Abstract
A reverse genetics approach was used to investigate the role of γ-aminobutyric acid metabolism in the wheat pathogenic fungus Stagonospora nodorum. The creation of mutants lacking Sdh1, the gene encoding succinic semialdehyde dehydrogenase, resulted in strains that grew poorly on γ-aminobutyric acid as a nitrogen source. The sdh1 mutants were more susceptible to reactive oxygen stress but were less affected by increased growth temperatures. Pathogenicity assays revealed that the metabolism of γ-aminobutyric acid is required for complete pathogenicity. Growth assays of the wild-type and mutant strains showed that the inclusion of γ-aminobutyric acid as a supplement in minimal media (i.e., not as a nitrogen or carbon source) resulted in restricted growth but increased sporulation. The addition of glutamate, the precursor to GABA, had no effect on either growth or sporulation. The γ-aminobutyric acid effect on sporulation was found to be dose dependent and not restricted to Stagonospora nodorum with a similar effect observed in the dothideomycete Botryosphaeria sp. The positive effect on sporulation was assayed using isomers of γ-aminobutyric acid and other metabolites known to influence asexual development in Stagonospora nodorum but no effect was observed. These data demonstrate that γ-aminobutyric acid plays an important role in Stagonospora nodorum in responding to environmental stresses while also having a positive effect on asexual development.
Introduction
Stagonospora nodorum is the causal agent of stagonospora nodorum blotch, a significant disease of wheat in many parts of the world [1]. It is has now been established that the wheat pathogen S. nodorum causes disease through the secretion of small secreted proteins (effectors) that interact with dominant susceptibility host genes leading to cell death and disease [2]. These studies have fundamentally advanced our understanding of the interaction and provided significant insight into how this necrotrophic pathogen causes disease [3].
Whilst this effector model has now been established as the means by which the pathogen initiates disease, there still remains much to learn about how the fungus grows, develops and reproduces in the host. Several studies to date have identified key pathogen metabolic pathways required to complete the infection cycle. For example, reverse genetics approaches have demonstrated that the metabolism of mannitol, particularly through the activity of mannitol 1-phosphate dehydrogenase (Mpd1) is essential for asexual sporulation [4], [5], [6]. Mutants lacking the Mpd1 gene were unable to sporulate, either in vitro or in planta, unless supplied with exogenous mannitol. A similar approach identified that trehalose also plays a key role in sporulation [7]. Disruption of a trehalose 6-phosphate synthase resulted in decreased sporulation that could be restored in the presence of added trehalose. The role of either of these primary metabolites on sporulation remains elusive although it was clearly demonstrated that the metabolism of these compounds, rather than simply their presence, was required for sporulation.
In a more recent study, a short-chain dehydrogenase, Sch1, was identified as being negatively regulated in S. nodorum strain lacking the Gα subunit Gna1 [8]. Subsequent disruption of the Sch1 gene resulted in a strain that was unable to produce pycnidiospores, although abnormal pycnidia were differentiated. Metabolite analysis of the mutant strain showed that neither mannitol nor trehalose levels were affected. Indeed primary metabolism appeared relatively unaffected by the Sch1 mutation. Metabolite profiling though did reveal that the secondary metabolite, alternariol, was present at very high levels in the mutant leading to speculation as to its role in asexual differentiation. Subsequent studies on S. nodorum sporulation-deficient strains have consistently identified changes in abundance in alternariol suggesting that it, and/or its associated pathway, play an integral role in sporulation [9].
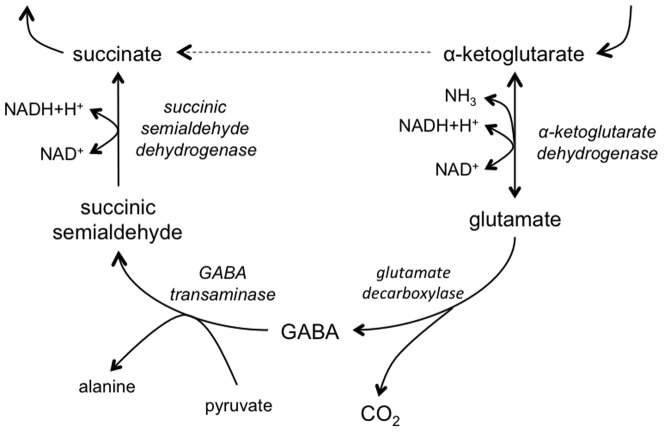
These studies highlight the critical role of specific primary metabolic processes in the development and differentiation of S. nodorum. However there are many other pathways whose roles aren't clear during either development or pathogenicity. One such pathway is the γ-aminobutyric acid (GABA) shunt (Figure 1). This pathway is a bypass of the TCA cycle from α-ketoglutaric acid to succinic acid via glutamate, GABA and succinic semialdehyde. In contrast to the TCA cycle (between α-ketoglutaric acid and succinic acid), the GABA shunt results in no net gain in NADH or ATP, leaving many to question its biological role.
Figure 1. The GABA shunt.
GABA itself is a well described negative neurotransmitter in the central and peripheral nervous system of vertebrates and some non-vertebrates [10]. However the role of GABA and its metabolism in other systems is less well understood. In plants, GABA has been shown to accumulate under different forms of stress [11], [12], [13], [14]. Mutational studies in Arabidopsis thaliana have demonstrated that the GABA pathway plays a role in responding to osmotic stress, as well as high temperature and light stress [15]. GABA has also been found to accumulate in tomato leaves during a compatible interaction with Cladosporium fulvum, leading to speculation that the pathogen was manipulating the host to produce a preferred nitrogen source [16]. However there has been no further evidence to support this.
The role of GABA and/or its metabolism in fungi is even less clear although it is well known that GABA is a suitable nitrogen source for many fungi [17]. A recent study in Magnaporthe oryzae identified a gene encoding succinic semialdehyde dehydrogenase (MoSSADH) as being important for pathogenicity [18]. Characterisation of the MoSSADH mutants found that the gene was required for appressorium-like penetration, invasive growth and normal development and conidiation. The mutants were also highly sensitive to H2O2 and displayed attenuated peroxidase and laccase activities. The bases of these phenotypes and the role of MoSSADH though were not elucidated.
In this study, we have focused on understanding the role of GABA catabolism on the pathogenicity and development of S. nodorum through the inactivation of the succinic semialdehyde dehydrogenase gene, Sdh1. The gene encoding succinic semialdehyde dehydrogenase was selected to allow a direct comparison of its role in a necrotrophic pathogen compared to the hemibiotrophic M. oryzae. This study has revealed various important roles for succinic semialdehyde dehydrogenase in S. nodorum, and like previous studies have done [7], has highlighted the significant differences that exist in the metabolic requirements of these different pathogens during infection. These data have also shed light on the novel role of GABA promoting asexual sporulation in S. nodorum.
Materials and Methods
Fungal strains and growth conditions
Stagonospora nodorum was maintained and grown as previously described [19]. Minimal media consists of 30 mM sucrose, 2 g/L NaNO3, 1 g/L NaH2PO4 and 10 mL trace stock solution, pH 6. For the supplementation assays, the chemicals (e.g. GABA) were added to media prior to pH adjustment. All strains were routinely grown under at 12 hr light/dark cycle at 22°C unless otherwise stated. Oxidative stress growth assays were undertaken by the addition of analytical grade H2O2 to agar plates to the final concentrations stated in the Results.
The dothideomycete Botryosphaeria sp. was kindly provided by Dr. Hugh Wallwork from the South Australian Research and Development Institute. The strain was grown on Botryosphaeria growth medium (BGM), which was prepared by adding 15 g of oats to 200 mL of deionised water and bringing to the boil in a microwave. To this, 40 g of fresh wheat leaves ground with a mortar and pestle in a small amount of water were added. The resulting liquid was then filtered and brought to 1 L with deionised water. 1.5% agar was added to the media prior to autoclaving. When used, 1 mM GABA was added prior to autoclaving.
Sporulation assays
For S. nodorum sporulation assays, agar plates were inoculated in the centre with 10 µL of 106 spores/mL, allowed to dry and then wrapped in micropore tape (3M). The plates were incubated for 20 days prior to being flooded with 5 mL sterile H2O and scraped using a 1 mL pipette tip. The plates were then allowed to rest for 10 min prior to a further 5 mL being applied. The liquid on the plates was then passed through a 10 mL syringe in which a small amount of sterile glass wool had been inserted into the barrel. The eluate from the syringe was collected in a 15 mL sterile plastic tube and centrifuged at 4000 g for 5 min. The pellet was resuspended in 5 mL sterile water and then diluted as required for subsequent counting using a haemocytometer.
The Botryosphaeria sp. pycnidia were counted by dividing the agar plates into eight and counting the pycnidia present on three representative sections from each plate. Three plates were used for each assay.
Sdh1 inactivation
The succinic semialdehyde dehydrogenase gene (Sdh1) disruption construct was created using the split-marker method [20]. p1 and p2 was used to amplify a 775 bp fragment 5′ of the Sdh1 gene whilst p3 and p4 were used to amplify a 760 bp 3′ fragment. These 5′ and 3′ flanks were fused to the overlapping fragments of hygromycin cassette creating the constructs 5′sdh1KO-YG and 3′sdh1KO-HY. These constructs were then further amplified using standard proof-reading PCR conditions followed by PCR purification. 3 µg of each construct was then co-transformed into S. nodorum SN15 as previously described [19]. The resulting transformants were screened by PCR for homologous recombination using primers SdhKOscreenF and SdhKOScreenR, with a 2360 bp band representing the wild-type locus and a 3870 bp band showing a homologous recombination event. The sdh1 complementation construct was created by amplifying the wild-type Sdh1 gene as well as 1 kb upstream and 500 bp downstream of the open reading frame using the Sdh1compF and Sdh1compR primers. The resulting amplicon was fused to a phleomycin cassette as previously described [20] and transformed into the sdh1-9 mutant. Gene copy number was determined by quantitative PCR as previously described [21]. The primer sequences are listed in Table S1.
In terms on nomenclature throughout the manuscript, Sdh1 refers to the wild-type gene whilst Sdh1 denotes the protein. The inactive gene is denoted by sdh1 (no capital ‘s’).
Pathogenicity assays
The wheat line Grandin was grown for two weeks under natural day/night cycle at approximately 22°C in small pots containing vermiculite and 5 g of Osmocote slow release fertiliser. After two weeks, the plants were inoculated with spores at a concentration of 1×106/mL containing 0.02% Tween 20 using an airbrush (Paasche, Chicago, USA). The plants were then covered for two days in the dark and scored visually as previously described at seven and 14 days post infection (dpi) [19].
Statistical analysis
All statistical analyses were undertaken using the JMP7 package (SAS Institute). Statistical significance was determined using the Tukey–Kramer analysis.
Results
The identification and disruption of Sdh1
To identify the gene encoding a succinic semialdehyde dehydrogenase in S. nodorum, the Uga2 protein sequence from S. cerevisiae was used to interrogate the S. nodorum annotated genome sequence [22]. This approach identified the gene SNOG_00899 as being 54% identical to Uga2 with an E-value of 0. A reciprocal blast of the SNOG_00899 protein sequence against yeast identified Uga2 as the best match. A subsequent BlastP analysis of the SNOG_00899 protein sequence against the non-redundant database identified multiple genes putatively encoding fungal succinic semialdehyde dehydrogenases. Analysis of the protein sequence revealed multiple matches with known protein motifs associated with succinic semialdehyde dehydrogenase proteins (TIGR01780 and cd07103). On this basis, SNOG_00899 was renamed Sdh1 and was chosen for subsequent analysis through targeted gene disruption.
The Sdh1 gene disruption construct was amplified as described above and transformed into the wild-type S. nodorum strain SN15. Greater than 50 transformants were recovered, of which 24 were chosen for screening by growth on GABA as a sole nitrogen source. Four of the 24 transformants showed little or no growth when GABA was supplied as the sole nitrogen source. PCR screening of these four transformants resulted in an enlarged PCR amplicon across the Sdh1 locus confirming the homologous integration of the disruption construct. These transformants were named S. nodorum sdh1-9, 12, 21 and 24. Quantitative PCR was used to confirm that each of the strains had only one copy of the hygromycin marker. Two of these homologous mutants, sdh1-9 and sdh1-21, as well as an ectopic transformant, S. nodorum Sdh1-2, were selected for further characterisation (Figure S1). To confirm that the resulting phenotypes were due to the disruption of the Sdh1 locus, a complementation construct was transformed back into the sdh1-9 background restoring the ability of that strain to grow on GABA as a sole nitrogen source.
Characterisation of S. nodorum sdh1 strains
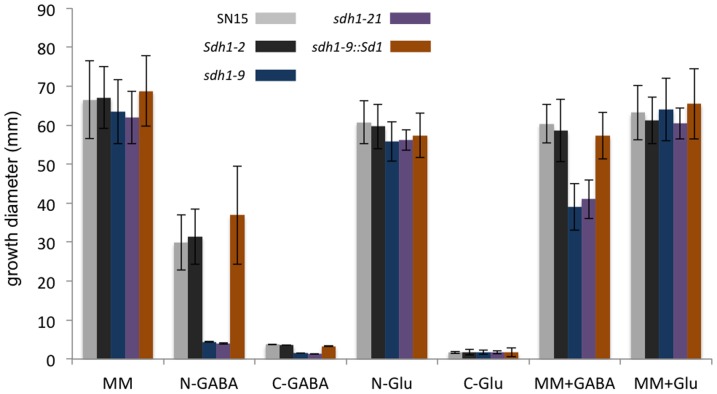
The role of Sdh1 was assessed during vegetative growth by measuring the growth of sdh1-9 and sdh1-21 strains on various defined media. The mutants displayed no difference in growth rates compared to Sdh1 strains when grown in minimal media in the absence of GABA (Figure 2; Figure S2). When GABA was supplied as a nitrogen source rather than nitrate, the growth of the sdh1 mutants was nearly 100-fold less compared to SN15 and the ectopic strain. None of the strains assayed grew strongly when GABA was supplied as a carbon source although the wild-type strain grew about 3-fold more than the sdh1 strains. All of the strains grew comparably to each other when grown on glutamate as a carbon or nitrogen source. The growth of the strains harbouring an intact Sdh1 gene was marginally affected by the inclusion of 1 mM GABA in complete minimal media. In contrast, the growth of the sdh1 mutants was reduced by approximately 30% when GABA was included in the media. No difference in growth was observed when the mutants were grown on complex V8PDA media (data not shown).
Figure 2. Plate growth assay.
Colony diameter was measured after 18 days growth with n = 3. Standard error bars are shown. MM, minimal media; N-GABA, minimal media with GABA as the sole nitrogen source; C-GABA, minimal media with GABA as the sole carbon source; N-Glu, minimal media with glutamate as the sole nitrogen source; C-Glu, minimal media with glutamate as the sole carbon source; MM+GABA, minimal media with 1 mM GABA; MM+Glu, minimal media with 1 mM glutamate.
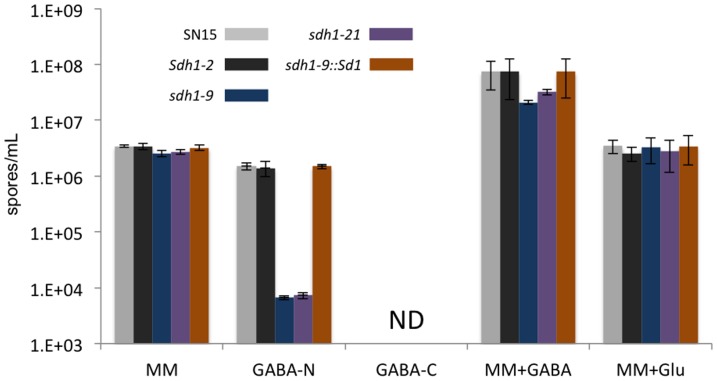
The strains were also assayed for their ability to sporulate in the media described above (Figure 3). There was no significant difference in the ability of strains to asexually sporulate on minimal media. The sporulation of the sdh1 mutants on GABA as either a nitrogen or carbon source showed a similar trend to that observed for the growth assays as GABA proved to be a poor nitrogen source whilst no spores were detected in any of the strains assayed with GABA as a carbon source. When 1 mM GABA was added to minimal media as a supplement, the sporulation of all strains increased by five to 10-fold compared to the absence of GABA. This increase in total spores was particularly surprising for the sdh1 strains given the negative impact on growth observed when GABA was included in the media as a supplement. Glutamate was also assayed as a supplement to determine whether or not the increased sporulation was due to more nitrogen being available. There was no increase in sporulation when glutamate was added to the media implying that the GABA sporulation effect was not due to nitrogen. Analyses of the resulting pycnidia from these sporulation assays revealed no phenotypic or viability differences when differentiated in the presence of GABA (data not shown).
Figure 3. Sporulation assay for each of the strains grown under different nutritional conditions.
MM, minimal media; N-GABA, minimal media with GABA as the sole nitrogen source; C-GABA, minimal media with GABA as the sole carbon source; MM+GABA, minimal media with 1 mM GABA; MM+GABA, minimal media with 1 mM GABA; MM+Glu, minimal media with 1 mM glutamate.
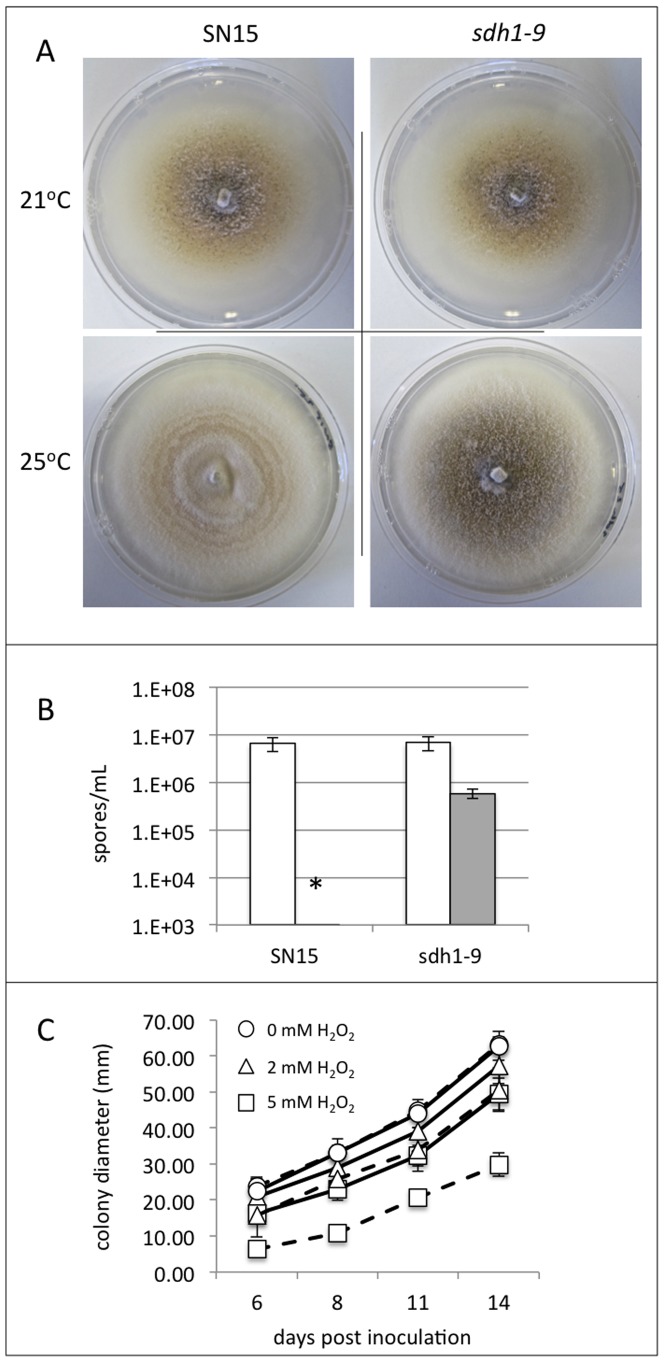
The sdh1 mutants were also tested for their resistance to a variety of stresses. The growth of the strains was examined during osmotic, pH, oxidative and temperature stresses. There was no difference on growth apparent in any of the strains when grown under different osmotic strengths or pH environments (data not shown). Similarly, the rate of growth was unchanged in the sdh1 strains compared to SN15 at different incubation temperatures. There was however a strong phenotypic difference between the wild-type and mutant isolates when grown at 25°C, a high growth temperature for S. nodorum (Figure 4A). Incubation of the wild-type strain at the higher temperature resulted in a fluffy white appearance with no evidence of pycnidia. In contrast, the sdh1 strains appeared much more similar to the phenotype observed at 21°C with pycnidia evident, although to a lesser degree at 25°C. Harvesting and counting of the spores on these plates revealed no significant difference in sporulation at 21°C when comparing the wild-type and sdh1 strains (Figure 4B). At 25°C, the wild-type did not sporulate whilst a 10-fold decrease in sporulation was observed for the sdh1 strains compared to growth at 21°C.
Figure 4. An analysis of the role of Sdh1 during temperature and reactive oxygen species (ROS) stress.
(A) Plate growth assays of the S. nodorum wild-type SN15 strain and the sdh1-9 mutant at 21°C and 25°C. (B) Spores collected and counted from the plate sporulation in (A). (C) Growth assays of the SN15 (solid lines) and sdh1-9 (broken lines) strains on different concentrations of H2O2.
The susceptibility of the sdh1 strains to reactive oxygen stress was assessed by including different concentrations of H2O2 in minimal media (Figure 4C). There was no significant difference in growth rate of the SN15 and sdh1 strains up to concentrations of 1 mM H2O2. There was a small but significant decrease in growth of the sdh1 strains compared to SN15 at 1 mM whilst the sdh1 strains only grew to approximately 60% of SN15 at 5 mM H2O2 demonstrating that Sdh1 has a role in detoxifying exogenous reactive oxygen species (ROS).
Sdh1 is required for complete pathogenicity
Spores generated from the sdh1 mutants were used to inoculate wheat seedlings to determine the involvement of the gene in virulence. Disease scores recorded at seven dpi revealed the sdh1 strains to be only half as pathogenic as the wild-type and ectopic isolates (Figure 5; Figure S3). A re-assessment of the disease after 14 dpi showed the disease scores of the sdh1 mutants was closer to that of the wild-type, but still significantly less proving that Sdh1 does have a role in disease development.
Figure 5. Pathogenicity scores for the wheat leaf infection assays.
N = 5 and standard error bars are shown.
GABA promotes sporulation of S. nodorum
In the course of characterising of the sdh1 strains, it was observed that the addition of GABA to minimal media (containing nitrate and sucrose) promoted sporulation. To investigate this further, S. nodorum SN15 was plated out onto varying concentrations of GABA to determine if the positive effect on sporulation was dose dependent. An equal number of spores were plated onto the centre of a standard minimal media plate containing 0, 0.1, 0.3, 1, 3 or 10 mM GABA (Figure 6). The plates were then allowed to grow for 18 days prior to the spores being harvested and counted.
Figure 6. Growth and sporulation assays of S. nodorum SN15 growing in increasing GABA concentrations.
(A) Images of S. nodorum growing at different GABA concentrations captured at eight days post-inoculation. Increasing levels of pycnidia (small dark spots) are clearly evident with higher concentrations of GABA. (B) Colony diameter; (C) Total number of spores per mL produced; (D) Number of spores produced divided by the area of colony growth. N = 6 and standard error bars are shown. The letters shown above each of the bars represent the statistical significance for that treatment with different letters representing treatments that are statistically different (p<0.05).
The first observation was that the increasing concentrations of GABA appeared to inhibit growth. There was no change in total growth observed up to 0.3 mM GABA, however higher concentrations restricted the rate of growth. Spore counts from the same plates revealed a positive correlation of GABA concentration and sporulation. The addition of 0.3 mM GABA was required to see a significant increase in sporulation with 10 mM resulted in an eight-fold increase in the number of spores produced per plate. When the reduced growth area of the cultures growing on higher levels of GABA was considered, the supplementation of the media with 10 mM GABA resulted in a 10-fold increase in sporulation.
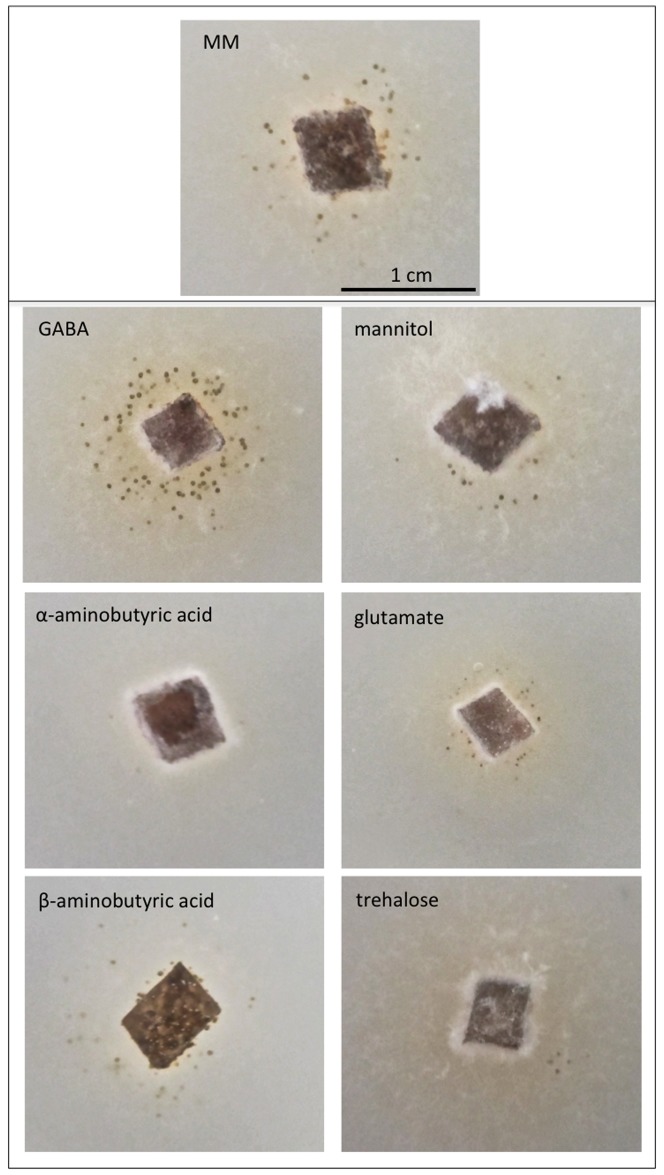
Minimal media was supplemented with isomers of GABA and other compounds known to induce sporulation in S. nodorum to see if this positive effect on sporulation was specific to GABA (Figure 7). With the exception of GABA, none of the compounds assayed resulted in any increase in sporulation. A decrease in sporulation was observed with the addition of α-aminobutyric acid to minimal media, probably reflecting the poor growth of S. nodorum in the presence of this GABA isomer.
Figure 7. Plate growth assays of S. nodorum SN15 growing on minimal media supplemented with different compounds.
All compounds were added to a final concentration of 1
The GABA effect on sporulation is not specific to S. nodorum
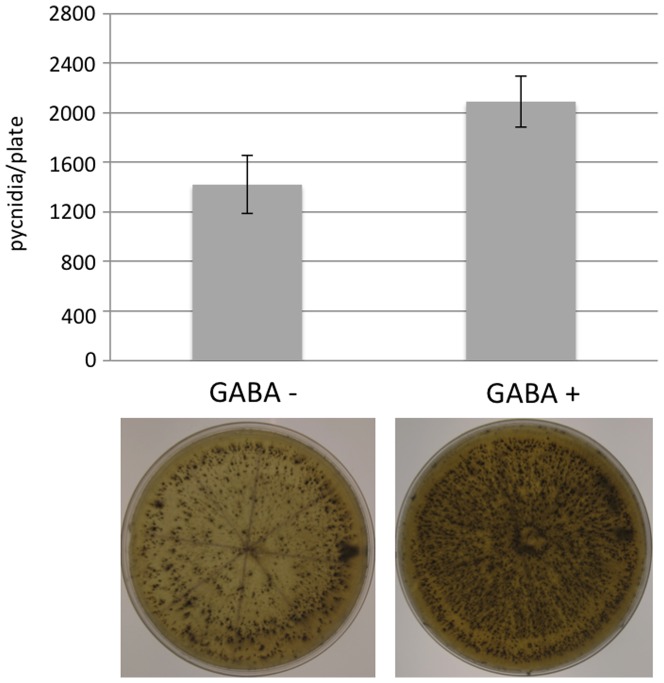
To determine if the positive effect of GABA on sporulation was specific to S. nodorum, we examined its effect on the ability of a related fungus Botryosphaeria sp. to asexually differentiate. In the absence of GABA, an average of 1400 pycnidia were counted per plate shortly after exposure to near-UV light (Figure 8). The addition of 1 mM GABA to the same growth media resulted in an approximate 47% increase in the number of pycnidia counted (average 2064 per plate). The data suggest that the effect of GABA on sporulation is not specific to S. nodorum.
Figure 8. Botryosphaeria sp. sporulation assays in the presence and absence of 1 mM GABA.
Discussion
Since the discovery of GABA metabolism in fungi, its role has been the subject of several hypotheses, but with little supporting evidence. Amongst these roles, it has been speculated that GABA metabolism in phytopathogenic fungi is important for disease. This idea was proposed by Solomon and Oliver [23] when they reported millimolar levels of GABA present in the tomato apoplast during a compatible interaction with the biotrophic fungus Cladosporium fulvum. In a subsequent study, the same authors demonstrated that the pathogen genes required to metabolise GABA were expressed during infection [24]. This lead to the suggestion that perhaps C. fulvum was able to manipulate the host in order to provide GABA as a nitrogen source during infection, although there has been no data published since to support this [16]. Consequently, to better understand the role of GABA in filamentous fungi, and in particular phytopathogens, we analysed its metabolism in the wheat pathogen S. nodorum.
The first observation from this study was that Sdh1, the gene encoding succinic semialdehyde dehydrogenase in S. nodorum, plays a role in stress response and pathogenicity. Under standard growth conditions, the growth rate and phenotype of S. nodorum mutants lacking Sdh1 appeared unaffected compared to SN15. When GABA was supplied as a nitrogen source, the sdh1 strains grew poorly as expected, as only a small amount of nitrogen would have been available for growth through alanine as a result of GABA transaminase. Comparable trends were observed when sporulation was assayed with the sdh1 mutant producing fewer spores than the wild-type when GABA was supplied as either a nitrogen or carbon source. These data fit the current dogma in that GABA can only be metabolised via a transamination and oxidation back to succinic acid.
It was apparent though from the growth assays that Sdh1 has a role in enabling S. nodorum to successfully respond to certain stresses. Upon exposure to 5 mM H2O2 the sdh1 mutants grew poorly compared to SN15 implying that the metabolism of GABA is involved in protection against ROS. Previous studies in S. cerevisiae and M. oryzae have demonstrated a role for GABA metabolism in resisting oxidative stress. In yeast, strains lacking either UGA5 (succinic semialdehyde dehydrogenase) or UGA1 (GABA transaminase) show increased susceptibility to higher concentrations of H2O2 directly implying that the catabolism of GABA has a role in ROS detoxification [25]. The authors speculated that the basis of this resistance is through the production of NADPH by UGA5 during the oxidation of succinic semialdehyde. In M. oryzae, MoSSADH was found to be regulated by a homologue of the YAP1 transcription factor, which is critical in regulating the response to ROS. Subsequent deletion of the MoSSADH gene revealed that the resulting mutants were susceptible to reactive oxygen stress. Whilst it is possible that decreased NADPH levels may be responsible for the susceptibility to ROS in MoSSADH mutants (although NADPH was not measured in the mutants), the mutants also displayed attenuated secreted peroxidase activity. One could simply speculate that reduced peroxidase activities were more likely to be responsible for the increased ROS susceptibility than reduced NADPH levels. It was unclear though how MoSSADH regulates peroxidase levels.
Another interesting phenotype displayed by the S. nodorum sdh1 mutants was their ability to better cope with higher than normal growth temperatures. At 21°C, the phenotypes of the sdh1 and SN15 strains appeared identical. At 25°C, the growth rate of the SN15 strain was comparable to that at 20°C, but appeared whiter in colour and more filamentous with no evidence of pycnidia or sporulation. In contrast the sdh1 strains displayed a more comparable phenotype to the growth observed at 20°C. Pycnidia were also evident, and although less than at 20°C, viable spores could be extracted. This is the first evidence that the GABA shunt plays a role in temperature stress in fungi, although the mechanism underlying this is unclear.
Perhaps not surprisingly, after demonstrating the increased susceptibility of the sdh1 strains to different stresses, the mutants were less pathogenic than wild-type isolates. At seven dpi, the sdh1 strains were about half as virulent as the wild-type, increasing to about 70% virulence at 14 dpi. It is highly likely that the pathogen would be subjected to a variety of stresses during infection. It has been previously established that elevated ROS levels are a result of the photosynthetic collapse caused by the effector SnToxA [26]. The increased susceptibility of the sdh1 mutants to H2O2 would suggest that the mutants may struggle to deal with the high in planta H2O2 levels resulting in slower growth. In contrast, deletion of MoSSADH in M. oryzae resulted in a completely non-pathogenic strain [18]. The M. oryzae mutants were unable to penetrate the cuticle effectively nor grow vegetatively within the leaf. Stagonospora nodorum and M. oryzae are contrasting pathogens with different penetration and infection mechanisms. There are multiple examples of genes that are involved in the pathogenicity of M. oryzae that do not share a similar role in S. nodorum [7], [27], [28]. The data presented in this study would suggest that Sdh1 maybe another.
Of particular interest was serendipitous observation of the effect of GABA on the growth and sporulation of S. nodorum when added to complete minimal medium (i.e. containing sucrose and nitrate). Increasing concentrations of GABA in the minimal medium clearly inhibited the growth of S. nodorum, both in wild-type and sdh1 strains. This impairment of growth is likely to be due to the increased levels of succinic semialdehyde which itself could be toxic [15].
Perhaps the more striking observation of GABA supplementation was that S. nodorum produced nearly 10-fold more spores on MM when supplemented with GABA. This was surprising when considering that sporulation was unaffected when GABA was supplied as a sole nitrogen source; the GABA effect was only evident when the compound was added to complete MM. GABA has been previously described to promote conidiation in Penicillium marneffei, although no further details were reported [29]. The positive effect of GABA on asexual sporulation was dose dependent, with increased GABA levels leading to higher spore production. Isomers of GABA and other compounds previously associated with sporulation in S. nodorum were also assayed with no effect observed other than a growth defect on β-aminobutyric acid (BABA). Previous reverse genetic studies have demonstrated the requirement for the mannitol and trehalose metabolic pathways for asexual sporulation [5], [7]. However, these compounds do not increase sporulation when included in complete minimal media suggesting the mechanisms behind GABA-induced sporulation are independent of mannitol and trehalose.
The obvious question from this data is why does the presence of GABA increase the rate of sporulation? Perhaps some light can be shed from the data on the sdh1 mutants. The inclusion of 1 mM GABA in minimal media significantly affected the growth of the sdh1 mutants, which as discussed above, was likely due to the increased accumulation of succinic semialdehyde. However, as for the wild-type strain, the presence of GABA also stimulated spore production in the sdh1 mutants. There are two possible reasons for this. Firstly, GABA could be metabolised via a route other than through succinic semialdehyde, and that this alternative pathway may contribute to sporulation. However the inability of the sdh1 strains to be able to grow on nitrogen as a sole nitrogen source would suggest this is unlikely.
Another possibility is that that whilst GABA is metabolised through the conventional pathway (and thus leading to the growth defect in the sdh1 strains), it may also have another role in inducing sporulation. Precisely how GABA would fulfil this alternative role is unclear but there is existing evidence that it can act as a signalling molecule. Chevrot and colleagues discovered that GABA stimulated the inactivation of the N-(3-oxootanoyl)homoserine lactone quorum-sensing signal secreted by Agrobacterium tumefaciens [30]. Quorum sensing is a mechanism of cell-to-cell communication predominantly undertaken by bacteria [31], [32]. Multiple studies have shown that quorum sensing is involved in the successful association of bacteria with eukaryotic organisms [33], [34]. The concept of small diffusible signalling molecules triggering sporulation in filamentous fungi is not without precedent. Studies by Adams and colleagues identified that FluG in Aspergillus nidulans is involved in the secretion of a small molecule that accumulates externally to induce sporulation [35]. Whilst it has been shown since that the molecule in A. nidulans is not GABA [36], our data does show that exogenous GABA does stimulate sporulation. Studies are underway to dissect this intriguing phenomenon further.
Supporting Information
(A) PCR amplification of Sdh1 locus using the SdhKOscreenF/R primers (Supplementary Table S1). A band of 2321 bp represents the wild-type locus and 4243 bp the sdh1 mutants. (B) Screening of the different strains for the presence of hygromycin (528 bp) and phleomycin (1998 bp). For both (A) and (B), lane 1 – 1 kb ladder, lane 2 – S. nodorum SN15, lane 3 – sdh1-9, lane 4 – sdh1-21, lane 5 – Sdh1-2 and lane 6 - sdh1-9::Sdh1. (C) A histogram representing the number of copies of the hygromycin and phleomycin relative to that of γ-actin. The primers for these listed in Supplementary Table S1. The S. nodorum strain mpdmdh102 was included as a positive control as it has been previously demonstrated to have one copy each of hygromycin and phleomycin (Solomon PS, Ipcho SVS, Hane JK, Tan K-C, Oliver RP (2008) A quantitative PCR approach to determine gene copy number. Fungal Genetics Reports 55: 5–8.).
(TIFF)
An example of the growth differences of the sdh1 strains growing on either nitrate (A) or GABA (B) as a nitrogen source.
(TIFF)
Representative pots infected with S. nodorum SN15 (left) or the sdh1-9 strain (right). Images were captured at 7 dpi. The red arrows show examples of heavily infected leaves.
(TIFF)
Primer sequences.
(DOCX)
Funding Statement
The work was supported by Australian Research Council and Grains Research and Development Corporation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Solomon PS, Lowe RGT, Tan KC, Waters ODC, Oliver RP (2006) Stagonospora nodorum: Cause of stagonospora nodorum blotch of wheat. Molecular Plant Pathology 7: 147–156. [DOI] [PubMed] [Google Scholar]
- 2. Oliver RP, Friesen TL, Faris JD, Solomon PS (2012) Stagonospora nodorum: From pathology to genomics and host resistance. Annual Review of Phytopathology 50: 23–43. [DOI] [PubMed] [Google Scholar]
- 3. Oliver RP, Solomon PS (2010) New developments in pathogenicity and virulence of necrotrophs. Current Opinion in Plant Biology 13: 415–419. [PubMed] [Google Scholar]
- 4. Solomon PS, Tan KC, Oliver RP (2005) Mannitol 1-phosphate metabolism is required for sporulation in planta of the wheat pathogen Stagonospora nodorum . Molecular Plant-Microbe Interactions 18: 110–115. [DOI] [PubMed] [Google Scholar]
- 5. Solomon PS, Waters ODC, Jörgens CI, Lowe RGT, Rechberger J, et al. (2006) Mannitol is required for asexual sporulation in the wheat pathogen Stagonospora nodorum (glume blotch). Biochemical Journal 399: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Solomon PS, Waters ODC, Oliver RP (2007) Decoding the mannitol enigma in filamentous fungi. Trends in Microbiology 15: 257–262. [DOI] [PubMed] [Google Scholar]
- 7. Lowe RGT, Lord M, Rybak K, Trengove RD, Oliver RP, et al. (2009) Trehalose biosynthesis is involved in sporulation of Stagonospora nodorum . Fungal Genetics and Biology 46: 381–389. [DOI] [PubMed] [Google Scholar]
- 8. Tan KC, Heazlewood JL, Millar AH, Thomson G, Oliver RP, et al. (2008) A signaling-regulated, short-chain dehydrogenase of Stagonospora nodorum regulates asexual development. Eukaryotic Cell 7: 1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan KC, Trengove RD, Maker GL, Oliver RP, Solomon PS (2009) Metabolite profiling identifies the mycotoxin alternariol in the pathogen Stagonospora nodorum . Metabolomics 5: 330–335. [Google Scholar]
- 10. Tillakaratne NJK, Medina-Kauwe L, Gibson KM (1995) Gamma-aminobutyric acid (GABA) metabolism in mammalian neural and nonneural tissues. Comparative Biochemistry and Physiology Part A: Physiology 112: 247–263. [DOI] [PubMed] [Google Scholar]
- 11. Shelp BJ, Bown AW, Faure D (2006) Extracellular γ-aminobutyrate mediates communication between plants and other organisms. Plant Physiology 142: 1350–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Allan WL, Peiris C, Bown AW, Shelp BJ (2003) Gamma-hydroxybutyrate accumulates in green tea and soybean sprouts in response to oxygen deficiency. Canadian Journal of Plant Science 83: 951–953. [Google Scholar]
- 13. Bouche N, Lacombe B, Fromm H (2003) GABA signaling: A conserved and ubiquitous mechanism. Trends in Cell Biology 13: 607–610. [DOI] [PubMed] [Google Scholar]
- 14. Bouche N, Fromm H (2004) GABA in plants: Just a metabolite? Trends in Plant Science 9: 110–115. [DOI] [PubMed] [Google Scholar]
- 15. Ludewig F, Hüser A, Fromm H, Beauclair L, Bouché N (2008) Mutants of GABA transaminase (POP2) suppress the severe phenotype of succinic semialdehyde dehydrogenase (ssadh) mutants in Arabidopsis. PLoS ONE 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oliver RP, Solomon PS (2004) Does the oxidative stress used by plants for defence provide a source of nutrients for pathogenic fungi? Trends in Plant Science 9: 472–473. [DOI] [PubMed] [Google Scholar]
- 17. Kumar S, Punekar NS (1997) The metabolism of 4-aminobutyrate (GABA) in fungi. Mycological Research 101: 403–409. [Google Scholar]
- 18. Guo M, Chen Y, Du Y, Dong Y, Guo W, et al. (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae . PLoS Pathogens 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Solomon PS, Thomas SW, Spanu P, Oliver RP (2003) The utilisation of di/tripeptides by Stagonospora nodorum is dispensable for wheat infection. Physiological and Molecular Plant Pathology 63: 191–199. [Google Scholar]
- 20. Solomon PS, Rybak K, Trengove RD, Oliver RP (2006) Investigating the role of calcium/calmodulin-dependent protein kinases in Stagonospora nodorum . Molecular Microbiology 62: 367–381. [DOI] [PubMed] [Google Scholar]
- 21. Solomon PS, Ipcho SVS, Hane JK, Tan K-C, Oliver RP (2008) A quantitative PCR approach to determine gene copy number. Fungal Genetics Reports 55: 5–8. [Google Scholar]
- 22. Hane JK, Lowe RGT, Solomon PS, Tan KC, Schoch CL, et al. (2007) Dothideomycete-plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum . Plant Cell 19: 3347–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Solomon PS, Oliver RP (2001) The nitrogen content of the tomato leaf apoplast increases during infection by Cladosporium fulvum . Planta 213: 241–249. [DOI] [PubMed] [Google Scholar]
- 24. Solomon PS, Oliver RP (2002) Evidence that γ-aminobutyric acid is a major nitrogen source during Cladosporium fulvum infection of tomato. Planta 214: 414–420. [DOI] [PubMed] [Google Scholar]
- 25. Coleman ST, Fang TK, Rovinsky SA, Turano FJ, Moye-Rowley WS (2001) Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae . Journal of Biological Chemistry 276: 244–250. [DOI] [PubMed] [Google Scholar]
- 26. Vincent D, Du Fall LA, Livk A, Mathesius U, Lipscombe RJ, et al. (2012) A functional genomics approach to dissect the mode of action of the Stagonospora nodorum effector protein SnToxA in wheat. Molecular Plant Pathology 13: 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Solomon PS, Tan KC, Sanchez P, Cooper RM, Oliver RP (2004) The disruption of a Gα subunit sheds new light on the pathogenicity of Stagonospora nodorum on wheat. Molecular Plant-Microbe Interactions 17: 456–466. [DOI] [PubMed] [Google Scholar]
- 28. Solomon PS, Waters ODC, Simmonds J, Cooper RM, Oliver RP (2005) The Mak2 MAP kinase signal transduction pathway is required for pathogenicity in Stagonospora nodorum . Current Genetics 48: 60–68. [DOI] [PubMed] [Google Scholar]
- 29. Borneman AR, Hynes MJ, Andrianopoulos A (2000) The abaA homologue of Penicillium marneffei participates in two developmental programmes: Conidiation and dimorphic growth. Molecular Microbiology 38: 1034–1047. [DOI] [PubMed] [Google Scholar]
- 30. Chevrot R, Rosen R, Haudecoeur E, Cirou A, Shelp BJ, et al. (2006) GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens . Proceedings of the National Academy of Sciences of the United States of America 103: 7460–7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: The LuxR-LuxI family of cell density- responsive transcriptional regulators. Journal of Bacteriology 176: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller MB, Bassler BL (2001) Quorum sensing in bacteria. pp. 165–199. [DOI] [PubMed]
- 33. Qazi S, Middleton B, Muharram SH, Cockayne A, Hill P, et al. (2006) N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus . Infection and Immunity 74: 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baker CJ, Mock NM, Whitaker BD, Roberts DP, Rice CP, et al. (2005) Involvement of acetosyringone in plant-pathogen recognition. Biochemical and Biophysical Research Communications 328: 130–136. [DOI] [PubMed] [Google Scholar]
- 35. Lee BN, Adams TH (1994) The Aspergillus nidulans FluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes and Development 8: 641–651. [DOI] [PubMed] [Google Scholar]
- 36. Rodriguez-Urra AB, Jimenez C, Nieto MI, Rodriguez J, Hayashi H, et al. (2012) Signaling the induction of sporulation involves the interaction of two secondary metabolites in Aspergillus nidulans . ACS Chemical Biology 7: 599–606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) PCR amplification of Sdh1 locus using the SdhKOscreenF/R primers (Supplementary Table S1). A band of 2321 bp represents the wild-type locus and 4243 bp the sdh1 mutants. (B) Screening of the different strains for the presence of hygromycin (528 bp) and phleomycin (1998 bp). For both (A) and (B), lane 1 – 1 kb ladder, lane 2 – S. nodorum SN15, lane 3 – sdh1-9, lane 4 – sdh1-21, lane 5 – Sdh1-2 and lane 6 - sdh1-9::Sdh1. (C) A histogram representing the number of copies of the hygromycin and phleomycin relative to that of γ-actin. The primers for these listed in Supplementary Table S1. The S. nodorum strain mpdmdh102 was included as a positive control as it has been previously demonstrated to have one copy each of hygromycin and phleomycin (Solomon PS, Ipcho SVS, Hane JK, Tan K-C, Oliver RP (2008) A quantitative PCR approach to determine gene copy number. Fungal Genetics Reports 55: 5–8.).
(TIFF)
An example of the growth differences of the sdh1 strains growing on either nitrate (A) or GABA (B) as a nitrogen source.
(TIFF)
Representative pots infected with S. nodorum SN15 (left) or the sdh1-9 strain (right). Images were captured at 7 dpi. The red arrows show examples of heavily infected leaves.
(TIFF)
Primer sequences.
(DOCX)