Abstract
The recent growth in single molecule studies of translation has provided an insight into the molecular mechanism of ribosomal function. Single molecule fluorescence approaches allowed direct observation of the structural rearrangements occurring during translation and revealed dynamic motions of the ribosome and its ligands. These studies demonstrated how ligand binding affects dynamics of the ribosome, and the role of the conformational sampling in large-scale rearrangements intrinsic to translation elongation. The application of time-resolved cryo-electron microscopy revealed new conformational intermediates during back-translocation providing an insight into ribosomal dynamics from an alternative perspective. Recent developments permitted examination of conformational and compositional dynamics of the ribosome in real-time through multiple cycles of elongation at the single molecule level. The zero-mode waveguide approach allowed direct observation of the compositional dynamics of tRNA occupancy on the elongating ribosome. The emergence of single molecule in vivo techniques provided insights into the mechanism and regulation of translation at the organismal level.
Introduction
The study of translation has been revolutionized over the past decade by the remarkable structural biology on the ribosome and its ligand complexes. These structures have revealed how the ribosomal particles assemble from RNA and proteins, how transfer RNA ligands are bound and suggested mechanisms for catalysis of peptide bond formation(Reviewed in [1–3]). Recent structures have shown how GTPase factors (EF-Tu, EF-G) interact with the 50S subunit to stimulate their enzymatic activity to enhance function in tRNA delivery or translocation of tRNA-mRNA complexes with respect to the ribosome[4–8]. Together with biochemical and mechanistic data, a coherent structural mechanism for the basic steps of translation can be outlined. Yet despite this progress, dynamic data are needed to link these structural pictures to a time-resolved pathway of translation.
Single-molecule fluorescence methods have provided rich detail on the dynamics of ligand-ligand, ligand-ribosome, and ribosomal particle dynamics. By using fluorescence resonance energy transfer (FRET) on single ribosomes, conformational dynamics have been probed on various ribosomal complexes containing fluorescent labels. Original experiments focused on dynamic pathway of tRNA delivery to the ribosome, and subsequent tRNA dynamics within the hybrid state after peptide bond formation. More recent experiments have mapped dynamics between ribosomal proteins and tRNA, ribosomal proteins and factors, and within the ribosome particle itself. Here we review recent progress in single-molecule fluorescence investigations of translation. We emphasize the interplay of structural and dynamic data through the application of electron microscopy, and recent experiments that allow tracking of translation in real time both in vitro and in vivo.
Structural approaches to resolve conformational dynamics
Cryo electron microscopy and X-ray crystallography have obtained static structural snapshots of ribosomes in a variety of functionally-relevant conformations [9–13], whereas molecular dynamics simulations have suggested the molecular motions involved in the transition of the ribosome between these distinct states [14–17]. Recently, several groups have combined time-resolved methods with single-particle EM to provide experimental conformational trajectories. Fischer et al. [18] investigated the dynamics of translocation using time-resolved electron microscopy. Their study characterized the global conformational changes associated with thermally-driven ribosomal back-translocation. Ribosomes assembled in a post-translocation state, with cognate deacylated E-site tRNAfMet and P-site fMet-Val-tRNAVal (E/P state), were allowed to convert spontaneously to their A/P states. The experimentally identified ribosome configurations included ones consistent with the previously observed “classical” and “hybrid” states. In addition, a number of intermediate and previously unobserved sub-states were identified (Figure 1 and 2). Kinetic analysis indicated that the rate-limiting step for back-translocation (measured at 0.8 min−1 under these conditions) is the conversion of the post-translocation complex to the pre-translocation complex.
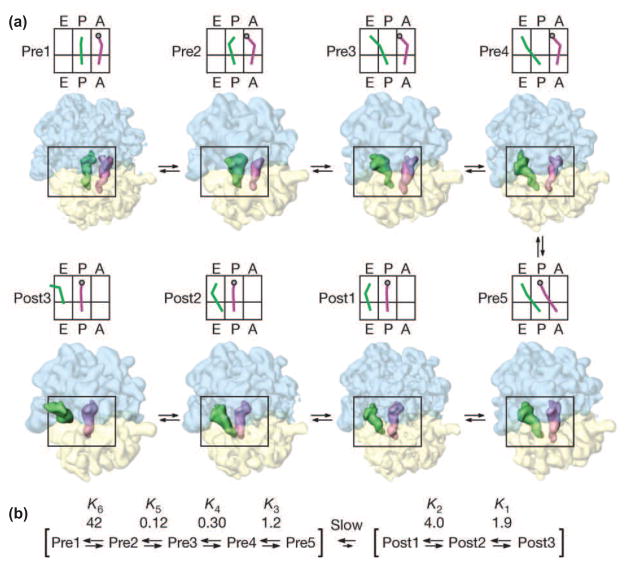
Figure 1.
(a) Conformations of the (retro-)translocating ribosome identified by cryo-EM. The individual complexes are classified by tRNA position. A reconstruction of a representative sub-state from the ensemble of sub-states for each pre- and post-translocational state (pre1 to pre5 and post1 to post3) is shown, along with a schematic representation of tRNA position. (b) Thermodynamics of translocation based on population analysis of the states identified by cryo-EM. The data indicate that conversion globally from the post- to pre-translocation state, while sub-states within the post- and pre- categories are in rapid equilibrium with each other. Reproduced with permission from Fischer et al., 2010.
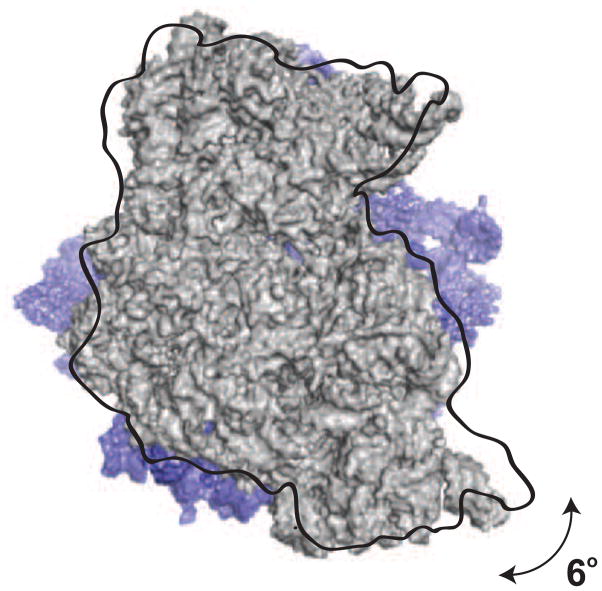
Figure 2. Intersubunit rotation.
Cryo-EM and X-ray crystallography methods revealed ribosomes in distinct conformations that differ by the relative orientation of the ribosomal subunits. 70S ribosome from is shown in top down view from the 30S subunit. 50S is shown in dark blue and 30S in un-rotated state is shown in light grey. 30S in rotated state is shown with the black outline. During elongation cycle 30S subunit rotates ~ 6° relative to the large subunit. Upon aminoacyl-tRNA selection and peptidyl transfer the 30S subunit rotates clockwise. The ribosome rotates counterclockwise upon translocation.
The study also illustrated that tRNA and ribosome motion, particularly 30S rotation, are coupled in the thermally-driven back-translocation process. The energy landscape for these coupled motions was shown to be relatively flat, leading to the conclusion that the overall process involves rapid sampling of the various conformational sub-states by the translocating ribosome. The data suggest that a number of parallel dynamic pathways exist by which translocation can occur. The study also showed that rotation of the 30S subunit, which impacts on tRNA motion, was more dynamic at 37 °C. The results were interpreted to provide further evidence that the ribosome acts as a Brownian ratchet. However, the influence of EF-G on the distribution and accessibility of the individual conformational sub-states was not investigated and awaits future study.
Williamson and co-workers used cryoEM to track the assembly of the 30S subunit from ribosomal RNA and proteins [19]. Using a method called discovery single particle analysis, they initiated assembly of 30S subunits, and rapidly trapped intermediates at different time points (1–100min) through negative staining. They could structurally classify the different assembly intermediates, and compare the observed conformations with their prior assembly kinetics and thermodynamics measurements. The results showed a heterogeneous, parallel mechanism of subunit assembly, and demonstrate the power of integrated biophysical and structural imaging of the translation machinery.
Probing conformational dynamics of the ribosome with smFRET
Conformational changes within the ribosome have been measured using single-molecule FRET. The efficiency of FRET is highly sensitive to the distance (1/R6, where R is inter-dye transition dipole distance) between the fluorophores in a range of 20–80Å within the >200Å ribosomal particle. FRET is thus well suited to monitoring the conformation and structural dynamics of the ribosome and factors during translation. The ability of single-molecule FRET methods to observe stochastic events provides a window onto dynamics otherwise invisible in bulk experiments. One powerful example involved experiments that measured rapid tRNA fluctuations in the A-site during tRNA selection [20,21]. The cognate tRNA has a reduced dissociation rate over near-cognate tRNAs from the A-site both before and after GTP hydrolysis by EF-Tu. Accordingly, rapid conformational sampling by tRNAs ensure that near-cognate tRNAs are quickly shuttled into unstable binding conformations, thereby only selecting the cognate tRNA for accommodation.
A variety of structural perspectives have explored conformational dynamics within the ribosome during the pre- and post-translocational phases of translation. There is general agreement that prior to translocation the ribosome shifts into an unlocked state characterized by rapid tRNA fluctuations between a classical and two hybrid tRNA configurations [22], and by dynamic movements of the subunits [23]. The translational machinery modulates the equilibria of these fluctuations to guide the ribosome and tRNAs into a desirable configuration for translocation. Wang et al. demonstrated that EF-G fluctuates between two binding conformations on the ribosome [24]. Such fluctuations only occur when the ribosome is in the pre-translocation state; this suggests that ribosome unlocking allows EF-G to access conformational states not otherwise available. There is additional evidence that EF-G binding to the unlocked ribosome shifts the equilibria of tRNA dynamics, perhaps with the help of L1 stalk movements, towards the hybrid conformations [25]. Furthermore, aminoglycosides, which shift the equilibrium of tRNA conformations back towards the classical state, decrease the EF-G-catalyzed translocation rate [26]. Taken together, these results suggest that EF-G, beyond driving translocation through GTP hydrolysis, also sets the stage for translocation by guiding the tRNAs into hybrid conformations favorable for translocation.
In the unlocked state, the ribosome becomes highly dynamic and these fluctuations must converge into the correct configuration to prepare for translocation[25,27,28]. Cornish et al. [29] observed 3 distinct conformational states of the L1 stalk and that the occupancy of the 50S tRNA E-site determines which of those 3 states are accessible to the L1 stalk. Such a finding suggests that at least some of these conformational changes are in fact linked and is consistent with the cryo-EM and X-Ray data and molecular dynamics simulations.
To investigate the conformational dynamics of translation termination, Sternberg et al. employed FRET between P-site tRNA and RF1 (release factor 1) to probe how release factors alter the equilibrium between two global ribosomal states that strongly resemble the locked and unlocked states during elongation [30]. Thus RF1 stabilizes the state analogous to a locked ribosome while RF3 binding favors the opposite state, suggesting that global conformational rearrangements of the ribosome are a central mechanism exploited throughout all phases of translation by different initiation, elongation, and release factors.
Real-time tracking of ribosomal via inter-subunit smFRET
Conformational rearrangements are a central theme in the dynamics of translation, underscoring the importance of a fluorescence signal that monitors the global conformational changes of the ribosome. Using specifically labeled 30S-Cy3 and 50S-Cy5 subunits [31], Marshall et al. characterized an inter-subunit FRET signal that reports on the global conformations of the ribosome and successfully applied that signal to study the role of GTP hydrolysis by IF2 upon subunit joining [32]. The authors demonstrated that IF2 accelerates subunit joining and that GTP hydrolysis by IF2 guides ribosomes joined in an unproductive low FRET state into an elongation-competent high FRET state.
The intersubunit FRET signal alternates between a high FRET state and a low FRET state on an elongating ribosome [33]. The transition from the high to the low FRET state occurs upon peptide-bond formation and is consistent with unlocking of the ribosome from the tightly locked state that preserves reading frame on the mRNA to a highly dynamic state in preparation for translocation [22,23,27,29,34]. The subsequent reverse transition from the low FRET state back to the high FRET state requires GTP hydrolysis by EF-G, but not EF-G dissociation, and likely corresponds to the re-locking of the ribosome upon translocation, where the local conformational dynamics of the ribosome are suppressed. Both FRET transitions do not occur spontaneously, thus indicating that the ribosome harnesses the free energies of peptidyl transfer and GTP hydrolysis respectively for two rearrangements to translocate during elongation [33]. Thus, the inter-subunit FRET signal provides a method to track global ribosomal conformation during the elongation cycle.
Aitken et al. used the inter-subunit FRET signal to monitor multiple rounds of elongation (Figure 3a and 3b) [35]. The maximum number of observed high-low-high FRET cycles produced by a single elongating ribosome corresponds to the number of codons in the translated mRNA (Figure 3c). Withholding a necessary tRNA ternary complex prevents progression past the cognate codon and arrests the FRET cycling. Therefore a cycle of high-low-high FRET transitions corresponds to a successful cycle of elongation, and multiple elongation cycles can be accurately tracked.
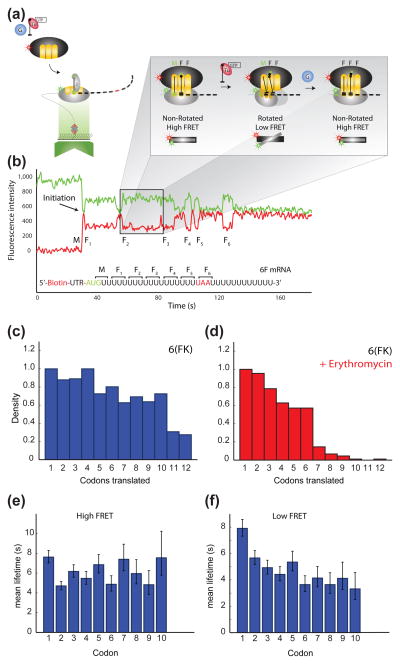
Figure 3. Intersubunit FRET.
(a) Single-molecule translation assay. Cy5-labeled 50S subunits, ternary complexes and EF-G are delivered to surface-immobilized Cy3-labeled 30S PICs. (b) Immobilization with an mRNA coding for six phenylalanines (6F) permits the observation of ribosome conformation during multiple rounds of elongation via the intersubunit FRET signal. The arrival of FRET corresponds to 50S subunit joining during initiation and is followed by multiple cycles of high-low-high FRET, each reporting on ribosome unlocking and locking during one round of elongation. (c) Number of FRET cycles observed on mRNAs encoding six alternating FK pairs. (d) Erythromycin stalls single translating ribosomes by blocking the nascent chain at codon 7. Mean lifetime estimates for the (e) high-FRET (locked) and (f) low-FRET (unlocked) states of the ribosome at the first ten codons of the 12F mRNA. Error bars denote 95% confidence intervals from single-exponential fits. There is an apparent increase in the translocation rate beyond initial codons, as evidenced by the decreasing low FRET (unlocked) state lifetime. Reproduced with permission from Aitkien et al., 2010.
The inter-subunit FRET signal provides a powerful method to monitor the global conformational dynamics of an elongating ribosome codon-by-codon in real-time. The authors showed that ribosomal translocation rates, as revealed by the low FRET lifetimes at each codon, increase as the ribosome translates the first several codons (Figure 3e and 3f). These data suggest improved processivity of translation as the ribosome moves from initiation to elongation, assuming a similar dissociation rate of translation complexes form mRNAs during translation. Thus, the early steps of ribosomal translocation are particularly slow, and could represent a point of potential regulation. The mechanistic origins for slow translocation at the initial codons is not due to disruption of Shine-Dalgarno pairing, but may be related to the length or chemical identity of the growing nascent chain. Further investigation is required.
The mechanisms of ribosome-targeting antibiotics have been explored by these single-molecule fluorescence methods. Fusidic acid, spectinomycin, and viomycin, effectively inhibited translation of multiple codons, and all showed distinct and codon-specific effects on the lifetimes of the FRET states [35]. The observed effects agree well with the expected mechanisms of the antibiotics and the cumulative evidence that these drugs act during elongation. Additionally, the authors observed a significant reduction in the number of ribosomes undergoing more than 6 FRET cycles in the presence of erythromycin, providing direct, real-time evidence that erythromycin blocks the peptide exit tunnel at a position where a polypeptide of 7 amino acids would reach (fMet plus 6 additional amino acids) (Figure 3c and 3d). The inter-subunit FRET signal will allow mechanistic exploration of many ribosome-directed antibiotics during real-time translation.
Full translation in zero-mode waveguides
New approaches have allowed direct tracking of translation under physiological conditions. Zero-mode waveguides (ZMWs) are an emerging platform for investigation of processes using single-molecule fluorescence [36,37]. Uemura et al. have employed new ZMW technology to monitor timing of arrival and departure of tRNAs in real time on the ribosome [38]. Traditional total internal reflection fluorescence (TIRF) microscopy limits fluorescently labeled tRNA concentrations to ~50nM due to background fluorescence from unbound labeled tRNA (Figure 4a). When tRNA is present at such low concentrations, the long delays between sequential tRNA arrival events can cause molecules to become photobleached, rendering them undetectable. The signals of interest consequently vanish prematurely during translation at rates supported by TIRF microscopy. Additionally, artificially long waiting times allow the ribosome to explore stable non-productive states.
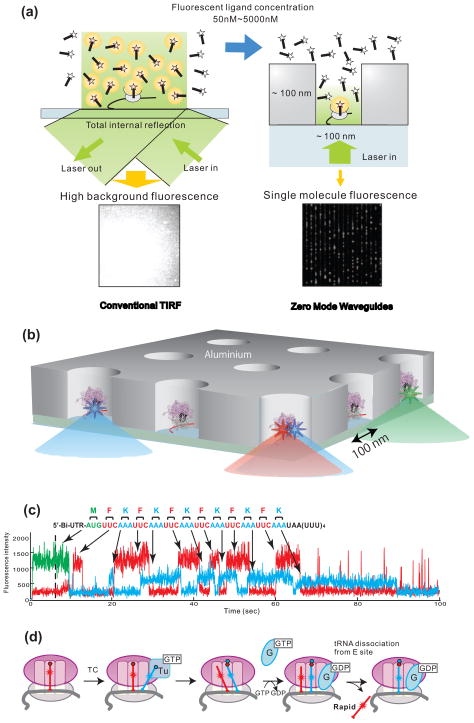
Figure 4. Visualization of single molecule translation in ZMWs.
(a) Comparison between conventional TIRF and use of zero-mode waveguides (ZMWs). At concentrations of fluorescently labeled ligands higher than ~50 nM, spots are not distinguishable due to high background fluorescence (left). ZMWs overcome this limitation (right) by decreasing illumination volume by three to four orders of magnitude compared to TIR excitation to an order of zeptoliters (10−21 L). This allows fluorescence detection in the μM range. (b) Schematic illustration of ZMW construction. Ribosomal complexes are specifically immobilized in the bottom of derivatized ZMWs using biotinylated mRNAs. Initial ribosome complex immobilized to the bottom surface of a ZMW contains Cy3-labeled fMet-tRNAfMet
(c) Fluorescence is excited by illumination at 488, 532 and 642 nm, and Cy2, Cy3 and Cy5 fluorescence are simultaneously detected. Heteropolymeric mRNAs encoding 13 amino acids were used: M(FK)6. Translation was observed in the presence of Phe-(Cy5)tRNAPhe, Lys-(Cy2)tRNALys complexed with EF-Tu and EF-G as a series of fluorescent pulses that mirror the mRNA sequence. A long Cy2 pulse is observed upon arrival of the ribosome at the stop codon. (d) Model for tRNA transit through the ribosome during translation. Blue-labeled tRNA arrives in the A-site, resulting in a 2-tRNA state with a red P-site-bound tRNA. This state forms an A/P hybrid state after peptide bond formation. Subsequent translocation of the 2 tRNA-mRNA complexes to the E and P sites is followed by rapid dissociation of red E-site tRNA. Reproduced with permission from Uemura et al., 2010.
ZMWs provides an elegant solution to these problems. Each ZMW consists of a ~50–200 nm diameter nanofabricated hole in metal film that restricts excitation light to a zeptoliter volume, making possible experiments with micromolar concentrations of fluorescently labeled ligands (Figure 4b). Recent advances in nanofabrication [39], surface chemistry [40], and detection instrumentation [41] have permitted direct monitoring of DNA polymerization in ZMWs [37]. Uemura et al. applied this approach to the study of translation. 70S initiation complexes were assembled containing Cy3 labeled N-formylmethionyl tRNAfMet and 5′-biotinylated mRNA, containing the Shine-Dalgarno sequence, an initiation codon followed by a coding sequence of 12 codons and terminated by a stop (UAA) codon. The complexes were immobilized in the ZMW via biotin-neutravidin interactions. Elongation factors and Cy5-labeled phenylalanyl-tRNAPhe and Cy2-labeled lysyl-tRNALys, which are cognate to the codons comprising the mRNA coding sequence were co delivered to the slide surface. At the start of the experiment, only the fMet-(Cy3)tRNA signal is detected. As successive codons are decoded by the ribosome the observed fluorescence signals report on the identity of the arriving tRNAs, the timing of events, and the number and identity of the tRNA occupancy on the immobilized ribosome (Figure 4c). The identity and order of arrival of elongator tRNAs illustrated directly the sequence-dependent behavior of translation. Detailed analysis also answered a long-standing question by demonstrating that tRNA release from the E site is uncoupled with the next tRNA arrival to the A site (Figure 4d) [38].
Another important conclusion from this ZMW work is how extensible the technology is to diverse biological systems. ZMW-based experiments are ideally tuned to explore the timing of labeled component binding and dissociation events while following the progress of a ribosome along a mRNA. The approach has been extended by us to prokaryotic translation initiation and eukaryotic translation, as well as to investigating the effects on translation of drugs targeting the ribosome. With the widespread availability of fluorescent reagents, we envisage that this technology can be applied effectively in a wide range of scientific fields.
in vivo Single molecule approaches
The emergence of in vivo single molecule techniques allows observation of unperturbed biological processes at the level of individual molecules. The Xie group developed methods of detecting individual molecules in living cells [18,42,43]. Observation of individual fluorophores in solution is challenged by diffusion of the molecules over distances greater than the size of a diffraction-limited spot during the observation time. This drawback was overcome using several approaches. First, YFP fusion with membrane protein Tsr was targeted to E. coli cell membrane, where decreased diffusion rates allowed observation of the single fluorophores on the millisecond time scale. Second, fluorescent molecules were observed during short intervals (300 μsec) [44]. Third, the problem could be circumvented by measuring the enzyme activity in bulk with precision that allows quantization of the enzyme copy number with single molecule precision [43]. Analysis of YFP-Tsr expression and measurements of the enzymatic activity of single β-galactosidase enzyme molecules demonstrated the stochastic nature of the protein production at transcription and translation levels in both E. coli and S. cerevisiae. The randomness in frequency and amplitude of the protein production results in ongoing variation of the proteome. When applied to the metabolic enzymes these observations demonstrate how individual cells differentially and continuously sample their environment and how cell populations fluidly adapt to the changes in conditions. These studies illustrated how random events visible only at the single-molecule level contribute to biological function.
Simultaneous genome-wide quantitation of the protein and mRNA levels revealed a lack of correlation between mRNA and protein abundance in the individual cells due to stochastic nature of the processes, fast mRNA degradation and accumulation of the protein over time. On the other hand, the correlation of the protein production in random gene pairs within a single cell demonstrated that relative protein abundance depends on global epigenetic factors such as global transcriptional and translational regulation [44].
Future Perspectives
Regulation of translation occurs predominantly during initiation phase (Reviewed in [45,46]). The mechanism of eukaryotic translational is highly complex and regulated. Recently the positions of ribosomes have been mapped on translating mRNAs, providing a static genome-wide snapshot of translation [47]. Translation initiation is governed by seven initiation factors and involves mRNA 5′-cap recognition, scanning and start codon selection. Alternative initiation pathways allow distinct translational responses. To allow application of the single molecule approaches to investigation of eukaryotic translation, Petrov & Puglisi successfully labeled small and large ribosomal subunits of the yeast ribosome and demonstrated their utility for single molecule fluorescence and force studies [48]. The future application of single-molecule methods will reveal molecular details of the eukaryotic translation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Agirrezabala X, Frank J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q Rev Biophys. 2009;42:159–200. doi: 10.1017/S0033583509990060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••4.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. Authors have determined the crystal structure of 70S:EF-Tu:Trp-tRNATrp:GDPCP complex to 3.2 Å resolution. This structure, and that of the 70S:EF-Tu:ribosomal complex after GTP hydrolysis [5] suggests a universal mechanism for ribosomal GTPase activation and GTP hydrolysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •5.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. Describes structure of the of 70S:EF-Tu:Trp-tRNATrp:GDP complex stalled with kirromycin and paromomycin to 3.6 and 3.8 Å resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci U S A. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Agirrezabala X, Lei J, Bouakaz L, Brunelle JL, Ortiz-Meoz RF, Green R, Sanyal S, Ehrenberg M, Frank J. Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM. EMBO J. 2008;27:3322–3331. doi: 10.1038/emboj.2008.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3. 5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- ••10.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. Upon EF-G binding to pre-translocating ribosomes 30S ribosomal subunit undergo ~ 6° rotational movement relative to 50S. The rotation is accompanied by ~ 20 Å movement of the L1 stalk. The observed motion provides insight into global ribosomal dynamics during elongation. [DOI] [PubMed] [Google Scholar]
- 11.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2. 4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 12.Agrawal RK, Penczek P, Grassucci RA, Burkhardt N, Nierhaus KH, Frank J. Effect of buffer conditions on the position of tRNA on the 70 S ribosome as visualized by cryoelectron microscopy. J Biol Chem. 1999;274:8723–8729. doi: 10.1074/jbc.274.13.8723. [DOI] [PubMed] [Google Scholar]
- 13.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5. 5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 14.Whitford PC, Geggier P, Altman RB, Blanchard SC, Onuchic JN, Sanbonmatsu KY. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA. 2010;16:1196–1204. doi: 10.1261/rna.2035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanbonmatsu KY. Energy landscape of the ribosomal decoding center. Biochimie. 2006;88:1053–1059. doi: 10.1016/j.biochi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Sanbonmatsu KY. Alignment/misalignment hypothesis for tRNA selection by the ribosome. Biochimie. 2006;88:1075–1089. doi: 10.1016/j.biochi.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci U S A. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••18.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. Dynamic snapshots of the 70S ribosome during retro-translocation were obtained by time-resolved electron cryo-microscopy, identifying a number of previously unobserved pre- and post-translocation sub-states. 30S-tRNA interactions were shown to be important for control of global translocation dynamics. [DOI] [PubMed] [Google Scholar]
- ••19.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. Authors employed time-resolved electron microscopy to elucidate pathways of 30S subunit assembly. The results showed that 30S assembly occurs via multiple compositional pathways. This study demonstrates the utility of structural imaging for resolving alternative pathways in translation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci U S A. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geggier P, Dave R, Feldman MB, Terry DS, Altman RB, Munro JB, Blanchard SC. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol. 2010;399:576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••23.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. The intersubunit rotation was observed by utilizing single molecule spectroscopy. After peptidyl transfer ribosomes undergo spontaneous oscillations between two states. The observed fluctuations correspond to the hybrid and classical states. EF-G binding stabilizes rotational conformation of the ribosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Qin H, Kudaravalli RD, Kirillov SV, Dempsey GT, Pan D, Cooperman BS, Goldman YE. Single-molecule structural dynamics of EF-G--ribosome interaction during translocation. Biochemistry. 2007;46:10767–10775. doi: 10.1021/bi700657d. [DOI] [PubMed] [Google Scholar]
- ••25.Fei J, Bronson JE, Hofman JM, Srinivas RL, Wiggins CH, Gonzalez RL., Jr Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci U S A. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. Authors observed dynamics of the L1 stalk fluctuation during translocation. Together with [10,29,27] this paper highlights the role of ribosomal dynamics during large-scale ribosomal movements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat Chem Biol. 2009;6:54–62. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2009;29:770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •29.Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci U S A. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. By employing smFRET authors demonstrated that L1 stalk could adopt three distinct conformational states. Findings suggest that L1 stalk movement modulates tRNA movement and dissociation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sternberg SH, Fei J, Prywes N, McGrath KA, Gonzalez RL., Jr Translation factors direct intrinsic ribosome dynamics during translation termination and ribosome recycling. Nat Struct Mol Biol. 2009;16:861–868. doi: 10.1038/nsmb.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorywalska M, Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall RA, Aitken CE, Puglisi JD. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell. 2009;35:37–47. doi: 10.1016/j.molcel.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •33.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. The authors observed global locking and unlocking of the ribosome via inter-subunit FRET. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HD, Puglisi JD, Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••35.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. The authors employed intersubunit ratcheting signal to monitor global ribosomal dynamics throughout initiation followed by multiple elongation cycles. Multiple continuous cycles of elongation were observed at the single molecule level in real-time. The signal was employed to distinguish the mechanisms of ribosome-targeting antibiotics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 37.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- ••38.Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. The authors utilized a ZMW approach to observe multiple rounds of tRNA transit on single translating ribosomes in real-time. The number of tRNA molecules simultaneously bound to each ribosome was directly determined by observing fluorescently labeled tRNAs. The codon identity and number of translated codons could be directly inferred from the tRNA signals. The authors demonstrated that E-site tRNA release is uncoupled from A-site tRNA binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foquet M, Samiee KT, Kong X, Chauduri BP, Lundquist PM, Turner SW, Freudenthal J, Roitman DB. Improved fabrication of zero-mode waveguides for single-molecule detection. Journal of Applied Physics. 2008;103:034301. [Google Scholar]
- 40.Korlach J, Marks PJ, Cicero RL, Gray JJ, Murphy DL, Roitman DB, Pham TT, Otto GA, Foquet M, Turner SW. Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures. Proc Natl Acad Sci U S A. 2008;105:1176–1181. doi: 10.1073/pnas.0710982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundquist PM, Zhong CF, Zhao P, Tomaney AB, Peluso PS, Dixon J, Bettman B, Lacroix Y, Kwo DP, McCullough E, et al. Parallel confocal detection of single molecules in real time. Opt Lett. 2008;33:1026–1028. doi: 10.1364/ol.33.001026. [DOI] [PubMed] [Google Scholar]
- 42.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 43.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- ••44.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. The authors quantitatively measured proteome-wide transcription and translation in vivo with single molecule precision. The authors demonstrated how stochastic biological processes contribute to the rapid cell response to the external stimuli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livingstone M, Atas E, Meller A, Sonenberg N. Mechanisms governing the control of mRNA translation. Phys Biol. 2010;7:021001. doi: 10.1088/1478-3975/7/2/021001. [DOI] [PubMed] [Google Scholar]
- 46.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••47.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. Ribosome position on mRNA was determined with nucleotide resolution. The genome-wide approach provides a snapshot of the ribosome position on actively translated mRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •48.Petrov A, Puglisi JD. Site-specific labeling of Saccharomyces cerevisiae ribosomes for single-molecule manipulations. Nucleic Acids Res. 2010;38:e143. doi: 10.1093/nar/gkq390. Large and small subunits of eukaryotic ribosome were specifically labeled. The authors demonstrated utility of the labeled ribosomes for single molecule spectrometry and force experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]