Abstract
To develop a global view of muscle transcriptional differences between older men and women and sex-specific aging, we obtained muscle biopsies from the biceps brachii of young and older men and women and profiled the whole-genome gene expression using microarray. A logistic regression-based method in combination with an intensity-based Bayesian moderated t test was used to identify significant sex- and aging-related gene functional groups. Our analysis revealed extensive sex differences in the muscle transcriptome of older individuals and different patterns of transcriptional changes with aging in men and women. In older women, we observed a coordinated transcriptional upregulation of immune activation, extracellular matrix remodeling, and lipids storage; and a downregulation of mitochondrial biogenesis and function and muscle regeneration. The effect of aging results in sexual dimorphic alterations in the skeletal muscle transcriptome, which may modify the risk for developing musculoskeletal and metabolic diseases in men and women.
Key Words: Aging, Sex, Sarcopenia, Transcription profile.
Degenerative changes in skeletal muscle are associated with an increased risk of physical disability and metabolic diseases such as type 2 diabetes in older individuals. The underlying molecular pathology may include mitochondrial dysfunction, insulin resistance, oxidative stress, proinflammatory state, and reduced neuronal stimulation (1,2). Many of these age-related changes in skeletal muscle appear to be influenced by sex. Although controversy exists regarding how sex influences each aspect of the aging process of skeletal muscle, sex differences have been reported in age-related loss of muscle mass and strength, that is, sarcopenia (3), decline of muscle insulin sensitivity (4,5), alterations in muscle fiber contractile properties and fatigue resistance (6), elevation in oxidative stress (7), the inflammatory response (8,9), as well as in the decline of mitochondrial content and activity (4,10). Thus, it is reasonable to postulate that the aging of muscle is regulated through different mechanisms in men and women (11). A better understanding of the potential sexually dimorphic response to the aging of skeletal muscle would help us gain a greater insight into the different susceptibilities to musculoskeletal and metabolic diseases for men and women, and could thereby facilitate the development of tailored countermeasures to ameliorate aging-related health problems.
The aging of skeletal muscle is complex involving multiple biological processes. Transcript profiling using microarrays is a powerful technique producing an unbiased global view of the molecular events occurring during various physiological states. It has been used to study sex-related differences in gene expression in human skeletal muscle (12,13); however, the impact of aging on sex differences in muscle has rarely been explored.
Therefore, the purpose of the present study was to use microarrays to establish a global transcriptional perspective of the aging process in skeletal muscle of men and women. In order to better differentiate the molecular events due to the biology of aging rather than changes in lifestyle, we chose to study arm muscle. Given that physical activity decreases with aging and most common activities (ie, walking, stair climbing) utilize lower body musculature, using arm muscle, rather than leg muscle may potentially reduce lifestyle-related bias. Indeed, it has been observed that muscle atrophy and loss of muscular strength with aging is greater in lower limbs than in upper limbs. As suggested by Janssen and colleagues (14), a greater influence of aging on lower limbs can be attributed to the age-associated reduction in physical activity. They also suggested that sex differences in skeletal muscle are more striking in the upper limbs (14). As such, studying arm muscle provided us more power to detect sex-specific biological processes associated with the aging of skeletal muscle.
By employing bioinformatic analytical methods that emphasize sensitivity, that is, an intensity-based Bayesian moderated t statistic (IBMT) (15) and a logistic regression-based method (LRpath) (16), our findings provide new evidence for the existence of a sexual dimorphism in the aging process of skeletal muscle and may provide additional insight into the pathological processes associated with the development of metabolic diseases during aging.
Methods
Participants
Twenty-two young participants were recruited from a large project, Functional SNPs Associated with Human Muscle Size and Strength (17,18). Fourteen young men (n = 7) and women (n = 7) provided resting muscle biopsies from the biceps brachii. Similarly, 20 older men (n = 10) and women (n = 10) were recruited according to the same exclusion criteria, which included: (i) no chronic diseases, (ii) no prior resistance training history, and (iii) no medications or dietary supplements, which may affect musculature. Among them, four older men and four older women volunteered to provide resting muscle biopsies from the biceps brachii. Individuals who had a job or daily activity that required repetitive use of their arms or had participated in weight lifting within 1 year before entering the study were also excluded. All older women were postmenopausal and not on hormone replacement therapy. Participant characteristics for the entire group (n = 42) are presented in Table 1. The human subjects committee approved the study protocol and an informed consent was obtained from all participants before joining the study.
Table 1.
Participant Characteristics
| Older Group | Young Group | |||||
| Women | Men | p | Women | Men | p | |
| n = 10 | n = 10 | n = 11 | n = 11 | |||
| Age (y) | 70.5±1.9* | 73.3±1.2* | ns | 22.7±0.8 | 24.7±1.5 | ns |
| Height (cm) | 157.4±1.8 | 172.7±2.0 | <.001 | 156.2±3.2 | 177.9±2.5 | <.001 |
| Weight (kg) | 66.3±2.6 | 88.6±5.0 | .001 | 60.0±5.2 | 76.9±4.6 | .02 |
| BMI (kg/m2) | 26.8±0.9 | 29.7±1.4* | ns | 23.7±1.7 | 24.2±1.4 | ns |
| Isometric muscle strength (kg) | 15.9±1.7* | 37.0±1.9* | <.001 | 25.3±2.9 | 55.4±4.6 | <.001 |
| n = 8 | n = 7 | n = 11 | n = 11 | |||
| Arm muscle CSA (cm2) | 62.7±5.0 | 69.1±6.5 | ns | 55.4±5.5 | 58.8±6.7 | ns |
| Arm CSA (cm2) | 65.1±5.1 | 72.6±6.5 | ns | 57.7±5.6 | 66.7±4.4 | ns |
| Muscle volume (cm3) | 561.5±42.1 | 620.8±53.7 | ns | 516.9±50.6 | 584.9±40.0 | ns |
| Arm volume (cm3) | 582.2±42.7 | 652.5±53.7 | ns | 537.7±51.4 | 613.1±40.3 | ns |
| Muscle quality (kg/cm2) | 0.25±0.17* | 0.56±0.05* | .001 | 0.47±0.05 | 0.89±0.07 | <.001 |
Note: Data presented as mean ± SE. BMI = body mass index; CSA = cross-sectional area; ns = not significant.
*Significantly different from young women or men.
p, significance value from t test for the comparison between women and men for each age group.
Measurement of Muscle Mass and Strength
Measurement of muscle cross-sectional area was obtained using magnetic resonance imaging and muscle volume was calculated. Details of this test can be found in a previously published article (18). Isometric strength of the elbow flexor muscles of the dominant arm was determined using a specially constructed, modified preacher bench and strain gauge (model 32628CTL, Lafayette Instrument Company, Lafayette, IN). The testing protocol has been previously published elsewhere (18). Briefly, the arm was positioned on the preacher bench with the elbow fixed at 90°. The lever arm of the preacher bench was aligned with the participant’s forearm, and the participant’s elbow was aligned with the center of rotation of the lever arm. The upper arm rested against a padded upper arm support producing an angle of approximately 45° between the trunk and upper arm. In order to isolate the elbow flexor, the participant was required to rest his or her chest against the preacher bench chest support with legs straight, heels on the floor, and the opposite arm resting on the legs. Participants pulled maximally against a fixed resistance. Three maximal contractions were assessed. Each contraction lasted 3 s, and a 1min rest was allowed between contractions. An average of the peak force produced during the three contractions was used as an indicator of the strength of the biceps muscle.
Muscle Collection and RNA Isolation
Muscle tissue samples were obtained from the biceps brachii of the dominant arm of 8 older and 14 young participants for the purpose of studying the muscle transcriptome. The tissue collection was conducted in the morning with participants in the postabsorptive resting state. The participants were instructed to refrain from any exercise using their dominant arm throughout the study. A percutaneous needle biopsy technique using a ¼ inch diameter University College Hospital (UCH) biopsy needle was applied. All biopsy samples were immediately weighed and snap frozen in liquid nitrogen-cooled isopentane, and stored at −80°C for subsequent analyses.
RNA preparation, quality control, and cDNA synthesis have been previously described (13). Briefly, total RNA was extracted from frozen tissue with a polytron homogenizer (Brinkman, Westbury, NY) and Trizol reagent (Invitrogen, Rockville, MD) and purified with an RNeasy kit (Qiagen, Santa Clara, CA). Total RNA was used as a template for double-stranded cDNA synthesis (Superscript Double-Stranded cDNA synthesis kit, Invitrogen, Carlsbad, CA). Biotin-labeled cRNA was generated for microarray hybridization (EnzoBioarray High Yield RNA transcription Labeling Kit, Affymetrix, Santa Clara, CA).
Microarray Analysis
Microarray preparation and data processing were previously described in detail (13). Briefly, biotin-labeled cRNA was hybridized to Affymetrix Human Genome U133 Plus 2.0 arrays according to the manufacturer’s instructions. Following hybridization, the probe arrays were washed and stained. The intensity of bound dye was measured with an argon laser confocal scanner (GeneArray Scanner, Agilent), and the stored images were analyzed using the GeneChip software Microarray Analysis Suite 5.0 (Affymetrix). Overall 54,675 probe sets representing 20,080 annotated genes were profiled.
Cell intensity files were generated using Microarray Analysis Suite 5.0. The array quality was confirmed by using the R package affyQCreport to generate quality control reports from the cell intensity files. Raw intensity values of all samples were preprocessed and normalized by robust multi-array average procedure using Bioconductor in R. Differentially expressed genes were tested with an IBMT (15). IBMT is similar to other methods that employ hierarchical Bayesian models to test the differential expression of genes obtained from microarray studies. These methods resolve the problem of poor estimation of the variance from gene expression due to a small sample size in most microarray studies. Rather than considering each gene/probe separately, these Bayesian modified approaches pool information across all expressed genes to achieve a more accurate and stable variance estimation, thus improving the results of the tests (15). IBMT is further strengthened, compared with other previously developed methods, by incorporating the well-documented information about the dependence of the variance of genes on expression intensity levels (ie, the larger variance often seen in genes with lower transcript expression levels). The advantages of IBMT in processing microarray data from a limited number of samples have been established in previous studies (13,15). The data discussed in this study have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) website and are accessible through Gene Expression Omnibus Series accession number GSE38718.
Microarray Validation
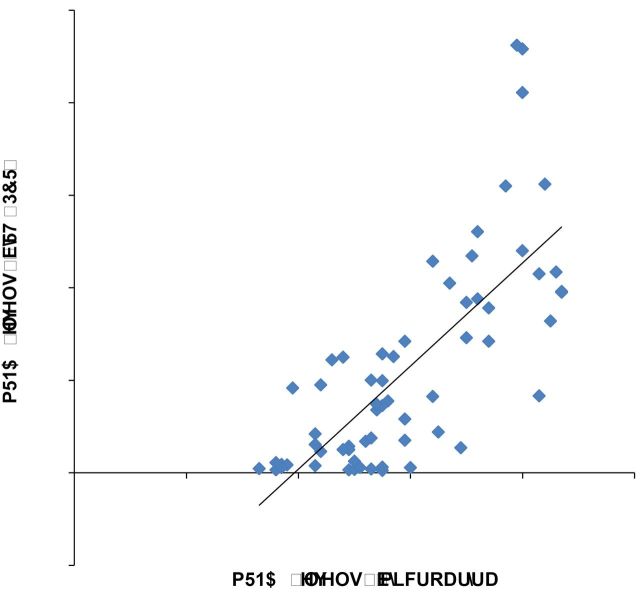
Our lab has been using the microarray technique to study human biceps brachii. We have completed projects related to sex differences, and the acute and chronic exercise responses, and have published the relevant data elsewhere (13,19). In those studies, we employed the same techniques and protocols in both the experimental procedures and data analysis as utilized within the current project. The validity of the microarray data has been confirmed using quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR) in a series of experiments (13,19). In the present study, the quantity of muscle from biopsy samples of the older adults was not sufficient for the additional qRT–PCR experiment. However, young muscle samples were previously analyzed using qRT–PCR for 10 genes including IGF1, IRS2, VEGFA, KDR, DUSP1, PPARGC1, FBXO40, SMAD3, ALDH2, and PFKFB3, and the consistency between microarray and qRT–PCR has been evidenced by the same pattern of differential expression of all selected genes between young men and women (13). We retrieved qRT–PCR data of the young muscle samples included in the present study for nine genes (PFKFB3 was excluded due to incomplete data). The consistency between qRT–PCR and microarray was analyzed by assessing the correlation between mRNA levels measured using two methods (Figure 1). Detailed experimental procedures used for qRT–PCR can be found in a previous publication (13).
Figure 1.
Correlation between quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR) and microarray data for transcript levels of nine genes. Each diamond represents one gene. mRNA levels (n = 7) were expressed as ratio relative to the mRNA levels of B2M, the internal control gene, for both qRT–PCR and microarray.
Functional Gene Networks and Pathway Analysis
Enriched Gene Ontology (GO) terms (20) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (21) were tested by a LRpath (13,16). Enriched GO terms/KEGG pathways were defined as those having a false discovery rate less than 0.01. We used a directional LRpath test to distinguish between upregulated and downregulated gene groups with −log(p value) calculated if the fold change is up and +log(p value) if the fold change is down.
Directly related GO terms that had considerable overlap in identified genes were considered redundant. The strategy to reduce redundancy is similar to that reported previously (13). Briefly, if only the child and parent term were enriched for a similar group of genes, the child term was retained; if the sibling terms and parent term were all involved, the more generalized parent term was retained.
Statistical Analysis
The physical characteristics of participants were compared between young and older groups for each sex and between men and women for each age group using an independent t test. Differentially expressed genes were tested using IBMT, and enrichment analysis for GO terms and KEGG pathways was conducted using LRpath. Multiple testing correction was performed using the Benjamini–Hochberg false discovery rate approach to calculate q values (22,23), and significantly enriched concepts (GO or KEGG) were defined as having a q value less than 0.01.
To validate the microarray data, a correlation was tested for mRNA levels measured by the microarray and qRT–PCR for nine genes using Pearson’s correlation. For the genes represented with multiple probes in the microarray, the probe with the highest average signal intensity (generally associated with lower error) was used. IBMT and LRpath were performed using R. All other analyses were conducted using SAS 9.1 (SAS Institute, Cary, NC).
Results
Participant Characteristics
Characteristics of all participants, including those who did and did not provide a muscle biopsy, are presented in Table 1. The biopsied participants, compared with those not biopsied, did not differ significantly in any measurement obtained. Compared with the women of the same age group, men were significantly taller and heavier, and had greater arm muscle strength and muscle quality (defined as muscle strength per square centimeter of muscle cross- sectional area). Compared with the young, older participants were significantly older, and had lower arm muscle strength and muscle quality. The body mass index of the older participants was higher than the young, but only reached significance in men.
Sex-Based Differential Gene Expression in Older Adults
Compared with older women, older men had higher transcript levels of genes involved in 81 GO terms and 8 KEGG pathways with false discovery rate less than 0.01, and they consistently represented four overriding biological themes including (i) mitochondrial structure and function (oxidative phosphorylation), (ii) muscle structure and movement, (iii) muscle protein catabolism, and (iv) gene transcription and translation (Supplementary Table 1). Genes with lower transcript levels in older men (or higher levels in older women) enriched 41 GO terms and 6 KEGG pathways, and these gene functional groups consistently reflected three overriding biological themes including (i) inflammatory response and immune function, (ii) lipids synthesis and storage, and (iii) extracellular matrix (ECM) and cytoskeleton development and organization (see Supplementary Table 1).
Age-Based Differential Gene Expression in Women
Compared with young women, older women had higher transcript levels for genes involved in 113 GO terms and 4 KEGG pathways, and these gene functional groups reflected six biological themes including (i) immune function, (ii) ECM remodeling, (iii) G-protein coupled receptor signaling, (iv) lipids storage, (v) tissue development, and (vi) neurological system process. Genes showing decreased expression in older women enriched 125 GO terms and 11 KEGG pathways, and they reflected four biological themes including (i) mitochondrial structure and function, (ii) muscle protein remodeling, (iii) gene transcription and translation, and (iv) muscle development. The full list of the significantly regulated GO terms and KEGG pathways are included in Supplementary Table 2.
Age-Based Differential Gene Expression in Men
Compared with young men, older men had higher transcript levels for genes involved in 38 GO terms and 2 KEGG pathways, and they reflected three biological themes including (i) gene transcription and translation, (ii) neurological system process, and (iii) ion channel activity. The downregulated genes in older men enriched 10 GO terms and 1 KEGG pathways, but no consistent biological themes were reflected. The full list of the significantly regulated GO terms and KEGG pathways are included in Supplementary Table 3.
Discussion
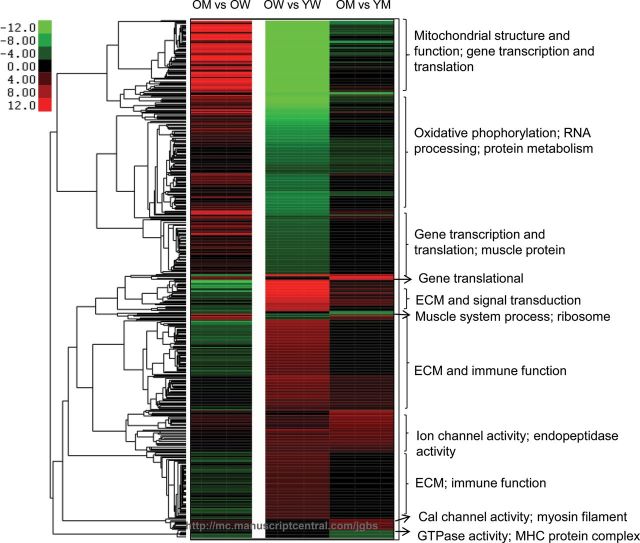
At present, reports on sex-based differences in the aging process of human skeletal muscle have been inconsistent, and a comprehensive investigation at the cellular and molecular level has been lacking. In the present study, we examined the global gene transcription profile of the biceps brachii of older men and women and identified extensive sex differences in the muscle transcriptome. The muscle transcriptional patterns associated with older women suggest that compared with their male counterparts, they are more predisposed to muscle degeneration including loss of mitochondrial content and mitochondrial dysfunction, muscle inflammation and fibrosis, and lipids infiltration. In order to understand the nature of the observed sex-based differential gene expression in the muscle of older adults, we further compared the muscle transcriptome of the older with that of young individuals within each sex. The within-sex analysis revealed that the majority of the sex-based differences were driven by the aging-associated transcriptional changes in women (Figure 2).
Figure 2.
Clustered Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways significantly enriched for differentially expressed genes in older men vs old women (OM vs OW), in older women vs young women (OW vs YW), and in older men vs young men (OM vs YM). The 456 GO terms/KEGG pathways significantly enriched in any of the three comparisons were included. The red color indicates upregulation in older men vs older women for the first column, and upregulation in older vs young muscle for the second and the third columns; the green color indicates downregulation in older men vs older women for the first column, and downregulation in older vs young muscle for the second and the third columns. The −log10(p value) was used as the value for strength of enrichment.
Sex differences in age-related muscle loss and degeneration have been documented in both humans and animals. Many studies (24,25), but not all (26), have reported that males experience a greater loss of muscle mass and strength with aging (ie, sarcopenia). However, data on the prevalence of functional limitation, disability, and frailty associated with sarcopenia indicate that women are more likely to suffer from these conditions (27,28). One explanation for the discrepancy between the sex preponderance in sarcopenia and the health conditions closely linked with sarcopenia is that the alteration of muscle “quality” during the aging process might be unfavorable to females. Baumgartner and colleagues (29) found among individuals aged 60 and older that fat-free mass decreased with age at nearly twice the rate in males as in females; however, body cell mass as a fraction of fat-free mass decreased significantly with age only in women, suggesting that active cell mass may be replaced by inert extracellular materials to a greater extent in females. There is also evidence indicating that with aging, females experience a greater decline in muscle force production and endurance (30). In particular, for the elbow flexor muscles, Valour and colleagues (31) reported that maximal power and maximal shortening velocity decreased with aging and was more prominent in women. The microarray data from the present study support these reports indicating that at the transcriptional level, in comparison with older men, older women exhibited more discernable aging-associated transcriptional changes in skeletal muscle including an upregulation of the immune response and ECM remodeling, and a downregulation of mitochondrial structure and oxidative phosphorylation, all of which have been identified as common signatures of aging across multiple tissue types and species including rats, mice, and humans (32,33).
Reduced muscle mitochondrial function plays a key role in age-related muscle dysfunction. Our data, specifically, indicated that older women, compared with older men, had lower transcript levels of genes involved in mitochondrial structure, function and biogenesis including genes encoding various subunits of ATP synthase, cytochrome c oxidase, mitochondrial ribosomal proteins, and NADH dehydrogenase (Table 2). We believe that these sex-related muscle transcriptome changes occur with aging because a previous study from our lab (13) did not reveal such a sex-related feature in young skeletal muscle. On the contrary, we observed that genes related to lipid beta-oxidation had elevated expression levels in young women, which is consistent with the findings of others (34). Notably, and in line with our findings, Boffoli and colleagues (10) found in orthopedic patients that the activity of mitochondrial electron transport complex III in skeletal muscle was significantly higher in young women than men, and declined sharply only in women with aging.
Table 2.
Genes With Higher Transcript Levels in Older Men Than in Older Women and Their Transcriptional Changes With Aging for Each Sex
| Gene Symbol | Older Men vs Older Women | Older Women vs Young Women | Older Men vs Young Men | |||
|---|---|---|---|---|---|---|
| Fold | p | Fold | P | Fold | p | |
| Mitochondrial structure and function | ||||||
| ATP5D | 1.62 | .023 | −2.01 | .001 | −1.37 | .018 |
| ATP5G1 | 1.43 | .040 | −1.48 | .004 | −1.03 | .712 |
| COX6B1 | 1.22 | .036 | −1.25 | .007 | 1.08 | .199 |
| COX8A | 1.34 | .023 | −1.46 | .001 | −1.08 | .307 |
| CYC1 | 1.45 | .047 | −1.63 | .001 | −1.27 | .004 |
| DHRS4 | 1.38 | .009 | −1.34 | .003 | 1.12 | .253 |
| MRPL24 | 1.26 | .007 | −1.39 | <.001 | −1.08 | .200 |
| MRPL34 | 1.36 | .048 | −1.41 | .001 | −1.19 | .171 |
| MRPS30 | 1.36 | .038 | −1.63 | <.001 | −1.39 | <.001 |
| MRPS7 | 1.36 | .012 | −1.41 | .002 | −1.05 | .591 |
| NDUFA11 | 1.27 | .004 | −1.44 | <.001 | −1.12 | .066 |
| NDUFS3 | 1.51 | .012 | −1.36 | .012 | 1.04 | .663 |
| NDUFV3 | 1.50 | .040 | −1.68 | <.001 | 1.03 | .793 |
| PDK2 | 1.43 | .025 | −1.53 | .004 | −1.17 | .031 |
| PGAM2 | 2.35 | .017 | −1.58 | .060 | 1.32 | .003 |
| PHKG1 | 2.01 | .023 | −1.63 | .074 | −1.08 | .320 |
| SLC25A11 | 1.44 | .025 | −1.65 | .003 | −1.10 | .441 |
| SLC25A12 | 1.85 | .007 | −1.59 | .012 | −1.02 | .805 |
| SLC25A25 | 1.70 | .004 | −2.06 | <.001 | −1.02 | .888 |
| UCP3 | 2.23 | .013 | −1.87 | .040 | −1.31 | .173 |
| PPARGC1A | 1.72 | .063 | −1.78 | .020 | −1.02 | .941 |
| Gene transcription and translation | ||||||
| RPLP0 | 1.32 | .002 | −1.15 | .022 | 1.08 | .257 |
| RPLP0-like | 1.44 | .002 | −1.25 | .008 | 1.09 | .306 |
| EEF1A2 | 1.54 | .040 | −2.00 | .001 | −1.48 | .002 |
| EEF1B2 | 1.23 | .054 | 1.02 | .806 | 1.53 | <.001 |
| FAU | 1.26 | .018 | −1.06 | .593 | 1.22 | .009 |
| BATF2 | 1.42 | .026 | −1.35 | .049 | 1.12 | .129 |
| HSF2 | 2.30 | .001 | −2.18 | .008 | 1.17 | .463 |
| IKBKB | 1.45 | .025 | −1.51 | .001 | −1.27 | .113 |
| GCN1L1 | 1.18 | .036 | 1.04 | .513 | 1.24 | .031 |
| MEX3B | 1.44 | .003 | −1.12 | .038 | 1.29 | .005 |
| MIF4GD | 1.26 | .041 | −1.32 | .003 | −1.09 | .266 |
| NCOA3 | 1.62 | .014 | 1.22 | .273 | 1.68 | .035 |
| RBM15 | 1.36 | .003 | −1.03 | .808 | 1.42 | .006 |
| RBM18 | 1.45 | .009 | −1.40 | .006 | 1.08 | .376 |
| RBM9 | 1.44 | .017 | 1.06 | .712 | 1.42 | .014 |
| RPL3L | 1.80 | .032 | −1.48 | .048 | 1.21 | .053 |
| RPL4 | 1.15 | .033 | 1.1 | .148 | 1.27 | .006 |
| RPLP0 | 1.32 | .002 | −1.15 | .022 | 1.08 | .257 |
Oxidative stress and lipid synthesis and storage also showed a female-specific transcriptional upregulation with aging in our study. A dramatic upregulation was observed for the gene encoding adiponectin, ADIPOQ, which increased over 20-fold in older women compared with their younger counterparts, but did not change with aging in men. Adiponectin is an adipokine that exhibits insulin-sensitizing and anti-inflammatory properties. In obesity and diabetes, lipid toxicity and oxidative stress in skeletal muscle can induce the upregulation of ADIPOQ, which may potentially serve as a cellular protective mechanism (35). Our finding that expression of ADIPOQ was drastically upregulated in older women might suggest a higher propensity to develop oxidative stress and ectopic lipid infiltration in muscle as they age. Interestingly, genes implicated in oxidative stress, such as caspase 6 and microsomal glutathione S-transferase 1 (MGST1), and genes involved in lipids storage, such as SCD, GPAM, and PPARG (Table 3), were significantly upregulated with aging in women, but only minor or nonsignificant changes were observed in men. In support of our results, Barreiro and colleagues (7) found significantly elevated oxidative stress levels in the external intercostal of older women but not men; and Goodpaster and colleagues (36) reported a higher fat infiltration in older women than men, as indicated by lower attenuation of skeletal muscle in women. The age-related upregulation of genes involved in lipid synthesis and storage in skeletal muscle may be directly linked with insulin resistance in older women. For example, we observed a dramatic increase in SCD in older women (~12-fold) but not in older men. SCD encodes stearoyl-CoA desaturase, which is a rate-liming enzyme catalyzing the synthesis of monounsaturated fatty acids. SCD plays an important role in the regulation of skeletal muscle metabolism affecting insulin sensitivity, mitochondrial fatty acid beta-oxidation, and ceramide de novo synthesis in oxidative myofibers (37,38).
Table 3.
Genes With Higher Transcript Levels in Older Women Than in Older Men and Their Transcriptional Changes With Aging for Each Sex
| Gene Symbol | Older Men vs Older Women | Older Women vs Young Women | Older Men vs Young Men | |||
|---|---|---|---|---|---|---|
| Fold | p | Fold | p | Fold | p | |
| Lipid synthesis | ||||||
| ADIPOQ | −22.72 | .024 | 21.34 | .003 | 1.03 | .681 |
| SCD | −13.53 | .036 | 12.38 | .006 | −1.00 | .991 |
| PPARG | −2.70 | .170 | 2.83 | .049 | 1.10 | .350 |
| FABP4 | −4.56 | .030 | 4.04 | .006 | 1.06 | .709 |
| CD36 | −3.11 | .009 | 2.13 | .021 | −1.34 | .266 |
| ACADL | −2.24 | .017 | 1.56 | .030 | −1.06 | .718 |
| CAV3 | 1.66 | .034 | −1.51 | .034 | 1.30 | .025 |
| CAV1 | −2.64 | .071 | 2.05 | .056 | −1.11 | .474 |
| CAV2 | −5.69 | .061 | 2.23 | .198 | −1.82 | .057 |
| PCK2 | −1.32 | .042 | 1.42 | .008 | 1.00 | .994 |
| Immune function | ||||||
| AOC3 | −5.99 | .038 | 5.11 | .010 | −1.01 | .926 |
| CLDN5 | −1.69 | .037 | 1.64 | .009 | −1.03 | .751 |
| JAM2 | −1.67 | .020 | 1.39 | .058 | −1.28 | .029 |
| LPL | −3.63 | .024 | 1.79 | .110 | −1.07 | .797 |
| MYH10 | −3.29 | .013 | 1.76 | .113 | −1.40 | .042 |
| PLAT | −2.88 | .021 | 2.16 | .020 | −1.28 | .110 |
| PLSCR4 | −3.16 | .032 | 2.45 | .020 | −1.40 | .043 |
| RAPGEF3 | −1.31 | .035 | 1.38 | .001 | −1.02 | .870 |
| SMAD3 | 1.87 | .001 | −1.28 | .038 | 1.74 | <.001 |
| TGFBR2 | −2.29 | .072 | 2.18 | .023 | −1.07 | .660 |
| TGFBR3 | −2.26 | .046 | 2.23 | .014 | 1.20 | .215 |
| ECM remodeling | ||||||
| FAT4 | −1.49 | .030 | 1.38 | .020 | −1.24 | .051 |
| FBLN2 | −1.53 | .028 | 1.82 | .013 | 1.08 | .515 |
| GADD45A | −2.01 | .001 | 1.63 | .034 | −1.33 | .245 |
| GJA1 | −2.72 | .032 | 1.89 | .053 | −1.46 | .112 |
| OFD1 | −1.73 | .003 | 1.26 | .041 | −1.28 | .005 |
| SDCBP | −1.88 | .015 | 1.44 | .035 | −1.07 | .574 |
| ACTN1 | −3.32 | .019 | 3.12 | .004 | −1.11 | .396 |
| TGFBR2 | −2.29 | .072 | 2.18 | .023 | −1.07 | .660 |
| TGFBR3 | −2.26 | .046 | 2.23 | .014 | 1.20 | .215 |
| COL1A1 | −2.90 | .019 | 1.03 | .947 | −1.96 | .117 |
| COL1A2 | −2.40 | .069 | 2.20 | .046 | −1.39 | .040 |
| COL3A1 | −3.18 | .059 | 1.28 | .622 | −1.74 | .127 |
| COL4A1 | −2.58 | .071 | 1.57 | .238 | −1.65 | .057 |
| COL5A1 | −2.02 | .024 | 1.39 | .313 | −1.52 | .055 |
| COL5A2 | −2.36 | .118 | 1.44 | .414 | −1.72 | .028 |
Consistent with transcriptional upregulation of oxidative stress and lipid deposition in the muscle of older women, genes involved in inflammation/immune function and ECM remodeling were also expressed at higher levels. Relevant to immune function, we observed over twofold increases in mRNA levels of C3, PLAT, TGFBR2, and TGFBR3 in older women and no change in older men compared with their younger counterparts. It has been suggested that women generally have lower levels of inflammation under various conditions and this state is maintained by estrogen levels before menopause; however, after menopause, due to the loss of estrogen protection, the disruption of the balance between proinflammatory and anti-inflammatory state manifests as a chronic inflammation in the body (8,9). The loss of estrogen in older women might also have a direct effect on the transcriptional regulation of ECM remodeling. In the skeletal muscle of monozygotic female twins discordant for hormone replacement therapy, hormone replacement therapy use has been associated with a lower expression level of genes involved in ECM–cell interaction (39). In our study, subunits of major collagen subtypes in skeletal muscle including collagen I, III, and IV were overexpressed in older women, which is due to an age-related upregulation in women but downregulation in men (Table 3). Reports on the impact of aging on specific muscle collagen fiber types are scarce. Nevertheless, it is interesting that a similar age and sex response has been reported on collagen fiber types in vascular smooth muscle in monkeys (40). The ECM and cytoskeleton are well known for their pivotal role in maintaining muscle structure and mechanical force transduction. In recent years, studies have indicated that the ECM also plays a central role in mediating resistance exercise-induced muscle hypertrophy (41), and may also be linked with muscle insulin activity (42). Overexpression of genes involved in immune activation and ECM remodeling, if translated into changes in proteins, might play a role in the declines of anabolic response and insulin activity in the skeletal muscle of older women.
Other remarkable transcriptional differences between aged skeletal muscle of men and women were also observed. In older men, overexpression of genes involved in gene transcription and translation, and muscle protein turn-over was observed. A similar finding has been previously made (12), in which genes involved in transcription and translation showed higher transcript levels in the vastus lateralis of men than in women; however, though both young and older participants were included in their study, the impact of age on this sex difference was not analyzed. Our investigation and the findings from a previous study (13) from our lab suggest that the sex-related differences in this gene functional group undergo a pattern reversal with aging, that is, genes involved in transcription and translation are overexpressed in young women but overexpressed in older men, which may be due to an age-associated downregulation in women and an upregulation in men. Transcriptional upregulation of gene transcription and translation machinery in men might represent a compensatory mechanism in the muscle for the declined muscle protein synthesis and escalated muscle breakdown with aging. It seems that such a compensatory mechanism fails in older women. Downregulated expression of transcriptional and translational machinery may work synergistically with downregulation of muscle protein turnover, leading to the accumulation of nonfunctional muscle proteins with aging in women.
Our study is not without limitations. First, our sample size was relatively small. However, to maximize the power of the study, we chose to use IBMT, a powerful analytic technique for use in studies with small sample sizes. A power analysis based on the comparison between older men and women indicated that we had enough power (85.8%) to detect a 1.5-fold change among 75% of all genes (α = 0.05). Higher power can be expected for the other within-sex analyses because the young groups had more samples (ie, n = 7 vs 4). Second, due to the limited amount of tissue biopsy collected, we were unable to measure protein markers for the relevant muscle phenotypes or examine muscle histological changes. Consequently, we were not able to directly relate age- and sex-based transcriptional changes with specific muscle phenotypes. However, transcriptional regulation represents the first and critical step in controlling gene expression and protein production, and we believe that studying global transcriptional changes is important to our understanding of the impact of aging and sex on muscle structure and function. Especially, when the transcript changes are observed for multiple genes in a coordinated fashion along a particular signaling pathway or across several interrelated biological processes, they likely represent mechanisms leading to overt phenotypic changes in the tissue. Similarly, after microarray experiments, we did not have sufficient samples left for all participants to run a complete confirmation analysis using qRT–PCR. However, we agree that the purpose of incorporating qRT–PCR in microarray study is to validate the overall dataset, and it is not feasible to experimentally evaluate our findings using individual genes. Consequently, using the samples from young participants, we demonstrated a high correlation between microarray and qRT–PCR. In addition, as mentioned earlier, we have utilized the same microarray methodology to study human biceps brachii and have observed a high consistency between microarray and qRT–PCR data in a series of experiments (13,19). Therefore, we are confident in the validity of our study methodology. Finally, due to the stringency of our study inclusion criteria, that is, recruiting “disease free” older participants for muscle biopsy, we chose not to exclude overweight but otherwise healthy (free from orthopedic injuries, cancer, thyroid disorders, arthritis, diabetes, and heart disease) participants from this study. As a result, we were unable to match older participants based on body mass index and subsequently our older men had slightly higher body mass index levels than older women. Nevertheless, this body compositional difference unlikely influenced the study results. Given that obesity is related to adverse changes in skeletal muscle, such as heightened oxidative stress and lipid deposition, one might expect that a higher body mass index in older men would result in more adverse skeletal muscle alterations in men than in women; however, this is contrary to what we observed. Although the influence of obesity may exist, the impact of sex and age on skeletal muscle appears to be more distinguishable. Therefore, we are confident that the transcriptional changes observed in this investigation are intrinsically related to sex-specific differences in the aging process of skeletal muscle.
Our study is the first to provide global evidence for the presence of extensive sex differences in the aging process of human skeletal muscle. A strength of this study lies in our use of arm muscle rather than commonly used leg muscle. Arm muscle can potentially differentiate a true aging effect from that induced by changes in physical activity with aging (14). In conclusion, our data suggest that transcriptional regulation likely plays a greater role in the aging process of skeletal muscle in women than in men, and older women may likely be more predisposed to dysfunctional declines in muscle with aging.
Funding
Funding for this study was provided by NIH R01 NS40606-01A1.
Supplementary Material
Supplementary material can be found at: http://biomedgerontology.oxfordjournals.org/
References
- 1. Petersen KF, Befroy D, Dufour S, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khamseh ME, Malek M, Aghili R, Emami Z. Sarcopenia and diabetes: pathogenesis and consequences. B J Diabetes Vascular Disease. 2011;11:230–234 [Google Scholar]
- 3. Doherty TJ. The influence of aging and sex on skeletal muscle mass and strength. Curr Opin Clin Nutr Metab Care. 2001;4:503–508 [DOI] [PubMed] [Google Scholar]
- 4. Karakelides H, Irving BA, Short KR, O’Brien P, Nair KS. Age, obesity, and sex effects on insulin sensitivity and skeletal muscle mitochondrial function. Diabetes. 2010;59:89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gómez-Pérez Y, Gianotti M, Proenza AM, Lladó I. Age-related decline of skeletal muscle insulin sensitivity in rats: effect of sex and muscle type. Rejuvenation Res. 2011;14:153–161 [DOI] [PubMed] [Google Scholar]
- 6. Wüst RC, Morse CI, de Haan A, Jones DA, Degens H. Sex differences in contractile properties and fatigue resistance of human skeletal muscle. Exp Physiol. 2008;93:843–850 [DOI] [PubMed] [Google Scholar]
- 7. Barreiro E, Coronell C, Laviña B, Ramírez-Sarmiento A, Orozco-Levi M, Gea J. Aging, sex differences, and oxidative stress in human respiratory and limb muscles. Free Radic Biol Med. 2006;41:797–809 [DOI] [PubMed] [Google Scholar]
- 8. Enns DL, Tiidus PM. The influence of estrogen on skeletal muscle: sex matters. Sports Med. 2010;40:41–58 [DOI] [PubMed] [Google Scholar]
- 9. Tiidus PM. Estrogen and gender effects on muscle damage, inflammation, and oxidative stress. Can J Appl Physiol. 2000;25:274–287 [DOI] [PubMed] [Google Scholar]
- 10. Boffoli D, Scacco SC, Vergari R, et al. Ageing is associated in females with a decline in the content and activity on the b-c1 complex in skeletal muscle mitochondria. Biochim Biophys Acta. 1996;1315:66–72 [DOI] [PubMed] [Google Scholar]
- 11. Iannuzzi-Sucich M, Prestwood KM, Kenny AM. Prevalence of sarcopenia and predictors of skeletal muscle mass in healthy, older men and women. J Gerontol A Biol Sci Med Sci. 2002;57:M772–M777 [DOI] [PubMed] [Google Scholar]
- 12. Welle S, Tawil R, Thornton CA. Sex-related differences in gene expression in human skeletal muscle. PLoS ONE. 2008;3:e1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu D, Sartor MA, Nader GA, et al. Skeletal muscle gene expression in response to resistance exercise: sex specific regulation. BMC Genomics. 2010;11:659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Janssen I, Heymsfield SB, Wang ZM, Ross R. Skeletal muscle mass and distribution in 468 men and women aged 18-88 yr. J Appl Physiol. 2000;89:81–88 [DOI] [PubMed] [Google Scholar]
- 15. Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;7:538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sartor MA, Leikauf GD, Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25:211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hubal MJ, Gordish-Dressman H, Thompson PD, et al. Variability in muscle size and strength gain after unilateral resistance training. Med Sci Sports Exerc. 2005;37:964–972 [PubMed] [Google Scholar]
- 18. Thompson PD, Moyna N, Seip R, et al. Functional polymorphisms associated with human muscle size and strength. Med Sci Sports Exerc. 2004;36:1132–1139 [DOI] [PubMed] [Google Scholar]
- 19. Gordon PM, Liu D, Sartor MA, et al. Resistance exercise training influences skeletal muscle immune activation: a microarray analysis. J Appl Physiol. 2012;112:443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Method). 1995;57:289–300 [Google Scholar]
- 23. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gallagher D, Visser M, De Meersman RE, et al. Appendicular skeletal muscle mass: effects of age, gender, and ethnicity. J Appl Physiol. 1997;83:229–239 [DOI] [PubMed] [Google Scholar]
- 25. Dey DK, Bosaeus I, Lissner L, Steen B. Changes in body composition and its relation to muscle strength in 75-year-old men and women: a 5-year prospective follow-up study of the NORA cohort in Göteborg, Sweden. Nutrition. 2009;25:613–619 [DOI] [PubMed] [Google Scholar]
- 26. Chan S, Head SI. Age- and gender-related changes in contractile properties of non-atrophied EDL muscle. PLoS ONE. 2010;5:e12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valentine RJ, Misic MM, Rosengren KS, Woods JA, Evans EM. Sex impacts the relation between body composition and physical function in older adults. Menopause. 2009;16:518–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Valentine RJ, McAuley E, Vieira VJ, et al. Sex differences in the relationship between obesity, C-reactive protein, physical activity, depression, sleep quality and fatigue in older adults. Brain Behav Immun. 2009;23:643–648 [DOI] [PubMed] [Google Scholar]
- 29. Baumgartner RN, Stauber PM, McHugh D, Koehler KM, Garry PJ. Cross-sectional age differences in body composition in persons 60+ years of age. J Gerontol A Biol Sci Med Sci. 1995;50:M307–M316 [DOI] [PubMed] [Google Scholar]
- 30. Hunter SK, Critchlow A, Enoka RM. Influence of aging on sex differences in muscle fatigability. J Appl Physiol. 2004;97:1723–1732 [DOI] [PubMed] [Google Scholar]
- 31. Valour D, Ochala J, Ballay Y, Pousson M. The influence of ageing on the force-velocity-power characteristics of human elbow flexor muscles. Exp Gerontol. 2003;38:387–395 [DOI] [PubMed] [Google Scholar]
- 32. de Magalhães JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zahn JM, Sonu R, Vogel H, et al. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maher AC, Fu MH, Isfort RJ, Varbanov AR, Qu XA, Tarnopolsky MA. Sex differences in global mRNA content of human skeletal muscle. PLoS ONE. 2009;4:e6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Delaigle AM, Senou M, Guiot Y, Many MC, Brichard SM. Induction of adiponectin in skeletal muscle of type 2 diabetic mice: In vivo and in vitro studies. Diabetologia. 2006;49:1311–1323 [DOI] [PubMed] [Google Scholar]
- 36. Goodpaster BH, Carlson CL, Visser M, et al. Attenuation of skeletal muscle and strength in the elderly: The Health ABC Study. J Appl Physiol. 2001;90:2157–2165 [DOI] [PubMed] [Google Scholar]
- 37. Dobrzyń A, Dobrzyń P. Stearoyl-CoA desaturase–a new player in skeletal muscle metabolism regulation. J Physiol Pharmacol. 2006;57(Suppl 10):31–42 [PubMed] [Google Scholar]
- 38. Hulver MW, Berggren JR, Carper MJ, et al. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab. 2005;2:251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ronkainen PH, Pöllänen E, Alén M, et al. Global gene expression profiles in skeletal muscle of monozygotic female twins discordant for hormone replacement therapy. Aging Cell. 2010;9:1098–1110 [DOI] [PubMed] [Google Scholar]
- 40. Qiu H, Tian B, Resuello RG, et al. Sex-specific regulation of gene expression in the aging monkey aorta. Physiol Genomics. 2007;29:169–180 [DOI] [PubMed] [Google Scholar]
- 41. West DW, Burd NA, Staples AW, Phillips SM. Human exercise- mediated skeletal muscle hypertrophy is an intrinsic process. Int J Biochem Cell Biol. 2010;42:1371–1375 [DOI] [PubMed] [Google Scholar]
- 42. Coletta DK, Mandarino LJ. Mitochondrial dysfunction and insulin resistance from the outside in: extracellular matrix, the cytoskeleton, and mitochondria. Am J Physiol Endocrinol Metab. 2011;301:E749–E755 [DOI] [PMC free article] [PubMed] [Google Scholar]