Abstract
Regulatory mechanisms to prevent centriole overduplication during the cell cycle are not completely understood. In this issue, FBXW5 is shown to control the degradation of the centriole assembly factor HsSAS-6. Moreover, the study proposes that FBXW5 is a substrate of both PLK4 and APC/C, two established regulators of centriole duplication.
Centrioles are fundamental for the assembly of microtubule-based structures, including cilia, flagella and the centrosome1. Abnormalities in centriole structure and function are associated with a number of diseases, including ciliopathies, obesity, primary microcephaly, male sterility and cancer. Not surprisingly, centriole duplication is exquisitely regulated and coordinated with the cell cycle. Each S phase, only one new centriole (termed a procentriole) is assembled from the lateral surface of the two pre-existing centrioles. The procentriole elongates until early mitosis and remains attached to its parental centriole until they disengage (that is, they lose their orthogonal configuration) in late mitosis. It is only after disengagement that centrioles are able to nucleate procentrioles2. In addition, to ensure centriole ‘copy number control’1, cells tightly regulate the levels and activity of centriole assembly factors.
PLK4, a divergent member of the Polo-like kinase family, is considered the master regulator of centriole copy number. Its levels and activity correlate with centriole number, both in model organisms and mammalian cells3. Depletion of PLK4 leads to the loss of centrioles with successive cell divisions, while, remarkably, overexpression of PLK4 produces multiple bona fide procentrioles around the parental centriole, in a rosette configuration3. Several assembly factors are required downstream of PLK4, including CPAP, CEP135, CEP152, CP110 and HsSAS-6 (ref. 4). Of these proteins, HsSAS-6 is recruited at the earliest stage of centriole formation5, and fascinating recent work suggests that it self-assembles into coiled-coil-containing oligomers that are critical for the initial steps of centriole structure formation6,7. Similarly to PLK4, HsSAS-6 is rate-limiting for centriole duplication, and its overexpression can also induce the formation of multiple procentrioles8.
The anaphase-promoting complex/cyclosome (APC/C) and the SKP1–CUL1–F-box protein (SCF) complex, two key ubiquitin ligase families involved in progression through the cell cycle, regulate the levels of several centrosome duplication factors. The APC/C is regulated by an interaction with either of two co-activating subunits, CDC20 or CDH1, which target substrates during M phase and G1 phase. APC/C–CDH1 induces the degradation of HsSAS-6 in G1 to restrict centriole duplication from occurring too early in the cell cycle8. Other notable APC/C substrates involved in the centrosome cycle include Cyclin A, Aurora A and PLK1. In contrast to the APC/C, which forms only two complexes, the SCF core scaffold can be used to assemble approximately 69 different complexes based on the recruitment, through SKP1, of a variable F-box protein that confers substrate specificity. A fundamental role for the SCF at the centrosome was initially highlighted by the localization of the core SCF components, SKP1 and CUL1, at interphase and mitotic centrosomes9. Moreover, interfering with SCF function by expression of a dominant-negative CUL1 mutant drives centrosome overduplication both in cell systems and a mouse model10. Specific F-box proteins implicated in the regulation of centrosome number include FBXW7, SKP2, Cyclin F (also called FBXO1) and βTrCP (see Fig. 1). FBXW7 and SKP2 regulate the levels and activity of Cyclin E, a critical coordinator of the centrosome duplication cycle. FBXW7 directly targets Cyclin E for degradation, whereas SKP2-based regulation of Cyclin E is indirect, through the degradation of p27, an inhibitor of Cyclin E–CDK (cyclin-dependent kinase) complexes. More recently, the F-box protein Cyclin F was shown to induce the degradation of the essential centriole duplication factor CP110, to prevent centriole overduplication in G2 phase11. Finally, βTrCP plays a critical role in the regulation of centriole number by promoting PLK4 turnover. PLK4 trans-autophosphorylation creates a phosphodegron recognized by SCF–βTrCP, establishing an important feedback mechanism to control PLK4 levels and limit centriole duplication to once per division12. Accordingly, βTrcp1−/− mouse embryonic fibroblasts display centrosome overduplication13.
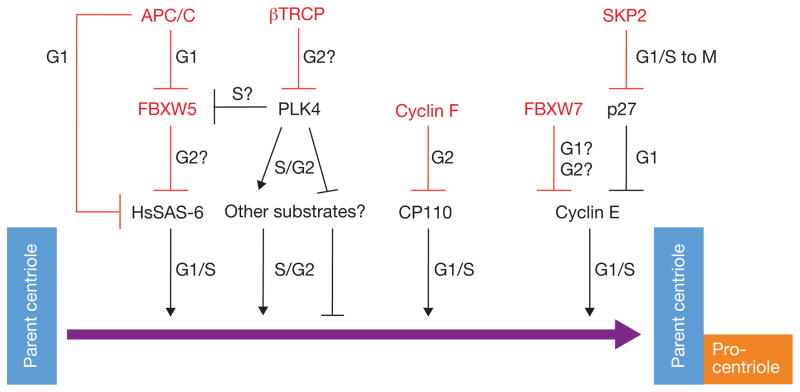
Figure 1.
Positive regulators of centriole duplication are regulated by degradation. In this issue, Malek and colleagues provide evidence for FBXW5-induced degradation of HsSAS-6. FBXW5 activity seems to be inhibited by PLK4-mediated phosphorylation of residue Ser 151 of FBXW5. FBXW5 levels in G1 are kept low by the APC/C, which also induces the degradation of HsSAS-6 in G1 (ref. 8). In addition to FBXW5 and itself, other targets of mammalian PLK4 remain unknown. Cyclin F induces CP110 degradation during G2 to prevent centriole overduplication. Cyclin E levels and activity are controlled directly by FBXW7 and indirectly by SKP2 through regulation of p27, an inhibitor of Cyclin E–CDK complexes. Red lines denote degradation processes. E3 ligases and F-box proteins are indicated in red.
On page 1004 of this issue, Malek and colleagues now show that SCF–FBXW5 is an important negative regulator of centrosome overduplication and propose that this occurs through its regulation of HsSAS-6 levels14. Cells depleted of FBXW5 accumulate supernumerary centrioles, multipolar spindles in mitosis and have increased levels of HsSAS-6. Conversely, the overexpression of FBXW5 inhibits centriole duplication and decreases the half-life of HsSAS-6. FBXW5 activity, and by inference the level of HsSAS-6, is regulated by two separate regulatory circuits. First, FBXW5 levels are kept low during mitosis and G1 by the APC/C. Second, to prevent premature FBXW5-mediated degradation of HsSAS-6 during S phase, the authors propose an intriguing mechanism whereby PLK4-mediated phosphorylation of FBXW5 on Ser 151 protects HsSAS-6 from degradation by inhibiting the ubiquitylating activity of SCF–FBXW5, although how this inhibition occurs is unclear.
FBXW5 depletion can only partially rescue the centriole phenotype caused by depletion of PLK4, so FBXW5 is likely to be one of several important PLK4 substrates involved in centriole duplication. Several other targets of PLK4 phosphorylation that are involved in centriole assembly have been proposed. Of relevance, in Caenorhabditis elegans it was recently found that ZYG-1 (the C. elegans orthologue of PLK4) phosphorylates SAS-6 at Ser 123, and this phosphorylation is critical for centriole duplication15. Perhaps PLK4 is involved in multiple layers of regulation of HsSAS-6, both directly and indirectly through the inhibition of FBXW5. SAS-6 Ser 123 is not a conserved residue in HsSAS-6, suggesting that if HsSAS-6 is a substrate of PLK4 in mammalian cells, the site required for regulation is different. Beyond SAS-6, other proposed substrates of PLK4 include CPAP and CEP152 (ref. 16), both of which are important centriole assembly factors required downstream of PLK4.
A number of important questions remain regarding the pathway proposed by Malek and colleagues. Firstly, how does FBXW5 target HsSAS-6 for degradation? Identification of the degron in HsSAS-6 would provide powerful information about how FBXW5 recognizes its substrates, and it would establish if HsSAS-6 recognition by FBXW5 is phosphorylation dependent, as substrate recognition by F-box proteins generally (although not always11) requires phosphorylation. Such details may reveal additional regulatory mechanisms, such as the kinase pathway for the phosphodegron, and thus information about the spatiotemporal regulation of HsSAS-6 degradation by FBXW5. Secondly, does FBXW5 reside exclusively at the centrosome, and if so, does it localize to a specific centrosomal substructure? The authors were unable to localize FBXW5 at the centriole by standard immunofluorescence microscopy analysis, although they showed that FBXW5 is present in centrosome-enriched fractions. The previously published substrates for FBXW5, TSC1 and TSC2, have no published role in centriole duplication and have not been shown to localize on the centro-some17, which may suggest regulated localization of FBXW5. Thirdly, it will be important for future work to establish the spatiotemporal dynamics of FBXW5 Ser 151 phosphorylation in vivo using phosphospecific antibodies. According to the authors’ model, Ser 151 phosphorylation should occur in the early stages of S phase to ensure that sufficient HsSAS-6 levels are protected for the initiation of centriole duplication. Lastly, it would be interesting to understand the mechanism by which phosphorylation inhibits the ligase activity of SCF–FBXW5 to shed light on the regulation of F-box proteins by post-translational modifications. Does phosphorylation influence binding between HsSAS-6 and FBXW5, or does it affect the catalytic activity of SCF–FBXW5? It will be important to investigate this mechanism in more detail, employing both phosphomimic and phosphodefective forms of FBXW5 in assays for its cell cycle levels, localization, substrate binding etc.
Clearly, tight regulation of the levels of centriole assembly factors is required to maintain the correct number of centrioles within a cell. Defects in this process lead to abnormal numbers of centrioles, a state that is associated with aneuploidy and is a hallmark of many cancer cells. Malek and colleagues have provided insight into a novel regulatory pathway controlling the levels of HsSAS-6 and centriole numbers. Increasing our understanding of the role of proteolysis pathways at the centrosome, such as the interplay between HsSAS-6, FBXW5 and PLK4, will undoubtedly enhance our understanding of the centriole duplication process.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
Contributor Information
Julia Pagan, Department of Pathology, NYU Cancer Institute, New York University School of Medicine, 522 First Avenue, SRB 1107, New York, New York 10016, USA.
Michele Pagano, Email: Michele.Pagano@nyumc.org, Department of Pathology, NYU Cancer Institute, New York University School of Medicine, 522 First Avenue, SRB 1107, New York, New York 10016, USA. Howard Hughes Medical Institute, 4000 Jones Bridge Road Chevy Chase, Maryland 20815-6789, USA.
References
- 1.Nigg EA. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Tsou MF, Stearns T. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 3.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 4.Kleylein-Sohn J, et al. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- 6.Kitagawa D, et al. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Breugel M, et al. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- 8.Strnad P, et al. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freed E, et al. Genes Dev. 1999;13:2242–2257. doi: 10.1101/gad.13.17.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piva R, et al. Mol Cell Biol. 2002;22:8375–8387. doi: 10.1128/MCB.22.23.8375-8387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Angiolella V, et al. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guderian G, Westendorf J, Uldschmid A, Nigg EA. J Cell Sci. 2010;123:2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- 13.Guardavaccaro D, et al. Dev Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 14.Puklowski A, et al. Nat Cell Biol. 2011;13:1004–1009. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- 15.Kitagawa D, Busso C, Fluckiger I, Gonczy P. Dev Cell. 2009;17:900–907. doi: 10.1016/j.devcel.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. J Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, et al. Genes Dev. 2008;22:866–871. doi: 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]