Abstract
UTX is known as a general factor that activates gene transcription during development. Here, we demonstrate an additional essential role of UTX in the DNA damage response, in which it upregulates the expression of ku80 in Drosophila, both in cultured cells and in third instar larvae. We further showed that UTX mediates the expression of ku80 by the demethylation of H3K27me3 at the ku80 promoter upon exposure to ionizing radiation (IR) in a p53-dependent manner. UTX interacts physically with p53, and both UTX and p53 are recruited to the ku80 promoter following IR exposure in an interdependent manner. In contrast, the loss of utx has little impact on the expression of ku70, mre11, hid and reaper, suggesting the specific regulation of ku80 expression by UTX. Thus, our findings further elucidate the molecular function of UTX.
Introduction
Maintaining genomic stability is crucial for ensuring the accurate cellular functioning of organisms ranging from bacteria to humans [1], [2]. During the course of evolution, cells have evolved multiple mechanisms, collectively known as the DNA damage response (DDR), that facilitate the cellular response to DNA damage [3], [4], [5]. These mechanisms include cell-cycle arrest, DNA repair and apoptosis [6], [7]. In addition, cells responding to DNA damage display a specific gene expression profile that facilitates DNA repair [8]. For example, CSA and HR23A are upregulated by the transcription factor USF-1 in response to UV damage [9]. In the normal diploid human lung fibroblast line MRC-5, exposure to ionizing radiation results in the upregulation of Ku70 via a p53/ATM-dependent mechanism [10]. DNA damage induces CRT1 transcription, which is downstream of DUN1 in the DNA damage pathway in yeast. In turn, CRT1 becomes hyperphosphorylated and dissociates from DNA, resulting in the transcriptional induction of three of the four RNR genes [11]. Over the last few years, a wealth of new information has been uncovered about the DDR, including the identification of many novel proteins involved in this process [12], but whether these proteins are regulated at the gene transcription level in response to DNA damage remains poorly understood.
The ubiquitously transcribed TPR gene on the X chromosome, or UTX, was first described as a gene that escapes from X chromosome inactivation [13], [14]. It is now clear that the UTX gene encodes a JmjC-domain-containing protein with histone lysine demethylase activity specific for the tri-methylated lysine 27 residues of histone H3 (H3K27me3) [15], [16], [17], [18], [19], [20], and it is officially referred to as KDM6A in the human genome. Several recent studies have found that UTX is a major component of the COMPASS complex, which includes myeloid/lymphoid or mixed-lineage leukemia (MLL), a SET-domain containing protein homologous to Drosophila Trithorax [21], [22], [23], [24], [25], and regulates transcription by coordinating the methylation of histone H3K4 and the demethylation of H3K27 [26]. In addition, based on the recently established link between a super elongation complex and MLL, UTX might play a role as a general factor that is involved in the activation of gene transcription [25], [27]. Interestingly, sporadic mutations and the abnormal expression of UTX have been linked to many types of human cancers, suggesting that UTX plays a role in tumorigenesis. However, the functional role of UTX in tumorigenesis remains elusive. Because the DDR is generally accepted as a crucial safeguard against cancer, we hypothesize that UTX is involved in the DDR and plays an important role in maintaining genome integrity.
In this study, we demonstrated that UTX plays an essential role in the DDR in Drosophila. UTX is specifically required for the p53-dependent expression of ku80 through mediating the demethylation of H3K27me3 upon exposure to ionizing radiation (IR). However, UTX is not required for the expression of other DNA repair genes, such as ku70 and mre11, or the apoptotic genes hid and reaper (rpr). UTX is physically associated with p53, and IR exposure induces the recruitment of both UTX and p53 to the ku80 promoter in an interdependent manner. These data favor a model in which UTX is a specific co-player in a p53-dependent cell survival response to DNA damage. Both UTX and p53 are functionally conserved from flies to humans. Therefore, our data demonstrate the role of UTX in the maintenance of genomic stability and might shed light on how UTX influences tumorigenesis.
Materials and Methods
Drosophila Genetics
All Drosophila lines were cultured in standard medium at 25°C. The P-element insertion mutant of utx, with a genotype of y1 w67c23; P{GSV6}GS10564/SM1, was obtained from the Drosophila Genetic Resource Center at the Kyoto Institute of Technology (http://kyotofly.kit.jp/cgi-bin/stocks). The P-element was mobilized using P [delta 2–3] as the source of P-element transposase according to standard protocols. A total of 176 independent white revertant lines were analyzed via PCR using genomic primers. One imprecise excision line, designated utxΔ95 (containing a 1,691 bp deletion from ggttatttgtatgtatgtat to taaaccaatcagtgggcaat), was recovered. The utx1 stock was kindly provided by Andreas Bergmann [28].
Kc Cell Culture, RNAi knockdown and Transfection
Kc167 (Kc) cells were ordered from DRSC (Drosophila RNAi Screening Center) and were routinely cultured in Schneider’s Drosophila medium (Life Technologies) containing 5% FBS (Life Technologies) at 25°C. RNAi-mediated gene knockdown experiments were performed essentially as described previously [29]. Double-stranded RNAs (dsRNAs) targeting control, utx and p53 sequences were synthesized as described elsewhere [26]. The following primer pairs were designed and used for the synthesis of the dsRNAs: utx forward, 5′-gaattaatacgactcactatagggagagagcaacaaaagttcggagc-3′; utx reverse, 5′-gaattaatacgactcactatagggagaatgaacagagggtgtgggag-3′; p53 forward, 5′- gaattaatacgactcactatagggagaatcgtgggacagcatgttat-3′; and p53 reverse, 5′- gaattaatacgactcactatagggagaaggctcagcaatttgttggc-3′; ku80 RNAi forward, 5′-gaattaatacgactcactatagggagacgtgctctcgtttcttcgga-3′ and ku80 RNAi reverse, 5′-gaattaatacgactcactatagggagaccgctctctttgacttctcc-3′. Five million cells were seeded in 25 cm2 flask with serum-free medium and incubated together with 37.5 µg of the dsRNAs (in equal amounts) for 30 min, and FBS was then supplemented at a 5% concentration. Total RNA was isolated two days later with the RNeasy mini kit (QIAGEN), and 2 µg of RNA was reverse transcribed using Superscript III reverse transcriptase (Life Technologies). We generated UTX mutant plasmid using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent technologies). The following primer pair is designed and used for the synthesis of utx mutant plasmid: utx-j mutant forward, 5′- cctggcgcgcaagcgaacaacaacttctgctcaatcaaca -3′ and utx-j mutant reverse, 5′-gttcgcttgcgcgccaggcgtcctacttccgggcaccttc-3′. For rescue experiment, we first treated the cells with dsRNA, and 2 days later re-plate the cells for transfection. Ten million cells were plated in 25 cm2 flask with medium containing 5% FBS and incubate at 25°C for 24 hours. Then each million cells transfected with 2 µg plasmid and X-tremeGENE Transfection Reagent (Roche). Incubated for 24 hours then following experiment.
Ionizing Radiation (IR) and Survival Assay
For the IR and qRT-PCR experiments using Kc cells, γ-ray irradiation was applied to the cells at 2 or 4 days after RNAi treatment, and the cells were then harvested for total RNA extraction using the RNeasy mini kit (QIAGEN). To assess cell survival, RNAi-treated Kc cells were irradiated with 4 or 8 Gy of IR, seeded at a density of 1 × 106 cells/ml into 6-well plates and counted at 2 and 4 days after irradiation. For hatching rate quantification, embryos were collected at 0–4 hours after embryo laying and irradiated with 10 Gy of IR, and the hatched larvae were counted after 3 days. For utxΔ95/utx1 embryos, utxΔ95/CyoGFP flies were crossed with utx1/CyoGFP flies, and the non-fluorescent embryos were collected. For the third instar larva experiments, the total irradiation dosage was 40 Gy. Total RNA was isolated at 2 hours after irradiation using the RNeasy mini kit (QIAGEN).
Real-time Quantitative RT-PCR Analysis
Real-time quantitative RT-PCR (qRT-PCR) assays were performed using the Applied Biosystems 7300 Real-Time PCR System (Life Technologies) and FastStart Universal SYBR Green Master Mix (Roche Applied Science). The following primer pairs were designed and employed for qRT-PCR: ku80 forward, 5′- aagtccgcaaaatgtgtggc-3′; reverse, 5′- atttcatcggtgtcgcaacc-3′; ku70 forward, 5′- cccatggtcgatgactttgac-3′; reverse, 5′- gaaaattgaacgccaaacagg-3′; mre11 forward, 5′- ccaaaacggaggctgtcaat-3′; reverse, 5′- cgatccactaactcctccacg-3′; hid forward, 5′- cccaccgaccaagtgctatac-3′; reverse, 5′- ggcggatactggaagatttgc-3′; rpr forward, 5′- ccagttgttaattccgaacg-3′; reverse, 5′- tcgcctgatcgggtatgtaga-3′; pnr forward, 5′- gcaaggaggagcatgatctca-3′; reverse, 5′- tggtgccgctcttcatatcc-3′. β-tubulin levels were used as an internal control as described [29].
Immunoprecipitation and Chromatin Immunoprecipitation (ChIP)
The immunoprecipitation experiments were performed with the PIERCE direct IP kit according to the manufacturer’s protocol using approximately 1×107 cells. The ChIP assays were conducted using the Upstate ChIP assay kit, also following the manufacturer’s protocol. Briefly, approximately 3×107 cells were collected, fixed and sonicated with a Bioruptor sonicator (Diagenode) to generate DNA fragments of approximately 500 bp in length. Next, immunoprecipitation was performed with either an antibody (3 µg) or normal rabbit IgG (3 µg), and the subsequent steps were performed as previously described [26]. The following primer pairs were used for qPCR: ku80 forward, 5′-gcaacgcggtgctagaaatat-3′; reverse, 5′-gcggcttactgacctaatgca-3′; ku70 forward, 5′-agcctgccgctgtaaaagtc-3′; reverse, 5′-accacctttcgatgacagcc-3′; mre11 forward, 5′-cggtctatgtgatggcgaaat-3′; reverse, 5′-tgtcgtggtgccattcatg-3′; hid forward, 5′-agcaaaacaaagcagcgaaga-3′; reverse, 5′-tgctggcttcctttttgtcct-3′; rpr forward, 5′-cggcgtgagagaaccaggt-3′; reverse, 5′-ttttttcgagatgcgttcgc-3′; pnr forward, 5′- gcgttagccagcacaaagtg-3′; reverse, 5′-tggtgagcgaaagagcaaga-3′.
Antibodies
Rabbit polyclonal anti-UTX antiserum was generated against a bacterially expressed GST-tagged UTX fragment (1–113aa). In the Western blot assays, the blots were first incubated with the proper concentrations of primary antibodies, followed by incubation with the indicated HRP-anti-rabbit IgG or HRP-anti-mouse IgG secondary antibodies (Sigma) and visualized using an ECL kit (Thermo Scientific). The antibodies used in the Western blot analysis were as follows: anti-UTX serum (1∶1,000), anti-β-Tubulin (1∶5,000, Sigma Cat. No. F1804) and anti-p53 (1∶1,000, DSHB Cat. No. 25F4); anti-UTX antiserum and anti-H3K27me3 (UPSTATE Cat. No. 07–449) were used for ChIP.
Results
The utx Gene is Essential for Cell Survival After IR Exposure
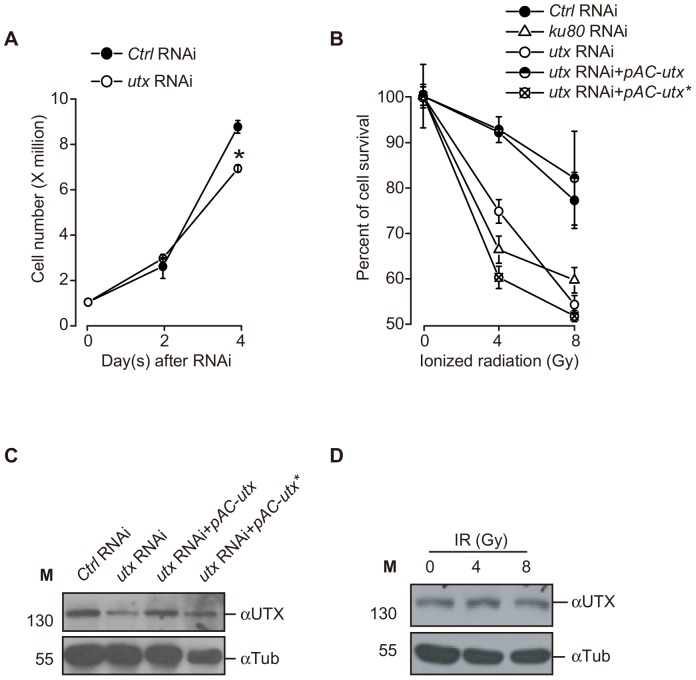
The UTX protein has been extensively studied regarding its function as a demethylase and its H3K27 demethylase-independent activity during development [15], [18], [28]. Previous studies indicate that UTX is also associated with human cancers [30], [31]. However, how UTX functions as a tumor suppressor is unclear. For this reason, we hypothesized that UTX is involved in the maintenance of genomic stability in response to DNA damage, as loss of UTX function results in genome instability and tumorigenesis. Under basal conditions, the RNAi-mediated depletion of utx did not affect the growth of Kc cells at 2 days after soaked with double-strend RNAs and caused only a slight slower growth at 4 days (Fig. 1A). We challenged 4 days RNAi-treated cells with IR at doses of 4 and 8 Gy and examined relative cell viability after 2 days. We found that both doses of IR induced a significant reduction in the viability of the cells treated with utx RNAi compared to controls RNAi cells. The reduction of cell survival is indeed due to the loss of utx, since over-expression of wild type UTX in RNAi cell could rescue the viability (Fig. 1B). UTX protein levels were significantly reduced after the RNAi-mediated knockdown of utx but remained steady after IR, suggesting that IR does not regulate the expression of UTX (Fig. 1C, 1D). These data indicate that UTX is required for cell survival/growth upon genotoxic insult in Kc cells.
Figure 1. UTX is required for cell survival following IR exposure in Kc cells.
(A) Cell counts at 2 and 4 days following RNAi treatment are indicated. Note that at 2 days, there is no difference in the growth rate of utx RNAi-treated compared to control (Ctrl) RNAi-treated cells. The asterisk indicates P<0.05 compared with the Ctrl RNAi group on the same day, as determined by two-way ANOVA followed by a post-hoc Tukey’s test for comparisons between groups. (B) utx RNAi-treated cells display significantly reduced cell counts compared to control RNAi-treated cells and over-expression wild type UTX could rescue RNAi effect but not mutant UTX after IR exposure. 4 days RNAi-treated cells were irradiated at doses of 4 and 8 Gy and examined relative cell viability after 2 days.(C) Western blot analysis of UTX expression confirming the efficiency of utx RNAi-mediated knockdown and over-expression of UTX. (D) IR treatment causes no detectable change in UTX protein levels. β-Tubulin (Tub) levels were used as loading controls. All of the data are representative of at least three independent experiments with similar results. Error bars indicate standard deviations from triplicate sets of the presented experiment.
UTX Upregulates ku80 Through Promoter Demethylation in Response to IR Exposure
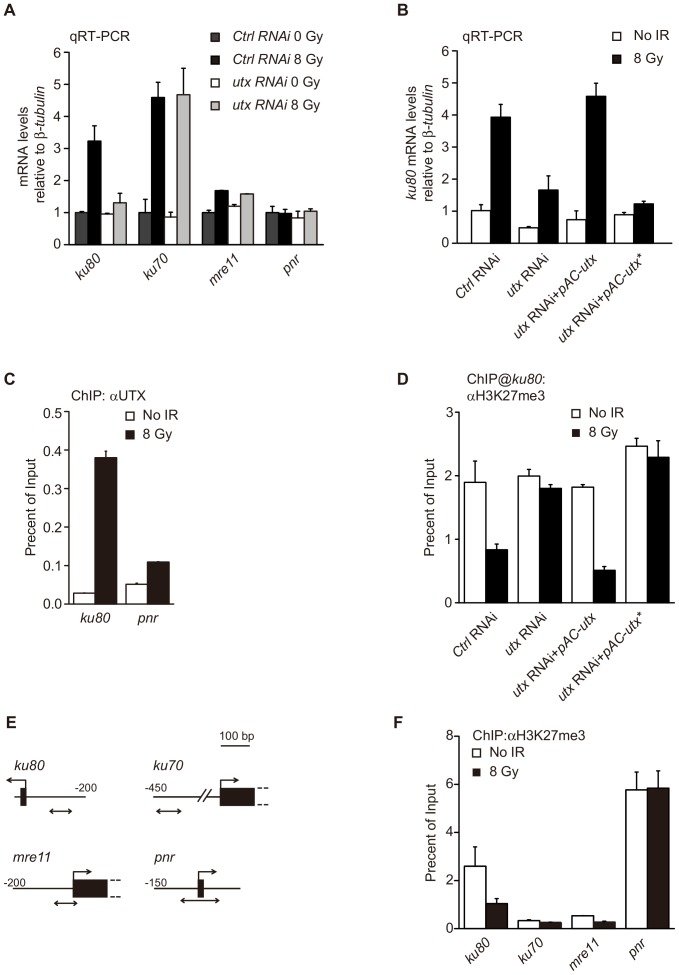
In a previous microarray analysis that conducted in our laboratory, we identified a list of genes that are upregulated following IR treatment in Drosophila Kc167 (Kc) cells (Table S1). This list included genes known for their roles in DSB repair, such as ku70, ku80 and mre11, similar to what has been previously reported in fly embryos [32]. To further determine the role of UTX in the DDR, we investigated whether UTX is involved in the regulation of these genes in response to IR exposure. Interestingly, we found that the RNAi-mediated knockdown of UTX expression significantly inhibited the upregulation of ku80, but not that of ku70 and mre11, following IR treatment (Fig. 2A). Over-expression of wild type UTX in utx RNAi cell restored the upregulation of ku80 expression (Fig. 2B). These data suggest that UTX regulates ku80 expression in a gene-specific manner in DDR. This notion is further supported by the fact that both utx and ku80 RNAitreated cells showed similar cell sensitivity to IR (Fig. 1B). Next, we explored whether UTX regulates directly or indirectly the expression of ku80 upon IR treatment. Chromatin immunoprecipitation (ChIP) assays revealed that UTX is recruited to the promoter region of ku80 upon IR treatment (Fig. 2C). Although UTX has predominantly been shown to regulate transcription by demethylating H3K27, it has also been found that UTX regulates the mesoderm differentiation of embryonic stem cells, independent of its H3K27 demethylase activity in mouse [33]. Therefore, we investigated whether the observed UTX-mediated ku80 expression was dependent on the demethylase activity of UTX.
Figure 2. UTX is required for the expression of ku80 after IR exposure via the demethylation of H3K27me3 in Kc cells.
(A) qRT-PCR analysis of the mRNA expression of the indicated genes before and after IR exposure in RNAi-treated Kc cells. The relative expression levels are normalized to β-tubulin levels. Note that ku80 is the only gene that requires utx for its expression. (B) qRT-PCT analysis of ku80 expression in different treatment cell. Over-expression WT UTX could rescue ku80 expression in UTX RNAi cell after IR but not mutant UTX. (C) ChIP assay with an anti-UTX antibody and the ku80 promoter with and without IR. Note the dramatic increase in the UTX occupancy of the ku80 promoter after IR. (D) ChIP assay for H3K27me3 at the ku80 promoter in different treatment Kc cells after IR. (E) The diagrams show PCR-amplified regions (double arrows) relative to the first exons (black box) in the ChIP analysis of the three DNA repair genes ku80, ku70 and mre11 and a control gene, panier (pnr). (F) Changes in H3K27me3 levels at the indicated genes 2 hours after IR treatment with a dose of 8 Gy. The H3K27me3 levels in the promoter regions of those genes were determined via ChIP assays and compared to the input genomic DNA.
Using ChIP assays, we found that IR treatment dramatically reduced the levels of H3K27me3 in the ku80, but not in utx depleted cells. Over-expression WT UTX could reduce the levels of H3K27me3 in utx RNAi cells after IR, but not JMJC domain mutant UTX which disrupt UTX enzyme activity (Fig. 2D) [28]. Furthermore, we found that over-expression JMJC domain mutant UTX could not rescue cell survival and ku80 expression compare with WT UTX in utx RNAi cell after IR (Fig. 1B, Fig. 2B). These data indicate that UTX functions as a histone demethylase in its regulation of ku80 upon IR exposure. To further assess whether DNA damage genes are activated in association with the altered levels of H3K27me3, we examined the promoter regions of these genes before and after IR treatment. We found that IR treatment induced a dramatic reduction of H3K27me3 levels in ku80, but not in ku70 or mre11 (Fig. 2E, 2F). As a control gene, we also examined a known Polycomb target gene, pannier (pnr), and we found that IR caused no apparent changes in either the expression or the recruitment of UTX to pnr [26]. These data indicate that during DNA damage, UTX specifically and directly regulates the expression of ku80 by demethylating histone H3K27me3 at the ku80 promoter.
UTX Coordinates with p53 to Directly Facilitate the Expression of ku80 Following IR Treatment
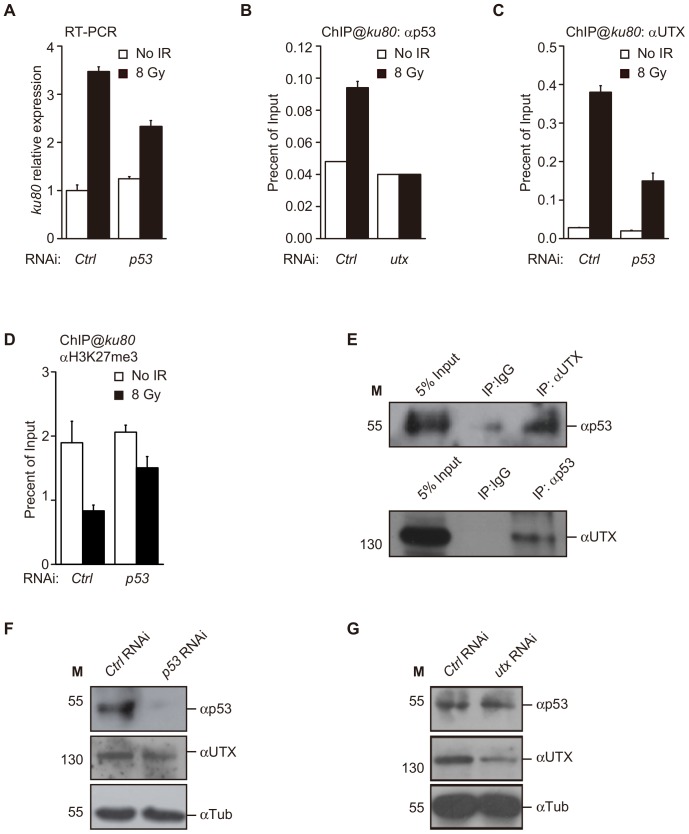
Previous studies have indicated that ku80, as well as ku70,is among the p53 target gene list in fly embryos [32] and larvae [34]. In Kc cells, we found that the expression of ku80 also requires p53, as the RNAi-mediated knockdown of p53 significantly reduced the expression of ku80 following IR treatment (Fig. 3A). In addition, using ChIP analysis, we showed that IR caused marked p53 enrichment in the ku80 promoter (Fig. 3B). Next, we asked whether UTX and p53 coordinate their activities to regulate the expression of ku80. The RNAi-mediated knockdown of UTX expression significantly inhibited the recruitment of p53 to the ku80 promoter, suggesting that UTX is required for the regulation of k80 expression by p53 (Fig. 3C). Intriguingly, we found both the UTX recruitment and the reduction of H3K27me3 levels in the ku80 gene were also prevented by the knock-down of p53 (Fig. 3C, 3D). These data indicate that both p53 and UTX directly regulate ku80 expression within the same pathway, thus requiring coordinated action between p53 and UTX. Further supporting this notion, we found that p53 coimmunoprecipitated with UTX, and UTX was similarly able to coimmuniprecipitate with p53, indicating a physical interaction between the two proteins (Fig. 3E). However, we did not observe a direct interaction in GST pull-down assays, suggesting that the interaction between p53 and UTX is indirect. The UTX protein level is not affected by the change of p53 level as confirmed by the Western blot (Fig. 3F). Similarly, UTX does not regulate p53 expression (Fig. 3G). These data exclude the possibility that UTX and p53 interacts in DDR by the mutual regulation of expression. Together, these data support a molecular model in which p53 and UTX form a complex to regulate ku80 expression and mediate the DDR following exposure to IR.
Figure 3. p53 and UTX are recruited in an interdependent manner to the ku80 promoter region.
(A) ku80 expression following IR exposure in Kc cells subjected to RNAi treatment, as indicated. (B, C) ChIP analysis of the physical occupancy of p53 and UTX at the ku80 promoter region. Note that knockdown of utx eliminates the increase in p53 binding, and knockdown of p53 reduces the binding of UTX to the ku80 promoter. (D) ChIP assay for H3K27me3 at the ku80 promoter in Kc cells treated with control or utx RNAi after IR. (E) Coimmunoprecipitation was performed using anti-p53 and anti-UTX antibodies and whole cell extracts of Kc cells. The immunoprecipitates were subjected to Western blot analysis with the indicated antibodies. (F, G) Western blot analysis to confirm the knockdown efficiency of p53 RNAi. β-Tubulin (β-Tub) levels were used as a loading control.
UTX Regulates ku80 Expression in Drosophila
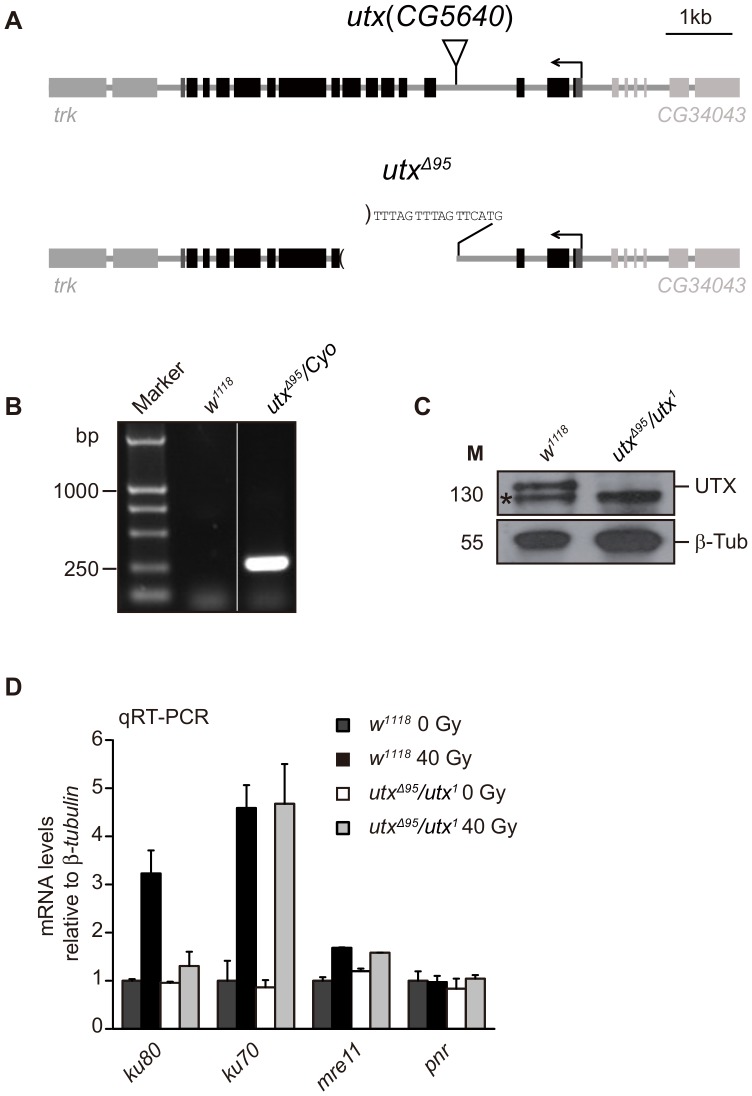
Using Drosophila as an in vivo model system, we next investigated whether the expression of ku80 is also regulated by UTX in Drosophila. To address this question, we generated a utx mutant allele, utxΔ95, through the imprecise excision of a P-element inserted into the utx locus (Fig. 4A). Genomic PCR analysis and sequencing data confirmed the existence of a deletion of five exons (1,691 base pairs) in utxΔ95, as indicated by FlyBase gene annotation (http://flybase.org/) (Figs. 4A & 4B). We found that animals homozygous for utxΔ95 only rarely survive to adults but utxΔ95/utx1 trans-heterozygotes can develop into adults and show no detectable morphological defects. Those results consistent with recently published article [35]. A reported EMS allele, utx1, bearing a nonsense mutation in the JmjC domain, has also been used [28]. Both utx alleles are null, as verified by the missing UTX band in a Western blot analysis of trans-heterozygous (utxΔ95/utx1) third instar larvae performed using an anti-UTX antiserum raised against the N-terminal 103 aa portion of the protein (Fig. 4C). As shown in Figure 4D, 8 Gy of IR dramatically upregulated the levels ku80, ku70 and mre11 in w1118 (wild type control) third instar larvae, similar to what was observed in Kc cells (Fig. 2A), indicating the expression of these genes upon DNA damage in Drosophila. However, IR treatment elicited a significantly reduced induction of ku80 expression in utxΔ95/utx1 third instar larvae compared to wild type larvae, whereas the expression of other genes remained relatively constant, suggesting an gene-specific requirement of UTX for ku80 expression during the DDR. We therefore conclude that UTX is essential for the expression of ku80 both in cell and Drosophila.
Figure 4. UTX is required for the expression of ku80 following IR exposure in Drosophila.
(A) Schematic illustration of the gene structure of wild type utx and a utx mutant allele (utxΔ95) generated via the imprecise excision of a P-element insertion. (B, C) Genomic PCR (B) and Western blot (C) analyses to verify the utxΔ95 genotype. (B) An approximately 250-bp band is detected in adult flies with a utxΔ95/Cyo genotype, but absent from the w1118 genotype. For details, please see the Materials and Methods section. (C) An approximately 130-kDa band indicated by an arrow in w1118 flies was not detected in the utxΔ95/utx1 third instar larvae. A non-specific band is indicated by an asterisk, and β-Tubulin (β-Tub) was used as a loading control. (D) qRT-PCR analysis of mRNA expression for the indicated genes before and after IR exposure in the w 1118 and utxΔ95/utx1 third instar larvae. The relative expression levels are normalized to β-tubulin levels. Note that ku80 is the only gene that requires utx for its expression.
Furthermore, to determine whether UTX is involved in DNA repair in Drosophila, we quantified the hatching rate of transheterozygous utx null (utxΔ95/utx1) and w1118 embryos treated with IR. The utx null embryos exhibited a markedly lower hatching rate of 69.6% compared to the wild type embryos, which displayed a hatching rate of 96.0% (Table 1). Treatment with 10 Gy of IR severely reduced the hatching rate for both genotypes. However, we conclude that the effect of IR was more significant for utx null embryos, as demonstrated by the statistically significant reduction of the normalized hatching rate (Table 1). These data suggest that utx mutant embryos are more sensitive to IR stress than wild type embryos. In addition, we found that the hatching rate of Drosophila embryo does not significantly change between wild type and mutant after UV irradiation (Table 2). Therefore, UTX might play an essential role in DNA repair through regulating ku80 expression both in cell and Drosophila.
Table 1. The hatching rate of Drosophila embryo.
| γ-ray irradiation | Normalized hatching rate | ||||
| Genotype | 0 Gy | 5 Gy | 10 Gy | 5 Gy | 10 Gy |
| w1118 | 499/520 (96.0%) | 98/185 (53.0%) | 28/166 (16.9%) | 55.3% | 17.6% |
| utxΔ95/utx1 | 188/270 (69.6%) | 42/127 (33.1%) | 7/170 (4.0%) | 47.7% a | 5.7% b |
P>0.05 and b P<0.01, as analyzed by Fisher’s Exact Test for normalized hatch rate upon IR treatment of 0–4 hrs w 1118 and utxΔ95/utx1 embryo.
Table 2. The hatching rate of Drosophila embryo.
| UV irradiation | Normalized hatching rate | ||||
| Genotype | 0 J/m2 | 10 J/m2 | 100 J/m2 | 10 J/m2 | 100 J/m2 |
| w1118 | 499/520 (96.0%) | 124/220 (56.4%) | 93/205 (45.4%) | 58.8% | 47.3% |
| utxΔ95/utx1 | 188/270 (69.6%) | 55/145 (37.9%) | 69/179 (38.5%) | 54.5% a | 55.3% a |
P>0.05, as analyzed by Fisher’s Exact Test for normalized hatch rate after 48 hrs upon UV treatment of 0–4 hrs w 1118 and utxΔ95/utx1 embryo.
UTX is Not Responsible for the Upregulation of Apoptotic Genes in Response to DNA Damage
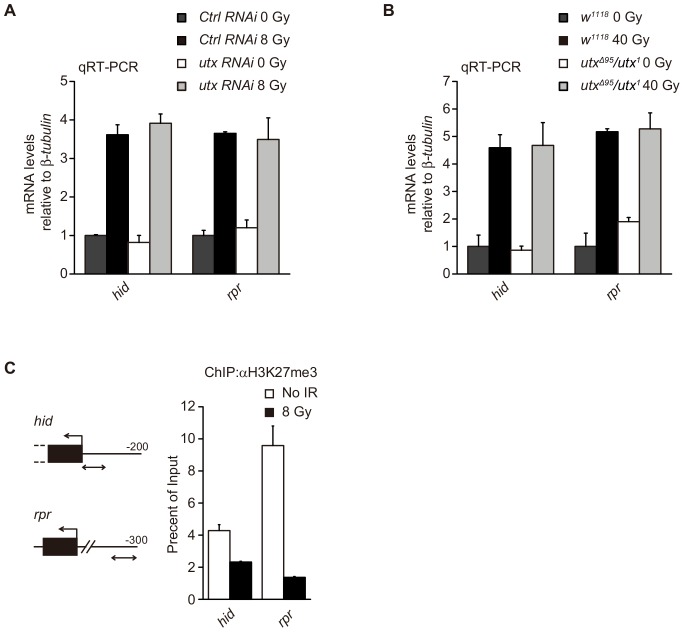
Previous studies have shown that p53 upregulates the apoptotic genes reaper and hid following treatment with IR [32]. Given that together with p53, UTX coordinately regulates the expression of ku80 upon IR exposure, we sought to determine whether UTX also participates in the regulation of apoptotic gene expression in response to DNA damage. To investigate this notion, we evaluated the changes in reaper and hid expression levels following IR treatment. Figures 5A and 5B show that independent of UTX, the expression levels of these apoptotic genes were upregulated to the same extent following IR exposure both in cell and Drosophila. These results were consistent with the findings of previous works showing that overexpression of UTX in primary human fibroblasts induces cell cycle arrest, but not apoptosis [36]. These data suggest that p53, but not UTX, is required for DNA damage-induced apoptosis. Interestingly, we found that the levels of H3K27me3 at the reaper and hid promoters were also reduced following IR treatment, similar to what was observed for the ku80 promoter (Fig. 5C). These data suggest that other demethylases might be responsible for the upregulation of apoptotic genes in response to DNA damage.
Figure 5. UTX is not responsible for the upregulation of apoptosis-related genes following IR exposure.
(A, B) qRT-PCR analysis of the mRNA expression of the indicated genes before and after IR treatment in RNAi-treated Kc cells, as shown for w1118 and utxΔ95/utx1 third instar larvae. The relative expression levels are normalized to β-tubulin levels. (C) The diagrams show PCR-amplified regions (double arrows) relative to the first exons (black box) in the ChIP analysis for two apoptosis-related genes, hid and rpr. The changes in H3K27me3 levels at the genes are indicated 2 hours after IR treatment with a dose of 8 Gy.
Discussion
To understand the mechanism underlying UTX function in tumorgenesis, we explored whether UTX is involved in DNA damage response in Drosophila. In this study, we found that UTX, play an essential role in DNA damage response by upregulation of ku80, which is uniquely required for p53 activated ku80 expression (Fig. 2–5). In addition, the gene activity of utx is correlated with loss of histone demethylation at H3K27 (Fig.2), suggesting that UTX could function as a histone demethylase and serve a gene-specific co-activator of p53 gene activation. We therefore provide an example that p53 target genes expression may be regulated at the level of histone modifications.
It is clear that p53 plays a pivotal role in the DNA damage response (DDR). One of the functions of p53 is to activate its target gene after DNA damage as transcription factor. For instance, p53 has been best characterized in regualting expression of cell cycle genes and apoptosis gene [37]. However, the precise reguation mechnism of p53 is still not clear. It is interesting that in Drosophila ku80 upregulation mediated by p53 requires UTX, but not other genes in related to DNA repair and apoptosis. However, we did observe reduced H3K27me3 levels in apoptotic genes (Fig.2), which raise the possibility that there could be additional histone demethylases participating in DDR pathways that coordinate with p53 regulating expression of hid and reaper after DNA damage, and remaining to be determined in further studies. In contrast, we did not detect reduced H3K27me3 levels in the ku70 promoter region following IR treatment. Further analysis revealed that the H3K27me3 level in the ku70 promoter region was lower than at the ku80 promoter. The expression of ku70 is independent of UTX, possibly due to the extremely low levels of H3K27me3 in the ku70 promoter region, which might not require demethylation for the expression of ku70 to occur (Fig. 2F). Thus, our data demonstrate the complexity of the function of p53 in the activation of target genes in response to DNA damage, particularly in terms of histone modification and the action of different demethylases (Fig. 6).
Figure 6. A model for the regulation of DNA damage response genes associated with DNA double-strand breaks in cells is suggested based on the results presented in Figures 2 – 5 .
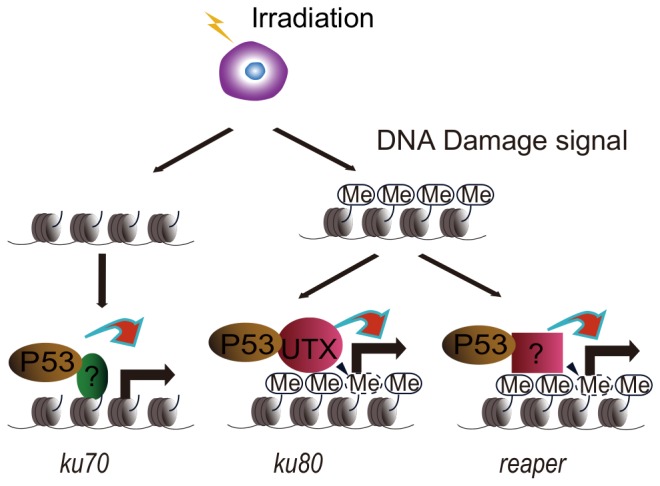
See text.
UTX has been reported to participate in many biological processes, including cell fate determination and animal development [15], [18], [28], [36], [38], largely depending on the transcriptional regulation of the target genes of UTX. UTX appears to play an important role in orchestrating several histone marker, including acetylation at H3K27 and ubiquitination at H2A [19], [39], [40], and mediates derepression of polycomb (Pc) target genes, such as HOX genes, by affecting Pc recruitment. These roles are consistent with UTX being a histone demethylase specific for H3K27 [20]. However, sporadic mutations of UTX have been linked to many types of human cancers [30], [41], [42] and it remains to be elucidated whether this is also sufficiently explained by its enzymatic activity. Indeed, several studies have proposed a role of UTX independent of its demethylase activity in chromatin remodeling and embryonic development [33], [43], [44]. In this study, we found UTX is also involved in DDR by upregulation of ku80 in Drosophila after IR. Although there are no available data demonstrating that ku80 mRNA levels are increased following DSBs in human cells, our data provide evidence that UTX functions to maintain genome stability and shed light on the mechanism underlying the function of UTX in human cancer. Recent studies suggest that loss of polycomb-mediated silencing might promote the upregulation of DNA repair genes [45] and facilitate the recovery of cells from genotoxic insults. UTX might therefore be required for various cell defense mechanisms under environmental stress, thereby contributing to tumor suppression.
Supporting Information
qRT-PCR analysis to confirm the knockdown efficiency of ku80 RNAi.
(TIF)
Contains partial of microarray data which shows fold change more than five times of genes expression up-regulated following IR.
(DOCX)
Acknowledgments
The authors thank Dr. Andreas Bergmann, Bloomington Stock Center and Developmental Studies Hybridoma Bank, for flies and antibodies.
Funding Statement
This work was supported in part by grants from the Natural Science Foundation of China (90919007 and 30900812), the New-Century Training Program Foundation for Talent of the State Education Commission and the Youth Foundation of Southeast University to Z.H. and grants from the National Basic Research Program of China (2012CB517904) and the Natural Science Foundation of China (30771068 and 31271321) to M.F. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745. [DOI] [PubMed] [Google Scholar]
- 2. Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374. [DOI] [PubMed] [Google Scholar]
- 3. Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mahaney BL, Meek K, Lees-Miller SP (2009) Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 417: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lord CJ, Ashworth A (2012) The DNA damage response and cancer therapy. Nature 481: 287–294. [DOI] [PubMed] [Google Scholar]
- 6. Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, et al. (2001) Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell 12: 2987–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Workman CT, Mak HC, McCuine S, Tagne JB, Agarwal M, et al. (2006) A systems approach to mapping DNA damage response pathways. Science 312: 1054–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jelinsky SA, Estep P, Church GM, Samson LD (2000) Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol 20: 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baron Y, Corre S, Mouchet N, Vaulont S, Prince S, et al. (2012) USF-1 is critical for maintaining genome integrity in response to UV-induced DNA photolesions. PLoS Genet 8: e1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown KD, Lataxes TA, Shangary S, Mannino JL, Giardina JF, et al. (2000) Ionizing radiation exposure results in up-regulation of Ku70 via a p53/ataxia-telangiectasia-mutated protein-dependent mechanism. J Biol Chem 275: 6651–6656. [DOI] [PubMed] [Google Scholar]
- 11. Huang M, Zhou Z, Elledge SJ (1998) The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605. [DOI] [PubMed] [Google Scholar]
- 12. Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lahn BT, Page DC (1997) Functional coherence of the human Y chromosome. Science 278: 675–680. [DOI] [PubMed] [Google Scholar]
- 14. Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, et al. (1998) The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet 7: 737–742. [DOI] [PubMed] [Google Scholar]
- 15. Agger K, Cloos PA, Christensen J, Pasini D, Rose S, et al. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734. [DOI] [PubMed] [Google Scholar]
- 16. De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, et al. (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130: 1083–1094. [DOI] [PubMed] [Google Scholar]
- 17. Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, et al. (2007) Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A 104: 18439–18444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, et al. (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694. [DOI] [PubMed] [Google Scholar]
- 19. Lee MG, Villa R, Trojer P, Norman J, Yan KP, et al. (2007) Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450. [DOI] [PubMed] [Google Scholar]
- 20. Swigut T, Wysocka J (2007) H3K27 demethylases, at long last. Cell 131: 29–32. [DOI] [PubMed] [Google Scholar]
- 21. Patel SR, Kim D, Levitan I, Dressler GR (2007) The BRCT-Domain Containing Protein PTIP Links PAX2 to a Histone H3, Lysine 4 Methyltransferase Complex. Dev Cell 13: 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, et al. (2007) Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 27: 1889–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cho YW, Hong T, Hong S, Guo H, Yu H, et al. (2007) PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, et al. (2011) The COMPASS family of H3K4 methylases in Drosophila. Molecular and Cellular Biology 31: 4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith E, Lin C, Shilatifard A (2011) The super elongation complex (SEC) and MLL in development and disease. Genes and Development 25: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fang M, Ren H, Liu J, Cadigan KM, Patel SR, et al. (2009) Drosophila ptip is essential for anterior/posterior patterning in development and interacts with the PcG and trxG pathways. Development 136: 1929–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luo Z, Lin C, Shilatifard A (2012) The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13: 543–547. [DOI] [PubMed] [Google Scholar]
- 28. Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, et al. (2010) The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol 30: 2485–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang L, Meng F, Ma D, Xie W, Fang M (2013) Bridging Decapentaplegic and Wingless signaling in Drosophila wings through repression of naked cuticle by Brinker. Development 140: 413–422. [DOI] [PubMed] [Google Scholar]
- 30. van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, et al. (2009) Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41: 521–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mar BG, Bullinger L, Basu E, Schlis K, Silverman LB, et al. (2012) Sequencing histone modifying enzymes identifies UTX mutations in acute lymphoblastic leukemia. Leukemia. [DOI] [PMC free article] [PubMed]
- 32. Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, et al. (2004) Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol 24: 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, et al. (2012) UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A 109: 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Bergeijk P, Heimiller J, Uyetake L, Su TT (2012) Genome-wide expression analysis identifies a modulator of ionizing radiation-induced p53-independent apoptosis in Drosophila melanogaster. PLoS One 7: e36539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Copur O, Muller J (2013) The histone H3-K27 demethylase Utx regulates HOX gene expression in Drosophila in a temporally restricted manner. Development 140: 3478–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang JK, Tsai MC, Poulin G, Adler AS, Chen S, et al. (2010) The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev 24: 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meek DW (2009) Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer 9: 714–723. [DOI] [PubMed] [Google Scholar]
- 38. Lee S, Lee JW, Lee SK (2012) UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell 22: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ (2012) Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Molecular and Cellular Biology 32: 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, et al. (2012) Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes and Development 26: 2604–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gui Y, Guo G, Huang Y, Hu X, Tang A, et al. (2011) Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 43: 875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Varela I, Tarpey P, Raine K, Huang D, Ong CK, et al. (2011) Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller SA, Mohn SE, Weinmann AS (2010) Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 40: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shpargel KB, Sengoku T, Yokoyama S, Magnuson T (2012) UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 8: e1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shaw T, Martin P (2009) Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep 10: 881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR analysis to confirm the knockdown efficiency of ku80 RNAi.
(TIF)
Contains partial of microarray data which shows fold change more than five times of genes expression up-regulated following IR.
(DOCX)