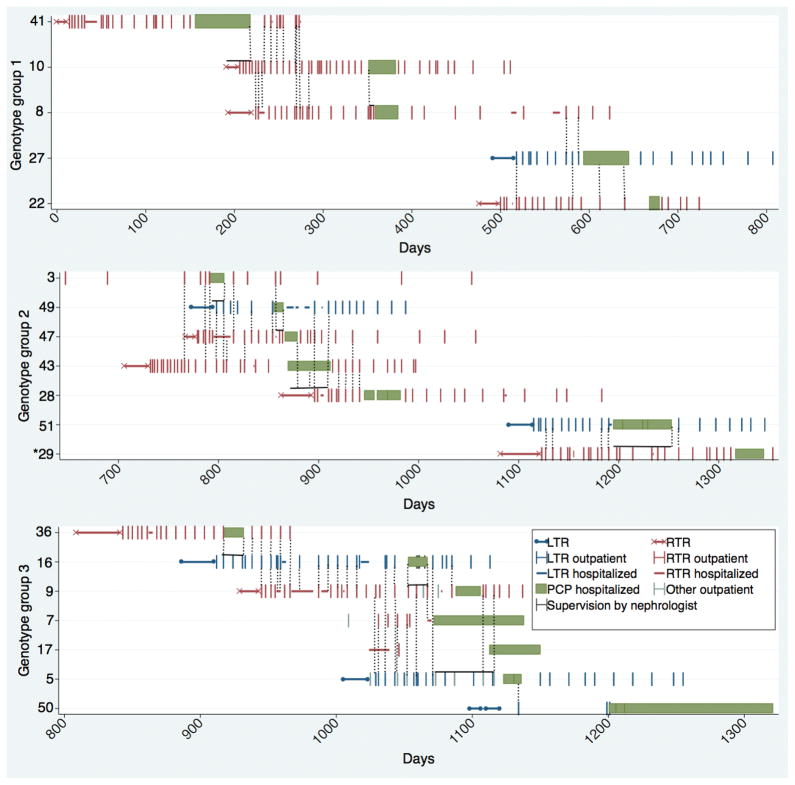
Transmission analysis
To assess possible cross-exposure, contacts between patients were investigated by a transmission map (figure 4). For many but not all cases, we were able to demonstrate exposure, during the period prior to diagnosis of PCP, to a PCP patient infected with the same genotype.
Figure 4.
Transmission map by case id. The transmission map is split according to genotype-group. Day “0” is April 9, 2007. The * indicates a colonized renal transplant recipient, the green bar indicates admission for pneumonia when PCP were suspected.
Group 1: Cases 2 and 3 developed PCP 19–20 weeks after the discharge of case 1. Cases 4 and 5 were transplanted a minimum of 13 weeks after the discharge of case 3.
Group 2: The interval from the discharge of case 10 to the transplantation of case 11 was 15 weeks.
Group 3: The interval between the discharge of case 13 to the transplantation of case 14 was 20 weeks.
Each major genotype in the outbreak affected both renal and liver recipients. The liver transplant ward is located on the floor above the renal transplant ward and direct contact between renal and liver recipients is limited to the possible brief contact at the entrance hall, elevators or the common out-patient blood sampling unit in the hospital.
Clinical features and risk factors
We conducted a matched case-control study and univariate analysis, to identify potential risk factors associated with development of PCP in the transplant setting. Previous studies have suggested that major risk factors, include high-dose immunosuppression, the use of specific immunosuppressive drugs, graft rejection, cytomegalovirus (CMV) infection and older age [7].
Renal transplant recipients
Four of 16 RTR patients (25%) were diagnosed more than one year after transplantation; the remaining 12 patients presented within ~6 months after transplantation (Table 1a). One case presented with PCP 72 days after cessation of a three month course of post-transplantation prophylaxis; no other patient received prophylaxis.
Current infection with CMV was associated with PCP (5/16 vs 2/32 for transplant-controls, P = 0.03). In addition, a significantly higher fraction of PCP cases received MMF compared to transplant-controls (16/16 vs 24/32, P = 0.04). Graft rejection within 6 months was not associated with PCP (4/16 vs 4/32, P = 0.4), nor was there a correlation between PCP and use of corticosteroids, exposure to other immunosuppressive regimens, the presence of comorbidity, CMV prior to PCP or cold graft ischemia (table 1a).
Liver transplant recipients
All 14 LTRs, including one diagnosed in 2002 who was not part of the outbreak, were diagnosed within 1 year of transplant. Four LTRs were diagnosed exclusively by a positive PCR, but had a clinical course consistent with PCP.
We again observed a statistically significant correlation between current CMV infection and PCP (4/14 vs 0/28 for transplant-controls, P = 0.009). Graft rejection within 6 months was not associated with PCP (3/14 vs 7/28, P = 1.0). No correlation between PCP and the use of corticosteroids, immunosuppressive regimens, CMV prior to PCP, co-morbidity, blood loss during surgery or biomarkers of liver and bile duct injury was observed (table 1b). In particular, the increased risk of PCP seen in RTRs receiving MMF was not observed in LTRs.
Discussion
This is the first reported outbreak of PCP occurring in both renal and liver transplant recipients. The outbreak was caused by three strains of Pneumocystis, as defined by genotyping; the same strains were found in only two cases of the large Pneumocystis-control population.
Our results differ from several recent studies which reported outbreaks of PCP among RTRs dominated by a single genotype [3, 8–15]. Surprisingly, LTRs were not infected with a distinct strain of Pneumocystis, but were part of clusters dominated by RTRs. This study provides the first molecular evidence linking Pneumocystis infection in LTRs to outbreak strains. In a previous report, LTRs were not infected during a single genotype outbreak of PCP among RTRs who attended the same outpatient clinic [12].
Outbreaks such as this one provide the opportunity to better understand the transmission dynamics of Pneumocystis infection. Genotyping studies provide strong evidence for recent transmission of Pneumocystis within clusters. Detection of P. jirovecii DNA by PCR in air samples obtained near PCP patients indicates possible aerosol spread [16]. In the current study, clustering of isolates by time and departments strongly suggests transmission directly between PCP cases or from a common source. If we assume that transmission may happen from 12 weeks prior to PCP symptoms to three weeks after initiation of treatment [17], direct transmission between the majority of our cases could have occurred. We were unable to identify potential exposure events for all cluster cases. However, seven cases were not genotyped and therefore not included in the transmission analysis; these cases may have contributed to transmission. Alternative sources of transmission may potentially include asymptomatic carriers of infection. Our genotype findings suggest that asymptomatic colonized transplant patients (e.g., patient 29) could have acted as one such reservoir. This hypothesis is supported by another recent study [14].
It has been hypothesized that more virulent strains of P. jirovecii may play a role in outbreaks: three previous outbreaks in RTRs were associated with a single genotype (ERT strain), but this strain was absent in our patients [13, 15]. The identification of multiple strains in this outbreak suggests that differential virulence is not playing an important role.
Health care workers (HCW) have been suggested as another possible reservoir, though at present there is no direct evidence to support this. Although serologic studies suggest greater exposure of HCW to Pneumocystis [18], no colonization of HCW could be demonstrated during one PCP outbreak [11]. In our hospital, different HCW care for renal and liver transplant recipients, and shared contacts by physicians are limited.
Analysis of Pneumocystis isolates in non-transplanted immunosuppressed patients usually does not show clustering of genotypes, but rather, reveals that broadly diversified strains are responsible for most infections [19, 20]. This suggests unique risk factors for RTRs and LTRs that could be related to exposure or to the underlying immunodeficiency. The reason for the sudden increase in cases of PCP cases in our clinic is unexplained by our risk factor analysis.
Several risk factors have been identified as predisposing transplant patients to PCP, most notably alteration in immunosuppressive therapy, graft rejection, CMV infection and older age [7]. The introduction of immunosuppressive drugs such as MMF and tacrolimus in the 1990s led to decreased rejection rates [21], but targeted T-cell suppressive drugs may predispose patients to PCP. We found a higher use of MMF in RTRs with PCP compared to matched transplant-controls, but this was not seen in LTRs. The association of MMF with PCP was previously reported in a large cohort study and two case-control studies [4, 22, 23]. In our center, MMF has been used as a key component of immunosuppression in RTRs for the past 14 years, yet no outbreaks of PCP were seen in the first 10 years after the introduction of MMF.
The use of corticosteroids is commonly recognized as a significant risk factor for PCP [23]. In one report, among LTRs managed with a corticosteroid-free regimen and no PCP prophylaxis, no PCP cases were observed [6]. Rejection therapy is also regarded as a prominent risk factor for PCP [24, 25]. However, while we observed a non-significant trend towards a higher rejection rate among RTRs with PCP, no clear correlation between corticosteroid exposure and PCP was found, in accordance with other studies [4].
Current infection with CMV was found to be strongly correlated with PCP in both renal and liver transplant recipients. This likely indicates a higher level of immunosuppression, but could also be caused by CMV re-activation secondary to PCP. These results should be interpreted with care, as the cases were suspected of pneumonitis and more likely to be tested. Prior CMV infection did not correlate with PCP.
The use and length of routine PCP prophylaxis for renal and liver transplant recipients has been debated. In our study, most patients developed PCP within 6 months after transplantation, in agreement with other studies [5, 25]. It is noteworthy, however, that 25% of PCP cases in RTRs occurred more than one year after transplantation, similar to the experience of others [4, 8, 9, 11]. In contrast, all LTRs with PCP were diagnosed within one year after transplantation.
Three months of trimethoprim/sulfamethoxazole prophylaxis has been suggested to adequately reduce PCP risk in renal and liver transplants [26–28]. However, studies of a three month prophylaxis should be interpreted with care, due to the risk of PCP outbreaks that have been identified with increasing frequency. Based on our findings, we suggest that PCP prophylaxis should be recommended for all patients 6 months after transplantation and prophylaxis should be considered for other susceptible patients, such as those undergoing rejection treatment, during outbreaks of PCP.
Our study has limitations. The point of contact between cases was difficult to establish, due to the retrospective design. Five cases, four of which were LTRs, were diagnosed solely by PCR; DFA was negative in two of these. However, the clinical course including response to therapy was consistent with a diagnosis of PCP.
Our study is not powered to rule out risk factors predisposing patients to PCP, in particular rejection therapy, which in other studies was an important risk factor for PCP. Immunosuppressive drugs, especially corticosteroids, could be an important risk factor for PCP that was not identified due to similar high frequency of use between cases and transplant-control subjects. Thus the entire transplant population could have been at risk, but only a minority may have developed it as a result of exposure to the infection.
A major strength of our study is the large number of Pneumocystis-control samples. For 2009 and 2010 the analyzed samples comprised almost all PCP infections at our hospital.
In conclusion, LTRs and RTRs were simultaneously infected in an outbreak of PCP that was predominantly caused by three unique strains of Pneumocystis. Our findings provide strong evidence of an outbreak profile caused by nosocomial transmission.
Materials and Methods
Study population
Rigshospitalet, Copenhagen University Hospital, is a tertiary care hospital in Denmark with a liver transplant program that covers the Danish population of 5.5 million, and a renal transplant program that covers a population of 2.5 million.
By database query, all renal or liver transplant recipients with a positive microscopy or PCR test for P. jirovecii between January 1st 2002 and December 31st 2010 were included in the study if the following criteria were met: 1) Symptoms compatible with PCP [29], 2) Radiographic findings and laboratory values compatible with PCP and 3) Detection of P. jirovecii by direct immunofluorescence microscopy or PCR. Samples examined for P. jirovecii included BAL (n=28), tracheal aspirations (n=1) and oral wash (n=1). Twenty-five of 27 samples tested by direct immunofluorescence microscopy were positive. In five cases a single-round touchdown PCR test, using previously described methods, was the only positive assay [30].
Based on these definitions, two RTRs, who were PCR positive but microscopy negative, and whose clinical course was not consistent with PCP, were excluded from the transplant-control analysis, but included as colonized RTRs in the genotype analysis.
One combined liver and renal transplant recipient is considered a liver transplant in our analysis. Guidelines of the Danish Health and Medicines Authority and the National Institutes of Health (NIH) were followed in the conduct of these studies.
Genotyping
Pneumocystis specimens from cases and Pneumocystis-controls were retrieved from stored samples at the Biobank of the Department of Clinical Microbiology and Statens Serum Instititut. Pneumocystis-controls were defined before genotyping as patients without renal or liver transplants who were diagnosed with PCP at Rigshospitalet during the same years as cases.
Blinded genotype analysis was done at the NIH, Bethesda, MD, USA. The method for RFLP analysis of the msg gene has been previously described [13, 19]. Briefly, DNA was extracted using the QIAamp DNA mini Kit (Qiagen), msg gene copy number was quantified by a real-time quantitative PCR, and a minimum of approximately 1000 msg gene copies per RFLP PCR reaction were amplified using semi-nested PCR with primers GK 472, GK 452 and GK 195, as described [19]. Purified PCR products were treated with restriction enzymes Dra1 or Hpy188I. Analysis of the digested fragments was done by agarose gel electrophoresis [13, 19].
MLST analysis was performed by amplifying and sequencing three regions of the P. jirovecii genome, ITS, 26S, and mt26S, as described [31]. Sequencing a region of the beta-tubulin gene was not found to have discriminatory utility. Regions with any polymorphism in any of the sequences were identified and utilized to compare the isolates. Isolates with one or more nucleotide differences were considered to be different strains.
Transmission maps
To investigate potential transmission events among patients, all admissions and outpatient visits for PCP cases between 2007 and 2010 were retrospectively recorded and analyzed.
Risk factor analysis - matched case-control selection
We used databases at the transplant units to match two control subjects without PCP (transplant-controls) to each case patient. Each transplant-control was matched by 1) type of organ, 2) age (+/− 10 years) and 3) the transplant-controls were matched according to transplantation date and duration of follow-up at the same hospital (Rigshospitalet); thus patients in the control group were at the same risk of exposure as the PCP cases.
Clinical data and definitions
Selected medical and microbiological patient data variables were recorded in an Epidata entry form [32]. Data were cross-validated by repeated recording. Follow up was complete for all patients through December 2010. The interval from transplantation to PCP (time of event), was defined as time to onset of PCP symptoms. For the transplant-controls, we calculated follow up intervals equivalent to the time of event for the matched case (risk date).
Immunosuppression
Standard immunosuppression included induction with basiliximab, daclizumab or thymoglobulin for RTRs and corticosteroids for LTRs, followed by maintenance therapy with calcineurin inhibitors, MMF or azathioprine and corticosteroids. Dosages were adjusted individually according to the unit’s protocol.
Rejection
Graft rejection was graded histologically using the BANFF criteria [33, 34]. Borderline rejections were considered as rejections if treated. In RTRs first-line graft rejection therapy consisted of 250–500 mg prednisolone for 3 days. In cases of steroid resistant rejection, anti-thymocyte globuline was subsequently added. If the rejection was antibody mediated, treatment with plasmapheresis and IVIG, and in some cases rituximab, was initiated. In LTRs, high dose pulse prednisolone (500 mg for 5 days) was used for rejection.
Infections and prophylaxis
Standard CMV prophylaxis consisted of valganciclovir for 3–6 months after transplantation. CMV infection was defined as detection of CMV-DNA by routine real time PCR in a blood sample [35]. For transplant-controls, current CMV infection was defined as infection within 2 months before the risk date; prior CMV infection was >2 months.
Co-morbidity
The Charlson co-morbidity index predicts the ten-year mortality for renal and liver transplant patients, based on a wide range of co-morbid conditions (total of 22 conditions) [36, 37]. Co-morbid conditions were scored according to severity and summed for a total Charlson co-morbidity index score. Postoperative complications were not included as co-morbidity. Diabetes was included in the score if it was diagnosed and treated prior to transplantation.
Other risk factors and definitions
Other risk factors analyzed included blood loss during the liver transplant procedure, cold ischemia time of renal grafts, neutropenia at the time of PCP diagnosis and among liver transplant recipients biomarkers of liver and bile duct injury.
Statistics
Student’s t-test and the Mann-Whitney test were used for comparison of quantitative data for normally and non-normally distributed data, respectively. Differences in proportions were compared using Fisher’s exact test. Statistical significance was defined by a two-sided p-value < 0,05. Statistical analyses were done in Stata (Version 11.2, StataCorp, College Station, Texas).
Acknowledgments
We would like to acknowledge and thank Jørgen Skov Jensen (Statens Serum Institut) for the work done by retrieving microbiology data and samples from PCP cases diagnosed by PCR.
This work was supported by grants from the Danish Kidney Association’s Research Foundation and The Helen and Ejnar Bjørnow Foundation. This research was supported in part by the Intramural Research Program of the National Institutes of Health Clinical Center.
Abbreviations
- CMV
Cytomegalovirus
- DFA
Direct fluorescens antibody
- LTR
Liver transplant recipient
- MLST
Multi locus sequence typing
- MMF
Mycophenolate mofetil
- NIH
National Institutes of Health
- PCP
Pneumocystis Pneumonia
- PCR
Polymerase chain reaction
- RFLP
Restriction fragment length polymorphism
- RTR
Renal transplant recipient
Footnotes
1 A.A.R.1,5: Participated in research design, clinical data collection, data analysis, manuscript preparation and editing. Supported by two independent grants from the Danish Kidney Association’s Research Foundation and The Helen and Ejnar Bjørnow Foundation.
M.S.2: Participated in research design, data analysis, and manuscript editing; performed laboratory research studies.
J.A.L.K.3: Participated in research design, laboratory diagnosis, sample biobank and manuscript editing.
S.S.S.4: Participated in research design, clinical data collection, contributed with a database of renal transplant recipients and manuscript editing.
A.R.5: Participated in research design, clinical data collection and manuscript editing.
C.R.5: Participated in research design, clinical data collection and manuscript editing.
E.G.2: Performed laboratory research studies and data analysis.
C.H.6: Performed laboratory research studies.
G.K.2: Performed laboratory research studies and data analysis.
J.A.K.2: Participated in research design, data analysis, and manuscript editing.
J.H.1: Participated in research design, clinical data collection, data analysis, manuscript preparation and editing. Corresponding author.
Department of Infectious Diseases, Rigshospitalet, Blegdamsvej 9, 2100 Copenhagen
Critical Care Medicine Department, NIH Clinical Center, Bethesda, MD 20892, USA
Centre for Medical Parasitology, Department of Clinical Microbiology, Rigshospitalet, Blegdamsvej 9, 2100 Copenhagen
Department of Nephrology, Rigshospitalet, Blegdamsvej 9, 2100 Copenhagen
Department of Surgical Gastroenterology, Rigshospitalet, Blegdamsvej 9, 2100 Copenhagen
Department of Laboratory Medicine, NIH Clinical Center, Bethesda, MD 20892, USA
None of the authors have any conflict of interest.
References
- 1.Huang L, Cattamanchi A, Davis JL, et al. HIV-associated Pneumocystis pneumonia. Proc Am Thorac Soc. 2011;8(3):294–300. doi: 10.1513/pats.201009-062WR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keely SP, Stringer JR, Baughman RP, Linke MJ, Walzer PD, Smulian AG. Genetic variation among Pneumocystis carinii hominis isolates in recurrent pneumocystosis. J Infect Dis. 1995;172(2):595–8. doi: 10.1093/infdis/172.2.595. [DOI] [PubMed] [Google Scholar]
- 3.Thomas S, Vivancos R, Corless C, Wood G, Beeching NJ, Beadsworth MB. Increasing frequency of Pneumocystis jirovecii pneumonia in renal transplant recipients in the United Kingdom: clonal variability, clusters, and geographic location. Clin Infect Dis. 2011;53(3):307–8. doi: 10.1093/cid/cir329. [DOI] [PubMed] [Google Scholar]
- 4.Neff RT, Jindal RM, Yoo DY, Hurst FP, Agodoa LY, Abbott KC. Analysis of USRDS: incidence and risk factors for Pneumocystis jiroveci pneumonia. Transplantation. 2009;88(1):135–41. doi: 10.1097/TP.0b013e3181aad256. [DOI] [PubMed] [Google Scholar]
- 5.Gordon SM, LaRosa SP, Kalmadi S, et al. Should prophylaxis for Pneumocystis carinii pneumonia in solid organ transplant recipients ever be discontinued? Clin Infect Dis. 1999;28(2):240–6. doi: 10.1086/515126. [DOI] [PubMed] [Google Scholar]
- 6.Orlando G, Tariciotti L, Manzia TM, et al. Ab initio calcineurin inhibitor-based monotherapy immunosuppression after liver transplantation reduces the risk for Pneumocystis jirovecii pneumonia. Transpl Infect Dis. 2010;12(1):11–5. doi: 10.1111/j.1399-3062.2009.00449.x. [DOI] [PubMed] [Google Scholar]
- 7.de Boer MG, de Fijter JW, Kroon FP. Outbreaks and clustering of Pneumocystis pneumonia in kidney transplant recipients: a systematic review. Med Mycol. 2011;49(7):673–80. doi: 10.3109/13693786.2011.571294. [DOI] [PubMed] [Google Scholar]
- 8.Yazaki H, Goto N, Uchida K, Kobayashi T, Gatanaga H, Oka S. Outbreak of Pneumocystis jiroveci pneumonia in renal transplant recipients: P. jiroveci is contagious to the susceptible host. Transplantation. 2009;88(3):380–5. doi: 10.1097/TP.0b013e3181aed389. [DOI] [PubMed] [Google Scholar]
- 9.Gianella S, Haeberli L, Joos B, et al. Molecular evidence of interhuman transmission in an outbreak of Pneumocystis jirovecii pneumonia among renal transplant recipients. Transpl Infect Dis. 2010;12(1):1–10. doi: 10.1111/j.1399-3062.2009.00447.x. [DOI] [PubMed] [Google Scholar]
- 10.Schmoldt S, Schuhegger R, Wendler T, et al. Molecular evidence of nosocomial Pneumocystis jirovecii transmission among 16 patients after kidney transplantation. J Clin Microbiol. 2008;46(3):966–71. doi: 10.1128/JCM.02016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phipps LM, Chen SC, Kable K, et al. Nosocomial Pneumocystis jirovecii pneumonia: lessons from a cluster in kidney transplant recipients. Transplantation. 2011;92(12):1327–34. doi: 10.1097/TP.0b013e3182384b57. [DOI] [PubMed] [Google Scholar]
- 12.de Boer MG, Bruijnesteijn van Coppenraet LE, Gaasbeek A, et al. An outbreak of Pneumocystis jiroveci pneumonia with 1 predominant genotype among renal transplant recipients: interhuman transmission or a common environmental source? Clin Infect Dis. 2007;44(9):1143–9. doi: 10.1086/513198. [DOI] [PubMed] [Google Scholar]
- 13.Sassi M, Ripamonti C, Mueller NJ, et al. Outbreaks of Pneumocystis pneumonia in 2 renal transplant centers linked to a single strain of Pneumocystis: implications for transmission and virulence. Clin Infect Dis. 2012;54(10):1437–44. doi: 10.1093/cid/cis217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Gal S, Damiani C, Rouille A, et al. A cluster of Pneumocystis infections among renal transplant recipients: molecular evidence of colonized patients as potential infectious sources of Pneumocystis jirovecii. Clin Infect Dis. 2012;54(7):e62–71. doi: 10.1093/cid/cir996. [DOI] [PubMed] [Google Scholar]
- 15.Pliquett RU, Asbe-Vollkopf A, Hauser PM, et al. A Pneumocystis jirovecii pneumonia outbreak in a single kidney-transplant center: role of cytomegalovirus co-infection. Eur J Clin Microbiol Infect Dis. 2012;31(9):2429–37. doi: 10.1007/s10096-012-1586-x. [DOI] [PubMed] [Google Scholar]
- 16.Choukri F, Menotti J, Sarfati C, et al. Quantification and spread of Pneumocystis jirovecii in the surrounding air of patients with Pneumocystis pneumonia. Clin Infect Dis. 2010;51(3):259–65. doi: 10.1086/653933. [DOI] [PubMed] [Google Scholar]
- 17.Manoloff ES, Francioli P, Taffe P, Van Melle G, Bille J, Hauser PM. Risk for Pneumocystis carinii transmission among patients with pneumonia: a molecular epidemiology study. Emerg Infect Dis. 2003;9(1):132–4. doi: 10.3201/eid0901.020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tipirneni R, Daly KR, Jarlsberg LG, et al. Healthcare worker occupation and immune response to Pneumocystis jirovecii. Emerg Infect Dis. 2009;15(10):1590–7. doi: 10.3201/eid1510.090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ripamonti C, Orenstein A, Kutty G, et al. Restriction fragment length polymorphism typing demonstrates substantial diversity among Pneumocystis jirovecii isolates. J Infect Dis. 2009;200(10):1616–22. doi: 10.1086/644643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helweg-Larsen J, Tsolaki AG, Miller RF, Lundgren B, Wakefield AE. Clusters of Pneumocystis carinii pneumonia: analysis of person-to-person transmission by genotyping. QJM. 1998;91(12):813–20. doi: 10.1093/qjmed/91.12.813. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro R, Young JB, Milford EL, Trotter JF, Bustami RT, Leichtman AB. Immunosuppression: evolution in practice and trends, 1993–2003. Am J Transplant. 2005;5(4 Pt 2):874–86. doi: 10.1111/j.1600-6135.2005.00833.x. [DOI] [PubMed] [Google Scholar]
- 22.Arichi N, Kishikawa H, Mitsui Y, et al. Cluster outbreak of Pneumocystis pneumonia among kidney transplant patients within a single center. Transplant Proc. 2009;41(1):170–2. doi: 10.1016/j.transproceed.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 23.Reichenberger F, Dickenmann M, Binet I, et al. Diagnostic yield of bronchoalveolar lavage following renal transplantation. Transpl Infect Dis. 2001;3(1):2–7. doi: 10.1034/j.1399-3062.2001.003001002.x. [DOI] [PubMed] [Google Scholar]
- 24.Radisic M, Lattes R, Chapman JF, et al. Risk factors for Pneumocystis carinii pneumonia in kidney transplant recipients: a case-control study. Transpl Infect Dis. 2003;5(2):84–93. doi: 10.1034/j.1399-3062.2003.00018.x. [DOI] [PubMed] [Google Scholar]
- 25.de Boer MG, Kroon FP, le Cessie S, de Fijter JW, van Dissel JT. Risk factors for Pneumocystis jirovecii pneumonia in kidney transplant recipients and appraisal of strategies for selective use of chemoprophylaxis. Transpl Infect Dis. 2011;13(6):559–69. doi: 10.1111/j.1399-3062.2011.00645.x. [DOI] [PubMed] [Google Scholar]
- 26.Branten AJ, Beckers PJ, Tiggeler RG, Hoitsma AJ. Pneumocystis carinii pneumonia in renal transplant recipients. Nephrol Dial Transplant. 1995;10(7):1194–7. [PubMed] [Google Scholar]
- 27.Kasiske BL, Zeier MG, Chapman JR, et al. KDIGO clinical practice guideline for the care of kidney transplant recipients: a summary. Kidney Int. 2010;77(4):299–311. doi: 10.1038/ki.2009.377. [DOI] [PubMed] [Google Scholar]
- 28.Trotter JF, Levi M, Steinberg T, Lancaster J. Absence of Pneumocystis jiroveci pneumonia in liver transplantation recipients receiving short-term (3-month) prophylaxis. Transpl Infect Dis. 2008;10(5):369–71. doi: 10.1111/j.1399-3062.2008.00318.x. [DOI] [PubMed] [Google Scholar]
- 29.Fujii T, Nakamura T, Iwamoto A. Pneumocystis pneumonia in patients with HIV infection: clinical manifestations, laboratory findings, and radiological features. J Infect Chemother. 2007;13(1):1–7. doi: 10.1007/s10156-006-0484-5. [DOI] [PubMed] [Google Scholar]
- 30.Helweg-Larsen J, Jensen JS, Benfield T, Svendsen UG, Lundgren JD, Lundgren B. Diagnostic use of PCR for detection of Pneumocystis carinii in oral wash samples. J Clin Microbiol. 1998;36(7):2068–72. doi: 10.1128/jcm.36.7.2068-2072.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nahimana A, Blanc DS, Francioli P, Bille J, Hauser PM. Typing of Pneumocystis carinii f. sp. hominis by PCR-SSCP to indicate a high frequency of co-infections. J Med Microbiol. 2000;49(8):753–8. doi: 10.1099/0022-1317-49-8-753. [DOI] [PubMed] [Google Scholar]
- 32.MLJB . EpiData Entry. Odense, Denmark: The EpiData Association; 2008. Version 3.1 ed. A comprehensive tool for validated entry and documentation of data. [Google Scholar]
- 33.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8(4):753–60. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- 34.Banff schema for grading liver allograft rejection: an international consensus document. Hepatology. 1997;25(3):658–63. doi: 10.1002/hep.510250328. [DOI] [PubMed] [Google Scholar]
- 35.Ljungman P, Griffiths P, Paya C. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin Infect Dis. 2002;34(8):1094–7. doi: 10.1086/339329. [DOI] [PubMed] [Google Scholar]
- 36.Jassal SV, Schaubel DE, Fenton SS. Baseline comorbidity in kidney transplant recipients: a comparison of comorbidity indices. Am J Kidney Dis. 2005;46(1):136–42. doi: 10.1053/j.ajkd.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Volk ML, Hernandez JC, Lok AS, Marrero JA. Modified Charlson comorbidity index for predicting survival after liver transplantation. Liver Transpl. 2007;13(11):1515–20. doi: 10.1002/lt.21172. [DOI] [PubMed] [Google Scholar]