Myofibrillogenesis is critical for muscle cell differentiation and contraction. This study shows that Smyd1b plays a key role in myofibrillogenesis in muscle cells. Knockdown of smyd1b results in up-regulation of hsp90α1 and unc45b gene expression, increased myosin degradation, and disruption of sarcomere organization in zebrafish embryos.
Abstract
Smyd1b is a member of the Smyd family that is specifically expressed in skeletal and cardiac muscles. Smyd1b plays a key role in thick filament assembly during myofibrillogenesis in skeletal muscles of zebrafish embryos. To better characterize Smyd1b function and its mechanism of action in myofibrillogenesis, we analyzed the effects of smyd1b knockdown on myofibrillogenesis in skeletal and cardiac muscles of zebrafish embryos. The results show that knockdown of smyd1b causes significant disruption of myofibril organization in both skeletal and cardiac muscles of zebrafish embryos. Microarray and quantitative reverse transcription-PCR analyses show that knockdown of smyd1b up-regulates heat shock protein 90 (hsp90) and unc45b gene expression. Biochemical analysis reveals that Smyd1b can be coimmunoprecipitated with heat shock protein 90 α-1 and Unc45b, two myosin chaperones expressed in muscle cells. Consistent with its potential function in myosin folding and assembly, knockdown of smyd1b significantly reduces myosin protein accumulation without affecting mRNA expression. This likely results from increased myosin degradation involving unc45b overexpression. Together these data support the idea that Smyd1b may work together with myosin chaperones to control myosin folding, degradation, and assembly into sarcomeres during myofibrillogenesis.
INTRODUCTION
Myofibrillogenesis, the process of sarcomere assembly, is critical for muscle cell differentiation and contraction. Myofibrillogenesis involves hundreds of sarcomeric proteins assembled into a highly organized structure called the sarcomere, the basic contractile unit in striated muscles. The sarcomere is divided into four major compartments: Z-line, I-band, A-band, and M-line. The Z-line anchors the actin thin filaments of the I-band. The M-line anchors the myosin thick filaments of the A-band. The assembly and disassembly of these multiprotein complexes follow ordered pathways that are highly regulated at the transcriptional, translational, and posttranslational levels. Disruption of these pathways leads to defective myofibril organization and skeletal and cardiac muscle diseases (Ehler and Gautel, 2008).
Recent studies show that Smyd1, a member of the Smyd family, plays a vital role in cardiogenesis and myofibrillogenesis (Gottlieb et al., 2002; Tan et al., 2006; Just et al., 2011). Smyd1, also known as skm-Bop, is a MYND and SET domain–containing protein that is specifically expressed in skeletal and cardiac muscles (Hwang and Gottlieb, 1995, 1997). Targeted deletion of Smyd1 in mice results in early embryonic lethality (Gottlieb et al., 2002). Embryos died around embryonic day 10.5, presumably due to the disruption of right ventricle formation and cardiomyocyte maturation (Gottlieb et al., 2002). The molecular mechanism by which Smyd1b functions is not well understood. Biochemical studies indicate that mouse Smyd1 and Smyd1b, one of the Smyd1 in zebrafish, can methylate histone H3 proteins in vitro (Tan et al., 2006; Sirinupong et al., 2010). However, the role of histone methylation in Smyd1b function is controversial (Just et al., 2011). In vitro studies have revealed that Smyd1 represses gene transcription in a histone deacetylase–dependent manner (Gottlieb et al., 2002). Consistent with its potential function in transcriptional regulation by histone modification, Smyd1 is initially localized in the nucleus of C2C12 myoblasts (Sims et al., 2002). However, it was reported that Smyd1 undergoes a nucleus-to-cytoplasm translocation during myoblast differentiation into myotubes (Sims et al., 2002). Moreover, recent studies show that Smyd1b binds to myosin (Just et al., 2011), suggesting that Smyd1b may have additional functions and targets in the cytoplasm. Consistent with the idea that Smyd1 may have additional targets, several recent studies demonstrated that Smyd2, another member of the Smyd family, can methylate nonhistone proteins, such as p53, RB1, and heat shock protein 90 (Hsp90; Huang et al., 2006; Saddic et al., 2010; Cho et al., 2012; Abu-Farha et al., 2011). Of interest, methylation of molecular chaperone Hsp90 by Smyd2 plays a critical role in sarcomere organization (Donlin et al., 2012; Voelkel et al., 2013).
A large body of evidence suggests that correct folding and assembly of myofibrillar proteins requires molecular chaperones and cochaperones (Barral and Epstein, 1999; Srikakulam and Winkelmann, 1999; Landsverk et al., 2007; Kim et al., 2008; Willis et al., 2009). It was shown that Hsp90 and Unc45 form a complex with newly synthesized myosin proteins and play vital roles in myosin folding and assembly (Srikakulam and Winkelmann, 2004). Genetic studies in Caenorhabditis elegans and biochemical analyses in vitro indicate that chaperone-mediated myosin folding is an integral part of myofibril assembly (Hutagalung et al., 2002). Knockdown or mutation of hsp90α1or unc45 in C. elegans or zebrafish embryos results in complete disruption of myofibril organization (Epstein and Thomson, 1974; Barral et al., 1998, 2002; Etard et al., 2007; Wohlgemuth et al., 2007; Du et al., 2008), a phenotype very similar to smyd1b knockdown in zebrafish embryos (Tan et al., 2006). However, the genetic and biochemical interactions among Smyd1, Hsp90α1, and Unc45b are not clear.
In this study, we characterize Smyd1b function in myofibril organization in skeletal and cardiac muscles of zebrafish embryos. We demonstrate that in addition to thick filament defects, knockdown of smyd1b causes significant disruption of thin and titin filaments, as well as M- and Z-lines in skeletal muscles of zebrafish embryos. Moreover, myofibril organization is also affected in cardiac muscles. Microarray and quantitative reverse transcription (qRT)-PCR analyses reveal that smyd1b knockdown significantly up-regulates hsp90α1 and unc45b gene expression in zebrafish embryos. Biochemical analysis by coimmunoprecipitation (coIP) shows that Smyd1b can be coimmunoprecipitated with Hsp90α1 and Unc45b. Moreover, knockdown of smyd1b results in dramatic reduction of myosin protein accumulation in zebrafish embryos without any effect on myosin mRNA expression. Together these data support the idea that Smyd1b may work together with myosin chaperone Unc45b to control sarcomere assembly during myofibrillogenesis.
RESULTS
Knockdown of smyd1b expression results in significant disruption of sarcomere organization in skeletal muscles
We previously demonstrated that Smyd1b plays an important role in the thick filament assembly in slow muscles of zebrafish embryos at 24 h postfertilization (hpf; Tan et al., 2006). However, a recent study showed that a loss-of-function Smyd1b mutant had a normal myofibril organization in slow muscles at 48 hpf, although the mutant embryos showed a clear fast muscle defect (Just et al., 2011). To better understand the function of Smyd1b in myofibril organization in the slow muscle, we characterized thin filaments and Z-line structures in slow muscles of Smyd1b-knockdown embryos at different times of development. When compared with the control embryo (Figure 1, A and G), the results show that knockdown of smyd1b significantly disrupts the myofibril organization of thin filaments and Z-lines in slow muscles of zebrafish embryos at 28 hpf (Figure 1, B and H). However, the myofibril defects at the thin filaments and Z-lines were partially recovered in smyd1b-knockdown embryos at 48 and 72 hpf, although the myofibers did not appear to be straight in the knockdown embryos (Figure 1, D, F, J, and L).
FIGURE 1:
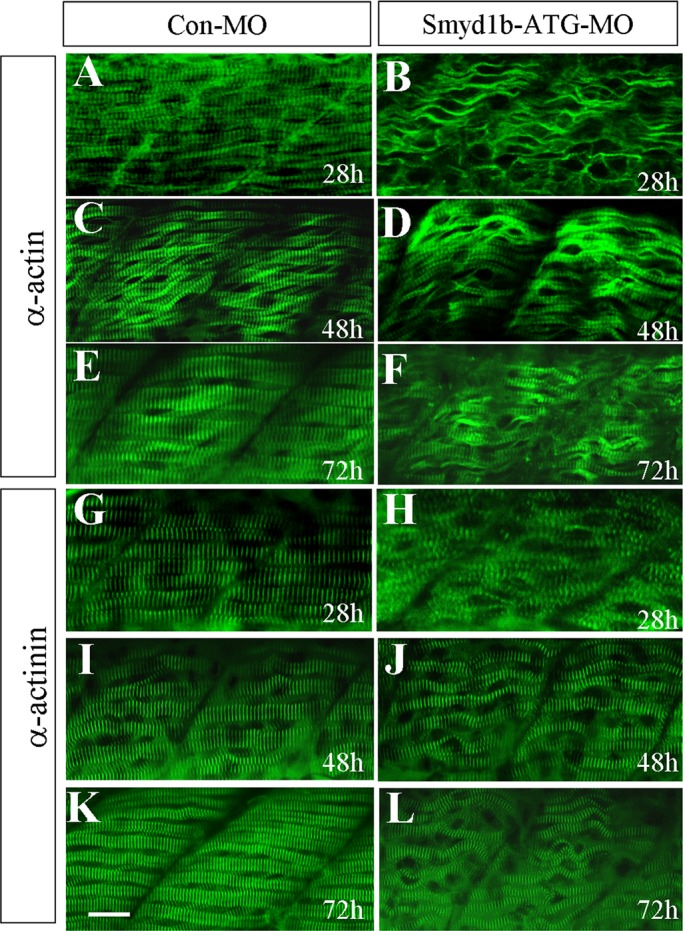
Knockdown of smyd1b expression results in defective thin filament and Z-line organization in slow muscles of early-stage zebrafish embryos. (A–F) Anti–α-actin antibody (Acl-20.4.2) staining shows organization of thin filaments in slow muscle fibers of control-MO (A, C, E) or Smyd1b-ATG-MO–injected (B, D, F) embryos at 28 (A, B), 48 (C, D), and 72 (E, F) hpf, respectively. (G–L) Anti–α-actinin antibody (EA-53) staining shows Z-line structure in control-MO (G, I, K) or Smyd1b ATG-MO–injected (H, J, L) embryos at 28 (G, H), 48 (I, J), and 72 (K, L) hpf, respectively. Scale bar, 25 μm.
To determine whether the M-line structure was also disrupted in early-stage embryos at 28 hpf and recovered at the later stages (48 and 72 hpf), we took advantage of the recently identified myomesin-3–red fluorescent protein (RFP) line generated from gene trapping (Clark et al., 2011). Myomesin-3-RFP fusion protein is specifically localized to the M-lines in slow muscles of zebrafish embryos (Clark et al., 2011; Xu et al., 2012). The myomesin-3–RFP localization was analyzed in smyd1b-knockdown embryos at 28, 48 and 72 hpf (Supplemental Figure S1). Compared with the control embryos (Supplemental Figure S1, A–C), the smyd1b-knockdown embryos showed no sarcomeric localization of myomesin-RFP at 28 hpf (Supplemental Figure S1D). The M-line localization of myomesin-RFP, however, became apparent at 48 and 72 hpf in the smyd1b-knockdown embryos (Supplemental Figure S1, E and F).
Recent studies demonstrated that zebrafish contains two smyd1 genes, smyd1a and smyd1b (Sun et al., 2008). Smyd1a shares high sequence similarity with Smyd1b and is specifically expressed in muscle cells of zebrafish embryos starting around 24 hpf (Sun et al., 2008). The recovery in smyd1b-knockdown embryos could be due to the redundant function of Smyd1a in later-stage embryos (48 and 72 hpf). To test this idea, we analyzed the phenotype of single and double knockdown of smyd1a and smyd1b. The results showed that knockdown of smyd1a and smyd1b together significantly disrupted the M-line organization of myomesin-3–RFP at all three stages analyzed (28, 48, and 72 hpf; Supplemental Figure S1). In contrast, knockdown of smyd1a or smyd1b alone had little or no effect at 48 and 72 hpf (Supplemental Figure S1). Collectively these data indicate that Smyd1a and Smyd1b are required for M-line organization and myofibril assembly in slow muscles of zebrafish embryos.
Zebrafish embryos contain two distinct types of muscles, slow and fast, which are located at different regions of the myotome (Devoto et al., 1996; Du et al., 1997). To analyze the effects of smyd1b knockdown on myofibril assembly in fast muscles, we examined the organization of thick and thin filaments, as well as M- and Z-lines, in fast muscles of smyd1b-knockdown zebrafish embryos. The results showed that knockdown of smyd1b completely disrupted the organization of thick and thin filaments in fast muscles of zebrafish embryos even at late stages at 72 hpf (Figure 2, B and D). Similarly, M- and Z-lines were also disrupted (Figure 2, F and H). It has been proposed that titin, the giant protein spanning from the Z-line to the M-line, is a key regulator of myofibrillogenesis. To determine whether titin organization was affected in smyd1b-knockdown embryos, we carried out immunostaining with the anti-titin antibody in zebrafish embryos. The results showed that titin filaments were also disrupted in smyd1b-knockdown embryos (Figure 2J). Collectively these data indicate that Smyd1b is required for sarcomere organization in both slow and fast muscles of zebrafish embryos. However, it is not clear whether Smyd1b has a direct role in the global organization of sarcomere assembly. It was reported that null mutants of the unc-54 that encodes myosin heavy chain in C. elegans also led to disruptions in sarcomere organization (Epstein and Thomson, 1974; MacLeod et al. 1977). Our recent studies show that knockdown of myosin heavy chain (MHC) not only disrupts thick filament organization, it also results in poor organization of M-lines (Xu et al., 2012). It remains to be determined whether the global disruption of sarcomere organization in smyd1b-knockdown embryos results directly from loss of Smyd1b function or indirectly from disruption of myosin thick filament organization.
FIGURE 2:
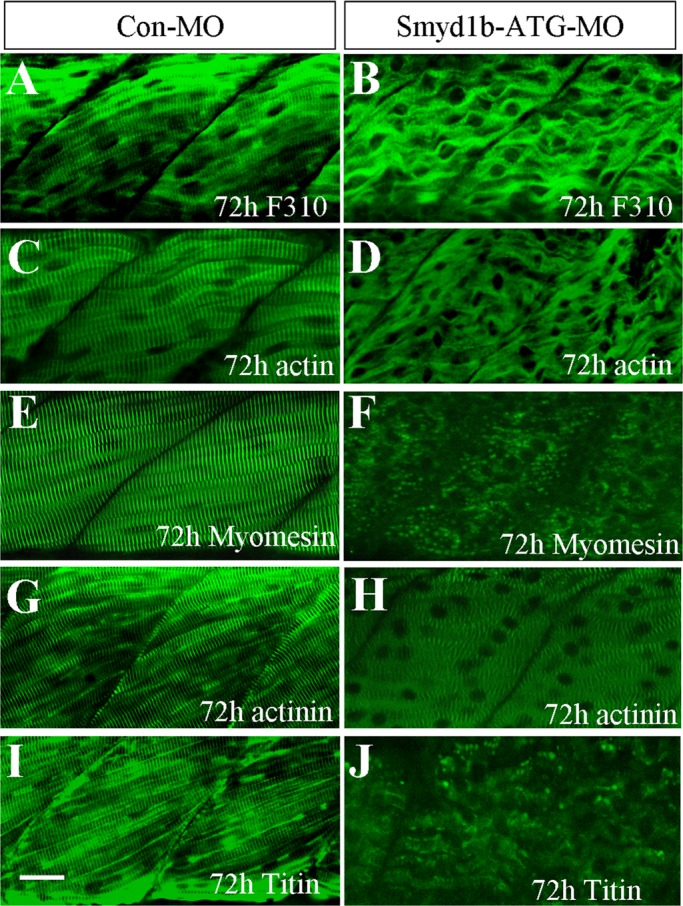
Knockdown of smyd1b expression results in defective sarcomere organization in fast skeletal muscles of zebrafish embryos. (A, B) Anti-MLC (F310) antibody staining (lateral view) shows thick filaments in fast muscle fibers of control (A) or Smyd1b-ATG-MO–injected (B) embryos at 72 hpf. (C, D) Anti–α-actin antibody (Acl-20.4.2) staining (lateral view) shows thin filaments in fast muscle fibers of control (C) or Smyd1b-ATG-MO–injected (D) embryos at 72 hpf. (E, F) Anti-myomesin antibody (mMaC myomesin B4) staining shows M-line structure in control-MO (E) or Smyd1b-ATG-MO–injected (F) embryos at 72 hpf. (G, H) Anti–α-actinin antibody (EA-53) staining shows Z-line structure in control-MO (G) or Smyd1b-MO–injected (H) embryos at 72 hpf. (I, J) Anti-titin (T11) antibody staining shows the sarcomeric localization of titin in fast muscle fibers in control-MO (I) or Smyd1b ATG-MO-injected (J) embryos at 72 hpf. Scale bar, 25 μm.
Smyd1b transgene rescues the skeletal muscle defect from knockdown
To confirm the specificity of the smyd1b-knockdown phenotype, we performed a rescue experiment using a Tg(Smyd1b:Smyd1bmyc) transgenic line expressing a myc-tagged Smyd1b driven by its own muscle-specific promoter (Du et al., 2006; Tan et al., 2006). The transgene was constructed using the Smyd1b cDNA, which does not need splicing for expression (Supplemental Figure S2). The Smyd1b splicing morpholino (MO; E9I9-MO) was microinjected into the Tg(Smyd1b:Smyd1bmyc) transgenic zebrafish embryos. Western blot showed that expression of the myc-tagged Smyd1b from the transgene was not affected by the E9I9 splicing MO (Supplemental Figure S2). The sarcomeric structures were analyzed in the E9I9-MO–injected embryos by immunostaining. The results showed that expression of the myc-tagged Smyd1b can rescue the myofibril defects in smyd1b-knockdown embryos (Figure 3). Compared with the nontransgenic control, transgenic embryos injected with the E9I9 spicing MO show normal organization of thin and titin filaments, as well as M- and Z-lines (Figure 3). Together these data confirm that muscle defects from smyd1b knockdown are gene specific, indicating that Smyd1b is essential for myofibrillogenesis in skeletal muscles.
FIGURE 3:
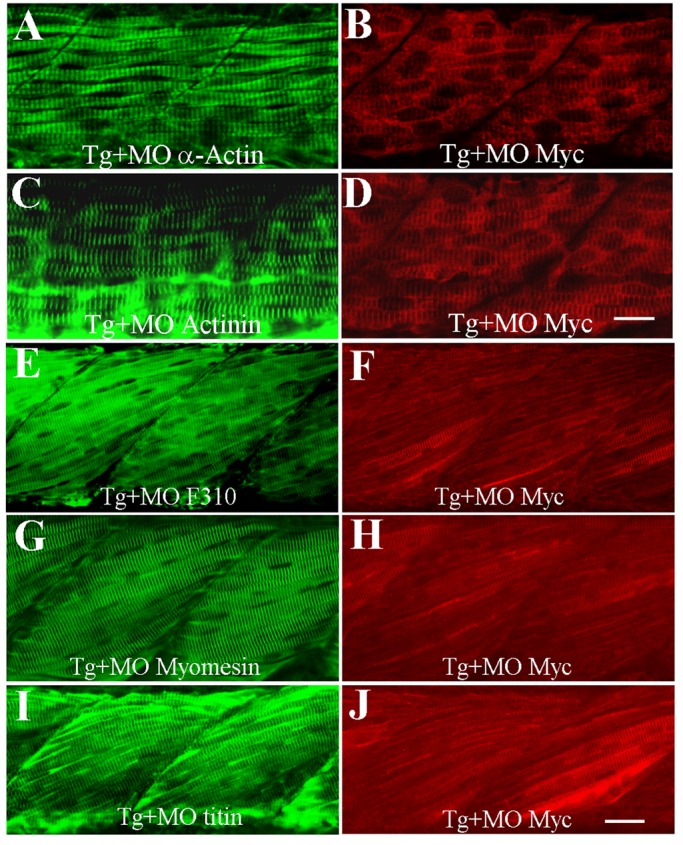
Rescue of myofibril defects by expression of myc-tagged Smyd1b. The Smyd1b E9I9-MO was injected into Tg(Smyd1b:Smyd1bmyc) transgenic zebrafish embryos expressing a myc-tagged Smyd1b at one- to two-cell stages. Expression of myc-tagged Smyd1b proteins was analyzed by Western blot and immunostaining with anti-myc antibodies. (A, B) Anti–α-actin and anti-myc (9E10) antibody double staining shows that expression of myc-tagged Smyd1b from the transgene (B) can rescue the knockdown effect on thin filament organization (α-actin) in Smyd1b MO–injected embryos at 24 hpf (A). (C, D) Anti-actinin and anti-myc (9E10) antibody double staining shows that expression of myc-tagged Smyd1b from the transgene (D) can rescue the knockdown effect on Z-line organization (α-actinin) in Smyd1b MO–injected embryos at 24 hpf (C). (E, F) Anti-MLC (F310) and anti-myc antibody double staining shows that expression of myc-tagged Smyd1b from the Tg(Smyd1b:Smyd1bmyc) transgene (F) can rescue thick filament defects in Smyd1b MO–injected embryos at 3 d postfertilization (dpf; E). (G, H) Anti-myomesin and anti-myc antibody double staining shows that expression of myc-tagged Smyd1b from the Tg(Smyd1b:Smyd1bmyc) transgene (H) can rescue the disruption of the M-line in Smyd1b MO–injected embryos at 3 dpf (G). (I, J) Anti-titin and anti-myc antibody double staining shows that expression of the Tg(Smyd1b:Smyd1bmyc) transgene (J) can rescue the disruption of titin assembly in Smyd1b MO–injected embryos at 3 dpf (I). Scale bars, 25 μm.
Knockdown of smyd1b disrupts sarcomere organization in cardiac muscles of zebrafish embryos
It was reported that targeted deletion of smyd1 results in defective cardiomyogenesis in mouse embryos that fail to form the right ventricle (Gottlieb et al., 2002). Our previous studies demonstrated that smyd1b-knockdown zebrafish embryos have no heartbeat and blood circulation (Tan et al., 2006). To better characterize the heart defects in smyd1b-knockdown embryos, we analyzed heart tube patterning in smyd1b-knockdown embryos by double staining using antibodies that recognize both ventricle and atrium (MF20) or atrium specifically (S46; Stainier and Fishman, 1992). The results show that heart tube patterning is normal in smyd1b-knockdown embryos (Figure 4, A–F).
FIGURE 4:
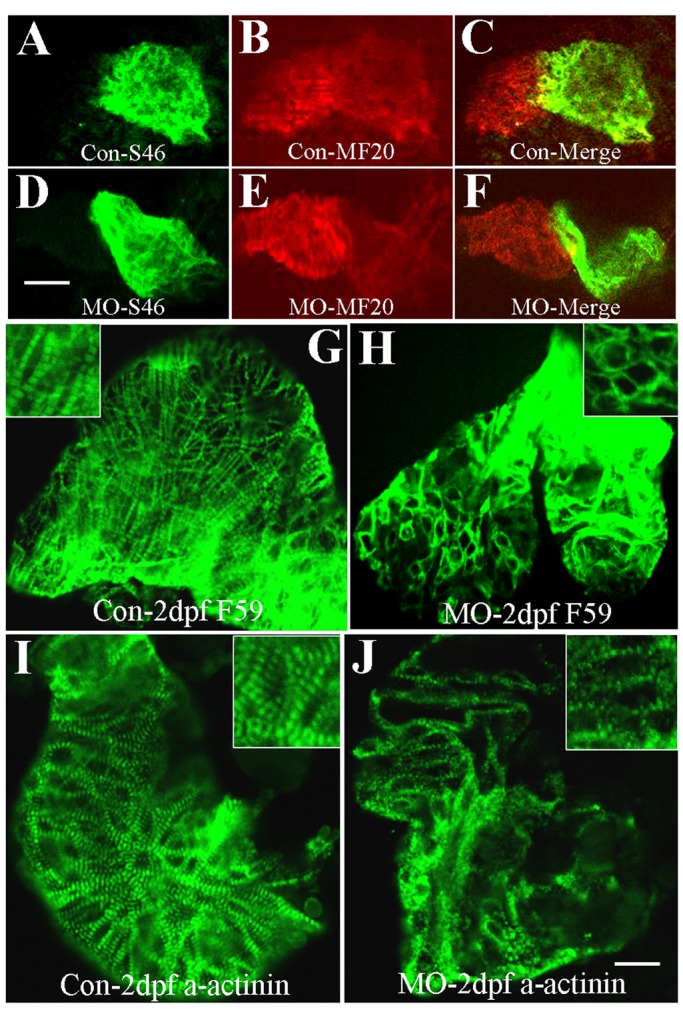
Immunostaining shows disorganized thick filaments and Z-line organization in cardiac muscles of smyd1b-knockdown embryos. (A–C) Double immunostaining with S46 (A) and MF20 (B) antibodies shows the atrium and ventricle formation in control-MO–injected embryos at 2 dpf. (C) Merged picture of A and B. Scale bars, 100 μm. (D–F) Double immunostaining with S46 (D) and MF20 (E) antibodies shows the atrium and ventricle formation in Smyd1b ATG-MO–injected embryos at 2 dpf. (F) Merged picture of D and E. Scale bar, 100 μm. (G, H) Anti-MHC (F59) antibody staining shows thick filament organization in control-MO (G) and Smyd1b ATG-MO–injected (H) embryos at 2 dpf. (I, J) Anti–α-actinin antibody staining shows Z-line structure in control-MO (I) and Smyd1b ATG-MO–injected (J) embryos at 2 dpf. Scale bar, 15 μm.
To further determine whether knockdown of smyd1b disrupts the sarcomere assembly in the cardiac muscles of zebrafish embryos, we examined thick filament and Z-line organization in cardiac muscles of smyd1b-knockdown embryos. Compared with the control (Figure 4G), few or no thick filaments could be detected in cardiac muscles of smyd1b-knockdown embryos (Figure 4H). In addition, the Z-line structure was also disrupted in cardiac muscles of Smyd1b-knockdown embryos (Figure 4J). Together these studies indicate that Smyd1b is also required for myofibril organization in cardiac muscles.
Knockdown of smyd1b results in up-regulation of hsp90α1 and unc45b gene expression
Members of the Smyd family are methyltransferases that are able to methylate histone proteins in vitro (Hamamoto et al., 2004; Brown et al., 2006; Tan et al., 2006; Sirinupong et al., 2010). Histone methylation has been implicated in regulation of gene expression. To test whether knockdown of smyd1b might alter gene expression involved in myofibrillogenesis, we carried out a microarray analysis comparing the expression profile of ∼20,000 genes in smyd1b-knockdown and control zebrafish embryos at 24 hpf. The data showed that the expression of 12 genes was up-regulated twofold to sixfold in smyd1b-knockdown embryos compared with the control. Strikingly, eight of them encode members of the heat shock protein family, including hsp70, hsp90, and hsp47 (Supplemental Table S1). In addition, >200 genes were down-regulated in smyd1b-knockdown embryos. However, no clear trend or common pathways were noted among those down-regulated genes.
Among these up-regulated hsp genes, hsp90α1 is especially interesting. hsp90α1 is specifically expressed in muscle cells of zebrafish embryos and plays a vital role in myofibril assembly (Sass et al., 1996, 1999; Du et al., 2008; Hawkins et al., 2008). It was shown that Hsp90 forms a complex with newly synthesized myosin protein and is involved in myosin folding and assembly (Srikakulam and Winkelmann, 2004). To validate the results from the microarray analysis, we analyzed hsp90α1 mRNA expression in smyd1b-knockdown embryos by in situ hybridization and qRT-PCR. The results confirmed that knockdown of smyd1b significantly up-regulated hsp90α1 gene expression (Figure 5B). qRT-PCR data indicated that the increase was approximately threefold (Figure 5G).
FIGURE 5:
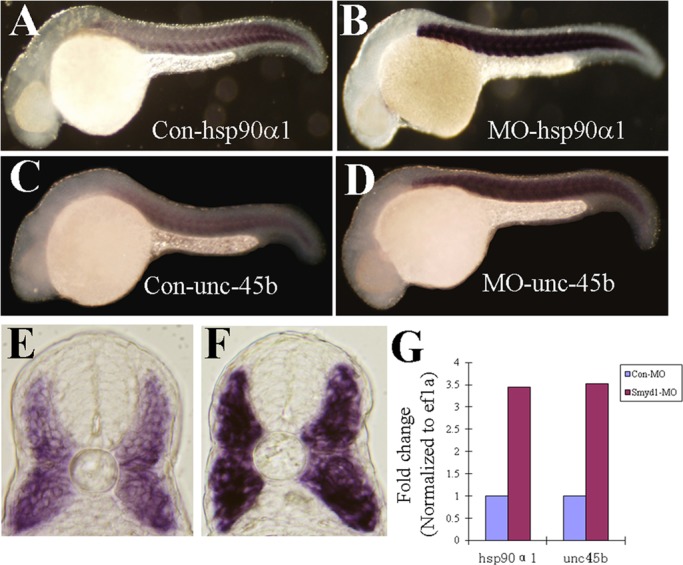
Whole-mount in situ hybridization and real-time PCR show the increased expression of hsp90α1 and unc45b in smyd1b-knockdown embryos. (A–D) In situ hybridization (side views) shows the expression of hsp90α1 (A, B) and unc45b (C, D) in control-MO (A, C) or Smyd1b ATG-MO–injected (B, D) embryos at 24 hpf. (E, F) Views of cross sections show the result of in situ hybridization of unc45b expression in control-MO (E) or Smyd1b ATG-MO–injected (F) embryos at 24 hpf. (G) Real-time PCR analysis of hsp90α1 and unc45b expression in control-MO or Smyd1b ATG-MO–injected embryos at 24 hpf.
It was reported that Unc45 associates with Hsp90 and functions as a myosin chaperone involved in myosin folding and assembly (Barral et al., 2002; Etard et al., 2007). To determine whether unc45b expression was also up-regulated in smyd1b-knockdown zebrafish embryos, we conducted in situ hybridization and quantitative PCR in the smyd1b-knockdown embryos. The result showed that unc45b expression was significantly increased in smyd1b-knockdown embryos (Figure 5, D, F, and G). Together these results indicate that Smyd1b knockdown could lead to up-regulation of heat shock protein and unc45b gene expression in skeletal muscles of zebrafish embryos.
The up-regulation of hsp90α1 gene expression by smyd1b knockdown raises the question of whether Smyd1b could be involved in stress response. To test this idea, we stressed zebrafish embryos with heat or cold shock. Expression of hsp90α1 and smyd1b was subsequently determined in the heat- or cold-stressed embryos. Although both heat and cold shock treatment significantly increased hsp90α1 expression (Figure 6, C and E), little or no change of smyd1b expression was detected in either heat- or cold-stressed embryos (Figure 6, D and F). This was further confirmed by qRT-PCR (Figure 6G). Together these data indicate that although knockdown of smyd1b induces heat shock protein gene expression, Smyd1b itself is not involved in cold- or heat-induced stress response.
FIGURE 6:
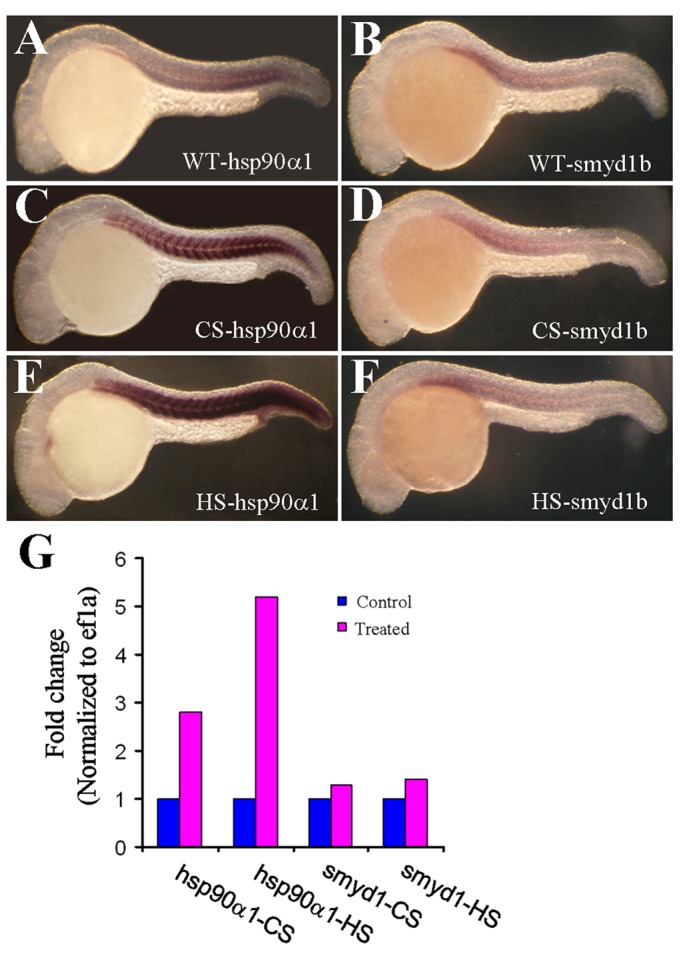
Cold and heat shock treatment has no effect on smyd1b gene expression in zebrafish embryos. Zebrafish embryos were subjected to cold or heat shock treatment. Expression of hsp90α1 and smyd1b was analyzed in these embryos by whole-mount in situ hybridization and real time RT-PCR. (A, B) Whole-mount in situ hybridization shows the expression of hsp90α1 (A) and smyd1b (B) in control embryos. (C, F) Whole-mount in situ hybridization shows the expression of hsp90α1 (C, E) and smyd1b (D, F) in cold-shocked (C, D) or heat-shocked (E, F) zebrafish embryos. (G) Real time RT-PCR shows the expression of hsp90α1 and smyd1b mRNA transcripts in cold-shocked (CS) or heat-shocked (HS) zebrafish embryos.
Smyd1 protein complex formation with Unc45b and Hsp90α1
Unc45b is a myosin chaperone that plays a pivotal role in myosin folding and assembly (Barral et al., 2002; Etard et al., 2007). Loss of Unc45b function results in defective myofibril organization in skeletal muscles of zebrafish embryos (Etard et al., 2007; Wohlgemuth et al., 2007; Hawkins et al., 2008), a phenotype very similar to smyd1b knockdown (Tan et al., 2006). To determine whether Smyd1b might be involved in myofibrillogenesis by forming a complex with myosin chaperone Unc45b, we performed coIP analysis using HEK293 cells expressing FLAG-tagged Unc45b and myc-tagged Smyd1b_tv1 or Smyd1b_tv2, encoded by two alternatively spliced isoforms of smyd1b (Li et al., 2011). The results show that Smyd1b_tv1 can be coimmunoprecipitated with FLAG-tagged Unc45b (Figure 7A). This association appears to be isoform specific (Figure 7A). The Smyd1b_tv2 isoform shows little or no interaction with Unc45b in the coIP assay (Figure 7A). The coIP results suggest that Smyd1b (Smyd1b_tv1) and Unc45b coexist in a protein complex. However, it is not clear whether Smyd1b and UNC-45b actually bind to one another. Recent studies show that Unc45b and Smyd1b can bind myosin heavy chain (Barral et al., 2002; Just et al., 2011). One possibility is that Smyd1b and Unc45b form a protein complex mediated by myosin heavy chain.
FIGURE 7:
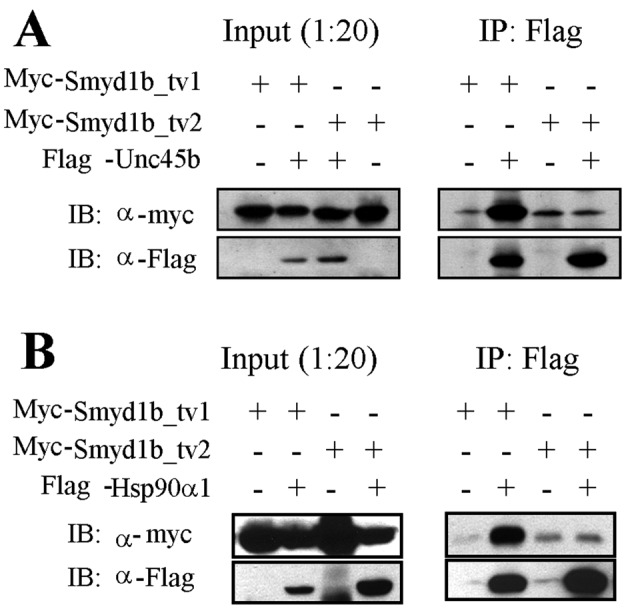
Coimmunoprecipitation shows Smyd1b-tv1, but not Smyd1b-tv2, interacts with myosin chaperone Hsp90α1 and Unc-45b. (A) coIP shows that myc-tagged Smyd1b-tv1 can be pulled down by FLAG-tagged Unc-45b coexpressed in HEK293 cells. (B) coIP shows that myc-tagged Smyd1b-tv1 can be pulled down by the FLAG-tagged Hsp90α1 coexpressed in HEK293 cells.
Recent studies show that Unc45b functions as a Hsp90α1 cochaperone in myosin folding and assembly (Barral et al., 1998; Etard et al., 2007; Du et al., 2008). To determine whether Smyd1b_tv1 is also able to form a protein complex with Hsp90α1, we performed a similar coIP assay with FLAG-tagged Hsp90α1. The results show that, similar to Unc45b, Smyd1b_tv1, but not Smyd1b_tv2, can be coimmunoprecipitated with FLAG-tagged Hsp90α1 (Figure 7B). Collectively these data indicate that Smyd1b_tv1 might associate directly or indirectly with Hsp90α1 and Unc45b, suggesting that they might work together to control myosin folding and assembly.
Knockdown of smyd1b reduces myosin protein accumulation in zebrafish embryos
Previous studies showed that loss of Hsp90α1 or Unc45b function significantly reduces myosin protein accumulation in zebrafish embryos (Du et al., 2008; Hawkins et al., 2008; Bernick et al., 2010). To determine whether knockdown of smyd1b can result in reduction of myosin protein accumulation in zebrafish embryos, we compared myosin protein levels in smyd1b-knockdown and control embryos at 24 hpf. Western blot analysis showed significant reduction of myosin heavy chain protein levels in smyd1b-knockdown embryos (Figure 8A). To test whether this reduced protein accumulation was due to decreased transcription of myosin gene or poor mRNA stability, we analyzed the myosin mRNA levels by in situ hybridization and quantitative RT-PCR in smyd1b-knockdown embryos. The result shows that there is little or no change in myosin mRNA levels in smyd1b-knockdown embryos compared with control (Figure 8, B–I). This was further confirmed by qRT-PCR (Supplemental Figure S3), indicating that the reduced myosin protein accumulation is not due to decreased myosin mRNA levels.
FIGURE 8:
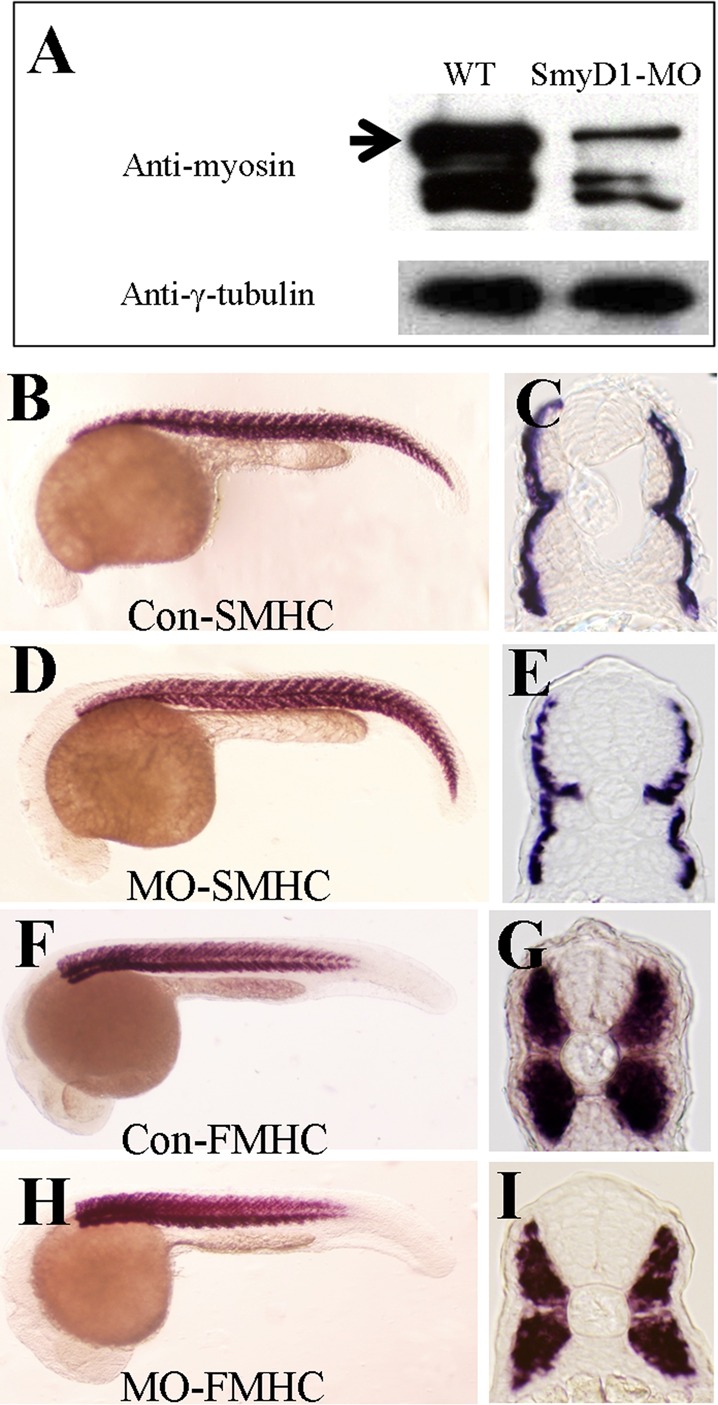
Knockdown of smyd1b results in decreased levels of myosin proteins but not their mRNAs. (A) Western blot using MF20 antibody shows decreased protein levels of myosin heavy chain in smyd1b-knockdown embryos. γ-Tubulin was used as loading control. (B–E) In situ hybridization shows the mRNA levels of slow myosin heavy chain in control-MO (B, C) or Smyd1b ATG-MO–injected (D, E) embryos at 24 hpf in different views. (F–I) In situ hybridization shows the mRNA levels of fast myosin heavy chain in control-MO (F, G) or Smyd1b ATG-MO–injected (H, I) embryos at 24 hpf in different views.
The reduced myosin accumulation in smyd1b-knockdown embryos is similar to previous findings in unc45b-knockdown embryos (Bernick et al., 2010). However, knockdown of smyd1b resulted in increased unc45b mRNA expression. Previous studies in C. elegans and zebrafish showed that overexpression of Unc45 results in thick filament defects and decreased myosin expression (Landsverk et al., 2007; Bernick et al., 2010; Ni et al., 2011). To better understand the interaction between Smyd1b and Unc45b in myosin folding and degradation, we directly expressed Unc45b in smyd1b-knockdown embryos by coinjecting a Tg(smyd1b:Unc45bFLAG) transgene encoding a FLAG-tagged Unc45b with the Smyd1b ATG-MO into zebrafish embryos. A typical mosaic pattern of Unc45b expression was detected in the injected embryos (Figure 9). Consistent with the idea that overexpression of Unc45 is detrimental to myosin thick filament organization, our data show that myofibers with ectopic Unc45b expression show no or little myosin expression and thick filament organization compared with neighboring myofibers without ectopic Unc45b expression (Figure 9, A–C). To test that FLAG-tagged Unc45b is bioactive, we coinjected the Tg(smyd1b:Unc45bFLAG) transgene with the Unc45b-ATG-MO into zebrafish embryos. In contrast to the effects in the smyd1b-knockdown embryos, ectopic expression of FLAG-tagged Unc45b rescued the thick filament defects in unc45b-knockdown embryos, suggesting that the FLAG-tagged Unc45b is biologically active (Figure 9, D–F). Collectively, these data suggest that Smyd1b and Unc45b might work together to control myosin folding and degradation. Moreover, up-regulation of unc45b expression might be involved in myosin degradation in smyd1b-knockdown embryos.
FIGURE 9:
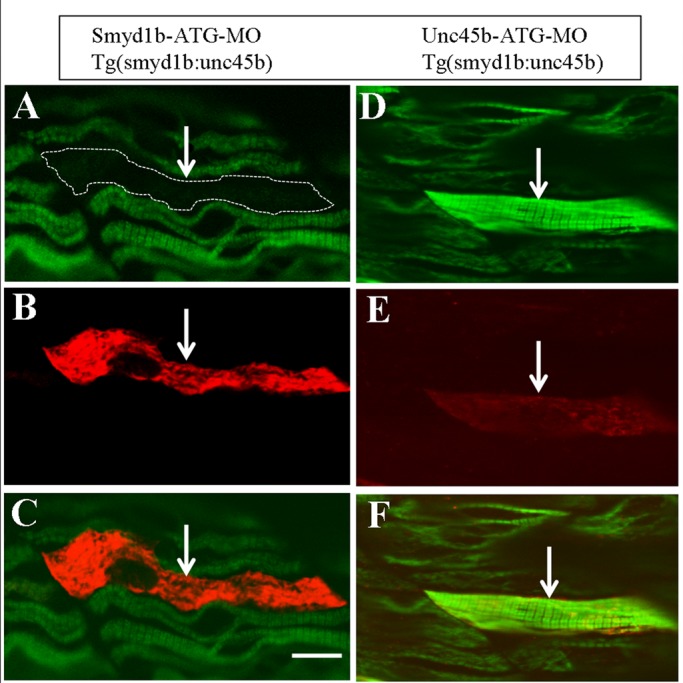
Overexpression of Unc45b increases myosin degradation in smyd1b-knockdown embryos. The DNA construct expressing FLAG-tagged Unc-45b under the control of smyd1 promoter was coinjected with Smyd1b-ATG-MO or Unc45b-ATG-MO into zebrafish embryos. The effect on myosin expression and thick filament organization was analyzed by double staining with anti-FLAG (red) and anti-myosin (F59, green) antibodies at 36 hpf. (A–C) Double staining shows the expression of FLAG-tagged Unc-45b (B) in a single fiber (indicated by an arrow) and its effect on myosin expression and thick filament organization (A, C). (C) Merged image of A and B, showing that the myofibril defect is restricted to the myofiber expressing the FLAG-tagged Unc-45b. (D–F) Double staining shows the rescue of myosin thick filament organization in Unc45b-knockdown zebrafish embryos coinjected with Unc45b-MO and the smyd1:unc45bFLAG DNA construct. A myofiber (arrow) expressing FLAG-tagged Unc45b (E) exhibited normal myosin expression and thick filament organization (D, F). Scale bar, 15 μm.
Protein methylation was suggested to play important roles in protein stabilization/destabilization (Yang et al., 2009; Egorova et al., 2010; Zhang et al., 2012). Several sarcomere proteins, such as myosin, actin, and creatine kinase, are methylated at their lysine residues (Tong and Elzinga, 1983; Iwabata et al., 2005). To test whether myosin heavy chain is methylated in zebrafish embryos, we carried out a Western blot analysis using anti–methyl lysine and anti-myosin antibodies. The data showed that several proteins, including myosin heavy chain, are indeed methylated in zebrafish embryos (Figure 10). Consistent with the reduced levels of myosin protein accumulation in smyd1b-knockdown embryos, the levels of methylated myosin were also significantly reduced in Smyd1-knockdown zebrafish embryos (Figure 10). Together these data indicate that myosin heavy chain proteins are methylated at lysine residues in zebrafish embryos. The significant reduction of myosin accumulation in smyd1b-knockdown may be directly or indirectly linked to myosin methylation, which may play an important role in protein stability and assembly into sarcomeres.
FIGURE 10:
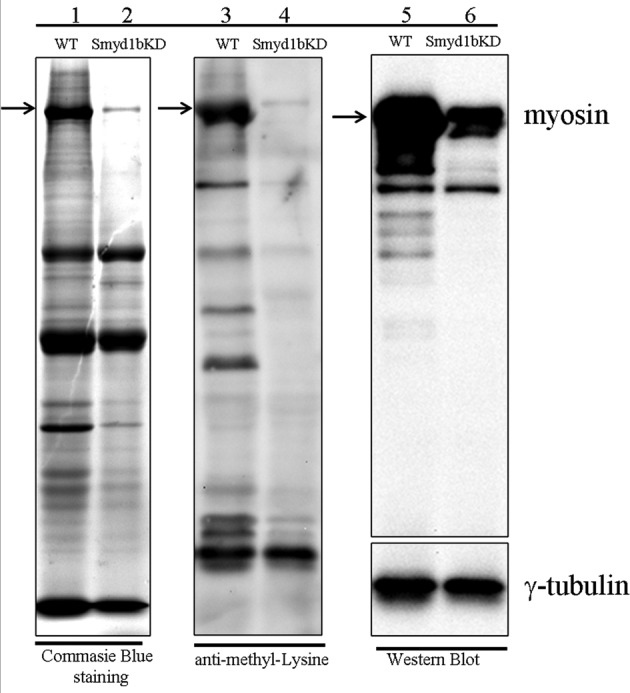
Analysis of protein methylation in smyd1b-knockdown embryos by Western blot. Lanes 1 and 2, Coomassie blue staining showing protein expression in wild-type (WT) control and smyd1b-knockdown embryos at 72 hpf. Lanes 3 and 4, protein methylation in WT control and smyd1b-knockdown embryos at 72 hpf by Western blot using anti–methyl lysine antibody. Lanes 5 and 6, myosin heavy chain protein levels in WT control and smyd1b-knockdown embryos at 72 hpf by Western blot using MF-20 antibody. γ-Tubulin was probed as a loading control.
DISCUSSION
In this study, we characterized the myofibril defects in skeletal and cardiac muscles of smyd1b-knockdown zebrafish embryos. We demonstrated that Smyd1b is required for myofibril organization in both skeletal and cardiac muscles. Knockdown of smyd1b expression resulted in complete disorganization of all key sarcomeric structures. including thick, thin, and titin filaments, as well as M- and Z-lines in skeletal and cardiac muscles of early-stage zebrafish embryos. We further demonstrated that knockdown of smyd1b resulted in up-regulation of unc45b and hsp90α1 gene expression, two important myosin chaperones involved in myosin folding and sarcomere assembly. Biochemical analysis by coIP revealed that Smyd1b_tv1 might associate directly or indirectly with Unc45b and Hsp90α1 in protein complexes. Moreover, knockdown of smyd1b expression resulted in a significant reduction of myosin methylation and protein accumulation without any significant effect on their mRNA expression. Together these data support the idea that Smyd1b is required for myofibril assembly in all striated muscles, and Smyd1b might work together with myosin chaperones to control sarcomere assembly during myofibrillogenesis.
Smyd1b is required for myofibril organization in slow muscles of early-stage zebrafish embryos
We previously demonstrated that Smyd1b is required for thick filament organization in slow muscles of zebrafish embryos (Tan et al., 2006). However, a recent report suggests that Smyd1b is not required for sarcomere organization in slow muscle of zebrafish embryos (Just et al., 2011). To test the hypothesis that the discrepancy between these two studies could be caused by different timing of the phenotypic analyses, we analyzed the sarcomere organization in slow muscles of Smyd1b-knockdown embryos at various stages during development. Consistent with this idea, we demonstrated that Smyd1b is required for myofibril assembly in slow muscles of early stages zebrafish embryos but becomes dispensable in late-stage embryos in slow muscles. This is likely due to the expression of another smyd1b-related gene, smyd1a, in muscle cells of zebrafish embryos starting around 24 hpf (Sun et al., 2008). Collectively data from these studies also suggest that Smyd1a and Smyd1b may not play an equal role in myofibrillogenesis. Smyd1b appears to play a more critical role than Smyd1a in sarcomere organization, especially in fast muscles and early-stage slow muscles of zebrafish embryos.
In addition to the thick filament defects, we showed that knockdown of smyd1b also disrupted the organization of thin and titin filaments, as well as of M-lines. This raises the question of whether M- and Z-line defects result directly from smyd1b knockdown or a secondary effect from myosin degradation and thick filament disorganization. Our recent studies from direct myosin knockdown in zebrafish embryos are consistent with a secondary effect (Codina et al., 2010; Xu et al., 2012). We showed that knockdown of myosin heavy chain 1 (myhc1) resulted in sarcomeric defects in both thick and thin filaments, as well as in M- and Z-lines (Codina et al., 2010; Xu et al., 2012). Together these studies indicate that the muscle phenotype from smyd1b knockdown could be due, at least in part, to disruption of myosin folding and assembly.
Protein complex formation with Unc45b and Hsp90α1
We showed by coIP assay that the longer isoform of Smyd1b (Smyd1b_tv1) could be coimmunoprecipitated with myosin chaperone Unc45b and Hsp90α1 expressed in HEK293 cells. The coIP results could only suggest that Smyd1b (Smyd1b_tv1) coexists with Unc45b or Hsp90α1 in a protein complex. They do not reveal whether Smyd1 actually binds to UNC-45b and Hsp90α1. Future pull-down studies using purified proteins will be required to demonstrate direct physical binding between Smyd1b and UNC-45b or Hsp90α1 proteins. Regardless of whether Smyd1b_tv1 can bind directly or indirectly with Unc45b and Hsp90α1, several pieces of evidence suggest that Smyd1b_tv1 may work together with Unc45b and Hsp90α1 to control myosin folding and sarcomere assembly during myofibrillogenesis. First, genetic analysis shows that knockdown or mutation of unc45b or Hsp90α1 results in similar muscle defects as Smyd1b knockdown in zebrafish embryos (Etard et al., 2007; Wohlgemuth et al., 2007; Du et al., 2008; Hawkins et al., 2008). Second, biochemical analysis reveals that knockdown or mutation of smyd1b results in significant reduction of myosin protein accumulation with no effect on mRNA levels, a phenotype very similar to that observed in Unc45b- or Hsp90α1-knockdown zebrafish embryos (Etard et al., 2007; Wohlgemuth et al., 2007; Du et al., 2008; Hawkins et al., 2008).
Up-regulation of hsp90α1 and unc45b gene expression in Smyd1b-knockdown embryos
In this study, we showed that knockdown of Smyd1b resulted in up-regulation of hsp90α1 and unc45b gene expression in zebrafish embryos. Up-regulation of hs90α1 and unc45b gene expression appears to be a common response in zebrafish embryos with myofibril disorganization. Disruption of myofibril organization by loss of Unc45b function also leads to up-regulation of hsp90α1 expression (Etard et al., 2007), and up-regulation of unc45b expression was noted in hsp90α1-mutant or -knockdown zebrafish embryos (Etard et al., 2007; Du et al., 2008; Hawkins et al., 2008). The mechanism underlying this up-regulation of gene expression is not clear. Disruption of myosin folding and assembly by smyd1b knockdown likely increases misfolding of myosin and other sarcomere proteins in muscle cells. The up-regulation of hsp90α1 and unc45b expression could result from increasing demand for refolding and degradation of misfolded sarcomeric proteins in smyd1b-knockdown embryos. Consistent with this, we found that other stress response genes, such as hsp70 and hsp47, are also up-regulated in smyd1b-knockdown zebrafish embryos (Supplemental Table S1).
Recent studies show that overexpression of Unc45 results in thick filament defects and decreased myosin expression in C. elegans and zebrafish (Landsverk et al., 2007; Bernick et al., 2010; Ni et al., 2011). Consistent with the idea that up-regulation of unc45 expression could be involved in myosin degradation in smyd1b-knockdown embryos, we demonstrated in this study that ectopic expression of Unc45b in smyd1b-knockdown embryos further enhanced myosin degradation and resulted in little or no thick filament organization compared with smyd1b knockdown alone. The molecular mechanism by which Unc45b regulates myosin degradation is not clear. It has been suggested that Unc45 functions as a myosin chaperone that regulates myosin accumulation and assembly by linkage to the ubiquitin proteasome system for degradation (Landsverk et al., 2007). Structural and functional analyses show that Unc45 contains an N-terminal TPR domain involved in interaction with Hsp90 and a C-terminal UCS domain required for interaction with myosin (Barral et al., 2002). Deletion of the C-terminal UCS domain abolished the disruptive effect of Unc45b overexpression in myosin thick filament organization. In contrast, deletion of the N-terminal TPR domain required for binding with Hsp90 had no effect (Bernick et al., 2010; Ni et al., 2011).
Smyd1b functions in cardiac muscle myofibrillogenesis
In this study we showed that knockdown of smyd1b does not affect heart tube patterning in zebrafish embryos. Heart tube with clear atrium and ventricle structures was observed in smyd1b-knockdown zebrafish embryos, consistent with a report by Just et al. (2011). The lack of heart tube patterning defects in fish embryos differs from the mouse smyd1b-knockout phenotype, in which the right ventricle fails to form in mouse embryos (Gottlieb et al., 2002). The reason for the observed discrepancy between these two studies is not clear. It could be due to the different heart patterning between fish and mammals. In contrast to mammalian heart, which has two separate ventricles, fish heart has only one ventricle.
Our studies, however, demonstrated that Smyd1b plays an important role in myofibril assembly in cardiac muscles. Knockdown of smyd1b resulted in disorganization of sarcomeric structures in heart muscle cells. Both myosin thick filaments and Z-lines were disrupted in cardiac muscles of smyd1b-knockdown zebrafish embryos. This is consistent with a report on smyd1b-mutant zebrafish embryos (Just et al., 2011). The myofibril defects in cardiac muscles resemble the skeletal muscle defects in smyd1b-knockdown embryos. Collectively these studies indicate that Smyd1b plays an important role in myofibril assembly in both skeletal and cardiac muscles.
Smyd1b and protein methylation
Previous studies showed that Smyd1b is a lysine methyltransferase that can methylate histone 3 in vitro (Tan et al., 2006). Recent studies (Just et al., 2011), however, demonstrated that the histone methyltransferase activity is not required for Smyd1b function in myofibrillogenesis. It has been increasingly recognized that members of the Smyd family are able to methylate nonhistone proteins. For example, Smyd2 can methylate Hsp90 and plays a critical role in sarcomere organization (Abu-Farha et al., 2011; Donlin et al., 2012; Voelkel et al., 2013). The in vivo targets of Smyd1 remain are not known. Yeast two hybrid and coIP analyses identified several tentative Smyd1-binding proteins, including skNAC, Mical2b, helicase, and FKBP8 (Park et al., 2010). It remains to be determined whether any of these proteins are methylation targets of Smyd1.
In this study, we analyzed the overall protein methylation in zebrafish embryos and showed that myosin heavy chain is methylated in zebrafish embryos. In addition, we found that the myosin methylation was significantly decreased in smyd1b-knockdown embryos. The significant reduction of myosin methylation could be due to the increased myosin protein degradation in smyd1b-knockdown embryos. Conversely, methylation of myosin by Smyd1b may be required for its folding and stability, and, hence, knockdown of smyd1b expression may result in poor methylation of myosin proteins, leading to increased degradation. Protein methylation has been suggested to play important roles in protein stabilization and destabilization (Yang et al., 2009; Egorova et al., 2010; Zhang et al., 2012). Because methylation and ubiquitination both occur at lysine residues, methylation of lysine residues could prevent them from ubiquitination. It has been suggested that methylation is a protein stable mark (Byvoet et al., 1972; Duerre and Lee, 1974; Rice and Allis, 2001). Given all of these considerations, one potential mechanism is that myosin methylation by Smyd1 may prevent myosin from ubiquitination, which leads to protein degradation. Consistent with the idea that myosin methylation may control its satiability, it was shown that myosin methylation markedly increased its ATPase activity in vitro (White and Rayment, 1993). Given that Smyd1b is a myosin-binding protein (Just et al., 2011), we speculate that Smyd1b may function as a muscle-specific methyltransferase involved in myosin methylation, which plays a critical role in myofibril assembly.
MATERIALS AND METHODS
Zebrafish lines and maintenance
Mature zebrafish were raised at the Zebrafish Facility of the Aquaculture Research Center, Institute of Marine and Environmental Technology (Baltimore, MD). The fish were maintained at 28°C with a photoperiod of 14 h light and 10 h dark in 8-gallon aquaria supplied with freshwater and aeration. The myomesin-RFP zebrafish line (GBT0067) was obtained from Stephen Ekker's laboratory at the Mayo Clinic (Rochester, MN). It carries the RFP insertion in the myomesin 3 gene (Clark et al., 2011; Xu et al., 2012; for additional information see www.zfishbook.org/index.php?topic=GBT0067#).
Morpholino and DNA microinjection into zebrafish embryos
Morpholino antisense oligos against zebrafish Smyd1a and Smyd1b were synthesized by Gene Tools (Philomath, OR). The Smyd1b translation blocker (Smyd1b ATG-MO) was based on the antisense sequence near the ATG start site. The Smyd1b splicing blocker (Smyd1b E9I9-MO) was based on the antisense sequence at the exon 9/intron 9 junction. The Smyd1a splicing blockers (Smyd1b E8I8-MO and Smyd1a E9I9-MO) were based on the antisense sequence at the exon 8/intron 8 and exon 9/intron 9 junctions, respectively. The standard control MO from Gene Tools was used as control. Morpholino antisense oligos were dissolved in Danieau buffer (Nasevicius and Ekker, 2000) to a final concentration of 0.5 and 1 mM. Zebrafish embryos were injected at the one- or two-cell stage with 2 nl of MO as described (Tan et al., 2006). DNA microinjection was carried out as described (Du et al., 1997)
Smyd1b ATG-MO: 5′-ACTTCCACAAACTCCATTCTGGATC-3′
Smyd1b E9I9-MO: 5′-CGTCACCTCTAGGTCTTTAGTGATG-3′
Smyd1a E8I8-MO: 5′-ATATCGCAACACTCACATGTATCCA-3′
Smyd1a E9I9-MO: 5′-GGTTGTACCTCCAGGTCTCTGCTGA-3′
Unc-45b ATG-MO: 5′-ATCTCCAATTTCTCCCATCGTCATT-3′
Tg(Smyd1b-Smyd1b) transgenic zebrafish and Tg(Smyd1b:Unc45bFLAG) transgene
The Tg(Smyd1b:Smyd1b-tv1myc)mb6 and Tg(Smyd1b:Smyd1b-tv2myc)mb7 transgenic zebrafish lines were generated as described previously (Tan et al., 2006). Tg(Smyd1b:Smyd1b-tv1myc) and Tg(Smyd1b:Smyd1b-tv2myc) minigenes were constructed by using cDNA encoding the myc-tagged Smyd1b-tv1 or Smyd1b-tv2 cloned after the 5.3-kb zebrafish smyd1b promoter and its 5′ flanking sequence (Du et al., 2006; Tan et al., 2006). The Tg(Smyd1b:Unc45bFLAG) transgene was constructed by using cDNA encoding the FLAG-tagged zebrafish Unc45b cloned after the 5.3-kb zebrafish smyd1b promoter and its 5′ flanking sequence (Bernick et al., 2010).
Immunostaining of whole-mount fish embryos
Immunostaining was carried out using whole-mount zebrafish embryos (24–72 hpf) as previously described (Tan et al., 2006), with the following antibodies: anti–myosin heavy chain for slow muscle (F59; Developmental Studies Hybridoma Bank [DSHB], Iowa City, IA), anti-myosin heavy chain (MF20; DSHB), anti-myosin heavy chain (S46; DSHB), anti–α-actinin (clone EA-53, #A7811; Sigma, St. Louis, MO), anti-MHC for fast muscles (F310; DSHB), anti-myomesin (mMaC myomesin B4; DSHB), anti–α-actin (Ac1-20.4.2; Progen, Heidelberg, Germany), and anti-titin (clone T11, #T9030; Sigma). Secondary antibodies were fluorescein isothiocyanate or tetramethylrhodamine isothiocyanate conjugates (Sigma).
Analysis of protein expression by Western blot
Wild-type or MO-injected zebrafish embryos were dechorinated manually at 24 or 72 hpf. The embryos (50 embryos each group) were washed with 1 ml of phosphate-buffered saline (PBS) and crushed gently to remove the yolk by pipetting with a glass pipette in 0.5 ml of PBS. The embryos were collected by a quick spin at 3000 rpm for 1 min. The embryo extract was washed once with 0.5 ml of PBS and solubilized in 100 μl of 2× SDS loading buffer (0.125 M Tris-Cl, pH 6.8, 4% SDS, 20% glycerol, 0.2 M dithiothreitol, 0.02% bromophenol blue) containing 1 mM phenylmethylsulfonyl fluoride and protease inhibitor (P8340; Sigma) by passage through a 21-gauge needle 10–15 times. The proteins were denatured by boiling for 3 min and analyzed on a 7.5% SDS–PAGE gel. Proteins from approximately five embryos were loaded on each lane of the SDS–PAGE gel. Proteins from the gel were transferred onto a polyvinylidene fluoride membrane (Immobilion-P; Millipore, Billerica, MA) by electrophoresis. Immunodetection of MHC, γ-tubulin, or myc-tagged proteins was carried out with their primary antibodies and followed by corresponding peroxidase-conjugated secondary antibodies. For Western blot to detect proteins with methylated lysine residues, the horseradish peroxidase–conjugated anti–methylated lysine antibody (TF1213; Enzo Life Sciences, Farmingdale, NY) was used. SuperSignal Western Blot Enhancer Kit (46640; Thermo Scientific, Waltham, MA) was used to enhance the sensitivity of the Western blot analysis according to the manufacturer's instructions. All blots were developed using the Pierce ECL Western Blotting Substrate (Thermo Scientific).
Real-time PCR analysis
Real-time PCR was carried out using 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA). The PCR was carried out using standard SYBR Green PCR Master Mix (Applied Biosystems). Standard curves of cDNA samples were constructed using 10-fold serial dilutions. The relative levels of gene expression were compared based on the normalized value of the endogenous control elongation factor 1-α (ef-1α). Real-time PCR was carried out using the following primers:
zfhsp90α1-P6: agccagacttcggtgaatcaa
zfhsp90α1-P7: ttctctctgtttctcaatgtaaa
zfmyhz2-P4: gctcacctaccagactgagga
zfmyhz2-P5: actcagcaatatcagcacgct
zfsmyhc1-P4: gctcacctaccagactgagga
zfsmyhc1-P5: catcttgttgacctgagattca
zfunc45b-P4: gctgcaaggaggtccaagaca
zfunc45b-P5: gatcatcagcatccagcatgt
zfef1a-P3: cttcaacgctcaggtcatcat
zfef1a-P4: acagcaaagcgaccaagagga
zSmyd1b-RT-F: atctgaacgtgtctgcaga
Smyd1b-P6: tcttccggcaccttgactccatcc
Whole-mount in situ hybridization
Whole-mount in situ hybridization was carried out using digoxigenin-labeled antisense probes as previously described (Du and Dienhart, 2001). The plasmid Hsp90α1-P was digested with NcoI and transcribed with Sp6 RNA polymerase to synthesize digoxigenin-labeled antisense RNA probe. Antisense probes against zebrafish slow myosin heavy chain 1 (smyhc1) or fast muscle myosin heavy chain 2 (myhz2) were synthesized by Sp6 RNA polymerase from NcoI or SphI linearized pGEM-smyhc1 or pGEM-myhz2 plasmid, respectively. The zebrafish unc-45b antisense probe was synthesized with Sp6 RNA polymerase from plasmid pGEM-unc45b linearized with BamHI.
Immunoprecipitation and immunoblotting
HEK293 cells were seeded at 2.5 × 105 per well in six-well plates 1 d before the transfection. Four micrograms of each indicated plasmid was transiently transfected by calcium phosphate precipitation. Cells were harvested 24 h after the transfection and lysed in 1× cell lysis buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM ethylene glycol tetraacetic acid, and 0.2% Nonidet P-40) with 1× protease inhibitor mixture (P2714; Sigma). For IP, 300 μg of total protein was incubated with 30 μl of prewashed Anti–FLAG M2 Affinity Gel (Sigma A2220) at 4°C for 2 h in a total volume of 600 μl containing 4% glycerol in 1× cell lysis buffer. The beads were washed three times in 1× cell lysis buffer before processing for SDS/PAGE and immunoblotting.
Heat shock and cold shock treatment of zebrafish embryos
For the heat shock experiment, 50 wild-type zebrafish embryos at 24 hpf were treated for 15 min from 28.5 to 38°C, then 38°C for 30 min, and then another 15 min from 38 to 28.5°C. For the cold shock experiment, 50 wild-type zebrafish embryos at 24 hpf were incubated for 30 min at 4°C. For real-time PCR, total RNA of the heat-shocked or cold-shocked embryos, as well of control embryos, was extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA was synthesized using the first-strand cDNA synthesis kit (Life Sciences). For in situ hybridization, the heat-shocked, cold-shocked, and wild-type embryos were fixed in 4% paraformaldehyde. Whole-mount in situ hybridization was performed with digoxigenin-labeled antisense probes as described (Tan et al., 2006).
Supplementary Material
Acknowledgments
This research was supported by research grants from the United States–Israel Binational Agricultural Research and Development Fund (IS-8713–08), the Maryland Stem Cell Research Fund, and a key research project grant from the Natural Science Foundation of China (31230076). We thank Vivien Xie and Nick Du for proofreading the manuscript.
Abbreviations used:
- hpf
hours postfertilization
- Hsp90
heat shock protein
- MO
morpholino
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-06-0352) on September 25, 2013.
REFERENCES
- Abu-Farha M, Lanouette S, Elisma F, Tremblay V, Butson J, Figeys D, Couture JF. Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J Mol Cell Biol. 2011;3:301–308. doi: 10.1093/jmcb/mjr025. [DOI] [PubMed] [Google Scholar]
- Barral JM, Bauer CC, Ortiz I, Epstein HF. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J Cell Biol. 1998;143:1215–1225. doi: 10.1083/jcb.143.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, Epstein HF. Protein machines and self assembly in muscle organization. Bioessays. 1999;21:813–823. doi: 10.1002/(SICI)1521-1878(199910)21:10<813::AID-BIES3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 2002;295:669–671. doi: 10.1126/science.1066648. [DOI] [PubMed] [Google Scholar]
- Bernick EP, Zhang PJ, Du S. Knockdown and overexpression of Unc-45b result in defective myofibril organization in skeletal muscles of zebrafish embryos. BMC Cell Biol. 2010;11:70. doi: 10.1186/1471-2121-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Sims RJ, 3rd, Gottlieb PD, Tucker PW. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer. 2006;5:26. doi: 10.1186/1476-4598-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byvoet P, Shepherd GR, Hardin JM, Noland BJ. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys. 1972;148:558–567. doi: 10.1016/0003-9861(72)90174-9. [DOI] [PubMed] [Google Scholar]
- Cho HS, et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia. 2012;14:476–86. doi: 10.1593/neo.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KJ, et al. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods. 2011;8:506–515. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codina M, Li J, Gutierrez J, Kao JP, Du SJ. Loss of Smyhc1 or Hsp90alpha1 function results in different effects on myofibril organization in skeletal muscles of zebrafish embryos. PLoS One. 2010;5:e8416. doi: 10.1371/journal.pone.0008416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto SH, Melançon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Donlin LT, et al. Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev. 2012;26:114–119. doi: 10.1101/gad.177758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SJ, Devoto SH, Westerfield M, Moon RT. Positive and negative regulation of muscle cell identity by members of the hedgehog and TGF-beta gene families. J Cell Biol. 1997;139:145–156. doi: 10.1083/jcb.139.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SJ, Dienhart M. Gli2 mediation of hedgehog signals in slow muscle induction in zebrafish. Differentiation. 2001;67:84–91. doi: 10.1046/j.1432-0436.2001.067003084.x. [DOI] [PubMed] [Google Scholar]
- Du SJ, Li H, Bian Y, Zhong Y. Heat-shock protein 90alpha1 is required for organized myofibril assembly in skeletal muscles of zebrafish embryos. Proc Natl Acad Sci USA. 2008;105:554–559. doi: 10.1073/pnas.0707330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SJ, Rotllant J, Tan X. Muscle-specific expression of the Smyd1b gene is controlled by its 5.3-kb promoter and 5’-flanking sequence in zebrafish embryos. Dev Dyn. 2006;235:3306–3315. doi: 10.1002/dvdy.20984. [DOI] [PubMed] [Google Scholar]
- Duerre JA, Lee CT. In vivo methylation and turnover of rat brain histones. J Neurochem. 1974;23:541–547. doi: 10.1111/j.1471-4159.1974.tb06057.x. [DOI] [PubMed] [Google Scholar]
- Egorova KS, Olenkina OM, Olenina LV. Lysine methylation of nonhistone proteins is a way to regulate their stability and function. Biochemistry (Mosc) 2010;75:535–548. doi: 10.1134/s0006297910050019. [DOI] [PubMed] [Google Scholar]
- Ehler E, Gautel M. The sarcomere and sarcomerogenesis. Adv Exp Med Biol. 2008;642:1–14. doi: 10.1007/978-0-387-84847-1_1. [DOI] [PubMed] [Google Scholar]
- Epstein HF, Thomson JN. Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature. 1974;250:579–580. doi: 10.1038/250579a0. [DOI] [PubMed] [Google Scholar]
- Etard C, Behra M, Fischer N, Hutcheson D, Geisler R, Strahle U. The UCS factor Steif/Unc-45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev Biol. 2007;308:133–143. doi: 10.1016/j.ydbio.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Gottlieb PD, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- Hawkins TA, et al. The ATPase-dependent chaperoning activity of Hsp90a regulates thick filament formation and integration during skeletal muscle myofibrillogenesis. Development. 2008;135:1147–1156. doi: 10.1242/dev.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Landsverk ML, Price MG, Epstein HF. The UCS family of myosin chaperones. J Cell Sci. 2002;115:3983–3990. doi: 10.1242/jcs.00107. [DOI] [PubMed] [Google Scholar]
- Hwang I, Gottlieb PD. Bop: a new T-cell-restricted gene located upstream of and opposite to mouse CD8b. Immunogenetics. 1995;42:353–361. doi: 10.1007/BF00179396. [DOI] [PubMed] [Google Scholar]
- Hwang I, Gottlieb PD. The Bop gene adjacent to the mouse CD8b gene encodes distinct zinc-finger proteins expressed in CTLs and in muscle. J Immunol. 1997;158:1165–1174. [PubMed] [Google Scholar]
- Iwabata H, Yoshida M, Komatsu Y. Proteomic analysis of organ-specific post-translational lysine-acetylation and -methylation in mice by use of anti-acetyllysine and -methyllysine mouse monoclonal antibodies. Proteomics. 2005;5:4653–4664. doi: 10.1002/pmic.200500042. [DOI] [PubMed] [Google Scholar]
- Just S, et al. The myosin-interacting protein SMYD1B is essential for sarcomere organization. J Cell Sci. 2011;124:3127–3136. doi: 10.1242/jcs.084772. [DOI] [PubMed] [Google Scholar]
- Kim J, Lowe T, Hoppe T. Protein quality control gets muscle into shape. Trends Cell Biol. 2008;18:264–272. doi: 10.1016/j.tcb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Landsverk ML, Li S, Hutagalung AH, Najafov A, Hoppe T, Barral JM, Epstein HF. The UNC-45 chaperone mediates sarcomere assembly through myosin degradation in Caenorhabditis elegans. J. Cell Biol. 2007;177:205–210. doi: 10.1083/jcb.200607084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu J, Bian YH, Rotllant P, Shen T, Chu W, Zhang J, Schneider M, Du SJ. Smyd1b_tv1, a key regulator of sarcomere assembly, is localized on the M-line of skeletal muscle fibers. PLoS One. 2011;6:e28524. doi: 10.1371/journal.pone.0028524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod AR, Waterston RH, Brenner S. An internal deletion mutant of a myosin heavy chain in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1977;74:5336–5340. doi: 10.1073/pnas.74.12.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene “knockdown” in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Ni W, Hutagalung AH, Li S, Epstein HF. The myosin-binding UCS domain but not the Hsp90-binding TPR domain of the UNC-45 chaperone is essential for function in Caenorhabditis elegans. J Cell Sci. 2011;124:3164–7313. doi: 10.1242/jcs.087320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Pierce SA, von Drehle M, Ivey KN, Morgan JA, Blau HM, Srivastava D. skNAC, a Smyd1-interacting transcription factor, is involved in cardiac development and skeletal muscle growth and regeneration. Proc Natl Acad Sci USA. 2010;107:20750–20755. doi: 10.1073/pnas.1013493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–73. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Saddic LA, West LE, Aslanian A, Yates JR, 3rd, Rubin SM, Gozani O, Sage J. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010;285:37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass JB, Martin CC, Krone PH. Restricted expression of the zebrafish hsp90alpha gene in slow and fast muscle fiber lineages. Int J Dev Biol. 1999;43:835–838. [PubMed] [Google Scholar]
- Sass JB, Weinberg ES, Krone PH. Specific localization of zebrafish hsp90 alpha mRNA to myoD-expressing cells suggests a role for hsp90 alpha during normal muscle development. Mech Dev. 1996;54:195–204. doi: 10.1016/0925-4773(95)00476-9. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Weihe EK, Zhu L, O'Malley S, Harriss JV, Gottlieb PD. m-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J Biol Chem. 2002;277:26524–26529. doi: 10.1074/jbc.M204121200. [DOI] [PubMed] [Google Scholar]
- Sirinupong N, Brunzelle J, Ye J, Pirzada A, Nico L, Yang Z. Crystal structure of cardiac-specific histone methyltransferase Smyd1b reveals unusual active site architecture. J Biol Chem. 2010;285:40635–40644. doi: 10.1074/jbc.M110.168187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikakulam R, Winkelmann DA. Myosin II folding is mediated by a molecular chaperonin. J Biol Chem. 1999;274:27265–27273. doi: 10.1074/jbc.274.38.27265. [DOI] [PubMed] [Google Scholar]
- Srikakulam R, Winkelmann DA. Chaperone-mediated folding and assembly of myosin in striated muscle. J Cell Sci. 2004;117:641–652. doi: 10.1242/jcs.00899. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Fishman MC. Patterning the zebrafish heart tube: acquisition of anteroposterior polarity. Dev Biol. 1992;153:91–101. doi: 10.1016/0012-1606(92)90094-w. [DOI] [PubMed] [Google Scholar]
- Sun XJ, et al. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS One. 2008;3:e1499. doi: 10.1371/journal.pone.0001499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Rotllant J, Li H, De Deyne P, Du SJ. Smyd1b, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci USA. 2006;103:2713–2718. doi: 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SW, Elzinga M. The sequence of the NH2-terminal 204-residue fragment of the heavy chain of rabbit skeletal muscle myosin. J Biol Chem. 1983;258:13100–13110. [PubMed] [Google Scholar]
- Voelkel T, Andresen C, Unger A, Just S, Rottbauer W, Linke WA. Lysine methyltransferase Smyd2 regulates Hsp90-mediated protection of the sarcomeric titin springs and cardiac function. Biochim Biophys Acta. 2013;1833:812–822. doi: 10.1016/j.bbamcr.2012.09.012. [DOI] [PubMed] [Google Scholar]
- White HD, Rayment I. Kinetic characterization of reductively methylated myosin subfragment 1. Biochemistry. 1993;32:9859–65. doi: 10.1021/bi00088a042. [DOI] [PubMed] [Google Scholar]
- Willis MS, Schisler JC, Portbury AL, Patterson C. Build it up-tear it down: protein quality control in the cardiac sarcomere. Cardiovasc Res. 2009;81:439–448. doi: 10.1093/cvr/cvn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth SL, Crawford BD, Pilgrim DB. The myosin co-chaperone UNC-45 is required for skeletal and cardiac muscle function in zebrafish. Dev Biol. 2007;303:483–492. doi: 10.1016/j.ydbio.2006.11.027. [DOI] [PubMed] [Google Scholar]
- Xu J, Gao J, Li J, Xue L, Clark KJ, Ekker SC, Du SJ. Functional analysis of slow myosin heavy chain 1 and myomesin-3 in sarcomere organization in zebrafish embryonic slow muscles. J Genet Genomics. 2012;39:69–80. doi: 10.1016/j.jgg.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XD, Lamb A, Chen LF. Methylation, a new epigenetic mark for protein stability. Epigenetics. 2009;4:429–33. doi: 10.4161/epi.4.7.9787. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wen H, Shi X. Lysine methylation: beyond histones. Acta Biochim Biophys Sin (Shanghai) 2012;44:14–27. doi: 10.1093/abbs/gmr100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


