Abstract
Drosophila colour vision is achieved by R7 and R8 photoreceptor cells present in every ommatidium. The fly retina contains two types of ommatidia, called ‘pale’ and ‘yellow’, defined by different rhodopsin pairs expressed in R7 and R8 cells. Similar to the human cone photoreceptors, these ommatidial subtypes are distributed stochastically in the retina. The choice between pale versus yellow ommatidia is made in R7 cells, which then impose their fate onto R8. Here we report that the Drosophila dioxin receptor Spineless is both necessary and sufficient for the formation of the ommatidial mosaic. A short burst of spineless expression at mid-pupation in a large subset of R7 cells precedes rhodopsin expression. In spineless mutants, all R7 and most R8 cells adopt the pale fate, whereas overexpression of spineless is sufficient to induce the yellow R7 fate. Therefore, this study suggests that the entire retinal mosaic required for colour vision is defined by the stochastic expression of a single transcription factor, Spineless.
The ability to discriminate between colours has evolved independently in vertebrates and invertebrates1,2. However, despite the obvious differences in eye development and design, both flies and humans have developed retinal mosaics where classes of photoreceptor cells (PRs) with different spectral sensitivity are randomly distributed3,4.
The compound eye of Drosophila consists of ~800 optical units (ommatidia), each containing eight PRs in addition to accessory cells5. In each ommatidium, the six ‘outer PRs’ (R1–R6) function like the vertebrate rod cells, as they are required for motion detection in dim light6,7. These cells express the broad-spectrum rhodopsin, Rh1 (ref. 8). The ‘inner PRs’ (R7 and R8) may be viewed as the equivalent of the colour-sensitive vertebrate cone cells, which express a range of different rhodopsin molecules9–13.
Ommatidial subset specification in Drosophila
The general rule of sensory receptor exclusion also applies to Drosophila ommatidia, where only one rhodopsin gene is expressed by a given PR14. The expression of inner PR rhodopsins can be used to distinguish three ommatidial subtypes15,16 (Supplementary Fig. 1a, b). Two of the subtypes are distributed randomly throughout the retina: ~30% of ommatidia express ultraviolet-sensitive Rh3 in R7 cells and blue-sensitive Rh5 in R8 cells, and therefore are specialized in the detection of short wavelengths (‘pale’ ommatidia, p; Fig. 1a, blue). The remaining ~70% express another ultraviolet-sensitive opsin (Rh4) in R7 and green-sensitive Rh6 in R8, making them more responsive to longer wavelengths (‘yellow’ ommatidia, y; Fig. 1a, yellow). The coupled expression of Rh3/Rh5 or Rh4/Rh6 within the same ommatidium results from communication between R7 and R8 (Supplementary Fig. 1b, c). In the dorsal rim area (DRA) (Fig. 1a, pink), a third type of ommatidia exists17 in which both R7 and R8 express ultraviolet-sensitive Rh3 (refs 18, 19). These ommatidia are used to detect the e-vector of polarized sunlight for orientation20,21. Spatially localized polarized light detectors and stochastically distributed colour-sensitive ommatidia therefore reflect two fundamentally different specification strategies that shape the retinal mosaic of Drosophila22.
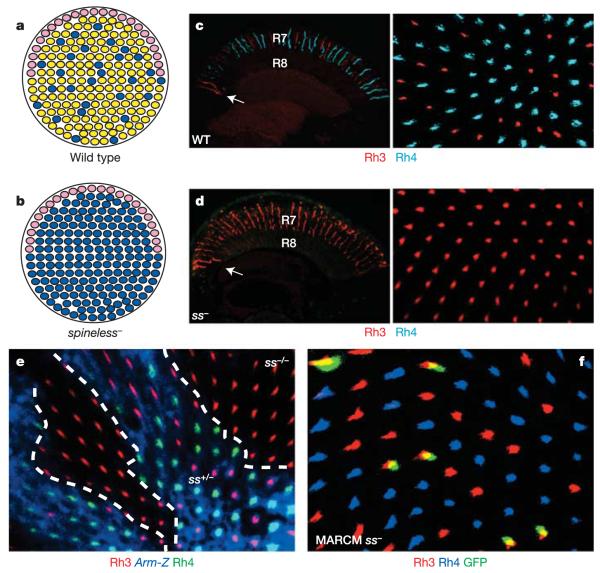
Figure 1. The yR7 subtype is lost in spineless mutants.
a, Three subtypes can be identified on the basis of molecular markers: ‘pale’ (blue), ‘yellow’ (yellow) and DRA (pink) ommatidia together form the wild-type retinal mosaic (schematic representation;dorsal is to the top). b, Schematic representation of the ss phenotype in R7 cells. c, Transverse section through a wild-type (WT) adult eye (left panel; dorsal is to the left). The arrow denotes the DRA. Ratio of R7 opsins in a wild-type whole-mount adult retina (right panel; dorsal is to the top) stained for Rh3 (red) and Rh4 (cyan). d, Transverse section through a ssD115.7 whole-mutant adult eye (left panel). Rh3 (red) is expanded and Rh4 (cyan) is completely lost. Opsin expression in a mutant whole-mount adult retina is also shown (right panel). e, Whole-mount retina with mitotic clones lacking Spineless (marked by the absence of expression of the armadillo-lacZ (Arm-Z) construct, blue). Rh4 (green) is always absent from mutant clones. Rh3 expression is shown in red. f, Whole-mount retina with MARCM clones lacking Spineless (marked by the presence of GFP, green). Every GFP-positive cell expresses Rh3 (red), whereas Rh4 (blue) is always absent from mutant cells.
The current model for specifying colour-sensitive ommatidia combines stochastic and instructive steps11. First, a subset of R7 (pale R7, pR7) stochastically chooses Rh3 expression over the ‘R7 default’, Rh4. Second, these cells then impose the p fate (Rh5) onto R8 (pale R8, pR8) of the same ommatidium (Supplementary Fig. 1c).
In this study, we report the identification of spineless (ss) as a key regulatory gene for establishing the retinal mosaic. ss encodes the Drosophila homologue of the human arylhydrocarbon (‘dioxin’) receptor, a member of the bHLH-PAS (basic helix–loop–helix-Period–Arnt–Single-minded) family of transcription factors23,24. At mid-pupation, ss is stochastically expressed in a majority of R7 that seem to correspond to the y subtype. ss is both necessary and sufficient to specify the yellow R7 (yR7) fate and subsequently the entire y ommatidia; pR7 cells are thus specified by default, and stochastic expression of ss represents the key regulatory event defining the retinal mosaic required for fly colour vision.
spineless is necessary for yellow ommatidia specification
We recently identified homothorax (hth) as the key regulatory gene necessary and sufficient for the specification of DRA ommatidia19. ss and hth cause similar homeotic phenotypes: that is, complete (hth) or partial (ss, ‘aristapedia’) transformation of antennae into legs25,26. Therefore, we tested for a potential role of ss in ommatidial subtype specification by generating whole-mutant eyes, as well as mitotic clones, lacking ss function using the null allele ssD115.7 and the ey-FLP/FRT technique27,28. Owing to ey-FLP expression in the antennal imaginal disc, ss mutant flies showed a strong aristapedia phenotype (Supplementary Fig. 2a), but lacked any obvious morphological eye phenotype. However, expression of rhodopsin genes was severely affected. In wild-type eyes, Rh3 is found in ~30% of R7 cells, as well as in both R7 and R8 of DRA ommatidia (Fig. 1c, arrow), whereas the remaining ~70% of R7 contain Rh4 (Fig. 1c). In ss mutant eyes, Rh4 was completely absent, whereas Rh3 was expanded into all R7 cells (Fig. 1d). The total number of ommatidia was not reduced, indicating that R7 cells were mis-specified into pR7, rather than yR7 being specifically eliminated. ss mutant mitotic clones were morphologically wild type (Supplementary Fig. 2b); however, Rh3 was always present in mutant R7 cells (marked by the absence of β-galactosidase (β-gal) expression), whereas Rh4 was always lost (Fig. 1e).
To test whether the R7 ss phenotype was cell autonomous, we generated individual mutant R7 cells using the MARCM technique29,30. All mutant R7 cells (marked by the presence of green fluorescent protein (GFP) expression) contained Rh3 and never Rh4, demonstrating that ss is required cell autonomously in R7 to induce Rh4 expression (Fig. 1f). DRA ommatidia were correctly specified in ss mutant eyes, as Rh3 was expressed normally in both DRA R7 and R8 cells (Fig. 1d, arrow). Therefore, ss is necessary for the establishment of the yR7 subtype without affecting PR fate specification (Fig. 1b and Supplementary Fig. 2c).
The ommatidial subtypes are first specified in R7, which then instruct R8 (ref. 16). Therefore, ss mutant eyes should exhibit a rhodopsin phenotype in R8. In wild types, ~30% of R8 cells contain Rh5, and the remaining ~70% contain Rh6 (Fig. 2c)11,12. In ss mutant eyes, the large majority (up to 95%) of R8 contained Rh5 (Fig. 2a, d), with some R8 still containing Rh6. However, most of these remaining yR8 cells were located in the dorsal third of the eye (Fig. 2a). In this part of the retina, instruction of pale R8 (pR8) by pR7 is less efficient, resulting in ommatidia with odd-coupled (Rh3/Rh6) rhodopsin expression (E.O.M. & C.D., manuscript in preparation). In ss mutants, the frequency of such ommatidia was significantly increased in the dorsal region (Fig. 2b).
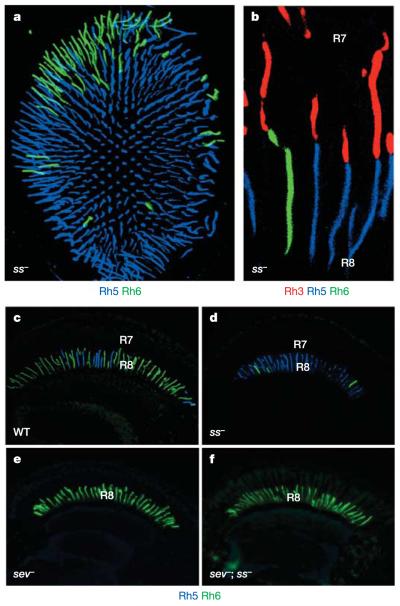
Figure 2. R8 phenotype of spineless mutant eyes.
a, Whole-mount retina from a ss mutant fly. The pR8 subtype (Rh5, blue) is expanded to almost all R8 cells (Rh6, green). b, Unusual mis-coupling of Rh3 (red) in R7 and Rh6 (green) in R8 of the same ommatidium is frequently observed in ss mutant retinas. Rh5 expression is shown in blue. c–f, Transverse sections stained for Rh5 (blue) and Rh6 (green): an adult wild-type eye (c), a ss mutant eye (d), a sev mutant eye (e), and a double-mutant (sev; ss) eye (f), which manifests the same R8 phenotype as sev mutants (expansion of yR8 cells and loss of pR8 cells).
To test whether the R8 opsin phenotype of ss mutants resulted from the inability of some mutant R7 cells to properly instruct R8, rather than from ss being directly required in R8, we generated sevenless; spineless (sev; ss) double-mutant eyes. These eyes, which lacked R7 cells, always exhibited the sev single-mutant phenotype (Fig. 2e), with virtually all R8 cells containing Rh6 (Fig. 2f). This indicates that ss is required in R7 for the formation of the yR7 subtype, and consequently for the formation of yR8, without being directly required in R8 PRs.
spineless induces the yellow ommatidial subtype
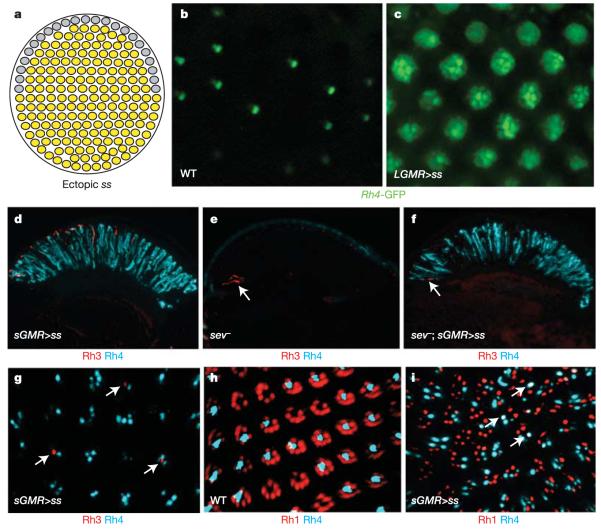
We tested whether ss was also sufficient to induce the y ommatidial subtype (Fig. 3a). Overexpression of ss in all developing PRs using a strong LGMR (long glass multiple reporter)-Gal4 driver31 and UAS-Ss (LGMR>ss flies) resulted in a rough eye phenotype, as well as a dramatic rhodopsin phenotype: Rh4 was activated in all PRs throughout the eye (R1–R6 as well as R7 and R8), as revealed by ectopic expression of an Rh4-GFP reporter in many PRs per ommatidium (Fig. 3c) compared with wild type (Fig. 3b). To avoid the strong phenotype in the eye, we misexpressed ss using the weaker, variegated GMR driver, sGMR (short GMR)-Gal431 (sGMR>ss flies). This led to strong ectopic induction of Rh4 in many PRs without severely affecting retinal morphology (Fig. 3d). This ectopic induction of Rh4 was also observed in sev mutants (Fig. 3f), and was thus independent of R7. Rh3 was still detected in some R7 in sGMR>ss flies, presumably due to the lack of variegated Gal4 expression in these cells (Fig. 3g, arrows), whereas Rh4 was expanded to some outer PRs. However, co-localization of Rh3 and Rh4 was never observed, confirming that gain of Rh4 in R7 cells always leads to the exclusion of Rh3. In contrast, gain of Rh4 in outer PRs did not lead to the exclusion of Rh1, as frequent coexpression of Rh1 and Rh4 was observed (Fig. 3i, arrows).
Figure 3. spineless is sufficient to induce the yR7 fate.
a, Summary of the ss gain-of-function phenotype. All R7 cells adopt the yR7 fate (yellow); the fate of DRA ommatidia is unclear (grey). b, Water immersion microscopy on living wild-type flies expressing the yR7-specific reporter Rh4-GFP. Expression is restricted to one inner PR in a large subset of ommatidia. c, Rh4-driven GFP expression is dramatically expanded in LGMR>ss flies, as visualized by water immersion. d, In sGMR>ss flies, Rh4 (cyan) is expanded through the whole ss-overexpressing retina (compare with Fig. 1c). e, Rh4 (cyan) is completely lost in sev mutants, although Rh3 (red) is present in DRA R8 cells (arrow). f, Overexpression of ss (sGMR>ss) leads to ectopic Rh4 (cyan) in sev mutants. Rh3 (red) is restricted to DRA R8 cells (arrow). g, In whole-mount retinas from sGMR>ss flies, variegated expression of ss leads to the expansion of Rh4 (cyan) into a variable number of PRs per ommatidium; coexpression with Rh3 (red) is never observed (arrows). h, In wild-type eyes, the outer-PR opsin Rh1 (red) and Rh4 (cyan) never co-localize in the same PR cell. i, Ectopic expression of ss in sGMR>ss flies leads to massive expansion of Rh4 (cyan), with some co-localization (arrows) with the outer-PR opsin Rh1 (red).
Using an Rh4-lacZ reporter construct in LGMR>ss flies, we found that β-gal-positive PR axons projected to both lamina and medulla, confirming the expansion of Rh4 into outer PRs (Supplementary Fig. 3a). However, Rh4-expressing outer PRs were not transformed into genuine R7 cells, as they maintained their lamina projections (Supplementary Fig. 3a). Notably, DRA inner PRs were the only cells not expressing Rh4 (Supplementary Fig. 3a), suggesting that the DRA fate19,32, specified by the gene hth, antagonizes ss function. Expression of Rh3 and Rh5 was completely lost (including in the DRA, where no rhodopsin was detected), while Rh6 expression was found in most R8 cells (Supplementary Fig. 3b). This resulted in R8 coexpressing Rh4 and Rh6, demonstrating that the ‘one sensory receptor per cell’ rule can be broken in Drosophila PRs, as has been shown in other insects14,33. Therefore, ectopic induction of the yR7 fate by ss specifically excludes the formation of pR7 cells. As a consequence, R8 cells expressing Rh5 are not induced16, with most R8 expressing Rh6. Rh6 was never found in outer PRs, supporting the hypothesis that ss is required only in R7 for the choice between Rh3 and Rh4, and not directly in R8 for the Rh6 choice (Supplementary Fig. 3b). In LGMR>ss flies, the specification of outer versus inner PRs (markers spalt and seven up; Supplementary Fig. 3c) or of R7 versus R8 (prospero and senseless; Supplementary Fig. 3d) was normal. Thus, ss acts by segregating ommatidial subtypes downstream of early PR specification events.
spineless can re-specify cell fate in R7 photoreceptor cells
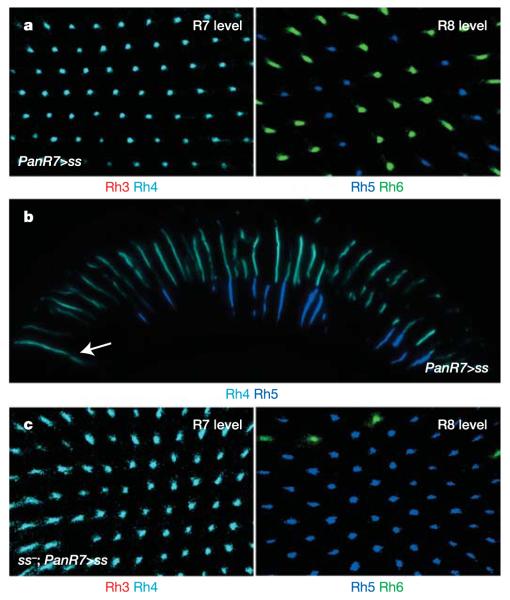
Colour PR cell fate determination seems to be a late event in PR differentiation. To test whether ss can transform the R7 fate at late stages of development, we used the PanR7-Gal4 driver (which is also expressed in DRA R8 cells). Late mis-expression of ss induced the y fate (Rh4) in all R7 cells, whereas Rh3 was absent (Fig. 4a, left panel). Opsin expression in the DRA was also altered, with Homothorax-positive cells (both R7 and R8) now expressing Rh4 (Supplementary Fig. 4). Hence, it is possible to reprogramme the R7 fate at later stages of differentiation, as PanR7-Gal4 becomes activated at the time of rhodopsin expression. Surprisingly, expression of R8 rhodopsins outside the DRA was not affected, as the distribution of Rh5 and Rh6 resembled the wild type (Fig. 4a, right panel). As a result, many ommatidia manifested the very unusual coupling of Rh4 in R7 and Rh5 in R8 (Fig. 4b). Therefore, although ss is able to reprogramme all R7 late in development, R8 cannot revert their fate once they have been instructed to become pR8, and they maintain Rh5. We have recently identified two antagonistic genes expressed in either of the two R8 subtypes, which act together as a molecular consolidation system responsible for this inertia of R8 (ref. 34). To confirm that late expression of ss exclusively in R7 was sufficient to transform R7, ss was mis-expressed in ssD115.7 mutants using PanR7-Gal4. This was sufficient to induce Rh4 and to repress Rh3 (Fig. 4c, left panel). R8 were again not reprogrammed and exhibited the ss mutant phenotype, with many R8 cells expressing Rh5 (Fig. 4c, right panel).
Figure 4. Control of PR cell fates by spineless.
a, In whole-mount retinas from PanR7>ss flies, all R7 cells express Rh4 (cyan), whereas Rh3 (red) is absent (left panel). Late expression of ss in R7 has no effect on the ratio between Rh5 (blue) and Rh6 (green) expression (right panel). b, Section of a PanR7>ss eye. As the fate of R8 is not affected by late ss expression in R7 cells, there is a great number of ‘odd-coupled’ Rh4 (cyan)/Rh5 (blue) ommatidia. The arrow indicates the DRA. c, In whole-mount retinas from PanR7>ss flies on a ss mutant background, Rh3 (red) is not expressed, while every R7 cell expresses Rh4 (cyan) (left panel). Late overexpression of ss in all R7 cells on a ss mutant background does not lead to a reprogramming of R8 cells as most R8 cells express Rh5 (blue) and few dorsal R8 express Rh6 (green) (right panel).
Transient expression of spineless precedes R7 specification
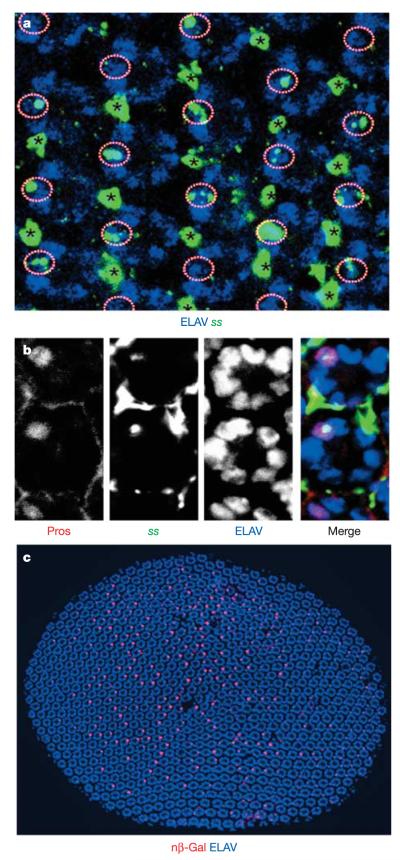
All of the results presented above strongly indicated that ss must be expressed in the y subtype of R7 at some point during pupal development. As several attempts to generate an anti-Spineless antibody had failed, we used in situ hybridization to detect ss messenger RNA in the retina at mid-pupation (Fig. 5a, b). At ~50% pupation, ss mRNA was detected in four neuronal cells per ommatidial cluster, one PR and three bristle cells (Fig. 5a, circles and asterisks). The PR was also labelled by anti-Prospero, confirming its identity as R7 (Fig. 5b). Although the expression levels of ss in bristle cells seemed uniformly high, levels of ss expression varied considerably among R7 cells, ranging from very faint to very strong in 60–80% of R7.
Figure 5. Mosaic expression of spineless in the developing retina.
a, In situ hybridization of a whole-mount pupal retina with an antisense ss probe (green) and an ELAV antibody (blue). ss expression can be observed in all bristle cells (asterisks), as well as in one PR (circles) per ommatidium in only 60–80% of all ommatidia. Note that ss levels vary from cell to cell. b, The PR cell positive for ss (green) is identified as an R7 cell by co-staining with the R7-specific marker Prospero (Pros, red). Neurons are marked with ELAV (blue). c, Nuclear β-gal (nβ-Gal, red) driven under the control of sseye-Gal4 reveals mosaic expression of ss in one cell per cluster. Neurons are marked with ELAV (blue). Dorsal is to the left in all panels.
We also identified a 1.6 kilobase ‘eye enhancer’ fragment (sseye) within the ss promoter that drives PR-specific expression. After crossing sseye-Gal4 to UAS-β-gal::NLS (nuclear localization sequence) reporters, PR-specific ss expression was first detected at mid-pupation—that is, approximately one day before rhodopsins are expressed, and before any visible molecular or morphological distinction between ommatidial subtypes (Fig. 5c). A single PR per ommatidium, which was identified as R7 through co-staining with Prospero, expressed ss (Supplementary Fig. 5a). Thus, the sseye enhancer recapitulates endogenous ss expression in PRs. ss expression was detected in 60–80% of R7, correlating well with the distribution of Rh4 in adult retina. Like Rh4-expressing ommatidia, β-gal-positive ommatidia were more abundant in the dorsal half of the eye, and no β-gal expression was detected in the DRA (marked by Homothorax), where Rh4 is also never expressed (Supplementary Fig. 5b). sseye-Gal4 expression was detectable for only ~2 h at midpupation. Although it was not possible to directly co-stain for ss and Rh4 (which starts to be expressed one day later during pupation), it seems that at mid-pupation a short pulse of ss is deployed in a large subset of R7, which will become yR7.
spineless levels are crucial for specifying yellow ommatidia
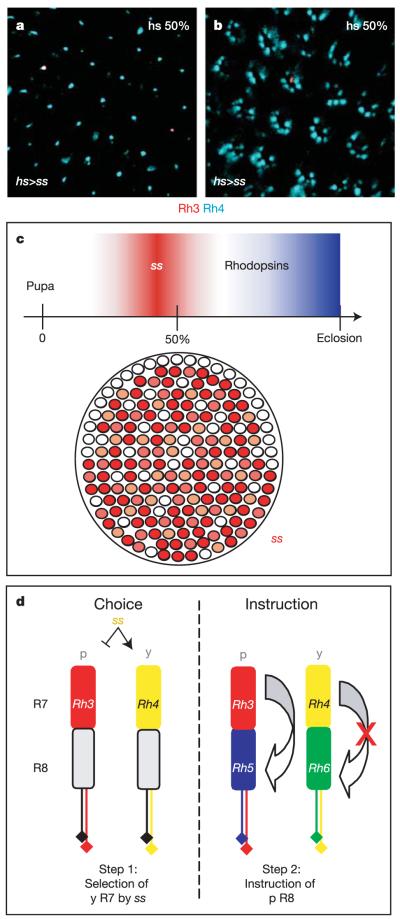
We tested whether a short pulse of ectopic ss expression was able to modify the entire retinal mosaic, using a heat shock-Gal4 driver (hs-Gal4) to temporally control ss expression (hs>ss flies). A 30-min heat shock at ~50% pupation indeed resulted in an increase of Rh4 expression with a concomitant reduction of Rh3 in adults. The phenotype varied extensively, from only R7 cells expressing Rh4 (~25% of the flies analysed had Rh4 in most R7) (Fig. 6a), to almost every PR expressing Rh4 (Fig. 6b). In contrast, a 30-min pulse of ss in one-day-old adult flies had no effect (data not shown). Heat shocks during larval or early pupal stages were lethal. Thus, PRs are extremely sensitive to a short pulse of ss during mid-pupation, at the time when endogenous ss is normally expressed.
Figure 6. Stochasticity of ss expression and a revised model for retinal patterning in Drosophila.
a, b, The effect of a 30-min pulse of ss expression at ~50% pupation leads to an almost 100% transformation of R7 to the y fate (a), or almost every PR (b). c, Top: transient expression of sseye-Gal4 (red) during pupation before the onset of opsin expression (blue). Bottom: variable expression of ss (different tones of red) in R7 cells. d, Left: the ss data suggest that ~70% of the R7 cells get promoted into the yR7 fate (Rh4, yellow) by expressing ss. The pR7 subtype (Rh3, red) therefore represents the R7 ‘default state’. Right: in ss-positive yR7 cells, the ability to communicate with the underlying R8 is abolished, resulting in y ommatidia as the R8 default state is expression of Rh6 (green). Only pR7 retain the competence to instruct the pR8 fate (Rh5, blue).
To further study the mechanism of the stochastic choice between p and y ommatidia, we analysed the retinal mosaic in different mutant backgrounds. Flies heterozygous for ssD115.7 had fewer Rh4-expressing R7 cells (control: 66.1 ± 3%; ss heterozygous: 53.8 ± 3.5%; P < 0.001; values are mean ± s.d.). As the ssD115.7 allele only affects the ss coding sequence, heterozygous flies have two functional promoters, only one of which produces a functional protein, suggesting that the non-productive promoter might sequester limiting factor(s) that regulate(s) the expression levels of ss. If this hypothesis is correct, addition of extra copies of the ss promoter should have a similar effect. Indeed, the addition of two functional copies of the ss eye enhancer (sseye-Gal4) in an otherwise wild-type background also caused a significant reduction of the yR7 subtype (50.2 ± 2.4%, P < 0.001). Therefore, the level of Spineless expression is important for the induction of the yR7 fate, which is less efficient in cells where the amount of Spineless is reduced.
Conclusions
Retinal patterning in Drosophila reveals an original mechanism for how PR mosaics can be generated: stochastic expression of a single transcription factor (Spineless) acts as a binary switch that transforms the seemingly homogeneous compound eye into a mosaic, distinguishing p and y subtypes. However, subtype specification and rhodopsin expression can be separated, as ss expression in yR7 has ceased well before the time of rhodopsin expression (Fig. 6c). Additional factors are therefore required downstream of ss to ensure expression of adult p- and y-specific markers such as rhodopsins and additional screening pigments4. We propose a revised two-step model for the stochastic specification of p and y ommatidia (Fig. 6d). First, R7 are stochastically divided into two subtypes by the induction of ss in yR7. ss-positive R7 express Rh4, whereas the remaining R7 choose the pR7 fate and express Rh3 by default (Fig. 6d, left). Second, only those R7 cells that did not express ss (pR7) retain the ability to induce the pR8 fate (Rh5), whereas yR8 express Rh6 by default (Fig. 6d, right). The ‘default states’ of R7 (Rh3) and R8 (Rh6) therefore belong to opposite subtypes. Expression of R8 rhodopsin genes is maintained by a bistable regulatory loop containing the genes warts and melted34. Notably, the localized specification of polarization-sensitive DRA ommatidia by hth antagonizes the stochastic choice executed by ss, placing these two genes into a new regulatory relationship during retinal patterning. Therefore, the role of the transcription factor Spineless is to generate the retinal mosaic required for fly colour vision by distinguishing yR7 from pR7 cell fates, and preventing R7 from instructing the underlying R8 cells.
Mosaic expression of sensory receptors has been described in detail for the olfactory system of both vertebrates35 and insects36, and random PR mosaics have been described for humans3 and amphibians37, as well as insects4,15,38–40. Two transcription factors have been shown to regulate the specification of blue versus red/green cone cell fates in mammals. Upon mutation of either—the human nuclear receptor NR2E3 (also known as PNR) or the rodent thyroid hormone β2 receptor—the number of blue cones is dramatically increased at the expense of green cones41,42, leading to ‘enhanced S-cone syndrome’. It should be noted that this retinal phenotype bears important similarity to the altered ommatidial mosaic in Drosophila ss mutants, where long wavelength-sensitive y ommatidia are lost at the expense of the short wavelength-sensitive p type.
The stochastic cell fate choice occurs at the level of the ss promoter: the very short pulse of ss expression at mid-pupation is not only controlled temporally, but its levels are also critical, and only ~70% of R7 receive enough Spineless to commit to the yR7 fate. Elucidating the mechanism that controls ss expression will shed some light into the fascinating process of stochastic gene expression, and the identification of its downstream targets will provide insights into consolidation and maintenance of cell fates.
METHODS
Drosophila strains and crosses, constructs, generation of transgenic flies by germ line transformation, antibody staining on mid-pupal and adult whole-mounted or cryo-sectioned retinas, in situ hybridization on pupal retinas, MARCM, cornea neutralization, and adult eye plastic sections were all performed by standard methods, and are described in detail in Supplementary Information.
Supplementary Material
Acknowledgements
We thank S. Britt, R. Carthew, S. Crews, R. Mann, G. Struhl, J. Treisman, the Hybridoma Bank, and the Bloomington Stock Center for flies and reagents. We also would like to thank J. Treisman and J. Blau for comments on the manuscript. We thank N. Franceschini, as well as all past and present members of the Desplan laboratory, for suggestions throughout the project and their comments on the manuscript. M.F.W. was supported by the Boehringer Ingelheim Fonds and the Studienstiftung des Deutschen Volkes, E.O.M. was supported by the Dean’s Dissertation Fellowship from NYU. This work was funded by a grant from the National Eye Institute/NIH to C.D. and conducted in a facility constructed with the support of a Research Facilities Improvement Grant from the NCRR, NIH.
Footnotes
Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Gegenfurtner KR, Kiper DC. Color vision. Annu. Rev. Neurosci. 2003;26:181–206. doi: 10.1146/annurev.neuro.26.041002.131116. [DOI] [PubMed] [Google Scholar]
- 2.Briscoe AD, Chittka L. The evolution of color vision in insects. Annu. Rev. Entomol. 2001;46:471–510. doi: 10.1146/annurev.ento.46.1.471. [DOI] [PubMed] [Google Scholar]
- 3.Roorda A, Williams DR. The arrangement of the three cone classes in the living human eye. Nature. 1999;397:520–522. doi: 10.1038/17383. [DOI] [PubMed] [Google Scholar]
- 4.Franceschini N, Kirschfeld K, Minke B. Fluorescence of photoreceptor cells observed in vivo. Science. 1981;213:1264–1267. doi: 10.1126/science.7268434. [DOI] [PubMed] [Google Scholar]
- 5.Wolff T, Ready DF. In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1993. pp. 1277–1325. [Google Scholar]
- 6.Heisenberg M, Buchner E. The role of retinula cell types in visual behavior of Drosophila melanogaster. J. Comp. Physiol. 1977;117:127–162. [Google Scholar]
- 7.Miller G, Hansen K, Stark W. Phototaxis in Drosophila: R1-R6 input and interaction among ocellar and compound eye receptors. J. Insect Physiol. 1981;27:813–819. [Google Scholar]
- 8.Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]
- 9.Montell C, Jones K, Zuker C, Rubin G. A second opsin gene expressed in the ultraviolet-sensitive R7 photoreceptor cells of Drosophila melanogaster. J. Neurosci. 1987;7:1558–1566. doi: 10.1523/JNEUROSCI.07-05-01558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fryxell KJ, Meyerowitz EM. An opsin gene that is expressed only in the R7 photoreceptor cell of Drosophila. EMBO J. 1987;6:443–451. doi: 10.1002/j.1460-2075.1987.tb04774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou WH, et al. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron. 1996;17:1101–1115. doi: 10.1016/s0896-6273(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 12.Papatsenko D, Sheng G, Desplan C. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development. 1997;124:1665–1673. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- 13.Huber A, et al. Molecular cloning of Drosophila Rh6 rhodopsin: the visual pigment of a subset of R8 photoreceptor cells. FEBS Lett. 1997;406:6–10. doi: 10.1016/s0014-5793(97)00210-x. [DOI] [PubMed] [Google Scholar]
- 14.Mazzoni EO, Desplan C, Celik A. ‘One receptor’ rules in sensory neurons. Dev. Neurosci. 2004;26:388–395. doi: 10.1159/000082281. [DOI] [PubMed] [Google Scholar]
- 15.Feiler R, et al. Ectopic expression of ultraviolet-rhodopsins in the blue photoreceptor cells of Drosophila: visual physiology and photochemistry of transgenic animals. J. Neurosci. 1992;12:3862–3868. doi: 10.1523/JNEUROSCI.12-10-03862.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou WH, et al. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development. 1999;126:607–616. doi: 10.1242/dev.126.4.607. [DOI] [PubMed] [Google Scholar]
- 17.Wada S. Spezielle randzonale ommatidien der fliegen (Diptera: Brachycera): Architektur und verteilung in den komplexaugen. Z. Morphol. Tiere. 1974;77:87–125. [Google Scholar]
- 18.Fortini ME, Rubin GM. Analysis of cis-acting requirements of the Rh3 and Rh4 genes reveals a bipartite organization to rhodopsin promoters in Drosophila melanogaster. Genes Dev. 1990;4:444–463. doi: 10.1101/gad.4.3.444. [DOI] [PubMed] [Google Scholar]
- 19.Wernet MF, et al. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 2003;115:267–279. doi: 10.1016/s0092-8674(03)00848-1. [DOI] [PubMed] [Google Scholar]
- 20.von Philipsborn A, Labhart T. A behavioral study of polarization vision in the fly, Musca domestica. J. Comp. Physiol. 1990;167:737–743. [Google Scholar]
- 21.Wolf R, Gebhardt B, Gademann R, Heisenberg M. Polarization sensitivity of course control in Drosophila melanogaster. J. Comp. Physiol. 1980;139:177–191. [Google Scholar]
- 22.Wernet MF, Desplan C. Building a retinal mosaic: cell-fate decision in the fly eye. Trends Cell Biol. 2004;14:576–584. doi: 10.1016/j.tcb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Emmons RB, et al. The Spineless-Aristapedia and Tango bHLH-PAS proteins interact to control antennal and tarsal development in Drosophila. Development. 1999;126:3937–3945. doi: 10.1242/dev.126.17.3937. [DOI] [PubMed] [Google Scholar]
- 24.Duncan DM, Burgess EA, Duncan I. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 1998;12:1290–1303. doi: 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess EA, Duncan I. Direct control of antennal identity by the spineless-aristapedia gene of Drosophila. Mol. Gen. Genet. 1990;221:347–357. doi: 10.1007/BF00259398. [DOI] [PubMed] [Google Scholar]
- 26.Casares F, Mann RS. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- 27.Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 29.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 30.Pignoni F, et al. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- 31.Moses K, Rubin GM. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- 32.Tomlinson A. Patterning the peripheral retina of the fly: decoding a gradient. Dev. Cell. 2003;5:799–809. doi: 10.1016/s1534-5807(03)00326-5. [DOI] [PubMed] [Google Scholar]
- 33.Tahayato A, et al. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev. Cell. 2003;5:391–402. doi: 10.1016/s1534-5807(03)00239-9. [DOI] [PubMed] [Google Scholar]
- 34.Mikeladze-Dvali T, et al. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 2005;122:775–787. doi: 10.1016/j.cell.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 35.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 36.Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 37.Sherry DM, Bui DD, Degrip WJ. Identification and distribution of photoreceptor subtypes in the neotenic tiger salamander retina. Vis. Neurosci. 1998;15:1175–1187. doi: 10.1017/s0952523898156201. [DOI] [PubMed] [Google Scholar]
- 38.White RH, Xu H, Munch TA, Bennett RR, Grable EA. The retina of Manduca sexta: rhodopsin expression, the mosaic of green-, blue- and UV-sensitive photoreceptors, and regional specialization. J. Exp. Biol. 2003;206:3337–3348. doi: 10.1242/jeb.00571. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt A, Vogt A, Friedmann K, Paulsen R, Huber A. Rhodopsin patterning in central photoreceptor cells of the blowfly Calliphora vicina: cloning and characterization of Calliphora rhodopsins Rh3, Rh5 and Rh6. J. Exp. Biol. 2005;208:1247–1256. doi: 10.1242/jeb.01527. [DOI] [PubMed] [Google Scholar]
- 40.Wakakuwa M, Kurasawa M, Giurfa M, Arikawa K. Spectral heterogeneity of honeybee ommatidia. Naturwissenschaften. 2005;92:464–467. doi: 10.1007/s00114-005-0018-5. [DOI] [PubMed] [Google Scholar]
- 41.Haider NB, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nature Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 42.Ng L, et al. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nature Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.