Abstract
Photonic DNA nanostructures are typically prepared by the assembly of multiple sequences of long DNA strands that are conjugated covalently to various dye molecules. Herein we introduce a non-covalent method for the construction of porphyrin-containing DNA nanowires and their networks that uses the programmed assembly of a single, very short, oligodeoxyribonucleotide sequence. Specifically, our strategy exploits a number of supramolecular binding modalities (including DNA base-pairing, metal-ion coordination, and β-cyclodextrin-adamantane derived host-guest interactions) for simultaneous nanowire assembly and porphyrin incorporation. Furthermore, we also show that the resultant DNA-porphyrin assembly can be further functionalized with a complementary “off-the-shelf” DNA binding dye resulting in photonic structures with broadband absorption and energy transfer capabilities.
Keywords: DNA nanostructures, photonic nanowires, supramolecular assembly
Introduction
DNA derived self-assembly is an attractive modality for the construction of organized nanostructures since simple base-pairing rules and hierarchical self-organization can be used to form high-fidelity architectures.1–7 Recent effort has been dedicated to the development of functional DNA nanostructures by incorporating functional entities (e.g., carbon nanotubes,8 polymers,9, 10 viruses,11 and nanoparticles12, 13) onto DNA structures. In this regard, DNA nanostructures containing multiple and well-placed chromophores are of particular interest with applications in light-harvesting, photonics, information transfer, and biomolecular sensing.14–22
The predominant method that is used to achieve DNA-based multi-chromophore nanostructures is to assemble multiple unique sequences of oligodeoxyribonucleotide (ODN) strands that are covalently tethered with dye molecules (or incorporate fluorescent nucleobase analogs).23–28 An alternative and elegant strategy to install chromophores onto DNA nanostructures, without the need for elaborate covalent synthesis, is to use non-covalent binding (e.g., via intercalation, groove-binding, or electrostatic interactions with the phosphate backbone).29–33 However, to the best of our knowledge, there has been no exploration of using orthogonal chromophore-initiated binding to a DNA scaffold as a trigger to facilitate the assembly of photonic DNA nanostructures. Nanostructures that combine DNA assembly and orthogonal supramolecular functionalization are salient because such systems can lead to diverse architectures and engender novel properties.34 Additionally, employing orthogonal supramolecular assembly modes for DNA nanostructure formation can minimize the quantity of the DNA domain utilized, in terms of both the number of unique ODN sequences used and their length.
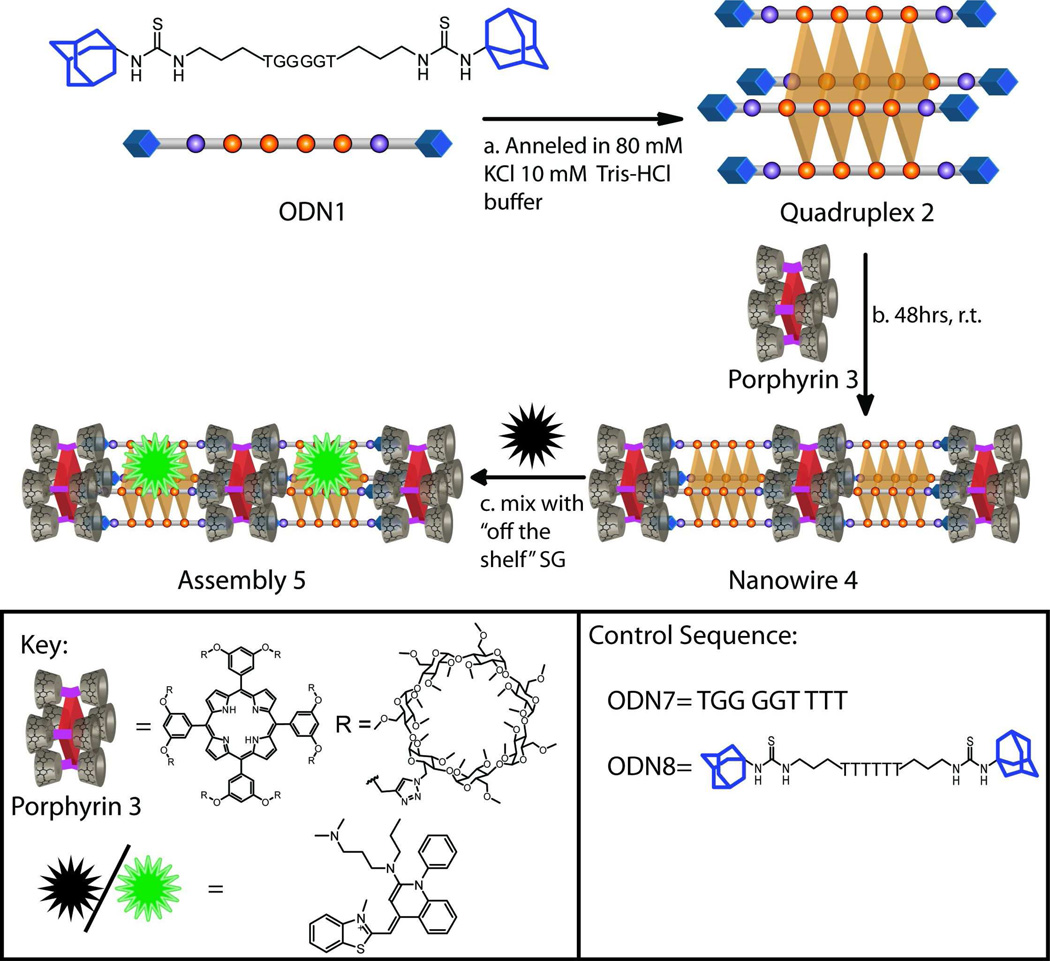
With these considerations in mind, we describe an approach that uses a range of supramolecular assembly modalities to construct chromophore-containing DNA-based nanowires and networks of wires. Importantly, in terms of the DNA domain, the nanowires are formed using only one, very short (6 nt long), ODN sequence (1). First, ODN 1, functionalized with two adamantane arms, self-assembles into a tetramolecular quadruplex 2 in the presence of templating potassium ions (Scheme 1). Second, incubation of quadruplex 2 with free-base porphyrin 3 decorated with eight permethylated β-cyclodextrin (PMβ-CD) arms leads to the formation of a porphyrin-containing DNA nanowire (and its higher-order networks) as a result of multivalent β-CD-adamantane based host-guest interactions. In addition, the assembly is further non-covalently functionalized with an “off-the-shelf” DNA-binding dye, resulting in novel photonic structures with broadband absorption properties in the visible spectrum (∼300 – 665 nm) and energy transfer capability from the DNA-binding dye donor to the free-base porphyrin acceptor.
Scheme 1.
Hierarchical construction of multi-chromophore containing DNA-based assembly 5.(a) Potassium ion induced self-assembly of bis-adamantane flanked ODN 1 into tetramolecular quadruplex 2. (b) Host-guest interactions derived assembly of quadruplex 2 and porphyrin 3 into multi-porphyrin containing nanowire 4 and its networks (see Scheme 2 for details). (c) Incorporation of a DNA-binding dye, SYBR green 1 (SG), to form assembly 5. Note: for exact chemical structures of the ODNs, see SI Scheme S1.
Our multi-chromophore containing DNA assembly design focused on guanine rich ODN 1 (d(AdTG4TAd), Ad = adamantyl head group). The core sequence contains four repetitive guanine residues because, in the presence of K+ ions, such a track of guanines form stable parallel guanine quadruplexes—a non-canonical DNA motif composed of repetitive stacks of guanine quartets (wherein the guanines are co-planar and are stabilized by hydrogen-bond driven base-pairing and metal ion-lone pair interactions).35 Furthermore, flanking thymine nucleobases and adamantane head-groups were added to the design to destabilize potential mismatched aggregation that can lead to conventional guanine wires.36–38 In addition, the adamantane moieties were specifically included since the self-assembled quadruplex 2 would then project four adamantane units from each of its two termini. Based on earlier work by our group and others on multivalent host-guest interactions on non-DNA derived supramolecular nanostructures,39–42 we envisioned that addition of a non-covalently “cross-linking” free-base porphyrin (3) that presents four PMβ-CD hosts from each face (and has dimensions that closely match the terminal face of a DNA quadruplex)43 should result in DNA-porphyrin array 4 as a result of repetitive multivalent host-guest interactions.
Results
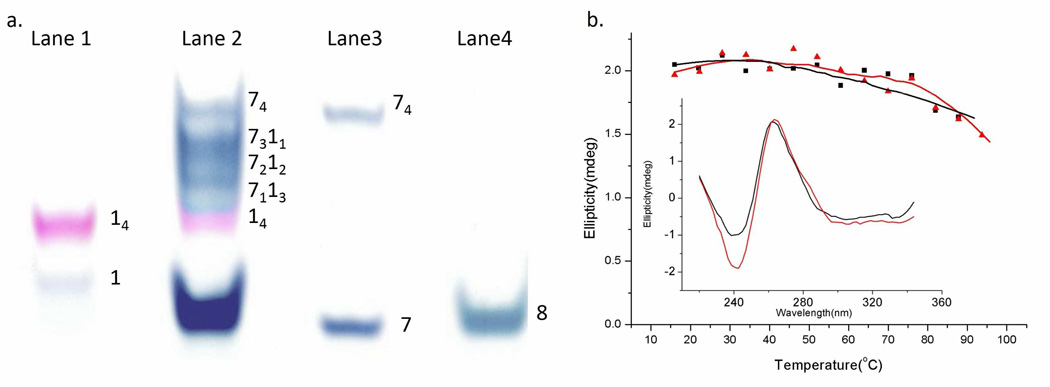
A key requisite of our self-assembly strategy is that ODN 1 first forms parallel tetramolecular quadruplex 2 (i.e., d(AdTG4TAd)4 or 14) in the presence of potassium ions (Scheme 1). Evidence for the formation of quadruplex 2 was obtained by (a) non-denaturing polyacrylamide gel electrophoresis (PAGE) and, (b) circular dichroism spectroscopy. When ODN 1 was incubated under standard quadruplex forming conditions44(see experimental section) in potassium containing buffer (80 mM KCl, 10 mM Tris-HCl, pH 7.5) and then loaded and run on a polyacrylamide gel, two distinct bands are observed (see Figure 1a, lane 1). The weak band that migrates fast is ascribed to a small amount of single-stranded ODN 1, as this migrates similar to a single stranded control (lane 4: ODN 8; d(AdT6Ad)), that has same length and terminal functionalization as ODN 1 but only contains thymine bases and thus cannot form a higher-order assembly. The intense slow-migrating band in lane 1 suggests the formation of a higher-order species i.e., tetramolecular quadruplex 2 (or 14). A well-established technique to probe whether tetramolecular quadruplexes are formed is to co-incubate two putative tetramolecular quadruplex forming strands containing the same track of guanines but are of varying length and see if a mixture of five unique quadruplexes are formed, each of distinct size and migration capacity.45 Thus, we incubated a long strand containing a track of four guanines ODN 7 (TG4T4: which is known to form a tetramolecular quadruplex in the presence of potassium cations)46 with ODN 1 under quadruplex forming conditions. As can be seen on lane 2, the resulting gel shows the presence of five slow migrating bands (corresponding to 74, 7311, 7212, 7113, and 14), verifying that both ODN 7 and ODN 1 form tetramolecular quadruplexes. It is also interesting to note that the homomeric tetramolecular quadruplex 2 band stains a unique purple color (when exposed to the DNA binding dye Stains-all) while the other quadruplexes stain a blue color.
Figure 1.
(a) 20% non-denaturing PAGE of ODNs after exposure to quadruplex forming conditions. Lane 1: ODN 1, lane 2: a mixture of ODN 1 and ODN 7, lane 3: ODN 7, lane 4: ODN 8. Gel was run at 150 V for 1 hr 20 min and visualized using Stains-all. (b) Thermal denaturation profile for quadruplex 2 (black squares) and quadruplex 74 (red triangles). Inset: Circular dichroism profile of quadruplex 2 (black line) and quadruplex 74 (red line) at 20°C. All circular dichroism experiments were conducted in 80 mM KCl, 10 mM Tris-HCl, pH 7.5 buffer and concentration of the quadruplexes were 4 µM.
Further evidence for quadruplex formation came from circular dichroism spectroscopy. In particular, upon incubation in quadruplex forming conditions, both ODN 1 and control ODN 7 exhibited characteristic profile for a parallel quadruplex structure, with a strong positive circular dichroism peak at 263 nm and a relatively weak negative peak at 242 nm (Figure 1b, inset).46, 47 In an effort to discern the thermal stability of the quadruplexes, we performed thermal denaturation experiments. As can be gauged from Figure 1b both quadruplex 2 (14) and quadruplex 74 are thermally very stable and show no significant denaturation.
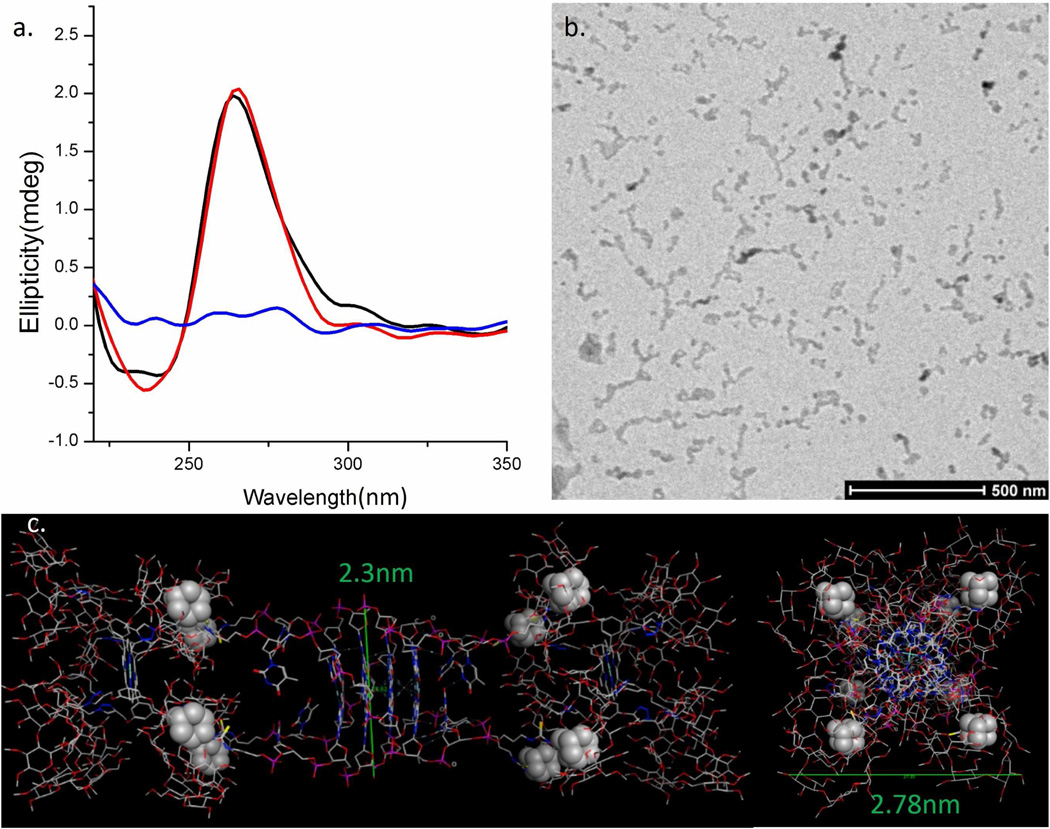
Since ODN 1 can form stable quadruplex 2, we subsequently incubated 2 for 48 hr (at room temperature) in the presence of water-soluble porphyrin 3 in a 1:1 ratio (see experimental section for assembly forming procedure). Circular dichroism spectroscopy of the resulting solution (shown in Figure 2a) clearly indicates that the parallel quadruplex structure of 2 is not destabilized by the presence of porphyrin 3. In order to probe whether an assembly was formed between quadruplex 2 and porphyrin 3 in conditions that mimic the solution phase, cryo- transmission electron microscopy (Cryo-TEM) was employed. As shown in Figure 2b, the Cryo-TEM image of a mixture of 2 and 3 (each diluted to 4 µM, after nanostructure forming conditions were employed) displayed wire-like structures from ∼100 nm to 200 nm in length and have a width between 7–20 nm. The high aspect ratio of these nanostructures suggests a directional assembly composed of consecutive host-guest interactions between the quadruplex and porphyrin units. However, given that a model for single nanowire 4 estimates the diameter to be 2.3–2.8 nm (Figure 2c), we postulated that the images seen in the Cryo-TEM are due to the higher-order association of single wires. Interestingly, some of these bundles display branching suggesting a flexible assembly where a single face of quadruplex 2 (or porphyrin 3) can bind to multiple partner molecules (i.e., via out-of-register host-guest interactions, see discussion section for more detail).
Figure 2.
(a) Circular dichroism spectra of quadruplex 2 (black), porphyrin 3 (blue), and a mixture of 2 and 3 (red) in potassium containing buffer (80 mM KCl, 10 mM Tris-HCl, pH 7.5). (b) Cryo-TEM image of assembly 4 in 160 mM KCl, 10 mM Tris-HCl, pH 7.5. Note: The concentrations of quadruplex 2 and porphyrin 3 for these studies were both 4 µM. (c) A representative model of a section of nanowire 4 (composed of two molecules of porphyrin 3 and one molecule of quadruplex 2). This model is based on the X-ray structure of the core quadruplex sequence (d[TGGGGT]4); PDB code 1S45. Note: the space-filled atoms are the adamantane binding units. The structure on the Left is a side view and structure on the Right is a top-view. The model was energy minimized (AMBER94 forcefield) using the MOE software. Note: This model is only a depiction and not a rigorous calculation.
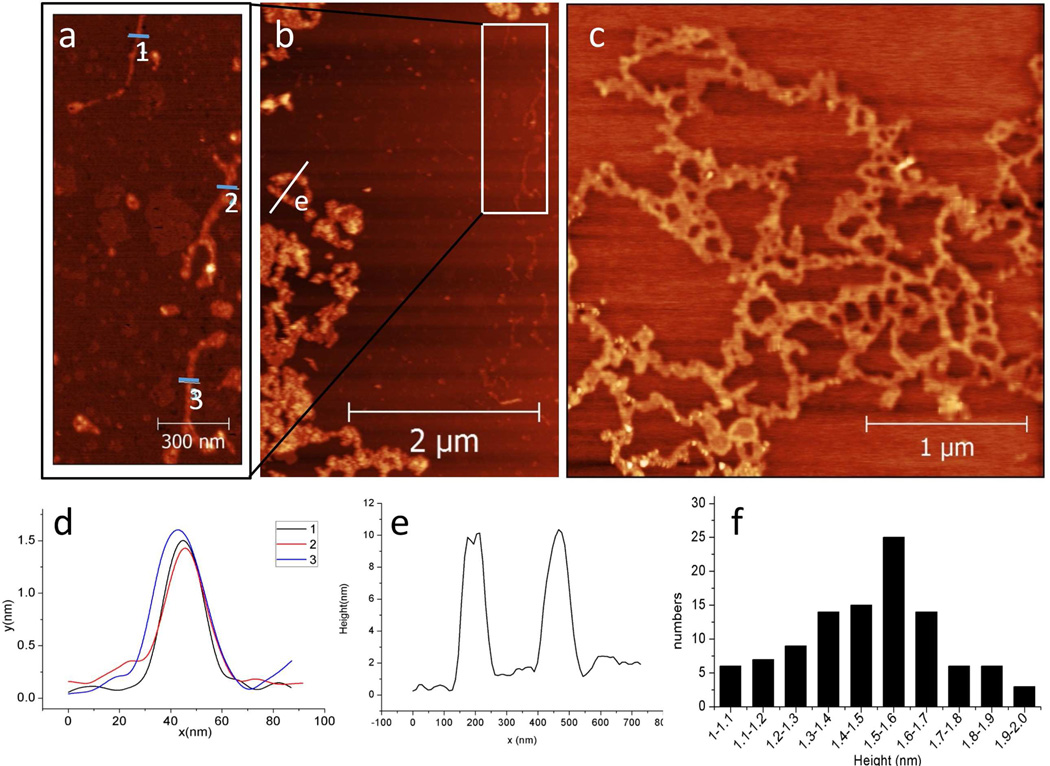
Atomic force microscopy (AFM) was also utilized to determine the structure of the DNA-porphyrin assembly. Since it is well established that Mg2+ ions can facilitate immobilization of ODN nanostructures onto mica, AFM measurements were conducted on Mg2+ treated, freshly cleaved, mica substrate under tapping mode in air.48–50 When AFM studies were conducted on a mixture of 2 and 3 (4 µM each), single nanowires are observed, as can be seen on the AFM image shown in Figure 3a (which is a zoom-in image from the white rectangle shown on Figure 3b). The apparent height of the single wires was found to be ca. 1.5 nm (see height profile on Figure 3d). This height—though shorter than expected—is not only in accord with previously reported STM images of porphyrin-only nanowires prepared from porphyrin 3 metalated with zinc,40 but is also in accord with previously reported AFM (∼1.6 nm)38 and STM (∼1.5 nm)51 data of conventional guanine wires. Furthermore, soft nanomaterials assembled from DNA and/or cyclodextrins typically exhibit lower measured heights when imaged on a surface due to various artifacts including high humidity, indentation of the assembly by the tip, and substrate induced flattening.51–53 In this case, the decreased height of nanowire 4 is thought to be due to a combination of the abovementioned artifacts and nanowire flexibility.
Figure 3.
(a) A 0.75×1.4 µm2 zoom-in image from rectangle shown in panel b (b) 3×5 µm2 and (c) 3×3 µm2 AFM images of assembly 4 on mica substrate. Note: The concentrations of quadruplex 2 and porphyrin 3 were both 4 µM. (d) Height profiles collected at the indicated white line (labeled 1,2,3,respectively) on panel a. (e) Height profile collected at the indicated white lines (labeled e) on panel b. (f) Histogram height distribution of the networks shown in panel c.
In addition, we also observed various aggregate states of the wires. For instance, networks of single wires are observed (Figure 3c and histogram on Figure 3f). Interestingly, these networks are composed of single wires with branching points that show the same height as other regions of the wires (see SI Figure S3 for further analysis of these height profiles). Taken together, these experiments suggest that the nanowire networks are formed by one face of the quadruplex or porphyrin units binding to more than one partner molecule, in a mismatched fashion. Larger networks composed of higher-order bundles of wires are also observed in these AFM images (see left hand portion of image shown in Figure 3b and height profile in Figure 3e).
The above-mentioned supramolecular strategy has thus far only utilized the DNA domain as a self-assembling scaffold to project the adamantane arms in a multivalent fashion. We were keen on investigating whether the core DNA domain can be used for further molecular recognition to rapidly incorporate complementary dyes within the DNA-porphyrin array. In particular, the fluorescence properties of assembly 4 was probed upon addition of a green fluorescent DNA binding dye, SYBR Green I (SG). This dye was specifically chosen because it exhibits significantly enhanced fluorescence upon binding (via stacking and groove-binding)54 to structured DNA assemblies but not to single stranded DNA. Furthermore, SG complements the porphyrin chromophore since it absorbs strongly between 450 and 540 nm (see SI Figure S8 for UV-Vis and fluorescence spectra of SG and porphyrin 3), a region of the visible spectrum where porphyrin 3 does not absorb significantly. Last, the emission spectrum of SG overlaps with the Q-absorption bands of porphyrin 3 thereby enabling us to investigate fluorescence resonance energy transfer.
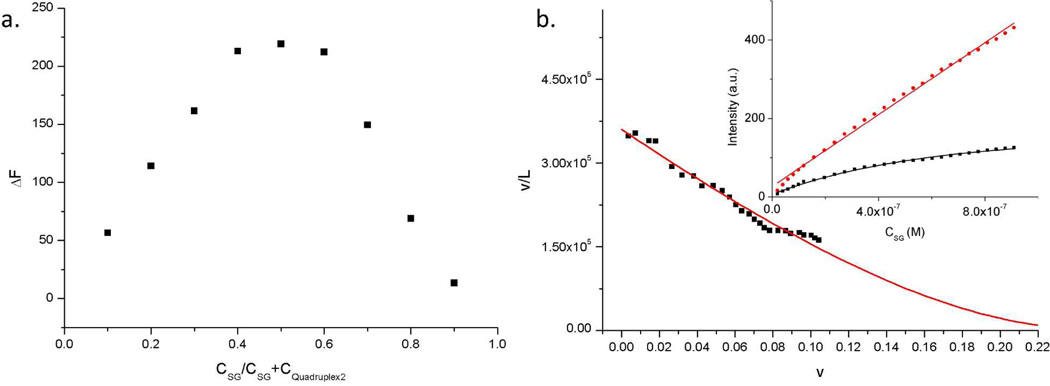
While the binding behavior of SG to duplex DNA has been described in the literature,54 the binding interaction of SG to tetramolecular quadruplexes has not been investigated. Preliminary UV-Vis studies suggested that SG exists as a monomer in our buffer conditions and that the absorption maximum of SG shifts from 493 nm to 500 nm upon addition of quadruplex 2 (See SI Figure S6 for absorption data). Further, we probed whether SG binds to quadruplex 2 via fluorescence spectroscopy. These studies suggested that the fluorescence of SG is drastically enhanced in the presence of quadruplex 2 thus giving a convenient spectroscopic signature for quadruplex-dye interaction. The binding stoichiometry of SG to 2 was investigated using a Job plot performed in the same quadruplex forming buffer, and following the fluorescence enhancement (ΔF) of SG when complexed with quadruplex 2. This plot (Figure 4a) clearly shows that under these concentrations a 1:1 binding stoichiometry is present for SG and quadruplex 2.
Figure 4.
(a) Job plot for complexation of SG with quadruplex 2, based on the fluorescence enhancement (AF) of SG when it binds to quadruplex 2. For the Job plot the total concentration of SG and quadruplex 2 were held constant at 4 µM. (b) Scatchard coordinates showing SG binding to quadruplex 2. The red line shows the non-linear curve-fit (R2 = 0.97). Inset: Isotherms showing the binding of SG to quadruplex 2. The fluorescence intensity of SG (Ex λ = 495 nm, Em λ = 525 nm) was measured upon the titration of SG into 0.6 µM quadruplex 2 (i.e., 2.4 µM in terms of guanine-quartets, black line) and 15 µM quadruplex 2 (60 µM G-quartets, red line) in 10 mM TrisHCl buffer and 80 mM KCl, pH 7.5.
In addition to the abovementioned fluorescence measurements, binding stoichiometry and binding constant (Ka) were obtained by measuring the dependence of SG fluorescence upon titration of SG into quadruplex 2 (Figure 4b, inset) and converting these results into Scatchard coordinates (see experimental section) followed by non-linear curve-fitting to equation 1 (Figure 4b).54
| (1) |
Equation 1 describes the conditional probability model for non-cooperative excluded site binding of ligand-lattice interactions, derived by McGhee and VonHippel.55 In equation 1, v is the number of bound SG per guanine quartet (a guanine quartet is the tetrad composed of four co-planar guanines), L is the concentration of free SG in solution at equilibrium, Ka is the observed association constant and n is the size of binding site in number of guanine quartets. Analyzing the titration data resulted in a Ka of 3.60 ± 0.06 × 105 M−1. Further, the binding site size of SG to quadruplex 2 in terms of guanine quartets was found to be n = 3.66 ± 0.08 (i.e., close to 4 guanine-quartets), which further suggests that SG binds to DNA quadruplex 2 in a 1:1 stoichiometry since quadruplex 2 has, in total, only 4 guanine quartets.
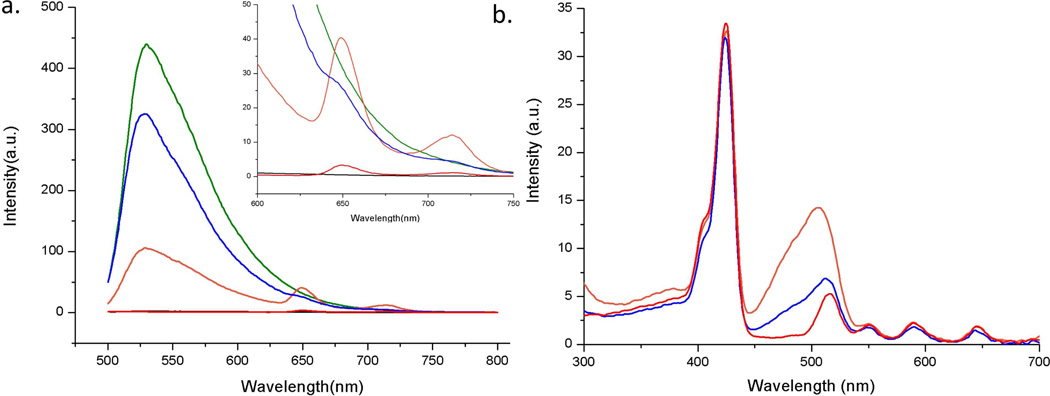
Upon determining that SG binds quadruplex 2 in a 1:1 fashion, we next set out to investigate the energy transfer capacity of SG to porphyrin 3 within the nanostructure. As shown in Figure 5a, SG displays no appreciable fluorescence when excited (at 480 nm) in the absence of quadruplex 2. In marked contrast, the fluorescence emission of SG is enhanced 200 fold upon addition to a solution of quadruplex 2 (SG/2 ratio = 1:1), again clearly suggesting binding of SG with the DNA quadruplex. Interestingly, when SG is added to assembly 4 (such that SG/2 ratio = 1:1) this enhancement in emission is diminished significantly and the presence of porphyrin-based emission bands at 650 and 715 nm is observed. The intensities of these porphyrin bands are ca. 10 fold higher than when only assembly 4 is excited at 480 nm (since porphyrin 3 has rather low absorption at 480 nm). Taken together, these experiments suggest that assembly 4 can be functionalized with SG resulting in a multi-chromophore array 5 and that an energy transfer process is operational from the SG donor to the porphyrin acceptor. Assuming that all of the quenching is due to FRET, an energy transfer efficiency of 75% was estimated based on the fluorescence emission profiles of SG:2 (donor) complex and assembly 5 (quenched state) according to the equation Ket = 1-F5/Fsg+2, where Ket is energy transfer efficiency, F5 and Fsg+2 are the areas of the emission profile from assembly 5 and the SG:2 complex, respectively.57
Figure 5.
(a) Solution based fluorescence (excitation λ = 480 nm) emission profile (inset shows the enlarged profiles around the porphyrin 3 emission region (from 600 nm – 750 nm) and (b) excitation profile (observed at emission λ = 715 nm,) of array 4 (red), SG only (black), SG + quadruplex 2 (green), assembly 5 (orange), and assembly 5 + excess β-CD (blue). Note: All solutions were 4 µM in quadruplex DNA, porphyrin 3, and SG. For excitation profiles a 500 nm cut off long pass filter was applied in front of the observation window
In order to verify that the energy transfer was due to a host-guest derived assembly, we also mixed assembly 5 with a large excess of free β-CD (250 equiv. per each PMβ-CD unit on porphyrin 3). The fluorescence of the resulting solution shows that the high intensity emission (at 530 nm) corresponding to the complex of SG and quadruplex 2 is substantially restored and the emission from the porphyrin peaks is diminished.
The excitation spectra (followed at 715 nm, where porphyrin 3 predominantly emits) of assemblies 4, 5, and 5 after incubation with excess β-CD were also investigated. The results are displayed in Figure 5b and clearly show that array 5 (i.e., assembly 4 + SG) possesses broadband excitation from 300 nm to 665 nm. This enhanced absorption profile is due to the transfer of excitation energy from SG to porphyrin 3 within assembly 5. Another feature that is clear from the excitation spectra shown in Figure 5b is that the SG-associated excitation bands are significantly diminished in intensity when the wire is dis-assembled (i.e., assembly 5 + excess β-CD). Taken together, these results further support the notion that the energy transfer from SG to porphyrin 3 is a result of the structure of self-assembled array 5 that is held together through host-guest interactions between porphyrin 3 and quadruplex 2.
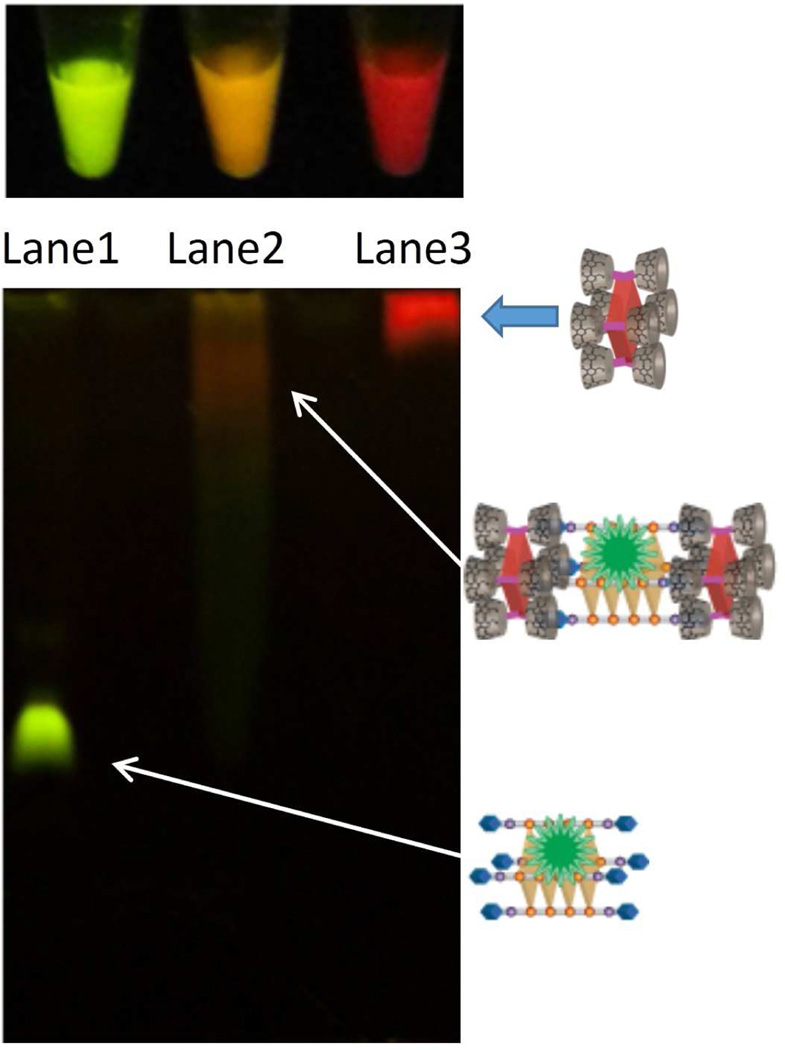
Further characterization of photonic supramolecular assembly 5 in terms of composition was obtained from non-denaturing gel-electrophoresis. Figure 6 (bottom) shows the result of a non-denaturing PAGE that visualizes the fluorescence emission (excitation using a broadband handheld UV-lamp; excitation λmax = 365 nm) of quadruplex 2, porphyrin 3, and assembly 4, each pre-mixed with excess SG. It is evident that DNA quadruplex 2 (lane 1) migrates fast and displays green fluorescence, as a result of binding to SG. On the other hand, porphyrin 3 (lane 3) hardly migrates and displays only porphyrin-based red fluorescence since SG does not bind to the porphyrin. Importantly, assembly 4 + SG (lane 2) displays a broad band that migrates much slower than quadruplex 2 indicating a distribution of high molecular weight species (which can be dis-assembled in the presence of excess free β-CD; see SI-Figure S12 for additional PAGE studies). Furthermore, this new band, ascribed to assembly 5, exhibits fluorescence that is orange in color, suggesting that both the porphyrin and SG-bound quadruplex units are present. Interestingly, these distinct fluorescence emission colors are consistent with those observed in solution by the naked eye (Figure 6, top) as well as by confocal microscopy (see SI-Figure S11 for images of fluorescent particles made up of assembly 4 or 5).
Figure 6.
(top) Fluorescent solutions (from left to right) composed of quadruplex 2 + SG, assembly 4 + SG, and porphyrin 3 + SG, respectively. The solutions were excited with a broadband UV-lamp, λmax = 365 nm. (bottom) 15% Non-denaturing PAGE. Lanes 1 through 3 correspond to SG pre-mixed with quadruplex 2, assembly 4, and porphyrin 3, respectively. The gel was imaged upon excitation with a broadband UV-lamp (λmax = 365 nm).
Discussion
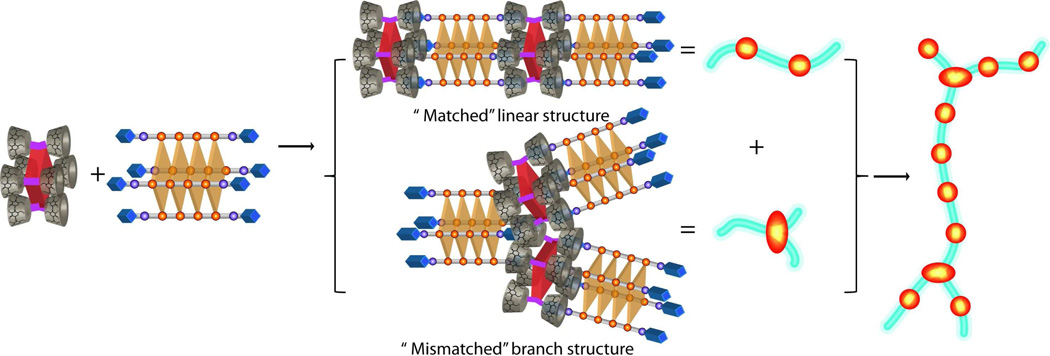
In previous studies we reported that the binding of the zinc coordinated version of tetraphenyl porphyrin 3 containing eight cyclodextrin (host) arms with a complementary tetraphenyl porphyrin projecting eight adamantane (guest) arms can lead to unidirectional nanowires as a result of good geometric matching (i.e., both supramolecular monomer cores are tetraphenyl porphyrins) and repetitive multivalent host-guest interactions.40 In the current manuscript, we provide new insight into how a self-assembled DNA quadruplex 2 projecting eight adamantane guest arms can assemble with porphyrin 3 to form nanowires and their networks. One rationale for using a DNA quadruplex scaffold was because the dimensions (∼1.1 nm × 1.1 nm)43 and four fold symmetry of the porphyrin macrocycle nicely match the geometry (∼1.1 nm × 1.1 nm)43 and four fold symmetry of a guanine quartet. Indeed, as can be seen in Figure 2c a linear nanowire should form when the self-assembling partners are properly aligned. However, these quadruplex-porphyrin assemblies exhibit regions that are unidirectional as well as regions containing junctions. Branching points were observed every 50–200 nanometers on the AFM image of the single wires (Figure 3a and 3c). The reason for this branching is thought to be mainly due to the flexibility of quadruplex 2. Unlike the covalently held porphyrin scaffold, quadruplex 2 is non-covalently assembled and the terminal thymine residues are not base-paired and thus are expected to be flexible. Further flexibility comes from the alkyl spacers that connect the adamantane thiourea arms to the terminal deoxyribose of the thymine residues (see SI Scheme S1 for the full chemical structure of ODN 1). Such flexibility may lead to out-of-register binding. For example, two adamantyl groups on one end of quadruplex 2 could bind to two cyclodextrin arms of one porphyrin and the remaining two adamantyl arms could bind to two cyclodextrin arms of another porphyrin (see Scheme 2). This type of mismatched binding mode can explain the presence of branching regions as well as to the formation of a network of single wires.
Scheme 2.
An illustration depicting the formation of linear wires, mismatched branched structures, and branched nanowire networks.
In addition to probing the formation of the quadruplex-porphyrin assembly, we have also investigated the possibility of non-covalently incorporating an additional complementary chromophore into the system, by using a dye, SG, which is known to bind to structured DNA assemblies. According to the fluorescence titration data, at the concentrations of SG used in our studies (< 4 µM) and the relatively large salt concentration (80 mM KCl), SG binds to quadruplex 2 in a 1:1 stoichiometry. Specifically, it was found that the binding site of SG to quadruplex 2 spans nearly all four guanine quartets (3.66 quartets). Furthermore, SG has been shown to bind to duplex DNA via a similarly large binding size (3.5 base-pairs)54.Detailed studies with duplex DNA has led to the proposal that SG undergoes intercalation via the benzo-thiazole and phenyl-quinilinium aromatic moieties while the propyl and dimethylaminopropyl arms span (and bind to) the minor groove. While the precise binding mode of SG to quadruplex DNA remains to be elucidated, since SG binds with a similar binding size and shows substantial enhancement in fluorescence when bound to either duplex DNA or quadruplex DNA, we speculate that the aromatic rings on SG are likely to intercalate between two guanine quartets of quadruplex 2 with the propyl and dimethylaminopropyl arms interacting with the quadruplex groove.
SG is a member of the monomethine family of cyanine dyes and is routinely utilized to detect double stranded DNA, and as we have now shown can also bind to quadruplex 2 in a 1:1 stoichiometry. Other cyanine dyes can also interact with quadruplex DNA in a 1:1 fashion. For instance, the Armitage group has observed that the carbocyanine dye DiSC(3) binds to a folded intramolecular quadruplex with a 1:1 stoichiometry.58 However, the nature of the cyanine dye is salient since another cyanine-based chromophore, DMSB, is also capable of binding to tetramolecular quadruplex DNA in a 2:1 fashion (where the dye forms a stacked homo-dimer)59. The incorporation of such cyanine based homodimers, or even chromophore tethered quadruplex DNA binding ligands (such as Distamycin A)60 that bind to quadruplex DNA with higher stoichiometry, may allow for further enhancement of the amount of light energy funneled into the porphyrin acceptor within the nanowire scaffold.
Analysis of the fluorescence properties of the porphyrin/quadruplex/SG assembly 5 shows that an energy transfer mechanism is operational leading to funneling of energy into the porphyrin macrocycle. Migration of energy onto a porphyrin chromophore precisely placed within a DNA nanostructure is of interest in terms of applications since such photoactive supramolecular complexes could lead to the development of self-assembled antenna structures for artificial light harvesting,26 or singlet oxygen generation (e.g., with potential in light-activated antibacterial applications)61.In addition, there is a need for the identification of quadruplex DNA specific binding agents62, thus such nanostructured assemblies incorporating quadruplex DNA could lead to high-throughput surface-based sensing platforms that can detect quadruplex binding agents via the disruption of an energy-transfer process.21 For instance, a small molecule or protein that binds to the quadruplex domain and displaces the dye will lead to loss of energy transfer to the porphyrin and thus can be detected.
From a basic science standpoint, in contrast to traditional covalent chemistry, the reported self-assembly strategy offers a straightforward route to investigate a variety of chromophore compositions by a simple mix and match protocol. In particular, a variety of metalated porphyrins (with unique absorption and emission properties) and other “off-the-shelf” quadruplex DNA binding dyes can be incorporated onto the nanowire networks leading to facile modulation of the absorption and energy transfer properties of the self-assembled nanostructures. Further, in a more general sense, this work is expected to serve as a stepping stone wherein better defined two- and three-dimensional DNA-based photonic nanostructures with directional FRET cascades can be constructed using an approach that minimizes the DNA domain utilized and maximizes orthogonal synthetic self-assembly in water.
Conclusions
In summary, we have developed a multifaceted supramolecular self-assembly strategy for the construction of photonic DNA-based nanowires and their networks. In contrast to traditional DNA-based photonic assemblies, this design starts from a single short ODN sequence without any chromophores covalently attached to it. It harnesses a number of important self-assembly modes in water including, DNA base-pairing, β-CD/adamantane derived host-guest interactions, and DNA-dye binding, to both facilitate nanostructure assembly and to incorporate complementary porphyrin and SG chromophores. Together these chromophores show broad spectrum absorption, from 300 nm to 665 nm. Further the SG unit is a commercially available DNA-binding dye. Thus this self-assembly strategy offers a simple route to investigate multi-chromophore containing photonic nanostructures by a simple mix and match protocol. We are currently investigating porphyrin-based DNA wires incorporating other “off-the-shelf” DNA binding dyes to modulate the absorption and energy transfer properties in the self-assembled nanostructures.
Experimental Section
1. General Experimental
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich or Acros Organics and solvents were purchased from Fischer Scientific. Sybr Green I (10,000x solution in DMSO, 1x = 0.68 µM) was purchased from Life Technologies. NMR spectra were recorded on a Varian 400 MHz spectrometer using CDCl3 as solvent. MALDI-TOF spectra were recorded on a Bruker Autoflex 3 Matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometer (MALDI-TOF MS). The matrices used were 2,5-dihydroxybenzoic acid (DHB) for oligodeoxyribonucleotide (ODNs) and α-cyano-4-hydroxycinnamic acid for the small molecules. UV-Vis studies were undertaken using a Hewlett Packard 8452A Diode Array Spectrophotometer. Fluorescence spectra were recorded on a Varian Cary Eclipse Fluorescence Spectrophotometer.
CD experiments
Circular dichroism spectra were taken on an Olis RSM 1000 CD using a cylindrical cuvette with 1 mm path length. The quadruplex formation of ODN 1 was monitored by CD and the data was subtracted from the spectra of a solution containing only buffer (10 mM Tris-HCl, 80 mM KCl, pH 7.5).
PAGE Studies
Non-denaturing polyacrylamide gel electrophoresis studies were conducted using a BioRad mini protean tetra cell equipped with BioRad PowerPac HC. For the gel shown in Figure 1a, in lanes 1, 3, and 4, ∼8 × 10−4 µmole of ssDNA was introduced, and in lane 2, ∼16 × 10−4 µmole of single strand ODN 1 and ∼16 × 10−4 µmole of single strand ODN 7 were used. All ODNs were applied to the gel after exposure to the standard quadruplex forming condition. The gel was stained using DNA binding dye Stains all, and imaged on a Nikon D200 camera.
For each lane of the gel shown in Figure 6, ∼2 × 10−4 µmole of quadruplex DNA and/or porphyrin 3 were premixed with 2 µL of 300x SybrGreen I (∼4 × 10−4 µmole ) for 15 min before applying to the gel. The running buffer was 1x TBE buffer containing 24 mM KCl at 110 V for 1.5 hrs. The gel was imaged on a Nikon D200 camera upon excitation with a broadband UV-lamp (λmax = 365 nm). A 520 nm cut-off filter (blocks light < 520 nm) was applied in front of the camera lens to remove any background/scattered light.
HPLC conditions
RP-HPLC purification was achieved using a Varian Prostar HPLC system, equipped with a Polymer Laboratories 100 Å 5 µm PLRP-S reverse phase column. The column was maintained at 65 °C for all runs. The flow rate was set at 0.75 mL/min. A gradient composed of two solvents (solvent A is 0.1 M TEAA in 5% acetonitrile and solvent B is 100% acetonitrile) was used.
Microscopy Protocols
Cryo-TEM experiments were performed on a FEI Tecnai G2 F30 Twin Transmission Electron Microscope Instrument (accelerating voltage = 120 kV). Sample was prepared on 200 mesh copper grids with lacey carbon film (purchased from Electron Microscopy Science). In order to facilitate the formation of higher order assemblies that show better contrast on TEM images, the sample was prepared in a 10 mM TrisHCl pH 7.5 buffer that contains 160 mM KCl.
AFM experiments were carried out on a Veeco Bioscope AFM (Digital Instruments) under tapping mode in air. Bruker OTESPA AFM probes with nominal frequency, tip diameter, and spring constants of 300 KHz, 7 nm, and 42 N/m, respectively were used. Mica (highest grade V1 Mica disc, 10 mm diameter, was purchased from TED PELLA, Inc.) was used as the substrate and the mica plate was freshly cleaved via scotch tape to achieve a flat surface before use. Prior to sample introduction, a 2 mM MgCl2 aqueous solution was applied for 5 minutes and then dried using a nitrogen gas flow. A solution of assembly 4 was subsequently applied on the mica substrate for 10 minutes, then washed 10 times with 50 µL aliquots of deionized water. After drying with nitrogen gas flow, the sample was imaged.
Confocal microscopy images were collected on a Nikon A1RSi equipped with a 32-channel multianode photomultiplier detector. 488 nanometer laser excitation was used. The spectral data was acquired in sequential bandwidths of 10 nm spanning the wavelength range from 492–742 nm using 26 PMT channels to generate a lambda stack.
Analysis of SG binding to quadruplex 2 via fluorescence spectroscopy
This procedure is adopted from ref 54. Varying concentrations (0.02–1µM) of SG was titrated into a solution containing quadruplex 2 (2.4 µM in terms of guanine-quartets) and the observed fluorescence intensity (F) of the SG:quadruplex 2 complex was used to generate an isotherm (black line, Figure 4b, inset). In order to determine the fluorescence intensity of 100% bound SG (Fb), another titration was conducted where varying concentrations of SG was titrated into a solution containing a large excess of quadruplex 2 (60 µM in terms of guanine-quartets; red line, Figure 4b, inset). Note: since the fluorescence of the free SG in solution is significantly lower than the fluorescence of the SG:quadruplex 2 complex (Fcomplex/Ffree> 200 fold), the contribution of the fluorescence of the free SG dye to the total fluorescence intensity is negligible. Accordingly, the fraction of SG dye bound to quadruplex 2 can be expressed as:
| (2) |
Where F is the observed fluorescence intensity of the SG:quadruplex 2 complex and Fb is the observed fluorescence intensity of 100% SG bound to quadruplex 2.
At equilibrium, the concentration of free SG can be expressed as:
| (3) |
and the SG binding density (which is the number of bound SG per guanine-quartet) can be expressed as:
| (4) |
where CSG and CQ correspond to the total concentrations of SG and guanine-quartets in solution, respectively.
After determining v and L, a Scatchard plot was constructed, as shown in Figure 4b, and nonlinear regression analysis was used to fit the data to equation 1.
2. ODN Synthesis
General Protocol. After synthesis (vide infra) the ODNs were first purified with sephadex resin Microspin G-25 columns (GE Healthcare) and then were chromatographed on a Varian Prostar reverse-phase HPLC complete with a MetaTherm column heater. Once purified the ODNs were characterized by MALDI-TOF in linear negative mode. Concentrations of stock solutions of ODNs were quantified based on their 260 nm electronic absorption at 85°C and their molar extinction coefficients were obtained by standard nearest neighbor calculations.
Synthesis of core ODNs. ODN 6 (structure shown in SI, Scheme S1) was synthesized at the Keck Foundation Biotechnology Resource Laboratory at Yale University using standard automated solid phase synthesis. Modified phosphoramidites (5’-Amino-Modifier C3 and 3’-PT Amino-Mod C3) were purchased from Glen Research. After HPLC purification, ODN 6 was analyzed by MALDI-TOF under negative linear mode (See SI Figure S1, bottom; Found m/z = 2138.18 Da, calculated mass [M-H]−= 2136.39 Da).
-
General Synthesis of bis-adamantane functionalized ODN 1 and ODN 8. The RP-HPLC purified core ODN 6 bearing primary amino groups on both the 3' and 5' ends, was reacted with excess 1-adamantyl isothiocyanate (10 mg in 1 mL of DMSO), DIPEA (50 µL) and 500 µL of 60 mM sodium carbonate buffer (pH = 8.5). The resulting solution was agitated overnight at 55°C and then unreacted 1-adamantyl isothiocyanate was precipitated by adding 2 mL of H2O. The white precipitate was filtered off and the solvents were removed using a Savant SPD IIIV speed rotorvap. The resulting crude residue (ODN 1) was dissolved in 0.1 M TEAA buffer, desalted using a G-25 spin column, purified by HPLC (see ESI Figure S1, top), and analyzed by MALDI-TOF under negative linear mode (SI Figure S1, bottom; Found m/z = 2523.11 Da, calculated mass [M-H]− = 2523.00 Da).
ODN 8 was synthesized using the same procedure discussed above, starting from a modified core sequence that contains six thymidines (TTTTTT) bearing primary amino groups on both the 3’ and 5’ ends. The final product ODN 8 was analyzed by MALDI-TOF under negative linear mode (SI Figure S1 bottom; Found m/z = 2422.24 Da calculated mass [M-H]− = 2422.95 Da)
3. Standard Incubation Process for Formation of Quadruplex DNA
A stock solution of single strand ODN (1 or 7) was diluted to 160 µM in potassium containing Tris-HCl buffer (10 mM Tris-HCl, 80 mM KCl, pH 7.5) and sealed in an eppendorf tube.The tube was heated to 90°C for 10 minutes and then cooled at 4°C for 48 hr. The resultant solution containing 40 µM tetramolecular quadruplex (2 or 74) was diluted in potassium containing Tris-HCl buffer to an appropriate concentration prior to use in the various spectroscopic and microscopic studies.
4. Synthesis of Porphyrin 3
The starting material, zinc Porphyin 3 (3-Zn) has been previously reported by our group39.To a stirring solution of 3-Zn (50 mg, 0.004 mmol) in 100 mL chloroform was added 5 drops of concentrated HCl, and the stirring was continued for 10 min. The reaction mixture was then neutralized with triethylamine and washed with saturated sodium bicarbonate. Subsequently, the organic layer was dried over anhydrous Na2SO4. The solvent was evaporated in vacuo and the final product was purified by silica gel column (first eluted with hexanes to remove non-polar impurities and then the product was collected by eluting with CH2Cl2). The solvents were removed in vacuo to give 3 as a purple solid (41 mg, 81%). 1H NMR (400 MHz, CDCl3): δ = 8.90 (br s, 8H), 7.81 (br s, 8H), 7.46 (br s, 8H), 7.06 (br s, 4H), 5.28 (m, 16H), 5.23–5.00 (m, 56H), 4.82 (m, 8H), 4.06–3.14 (m, the rest of PMβ-CDs protons ca. 808H), −2.91(br, 2H). MALDI-TOF MS (reflective positive mode): calculated for C564H918N28O280K m/z = 12602.8 Da, Found m/z = 12599.6 Da (M+K+). UV-Vis (H2O): 422 nm (ε 203,489 dm3 mol−1 cm−1), 518 nm (ε 15,586 dm3 mol−1 cm−1), 548 nm (ε 8,787 dm3 mol−1 cm−1), 588 nm (ε 5,300 dm3 mol−1 cm−1), 650 nm (ε 5,326 dm3 mol−1 cm−1).
5. Preparation of Assembly 4
100 µL of 40 µM quadruplex 2 in Tris-HCl buffer (10 mM Tris-HCl, 80 mM KCl, pH 7.5) was mixed with 40 µL of 100 µM porphyrin 3 in Tris-HCl buffer(10 mM Tris-HCl, 80 mM KCl, pH 7.5), to give an equal concentration of porphyrin 3 and quadruplex 2 (28.6 µM each). The mixture was covered with aluminum foil and kept at room temperature for 48 hr to form the assembly. This solution was diluted with Tris-HCl buffer to an appropriate concentration (for most spectroscopic and microscopic experiments 4 µM solution of the assembly, in terms of the concentration of porphyrin 3 or quadruplex 2, was used).
6. Preparation of Assembly 5
Assembly 5 was prepared by adding 1 µL of 300x SG in Tris-HCl buffer (10 mM Tris-HCl, 80 mM KCl, pH 7.5) to 50 µL of 4 µM solution of nanowire 4 (final SG: quadruplex 2 ratio ≈1:1) and mixed for 5 minutes. As can be seen from inspection of the solid black line in SI Figure S8, assembly 5 shows broadband absorption.
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Jibao He, Michael Johnson, and Ashish Aryal (from Nikon) for their assistance with Cryo-TEM, AFM, and confocal microscopies, respectively. We are also grateful to Russ Schmehl for helpful discussions.
Funding Sources
Acknowledgements are made to the National Science Foundation (CHE-1112091), the National Institutes of Health (R01GM097571) and the American Chemical Society Petroleum Research Fund (50272-DNI4), for supporting this research.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Electronic Supplementary Information (ESI) available: Synthetic scheme and characterization of porphyrin 3, supplementary AFM images, UV-Vis, Circular dichroism and fluorescence spectra, PAGE, and confocal microscopy are available in the supplementary information. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Seeman NC. DNA in a material world. Nature. 2003;421(6921):427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 2.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452(7184):198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 3.Wengel J. Nucleic acid nanotechnology-towards Angstrom-scale engineering. Organic & Biomolecular Chemistry. 2004;2(3):277–280. doi: 10.1039/b313986g. [DOI] [PubMed] [Google Scholar]
- 4.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440(7082):297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 5.Endo M, Sugiyama H. Chemical Approaches to DNA Nanotechnology. ChemBioChem. 2009;10(15):2420–2443. doi: 10.1002/cbic.200900286. [DOI] [PubMed] [Google Scholar]
- 6.Feldkamp U, Niemeyer CM. Rational Design of DNA Nanoarchitectures. Angew. Chem. Int. Ed. 2006;45(12):1856–1876. doi: 10.1002/anie.200502358. [DOI] [PubMed] [Google Scholar]
- 7.Dietz H, Douglas SM, Shih WM. Folding DNA into Twisted and Curved Nanoscale Shapes. Science. 2009;325(5941):725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maune HT, Han S-p, Barish RD, Bockrath M, Goddard IIA, RothemundPaul WK, Winfree E. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat Nano. 2010;5(1):61–66. doi: 10.1038/nnano.2009.311. [DOI] [PubMed] [Google Scholar]
- 9.McLaughlin CK, Hamblin GD, Hänni KD, Conway JW, Nayak MK, Carneiro KMM, Bazzi HS, Sleiman HF. Three-Dimensional Organization of Block Copolymers on “DNA-Minimal“ Scaffolds. J. Am. Chem. Soc. 2012;134(9):4280–4286. doi: 10.1021/ja210313p. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, TØrring T, Dong M, Rosen CB, Besenbacher F, Gothelf KV. DNA-Templated Covalent Coupling of G4 PAMAM Dendrimers. J. Am. Chem. Soc. 2010;132(51):18054–18056. doi: 10.1021/ja109677n. [DOI] [PubMed] [Google Scholar]
- 11.Stephanopoulos N, Liu M, Tong GJ, Li Z, Liu Y, Yan H, Francis MB. Immobilization and One-Dimensional Arrangement of Virus Capsids with Nanoscale Precision Using DNA Origami. Nano Lett. 2010;10(7):2714–2720. doi: 10.1021/nl1018468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stearns LA, Chhabra R, Sharma J, Liu Y, Petuskey WT, Yan H, Chaput JC. Template-Directed Nucleation and Growth of Inorganic Nanoparticles on DNA Scaffolds. Angew. Chem. Int. Ed. 2009;48(45):8494–8496. doi: 10.1002/anie.200903319. [DOI] [PubMed] [Google Scholar]
- 13.Lo PK, Altvater F, Sleiman HF. Templated Synthesis of DNA Nanotubes with Controlled, Predetermined Lengths. J. Am. Chem. Soc. 2010;132(30):10212–10214. doi: 10.1021/ja1017442. [DOI] [PubMed] [Google Scholar]
- 14.Su W, Schuster M, Bagshaw CR, Rant U, Burley GA. Site-Specific Assembly of DNA-Based Photonic Wires by Using Programmable Polyamides. Angew. Chem. Int. Ed. 2011;50(12):2712–2715. doi: 10.1002/anie.201006735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heilemann M, Kasper R, Tinnefeld P, Sauer M. Dissecting and Reducing the Heterogeneity of Excited-State Energy Transport in DNA-Based Photonic Wires. J. Am. Chem. Soc. 2006;128(51):16864–16875. doi: 10.1021/ja065585x. [DOI] [PubMed] [Google Scholar]
- 16.Hannestad JK, Sandin P, Albinsson B. Self-Assembled DNA Photonic Wire for Long-Range Energy Transfer. J. Am. Chem. Soc. 2008;130(47):15889–15895. doi: 10.1021/ja803407t. [DOI] [PubMed] [Google Scholar]
- 17.Kienzler A, Flehr R, Kramer RA, Gehne Sr, Kumke MU, Bannwarth W. Novel Three-Color FRET Tool Box for Advanced Protein and DNA Analysis. Bioconjugate Chem. 2011;22(9):1852–1863. doi: 10.1021/bc2002659. [DOI] [PubMed] [Google Scholar]
- 18.Stein IH, Steinhauer C, Tinnefeld P. Single-Molecule Four-Color FRET Visualizes Energy-Transfer Paths on DNA Origami. J. Am. Chem. Soc. 2011;133(12):4193–4195. doi: 10.1021/ja1105464. [DOI] [PubMed] [Google Scholar]
- 19.Dutta PK, Varghese R, Nangreave J, Lin S, Yan H, Liu Y. DNA-Directed Artificial Light-Harvesting Antenna. J. Am. Chem. Soc. 2011;133(31):11985–11993. doi: 10.1021/ja1115138. [DOI] [PubMed] [Google Scholar]
- 20.Kumar CV, Duff MR. DNA-Based Supramolecular Artificial Light Harvesting Complexes. J. Am. Chem. Soc. 2009;131(44):16024–16026. doi: 10.1021/ja904551n. [DOI] [PubMed] [Google Scholar]
- 21.Su W, Bonnard V, Burley GA. DNA-Templated Photonic Arrays and Assemblies: Design Principles and Future Opportunities. Chemistry – A European Journal. 2011;17(29):7982–7991. doi: 10.1002/chem.201100924. [DOI] [PubMed] [Google Scholar]
- 22.Feng X, Duan X, Liu L, Feng F, Wang S, Li Y, Zhu D. Fluorescence Logic-Signal-Based Multiplex Detection of Nucleases with the Assembly of a Cationic Conjugated Polymer and Branched DNA. Angew. Chem. Int. Ed. 2009;48(29):5316–5321. doi: 10.1002/anie.200901555. [DOI] [PubMed] [Google Scholar]
- 23.Teo YN, Kool ET. DNA-Multichromophore Systems. Chem. Rev. 2012;112(7):4221–4245. doi: 10.1021/cr100351g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varghese R, Wagenknecht H-A. DNA as a supramolecular framework for the helical arrangements of chromophores: towards photoactive DNA-based nanomaterials. Chem. Commun. 2009;0(19):2615–2624. doi: 10.1039/b821728a. [DOI] [PubMed] [Google Scholar]
- 25.Malinovskii VL, Wenger D, Haner R. Nucleic acid-guided assembly of aromatic chromophores. Chem. Soc. Rev. 2010;39(2):410–422. doi: 10.1039/b910030j. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz E, Le Gac S, Cornelissen JJLM, Nolte RJM, Rowan AE. Macromolecular multi-chromophoric scaffolding. Chem. Soc. Rev. 2010;39(5):1576–1599. doi: 10.1039/b922160c. [DOI] [PubMed] [Google Scholar]
- 27.Wagenknecht H-A. Helical Arrangement of Porphyrins along DNA: Towards Photoactive DNA-Based Nanoarchitectures. Angew. Chem. Int. Ed. 2009;48(16):2838–2841. doi: 10.1002/anie.200900327. [DOI] [PubMed] [Google Scholar]
- 28.Stadler AL, Delos Santos JO, Stensrud ES, Dembska A, Silva GL, Liu S, Shank NI, Kunttas-Tatli E, Sobers CJ, Gramlich PME, Carell T, Peteanu LA, McCartney BM, Armitage BA. Fluorescent DNA Nanotags Featuring Covalently Attached Intercalating Dyes: Synthesis, Antibody Conjugation, and Intracellular Imaging. Bioconjugate Chem. 2011;22(8):1491–1502. doi: 10.1021/bc100485f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssen PGA, Vandenbergh J, van Dongen JLJ, Meijer EW, Schenning APHJ. ssDNA Templated Self-Assembly of Chromophores. J. Am. Chem. Soc. 2007;129(19):6078–6079. doi: 10.1021/ja0711967. [DOI] [PubMed] [Google Scholar]
- 30.Lubitz I, Borovok N, Kotlyar A. Interaction of Monomolecular G4-DNA Nanowires with TMPyP: Evidence for Intercalation†. Biochemistry. 2007;46(45):12925–12929. doi: 10.1021/bi701301u. [DOI] [PubMed] [Google Scholar]
- 31.Benvin AL, Creeger Y, Fisher GW, Ballou B, Waggoner AS, Armitage BA. Fluorescent DNA Nanotags: Supramolecular Fluorescent Labels Based on Intercalating Dye Arrays Assembled on Nanostructured DNA Templates. J. Am. Chem. Soc. 2007;129(7):2025–2034. doi: 10.1021/ja066354t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.OzhalUnal H, Armitage BA. Fluorescent DNA Nanotags Based on a Self-Assembled DNA Tetrahedron. ACS Nano. 2009;3(2):425–433. doi: 10.1021/nn800727x. [DOI] [PubMed] [Google Scholar]
- 33.Fechter EJ, Olenyuk B, Dervan PB. Sequence-Specific Fluorescence Detection of DNA by Polyamide-Thiazole Orange Conjugates. J. Am. Chem. Soc. 2005;127(47):16685–16691. doi: 10.1021/ja054650k. [DOI] [PubMed] [Google Scholar]
- 34.McLaughlin CK, Hamblin GD, Sleiman HF. Supramolecular DNA assembly. Chem. Soc. Rev. 2011;40(12):5647–5656. doi: 10.1039/c1cs15253j. [DOI] [PubMed] [Google Scholar]
- 35.Davis JT. G-Quartets 40 Years Later: From 5′-GMP to Molecular Biology and Supramolecular Chemistry. Angew. Chem. Int. Ed. 2004;43(6):668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 36.Marsh TC, Henderson E. G-Wires: Self-Assembly of a Telomeric Oligonucleotide, d(GGGGTTGGGG), into Large Superstructures. Biochemistry. 1994;33(35):10718–10724. doi: 10.1021/bi00201a020. [DOI] [PubMed] [Google Scholar]
- 37.Marsh TC, Vesenka J, Henderson E. A new DNA nanostructure, the G-wire, imaged by scanning probe microscopy. Nucleic Acids Res. 1995;23(4):696–700. doi: 10.1093/nar/23.4.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotlyar AB, Borovok N, Molotsky T, Cohen H, Shapir E, Porath D. Long, Monomolecular Guanine-Based Nanowires. Adv. Mater. 2005;17(15):1901–1905. [Google Scholar]
- 39.Fathalla M, Li S-C, Diebold U, Alb A, Jayawickramarajah J. Water-soluble nanorods self-assembled via pristine C60 and porphyrin moieties. Chem. Commun. 2009;0(28):4209–4211. doi: 10.1039/b908050c. [DOI] [PubMed] [Google Scholar]
- 40.Fathalla M, Neuberger A, Li S-C, Schmehl R, Diebold U, Jayawickramarajah J. Straightforward Self-Assembly of Porphyrin Nanowires in Water: Harnessing Adamantane/β-Cyclodextrin Interactions. J. Am. Chem. Soc. 2010;132(29):9966–9967. doi: 10.1021/ja1030722. [DOI] [PubMed] [Google Scholar]
- 41.Li Z-Q, Zhang Y-M, Guo D-S, Chen H-Z, Liu Y. Supramolecular Assembly with Multiple Preorganised π-Electronic Cages. Chemistry – A European Journal. 2013;19(1):96–100. doi: 10.1002/chem.201203575. [DOI] [PubMed] [Google Scholar]
- 42.Böhm I, Isenbügel K, Ritter H, Branscheid R, Kolb U. Fluorescent Nanowires Self-Assembled through Host–Guest Interactions in Modified Calcein. Angew. Chem. Int. Ed. 2011;50(32):7407–7409. doi: 10.1002/anie.201008145. [DOI] [PubMed] [Google Scholar]
- 43.Tagore DM, Sprinz KI, Fletcher S, Jayawickramarajah J, Hamilton AD. Protein Recognition and Denaturation by Self-Assembling Fragments on a DNA Quadruplex Scaffold. Angew. Chem. Int. Ed. 2007;46(1–2):223–225. doi: 10.1002/anie.200603479. [DOI] [PubMed] [Google Scholar]
- 44.Jayawickramarajah J, Tagore DM, Tsou LK, Hamilton AD. Allosteric Control of Self-Assembly: Modulating the Formation of Guanine Quadruplexes through Orthogonal Aromatic Interactions. Angew. Chem. 2007;119(40):7727–7730. doi: 10.1002/anie.200701883. [DOI] [PubMed] [Google Scholar]
- 45.Chaput JC, Switzer C. A DNA pentaplex incorporating nucleobase quintets. Proceedings of the National Academy of Sciences. 1999;96(19):10614–10619. doi: 10.1073/pnas.96.19.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu M, Guo Q, Kallenbach NR. Structure and stability of sodium and potassium complexes of dT4G4 and dT4G4T. Biochemistry. 1992;31(9):2455–2459. doi: 10.1021/bi00124a003. [DOI] [PubMed] [Google Scholar]
- 47.Petraccone L, Martino L, Duro I, Oliviero G, Borbone N, Piccialli G, Giancola C. Physico-chemical analysis of G-quadruplex containing bunch-oligonucleotides. Int. J. Biol. Macromol. 2007;40(3):242–247. doi: 10.1016/j.ijbiomac.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 48.Vesenka J, Bagg D, Wolff A, Reichert A, Moeller R, Fritzsche W. Auto-orientation of G-wire DNA on mica. Colloids and Surfaces B: Biointerfaces. 2007;58(2):256–263. doi: 10.1016/j.colsurfb.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 49.Lyubchenko YL, Shlyakhtenko LS, Ando T. Imaging of nucleic acids with atomic force microscopy. Methods. 2011;54(2):274–283. doi: 10.1016/j.ymeth.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neaves KJ, Huppert JL, Henderson RM, Edwardson JM. Direct visualization of G-quadruplexes in DNA using atomic force microscopy. Nucleic Acids Res. 2009;37(18):6269–6275. doi: 10.1093/nar/gkp679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shapir E, Sagiv L, Borovok N, Molotski T, Kotlyar AB, Porath D. High-Resolution STM Imaging of Novel Single G4-DNA Molecules. The Journal of Physical Chemistry B. 2008;112(31):9267–9269. doi: 10.1021/jp803478f. [DOI] [PubMed] [Google Scholar]
- 52.Cacialli F, Wilson JS, Michels JJ, Daniel C, Silva C, Friend RH, Severin N, Samori P, Rabe JP, O'Connell MJ, Taylor PN, Anderson HL. Cyclodextrin-threaded conjugated polyrotaxanes as insulated molecular wires with reduced interstrand interactions. Nat Mater. 2002;1(3):160–164. doi: 10.1038/nmat750. [DOI] [PubMed] [Google Scholar]
- 53.Samorí P, Ecker C, Gössl I, de Witte PAJ, Cornelissen JJLM, Metselaar GA, Otten MBJ, Rowan AE, Nolte RJM, Rabe JP. High Shape Persistence in Single Polymer Chains Rigidified with Lateral Hydrogen Bonded Networks. Macromolecules. 2002;35(13):5290–5294. [Google Scholar]
- 54.Dragan AI, Pavlovic R, McGivney JB, Casas-Finet JR, Bishop ES, Strouse RJ, Schenerman MA, Geddes CD. SYBR Green I: Fluorescence Properties and Interaction with DNA. Journal of Fluorescence. 2012;22(4):1189–1199. doi: 10.1007/s10895-012-1059-8. [DOI] [PubMed] [Google Scholar]
- 55.McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: Cooperative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 56.This decrease in emission of the SG:2 complex upon binding to porphyrin 3 provided a practical tool to probe the stoichiometry of quadruplex 2 and porphyrin 3 in the nanowire complex. A Jobs plot (see SI Figure-10) gave evidence for a 1:1 stoichiometry of binding (with a maximum at 0.5)
- 57.The excitation wavelength was 480 nm and the absorption intensity of the SG:2 complex and assembly 5 at this wavelength were closely matched (˜10 % difference). see SI Figure-9
- 58.Mohammed HS, Delos Santos JO, Armitage B. Noncovalent binding and fluorogenic response of cyanine dyes to DNA homoquadruplex and PNA-DNA heteroquadruplex structures. Artificial DNA: PNA & XNA. 2011;2(2):43–49. doi: 10.4161/adna.2.2.16339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gai W, Yang Q, Xiang J, Jiang W, Li Q, Sun H, Guan A, Shang Q, Zhang H, Tang Y. A dual-site simultaneous binding mode in the interaction between parallel-stranded G-quadruplex [d(TGGGGT)]4 and cyanine dye 2,2′-diethyl-9-methyl-selenacarbocyanine bromide. Nucleic Acids Res. 2013;41(4):2709–2722. doi: 10.1093/nar/gks1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martino L, Virno A, Pagano B, Virgilio A, Di Micco S, Galeone A, Giancola C, Bifulco G, Mayol L, Randazzo A. Structural and Thermodynamic Studies of the Interaction of Distamycin A with the Parallel Quadruplex Structure [d(TGGGGT)]4. J. Am. Chem. Soc. 2007;129(51):16048–16056. doi: 10.1021/ja075710k. [DOI] [PubMed] [Google Scholar]
- 61.Xing C, Xu Q, Tang H, Liu L, Wang S. Conjugated Polymer/Porphyrin Complexes for Efficient Energy Transfer and Improving Light-Activated Antibacterial Activity. J. Am. Chem. Soc. 2009;131(36):13117–13124. doi: 10.1021/ja904492x. [DOI] [PubMed] [Google Scholar]
- 62.Monchaud D, Teulade-Fichou M-P. A hitchhiker's guide to G-quadruplex ligands. Organic & Biomolecular Chemistry. 2008;6(4):627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.