Abstract
RNA helicases are enzymes that are thought to unwind double-stranded RNA molecules in an energy-dependent fashion through the hydrolysis of NTP. RNA helicases are associated with all processes involving RNA molecules, including nuclear transcription, editing, splicing, ribosome biogenesis, RNA export, and organelle gene expression. The involvement of RNA helicase in response to stress and in plant growth and development has been reported previously. While their importance in Arabidopsis and Oryza sativa has been partially studied, the function of RNA helicase proteins is poorly understood in Zea mays and Glycine max. In this study, we identified a total of RNA helicase genes in Arabidopsis and other crop species genome by genome-wide comparative in silico analysis. We classified the RNA helicase genes into three subfamilies according to the structural features of the motif II region, such as DEAD-box, DEAH-box and DExD/H-box, and different species showed different patterns of alternative splicing. Secondly, chromosome location analysis showed that the RNA helicase protein genes were distributed across all chromosomes with different densities in the four species. Thirdly, phylogenetic tree analyses identified the relevant homologs of DEAD-box, DEAH-box and DExD/H-box RNA helicase proteins in each of the four species. Fourthly, microarray expression data showed that many of these predicted RNA helicase genes were expressed in different developmental stages and different tissues under normal growth conditions. Finally, real-time quantitative PCR analysis showed that the expression levels of 10 genes in Arabidopsis and 13 genes in Zea mays were in close agreement with the microarray expression data. To our knowledge, this is the first report of a comparative genome-wide analysis of the RNA helicase gene family in Arabidopsis, Oryza sativa, Zea mays and Glycine max. This study provides valuable information for understanding the classification and putative functions of the RNA helicase gene family in crop growth and development.
Introduction
Helicases have been identified in organisms ranging from Escherichia coli to humans, viruses and plants. The helicases from all of these organisms represent a large gene family that may have a predominant role in modulating environmental responses. These proteins can be grouped into families based on sequence homologies [1], [2]. The RNA helicases are enzymes that use energy derived from the hydrolysis of a nucleotide triphosphate to unwind double-stranded RNAs [3]. The majority of RNA helicases belong to superfamily 2 (SF2), which consists of three subfamilies, known as DEAD, DEAH and DExD/H [4], [5], [6]. RNA helicases have been shown to be involved in every step of RNA metabolism, including nuclear transcription, pre-mRNA splicing, ribosome biogenesis, nucleocytoplasmic transport, translation, RNA decay, and organellar gene expression [3], [4], [7]. Based on the multiple functions of these genes in cellular RNA metabolism, the fact that the RNA helicases are also involved in plant growth and development and the response to abiotic stress is not surprising.
The RNA helicase family in plants is larger and more diverse than in other systems [8]. In fact, the expression level of several DEAD-box helicases has been shown to be regulated in response to changes in specific environmental conditions, including salt stress, oxygen levels, light or temperature [2], [9], [10]. Initially, three Arabidopsis DEAD-box RNA helicases, LOS4, STRS1 and STRS2, were shown to be involved in the stress responses to various abiotic stresses [11], [12], [13]. HUA ENHANCER2, encoding a putative DExH-box RNA helicase, was shown to be active in both B and C pathways in the flower and to affect vegetative and inflorescence development in Arabidopsis [14]. A plastid DEAD-box RNA helicase VDL gene in tobacco was reported to play an important role in chloroplast differentiation and plant morphogenesis [15]. The Arabidopsis DExH box helicase CAF/DICER-LIKE 1 has been shown to be critical for the biogenesis of microRNAs and plant development [16], [17]. In Zea mays, MA16, fibrillarin, and ZmDRH1 may be part of a ribonucleoprotein complex involved in ribosomal RNA metabolism [18]. Arabidopsis TEBICHI has been shown to be required for the regulation of cell division and differentiation in meristems [19], and ISE2, which is localised to cytoplasmic granules, has been shown to be involved in plasmodesmata function during embryogenesis in Arabidopsis [20]. OsBIRH1, the rice homolog of RH50, exhibited RNA helicase activities in vitro and helped to confer plant resistance against various stresses [21]. Another RNA helicase, PUTATIVE MITOCHONDRIAL RNA HELICASE2, is involved in group II intron splicing in mitochondria [22]. Additionally, SLOW WALKER3 has been shown to be essential for female gametogenesis as a putative DEAD-box RNA helicase in Arabidopsis [23]. Previously, we reported that the DEVH-box RNA helicase AtHELPS participates in the regulation of potassium-deprivation tolerance [24]. Recently, rice API5 has been shown to couple with two DEAD-box RNA helicases (API1 and API2) in regulating PCD during tapetum degeneration in rice [25]. A DExH-box RNA helicase, ABO6, was shown to mediate mitochondrial reactive oxygen species production and also mediate crosstalk between ABA and Auxin signalling [26]. Cumulatively, these investigations indicate that the RNA helicases may play an important role in building resistance to abiotic stresses and in plant growth and development.
Despite the diversity of their biological functions and the wide range of organisms in which these proteins have been identified, high sequence conservation has been maintained in the large group of helicases, suggesting that all the helicase genes evolved from a common ancestor. Hence, signature sequences can be used efficiently for the detection and the prediction of new helicases in the genome databases [27]. After a thorough analysis of all the sequence data, 32 different DEAD-box RNA helicases have been identified and named from AtRH1 to AtRH32 [27]. Subsequently, the analysis of RNA helicase genes in Escherichia coli have been identified and studied extensively, including 5 DEAD-box and 13 DExH-box RNA helicase genes [28], [29]. Recently, the complete analysis and classification of the RNA helicase gene family in Arabidopsis and Oryza sativa, which contain 113 and 115 RNA helicase genes, respectively, has been reported [2].
These studies added more evidence of the role of RNA helicase genes in development and response to abiotic stresses in different species, but related genome-wide resources are limited in two other important crops, maize and soybean. In addition, several whole-genome analysis studies in Arabidopsis and rice have been performed in the past ten years, focused on categories such as the RING-finger, receptor-like kinase gene family, MAPK and MAPKK gene family, KT/HAK/KUP potassium transporters gene family, NAC proteins and siRNAs [30], [31], [32], [33], [34], [35], [36], [37]. Recently, genome-wide analysis in maize has identified and analysed most of the cyclins, MuDR-related transposable elements, beta-glucosidase gene family, auxin response factor (ARF) gene family and two-component signal system (TCS) genes [38], [39], [40], [41], [42]. So far, no genome-wide information on the RNA helicase gene family is currently available in Zea mays or Glycine max. Extensive genome-wide comparative in silico analysis of the RNA helicase gene family in the completed genomes of Zea mays and Glycine max could identify numerous known or novel gene families associated with defence, photomorphogenesis, gene regulation, development, metabolism, transportation and/or stress tolerance.
In this study, we identified a total 161, 149, 136 and 213 RNA helicase genes in the Arabidopsis, Oryza sativa, Zea mays and Glycine max genomes, respectively, by genome-wide comparative in silico analysis. Each of the different subfamilies, such as DEAD-box, DEAH-box or DExD/H-box, has different rate of alternative splicing in each species. The chromosome location analysis showed that the RNA helicase protein genes were distributed across all chromosomes with different densities in the four species. The phylogenetic tree analyses identified the relevant homologs of DEAD-box, DEAH-box and DExD/H-box RNA helicase proteins in each of the four species. Additionally, microarray expression data showed that many of these predicted RNA helicase genes were expressed in different developmental stages and different tissues under normal growth conditions. Finally, real-time quantitative PCR analysis showed that the expression levels of 10 RNA helicase genes in Arabidopsis and 13 RNA helicase genes in Zea mays were in close agreement with the microarray expression data. To our knowledge, this is the first report of a comparative genome-wide analysis of the RNA helicase gene family in Arabidopsis, Oryza sativa, Zea mays and Glycine max. The comparative genome-wide analysis provides valuable information for understanding the classification and putative functions of the RNA helicase gene family and new insight into the organisation, evolution and functions of the RNA helicase gene family in crop growth and development.
Results
The Identification of the RNA Helicase genes in Arabidopsis, Oryza sativa, Zea Mays and Glycine Max
To identify the members of the RNA helicase gene family in Arabidopsis, Oryza sativa, Zea mays and Glycine max, we used bioinformatic methods to gather extensive information regarding this family. A total of 161 genes that encode 217 RNA helicase proteins were identified as potential members of the RNA helicase superfamily within the Arabidopsis genome (http://www.tair.org/), whereas 149 genes encoded 199 RNA helicase proteins were identified in the Oryza sativa genome (http://www.phytozome.net/) (Table S1). Our predicted number of RNA helicase proteins in the two species was greater than the number found in Arabidopsis (113) or rice (115) [2]. So far, predicted members of the RNA helicase gene family in Glycine max and Zea mays have not reported in detail. In our results, we identified a total of 136 and 213 RNA helicase genes in the Zea mays and Glycine max genome (http://www.phytozome.net/), respectively (Table S1).
Based on the characteristics of the conserved motifs, the RNA helicase genes were classified into three subfamilies: DEAD-box (50/51/57/87 genes), DEAH-box (40/33/31/48 genes) and DExD/H-box (71/65/50/78 genes) in the four species Arabidopsis, Oryza sativa, Zea mays and Glycine max (Table 1), respectively for each subfamily. In addition, the results revealed that Arabidopsis, Oryza sativa, Zea mays and Glycine max have 56, 50, 79 and 35 alternative splicing in whole RNA helicase gene family, respectively (Table 1). In Zea mays, there are two particular genes whose different alternative splicing products belong to different RNA helicase subfamilies. GRMZM2G010085_T01 and GRMZM2G010085_T02 belong to the DEAH-box and DExD/H-box subfamilies, respectively. Moreover, GRMZM2G420865_T02 and GRMZM2G420865_T03 belong to DEAD-box and DExD/H-box, respectively.
Table 1. The number of the DEAD-box, DEAH-box and DExD/H-box RNA helicase genes in Arabidopsis, Oryza sativa, Zea mays, Glycine max.
| Species | DEAD-box | DEAH-box | DExD/H-box | Total | Alternative Splicing | Alternative Splicing (%) |
| Arabidopsis | 50/66 | 40/52 | 71/99 | 161/217 | 56 | 25.81 |
| Oryza sativa | 51/79 | 33/41 | 65/79 | 149/199 | 50 | 25.12 |
| Zea mays | 57/86 | 31/44 | 50/85 | 136*/215 | 79 | 36.74 |
| Glycine max | 87/101 | 48/55 | 78/92 | 213/248 | 35 | 14.11 |
Note: In front of diagonal not include the number of alternative splicing; in the back of diagonal include the number of alternative splicing.
In Zea mays, there are two especial genes with different alternative splicing belongs to different RNA helicase subfamilies.
The Chromosome Localization of the RNA Helicase Gene Family in Arabidopsis, Oryza sativa, Zea Mays and Glycine Max
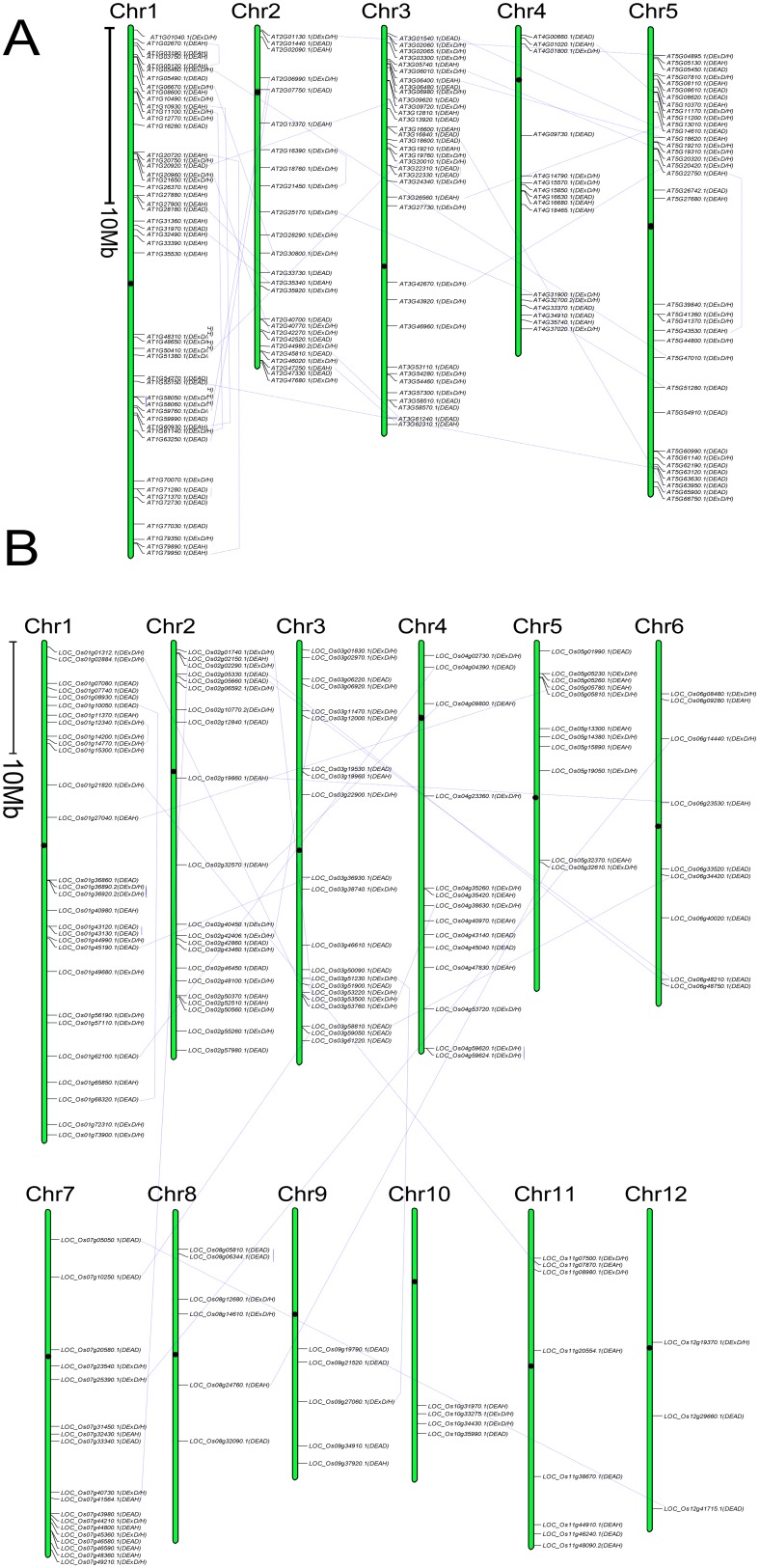
Using the Perl-based program MapDraw and Photoshop tools, the RNA helicase genes were then mapped onto the chromosomes of different species and named with their own Gene ID. In all four species, all predicted RNA helicase genes could be conclusively matched to a chromosome (Figure 1 and Figure 2). The chromosomal locations of 161 Arabidopsis RNA helicase protein genes were analysed first in our study. The chromosomes locations analysis showed that the Arabidopsis RNA helicase protein genes were distributed across all 5 chromosomes with different densities from 9.9% (chromosome 4) to 30.4% (chromosome 1) (Figure 1A). Second, we found a similar distribution pattern on Oryza sativa chromosomes from 2.0% (chromosome 12) to 19.5% (chromosome 1) (Figure 1B). Only 3 genes were mapped on chromosome 12. In addition, the Zea mays and Glycine max RNA helicase protein genes were mapped on the chromosomes from chromosomes 1 to 10 and from chromosomes 1 to 20, respectively. In Zea mays, chromosome 5 encompassed the most RNA helicase protein genes with 23 (16.9%), while chromosomes 6 and 9 contained 7 RNA helicase protein genes (5.1%) (Figure 2A). Compared with the preceding three species, relatively low densities of RNA helicase protein genes were observed on the 20 Glycine max chromosomes, with the densities from 1.9% (chromosome 6 ) to 10.8% (chromosome 8) (Figure 2B). To detect possible relationships between RNA helicase genes and potential genome duplication events, we mapped 35, 27, 25 and 62 paralogous gene pairs of RNA helicase genes in Arabidopsis, Oryza sativa, Zea mays and Glycine max, respectively (Figure 1 and Figure 2). It is noteworthy that the percentage of the paralogous gene pairs of RNA helicase genes in Glycine max was higher than the other three species, indicating that segmental and/or tandem duplications might more frequently occurred in Glycine max.
Figure 1. The chromosomal mapping of the RNA helicase gene family.
(A) Arabidopsis, (B) Oryza sativa. The scale bar represents a 10.0 Mb chromosomal distance. The chromosome number is indicated at the top of each chromosome. To simplify the presentation, we named the putative RNA helicase genes with their own Gene ID. The subfamily of each RNA helicase gene was shown in parentheses. The paralogous sister pairs of RNA helicase genes were connected by light blue line, which had very strong bootstrap support (>90%). The positions of centromeres were marked with the black dot.
Figure 2. The chromosomal mapping of the RNA helicase gene family.
(A) Zea mays, (B) Glycine max. The scale bar represents a 10.0 Mb chromosomal distance. The chromosome number is indicated at the top of each chromosome. To simplify the presentation, we named the putative RNA helicase genes with their own Gene ID. The subfamily of each RNA helicase gene was shown in parentheses. The paralogous sister pairs of RNA helicase genes were connected by light blue line, which had very strong bootstrap support (>90%). The positions of centromeres were marked with the black dot.
The Phylogenetic Tree Analysis of the RNA Helicase Gene Family in Arabidopsis, Oryza Sativa, Zea Mays and Glycine Max
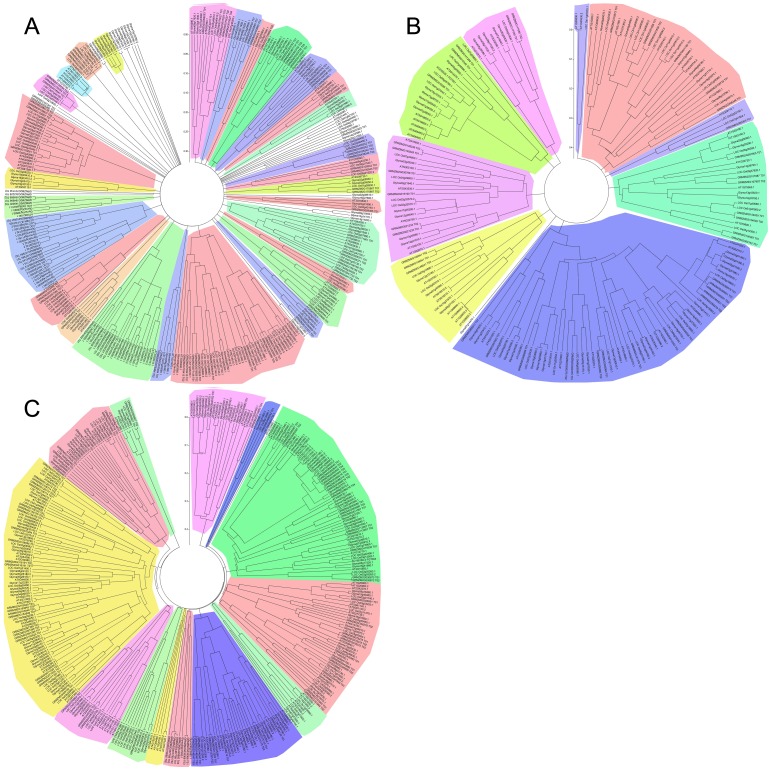
To determine their evolutionary relationship, the phylogenetic relationship of each subfamily of the RNA helicase proteins was examined by aligning their amino acid sequences and implementing the neighbour-joining method in MEGA 5.0. The phylogenetic tree analyses showed that the whole of DEAD-box (332), DEAH-box (192) and DExD/H-box (355) RNA helicase proteins in four species, respectively (Figure 3). We also performed the orthologs/paralogs relationship among the RNA helicase genes family in four species (Figure S1, Figure S2, Figure S3 and Figure S4). Three Arabidopsis DEAD-box RNA helicases, LOS4, STRS1 and STRS2, were shown to be involved in responses to multiple abiotic stresses [11], [12], [13]. These investigations indicate that DEAD-box RNA helicases may play an important role in building resistance to abiotic stress during plant growth and development. Figure 3A shows that Glyma18g32190, Glyma19g03410 and LOC_Os03g6220 have high homology to Arabidopsis LOS4 (At3G53110). In addition, the phylogenetic tree analyses showed that STRS1 (At1G31970) has a high level of identity to Glyma01g01390, Glyma09g34390, LOC_Os07g20580 and GRMAZM2G076484. An analysis of the DEAH-box RNA helicase proteins showed that this subfamily could be further classified into nine subgroups (Figure 3B), whereas the other two subfamilies, DEAD-box and DExD/H-box RNA helicase proteins, could be further classified into more than ten subgroups (Figure 3A and Figure 3C). ISE2 and AtHELPS, encoding DExD/H-box RNA helicases, were shown to be involved in plasmodesmata function during embryogenesis and potassium deprivation responses and tolerance in Arabidopsis thaliana, respectively [20], [24]. As shown in Figure 3C, we identified the relevant homologs of ISE2 (AT1G70070) and AtHELPS (AT3G46960) as LOC_Os02g50560 and Glyma18g07510, respectively. At the same time, the phylogenetic tree analyses revealed that OsBIRH1 (LOC_Os03g01830) has a higher homology to Arabidopsis AT3G06980, which is in accord with preceding research [21].
Figure 3. Phylogenetic tree analysis of RNA helicase in Arabidopsis, Oryza sativa, Zea mays and Glycine max.
(A) The DEAD-box RNA helicase proteins in four species. (B) The DEAH-box RNA helicase proteins in four species. (C) The DExD/H-box RNA helicase proteins in four species. The amino acid sequences of the RNA helicase proteins were aligned with ClustalX, and the phylogenetic tree was constructed using the neighbour-joining method in MEGA 5.0 software. Each node is represented by a number that indicates the bootstrap value for 100 replicates. The scale bar represents 0.05, 0.1 and 0.1 substitutions per sequence position, respectively.
The Expression Profile During Different Development Stage and Different Tissues of the RNA Helicase Genes in Four Species Under Normal Growth Conditions
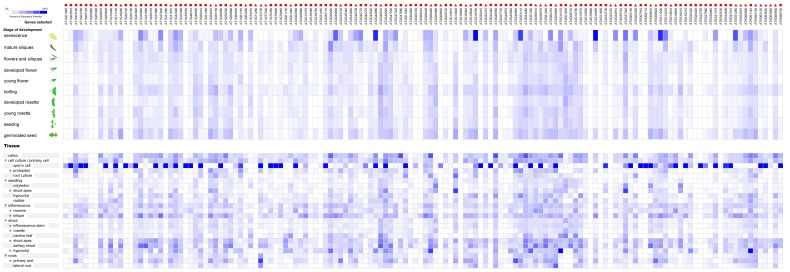
To investigate the potential functions of the RNA helicase proteins in crop development, we analysed microarray expression data from various datasets in the Gene Chip platform of Genevestigator. We found that not all of the predicted genes were expressed in different plant developmental stages and different tissues under normal growth conditions. As shown in Figure 4, among the 161 predicted genes in Arabidopsis, 144 genes (89.4%) were expressed in at least one of the development stages tested. More than half of the predicted RNA helicase genes were expressed in ten different development stages with various expression levels, including senescence, mature siliques, flowers and siliques, developed flower, young flower, bolting, developed rosette, young rosette, seedling and germinated seed. AT2G44980, AT3G19760, AT3G53110 (LOS4), AT4G18465, AT5G11200 and AT5G51280 were highly expressed in senescence stage. Our results showed that the most RNA helicase genes were expressed in more than 20 tissues in Arabidopsis with different expression levels and many predicted Arabidopsis RNA helicase genes were expressed in primary cell, seedling, inflorescence, silique, shoot and roots. Forty-four genes were highly expressed in sperm cells and AT5G10370 and AT5G61140 were only highly expressed in primary root tissue (Figure 4).
Figure 4. The expression profiles of 145 RNA helicase genes in Arabidopsis.
The expression profiles of 145 RNA helicase genes in different development stages (above) and tissues (below) in Arabidopsis. The deep and light blue shading represents the relative high or low percent potential expression levels, respectively, of the helicase genes in different development stages and different tissues. The DEAD-box, DEAH-box and DExD/H-box RNA helicase genes are indicated by the red triangle, circle and square, respectively.
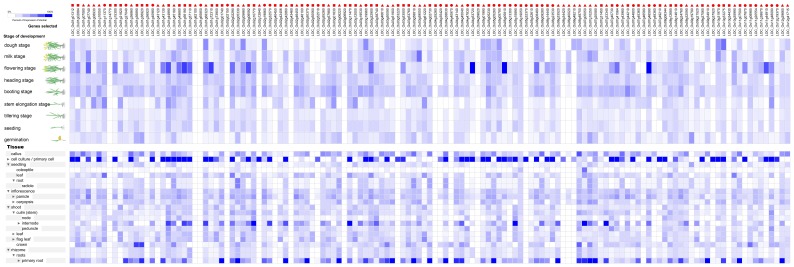
During the nine Oryza sativa development stages, including the dough stage, milk stage, flowering stage, heading stage, booting stage, stem elongation stage, tillering stage, seedling and germination, 135 genes (90.6%) were expressed in at least one of the development stages tested and several genes had high expression levels in the flowering stage (Figure 5). Notably, the expression levels of the RNA helicase in all tested Oryza sativa tissues were higher in primary cell, internode and primary root (Figure 5).
Figure 5. The expression profiles of 138 RNA helicase genes in Oryza sativa.
The expression profiles of 138 RNA helicase genes in different development stages (above) and different tissues (below) in Oryza sativa. The deep and light blue shading represents the relative high or low percent potential expression levels, respectively, of the helicase genes in different development stages and different tissues. The DEAD-box, DEAH-box and DExD/H-box RNA helicase genes are indicated by the red triangle, circle and square, respectively.
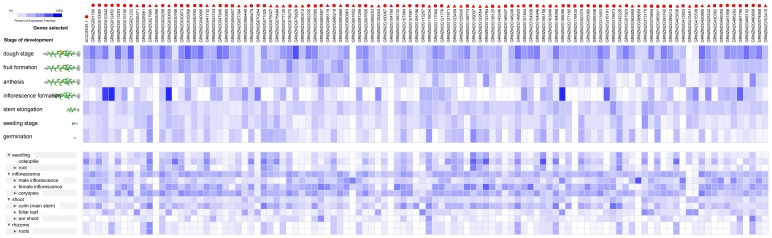
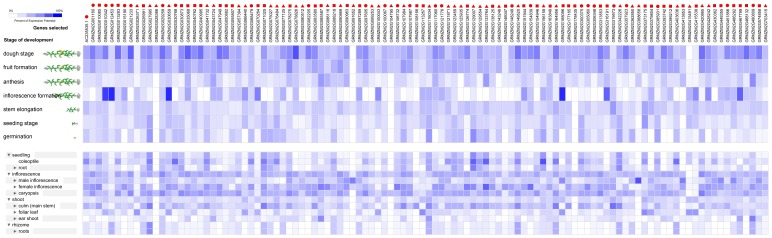
Approximately 109 Zea mays genes were expressed with various expression levels in the tested development stages, including dough stage, fruit formation, anthesis, inflorescence formation, stem elongation, seedling stage and germination (Figure 6). In addition, the expression of these genes exhibited similar profiles and showed higher expression level than in Arabidopsis and Oryza sativa development stage (Figure 6).
Figure 6. The expression profiles of 54 RNA helicase genes in Zea mays.
The expression profiles of 54 RNA helicase genes in different development stages (above) and different tissues (below) in Zea mays. The deep and light blue shading represents the relative high or low percent potential expression levels, respectively, of the helicase genes in different development stages and different tissues. The DEAD-box, DEAH-box and DExD/H-box RNA helicase genes are indicated by the red triangle, circle and square, respectively.
Approach to half of the Glycine max RNA helicase genes were expressed highly in fruit formation and bean development stage. For another, about half of the genes were expressed in main shoot growth and germination at lower levels. Moreover, approximately 15 genes were most highly expressed in the flowering stage (Figure 7). Many genes exhibited higher expression levels in the primary cell, leaf cell, shoot apex, axillary meristem, shoot apex and unspecified root type of soybean (Figure 7). Taken together, these data suggest that the highly expressed RNA helicase genes may play an important role in the regulation of four species’ growth and development, and these analyses further aid the understanding of the basal functions of many RNA helicase proteins in crop growth and development.
Figure 7. The expression profiles of 99 RNA helicase genes in Glycine max.
The expression profiles of 99 RNA helicase genes in different development stages (above) and different tissues (below) in Glycine max. The deep and light blue shading represents the relative high or low percent potential expression levels, respectively, of the helicase genes in different development sages and different tissues. The DEAD-box, DEAH-box and DExD/H-box RNA helicase genes are indicated by the red triangle, circle and square, respectively.
The Expression Profile of the RNA Helicase Genes in Various Tissues as Determined by qRT-PCR Analyses in Arabidopsis and Zea Mays
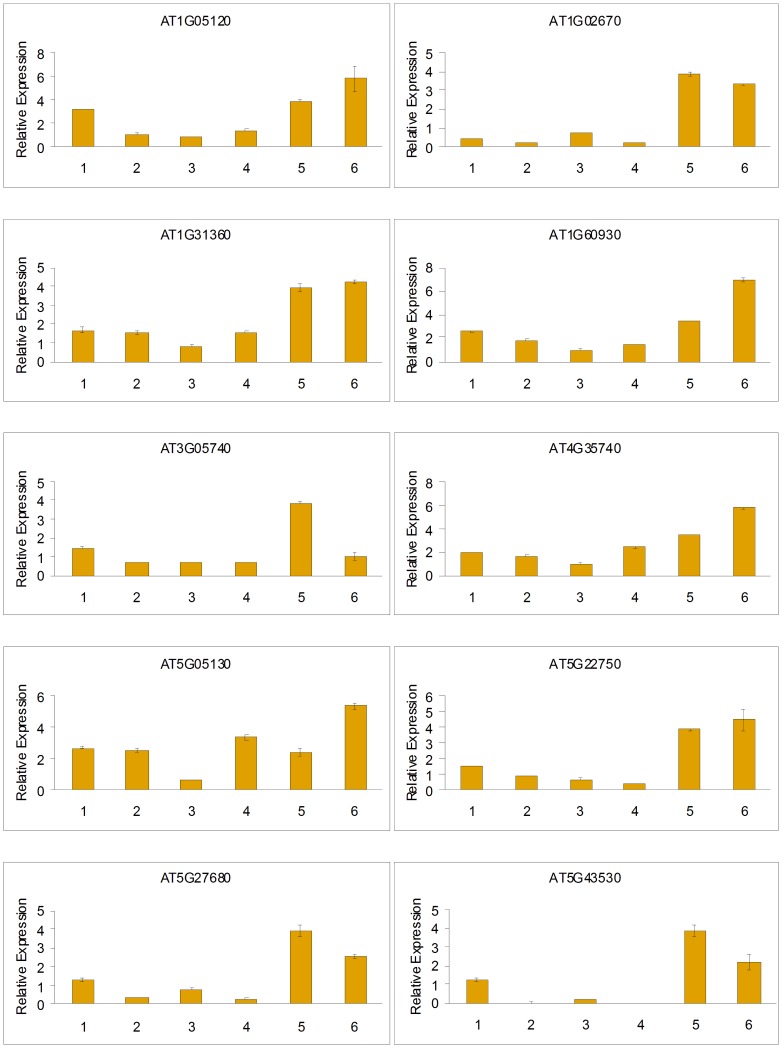
We performed the expression analysis of 10 RNA helicase genes in Arabidopsis under normal growth conditions in six different tissues: root, rosette leaf, stem, cauline leaf, flower and silique. Basically in accord with Genevestigator analysis, qRT-PCR results showed that 9 predicted genes were expressed in all of the tested tissues in Arabidopsis (Figure 8). Only one gene (AT5G43530) displayed tissue-specific expression patterns, which was not detected in rosette leaf and cauline leaf (Figure 8). Intriguingly, relatively higher expression levels of these helicase genes were observed in flower and silique, indicating that RNA helicase activities might be closely related with reproductive processes in Arabidopsis (Figure 8).
Figure 8. QRT-PCR analysis of 10 helicase genes in different tissues of Arabidopsis.
Error bars indicate standard deviations (n = 3). 1, root; 2, rosette leaf; 3, stem; 4, cauline leaf; 5, flower; 6, silique.
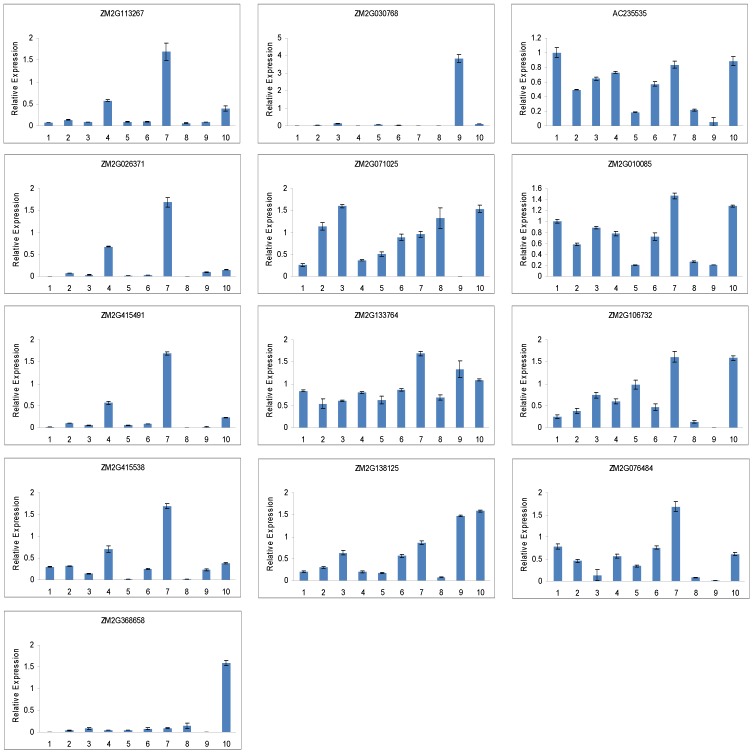
We also analysed the expression of 13 RNA helicase genes in maize under normal growth conditions in ten different tissues: primary root, pericarp, internode, adult leaf, silk, culm, seedling, endosperm, embryo and tassel. All of the 13 predicted genes were expressed in at least one of the ten tissues in maize (Figure 9). The results showed that ZM2G113267, ZM2G026371, ZM2G415491 and ZM2G415538 were primarily expressed in the seedlings and adult leaf, while they were barely expressed in the other eight tissues. ZM2G368658 and ZM2G030768 were especially abundant in the embryo and tassel, respectively, and were expressed at relatively low levels in other nine tissues. ZM2G071025, ZM2G133764, ZM2G138125, AC235535, ZM2G010085, ZM2G106732 and ZM2G076484 were detected in all tested tissues, whereas ZM2G071025 and AC235535 in the embryo, ZM2G138125 in the endosperm, ZM2G010085, ZM2G106732 and ZM2G076484 in the endosperm and embryo were weakly expressed (Figure 9). These results, also in accord with Genevestigator analysis, suggest that the tested 13 RNA helicase genes might be involved in tissue development in maize. Taken together, these results imply that the tested RNA helicase genes might play roles in regulating the development of different tissues.
Figure 9. QRT-PCR analysis of 13 helicase genes in different tissues of Zea mays.
Error bars indicate standard deviations (n = 3). 1, primary root; 2, pericarp; 3, internode; 4, adult leaf; 5, silk; 6, culm; 7, seedling; 8, endosperm; 9, embryo; 10, tassel.
Discussion
RNA helicases are found in various organisms, ranging from prokaryotes to mammals, and have become a focus of interest in recent years due to their participation in diverse cellular processes [8], [43], [44]. In the past ten years, although the RNA helicases have been intensively studied in plant growth and development and response to various stresses [8], [10], [11], [12], [13], [16], [17], [19], [20], [24], [45], [46], [47], only a few members have been identified in the regulation of crop plant growth and development. While Arabidopsis and Oryza sativa RNA helicase families have been partially predicted from The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org/) and the Rice Genome Annotation Project (RAP) database (http://rice.plantbiology.msu.edu/) [2], [48], [49], the characteristics of the gene family and the function of RNA helicase proteins is poorly understood in Zea mays and Glycine max. Therefore, the biological functions of a majority of the crop RNA helicases require further investigation.
In this study, we presented a complete analysis of the RNA helicase gene family in Arabidopsis and other crop species genomes by genome-wide comparative in silico analysis, including the gene classification, chromosomal locations, phylogenetic tree and expression profiles in different tissues and development stages under normal growth conditions.
The RNA helicase gene family has 113 members in Arabidopsis and 115 members in rice [2]. In this study, we identified a total of 161 and 149 RNA helicase genes in Arabidopsis and rice, respectively. We speculate that this phenomenon may be due to continual updates of the TAIR and RAP database. In addition, we also identified a total of 136 RNA helicase genes in maize and 213 members in soybean. Compared with Arabidopsis (genome size 125 Mb) [50] and rice (genome size 480 Mb) [51], the size of the RNA helicase gene family is smaller in maize (genome size 2500 Mb) [52], [53] and soybean (genome size 1115 Mb) [54]. But compared with the number of all genes in four species genome (25,500 genes in Arabidopsis genome, 37,500 genes in rice genome, 50,000 genes in maize genome and 66,000 genes in soybean genome), the percentage of the RNA helicase gene family is similar in four species (0.631% in Arabidopsis, 0.397% in rice, 0.272% in maize and 0.323% in soybean), except higher in Arabidopsis. Although the genome and the total genes number in four species is very different, it still not show differ greatly in the size of RNA helicase gene family. The presence of a large helicase gene family in four species suggests the RNA helicases play important roles in diverse processes. We further compared the number of RNA helicase genes in different subfamilies among Arabidopsis, Oryza sativa, Zea mays and Glycine max (Table 1). As showed in Table 1, we founded that the number of the DEAD-box and DExD/H-box RNA helicase genes have many more members than the DEAH-box RNA helicase genes in the four species. The key difference is that the number of the alternative splicing products (35) was much smaller in Glycine max than in Arabidopsis (56), Oryza sativa (50), and Zea mays (79), rather than the number of different genes in each of the subfamilies.
Among the total RNA helicase genes in the four species, the percentage of alternative splicing is lowest in soybean (14.11%) and highest in maize (35.81%). The data showed that the phenomenon of alternative splicing of RNA helicase genes is more common in dicot than in monocot. The regulation of alternative splicing is a key step in the control of gene expression, as splicing variants have different biological functions and regulatory features. Alternative splicing is one of the most complex cellular processes in eukaryotes, where information must be processed differently at different times (such as different development stages) or a very high level of diversity is required. Only a small number of alternative splicing events have been reported in plants. Recent progress has occurred in characterising the splicing signals in plant pre-mRNAs, in identifying the mutants affected in splicing and in discovering new examples of alternatively spliced mRNAs. Furthermore, although data from both animals and plants suggest tissue-specific and temporal regulation of alternative splicing [55], [56], [57], [58], the mechanisms that regulate alternative splicing in plants remain unknown. Alternative splicing can result in the production of different protein isoforms, thereby affecting transcriptome and proteome diversity, and, ultimately, the regulation of protein function and gene expression [59], [60], [61]. Recent genome-wide experiments have shown that >40% of Arabidopsis thaliana and rice genes can produce multiple diverse mRNA molecules by alternative splicing [62], [63], [64], [65]. In our results, the percentage of alternative splicing of RNA helicase genes in Arabidopsis thaliana and rice (>25%) was less than the average alternative splicing frequency of whole genome. Our data concerning the alternative splicing frequency of RNA helicase genes in different crop species will not only provide information on mechanisms of gene regulation through alternative splicing in future but also facilitate our understanding of the regulation of RNA helicase genes in crop growth and development. To our knowledge, this is the first report of a genome-wide analysis of the crop RNA helicase gene family. The different gene subfamilies and alternative splicing frequency of the RNA helicases might mirror the diverse functions of these genes in RNA metabolism.
We also utilised a Genevestigator analysis to gain insight into the expression profiles of the RNA helicase genes during different development stages and in different tissues under normal growth conditions. We found that under normal growth conditions, among the all of the predicted genes in Arabidopsis, rice and maize, more than 80% RNA helicase genes were expressed in at least one of the development stages and tissues tested (Figure 4, 5, 6). In addition, about half of the predicted RNA helicase genes were expressed in at least one of the development stage and tissues tested in soybean (Figure 7). Therefore, we speculated that the highly expressed RNA helicase genes may play a role in the regulation of crop growth and development. However, more research will be needed to determine the functions of the RNA helicase genes in these four species. In addition, the results also showed that the percentage of different subfamilies in different development stage and tissues changed dissimilarly. The DEAH-box RNA helicase genes higher proportion of the development stages and tissues in Arabidopsis, Oryza sativa and Zea mays (Figure 4, 5, 6). Taken together, we speculate that the RNA helicase proteins, specifically the DEAH-box RNA helicases, might play an important role in different development stages of crops and the growth of different tissues.
To our knowledge, this is the first report of a comparative genome-wide analysis of the RNA helicase gene family in Arabidopsis and Oryza sativa, Zea mays and Glycine max. This study provides valuable information for understanding the classification and putative functions of the RNA helicase gene family in crop growth and development.
Materials and Methods
The Identification of the Helicase Genes in Arabidopsis, Oryza Sativa, Zea Mays and Glycine Max
To identify the members of the helicase gene family in Arabidopsis, Oryza sativa, Zea mays and Glycine max, two different approaches were performed [66]. First, the genome of the four species were downloaded from the database with different genome sizes, including Arabidopsis (genome size 125 Mb, 25,500 genes), rice (genome size 480 Mb, 37,500 genes), maize (genome size 2500 Mb, 50,000 genes) and soybean (genome size 1115 Mb, 66,000 genes) [51]–[54]. All known Arabidopsis helicase gene sequences, which were downloaded from the Arabidopsis genome TAIR 9.0 release (http://www.arabidopsis.org/), were used as query sequences to perform multiple database searches against the proteome and genome files downloaded from the Phytozome database (http://www.phytozome.net/). Stand-alone versions of BLASTP and TBLASTN (http://blast.ncbi.nlm.nih.gov), which are available from the NCBI, were used with an e-value cutoff set to 1e-003 [67]. All of the protein sequences derived from the collected candidate helicase genes were examined using the domain analysis programs PFAM (http://pfam.sanger.ac.uk/) and SMART (http://smart.embl-heidelberg. de/) with the default cutoff parameters [68], [69]. Second, we analysed the domains of all of the peptide sequences using a Hidden Markov Model (HMM) analysis with protein family (Pfam) searching (http://pfam.sbc.su.se/). Then, we obtained the sequences with the PF00271 Pfam number, which contained a typical helicase domain, from the genome sequences using a Perl-based script. Finally, all of the protein sequences were compared with known helicase sequences using ClustalX (http://www.clustal.org/) to verify the sequences were candidate helicases [70].
The isoelectric points and molecular weights of the proteins were obtained with the help of the proteomics and sequence analysis tools on the ExPASy Proteomics Server (http://expasy.org/) [71]. The chromosomal locations and the exon/intron information were obtained from the Phytozome database using a Perl-based program.
The Chromosomal Location of the Helicase Genes
The chromosomal locations were retrieved from the genome data downloaded from the Phytozome database (http://www.phytozome.net/) using a Perl-based program and mapped to the chromosomes using the MapDraw and Photoshop tools.
Sequence Alignment and Phylogenetic Analysis
The helicase sequences were aligned using the ClustalX program with BLOSUM30 as the protein-weight matrix. The MUSCLE program (version 3.52) was also used to perform multiple sequence alignments to confirm the ClustalX results (http://www.clustal.org/) [72]. Phylogenetic trees of the helicase protein sequences were constructed using the neighbour-joining (NJ) method of the MEGA5 program (http://www.megasoftware.net/) using the p-distance and complete deletion option parameters [73]. The reliability of the obtained trees was tested using a bootstrapping method with 1000 replicates. The images of the phylogenetic trees were drawn using MEGA5.
Expression Analyses of the Helicase Genes in Arabidopsis, Oryza Sativa, Zea Mays and Glycine Max
Microarray expression data from various datasets were obtained using Genevestigator (https://www.genevestigator.com/gv/) with the Arabidopsis (ATH1∶22 k array), Oryza sativa (OS_51 k: Rice Genome 51 k array), Zea mays (ZM_84 k: Nimblegen Maize 385 k) and Glycine max (GM_60 k: Soybean Genome Array) Gene Chip platform. Then, the identified helicase-containing gene IDs were used as query sequences to perform searches in the Gene Chip platform of Genevestigator.
Plant Materials and Growth Conditions
Arabidopsis thaliana (Col-0) seeds were surface-sterilized and sown on MS plates. Seeds were stratified at 4°C for 2 days and then transferred to 22°C for 2 weeks. One month-old plants grown under a 16-h-light/8-h-dark photoperiod at 22°C with cool white light (120 mmol photons m−2 s−1) were used for sampling. For RNA extraction, the different tissues were frozen and stored in liquid nitrogen immediately after harvest.
For maize inbred line Qi 319 (from Shandong Academy of Agricultural Sciences), embryos 25 days after pollination was harvested from greenhouse-grown plants in sand under 16 h of light (25°C) and 8 h of dark (20°C), and eight-week-old seedling tissues and organs were harvested for expression analysis. Samples were collected and were immediately frozen in liquid nitrogen for further use. Two biological replicates were performed for each sample.
RNA Isolation and Real-time Quantitative RT-PCR Expression Analysis
Total RNAs were extracted using Trizol according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA) from leaves of maize seedlings with different treatments. The first strand cDNAs were synthesised using First Strand cDNA Synthesis kit (Fermentas, USA).
Real-time quantification RT-PCR reactions were performed in Bio-RAD MyiQ™ Real-time PCR Detection System (Bio-Rad, USA) using the TransStart Top Green qPCR SuperMix (TransGen, China) according to the manufacturer’s instructions. Each PCR reaction (20 µL containing 10 µL 2×real-time PCR Mix (containing SYBR Green I), 0.5 µL of each primer, and appropriately diluted cDNA. The thermal cycling conditions were 95°C for 30 s followed by 45 cycles of 95°C for 15 s, 55°C −60°C for 30 s, and 72°C for 15 s. The Zmactin gene was used as an internal reference for all the qRT-PCR analysis. Each treatment was repeated three times independently. Relative gene expression was calculated according to the delta-delta Ct method of the system. The primers used are described in Table S2.
Supporting Information
Phylogenetic tree analysis of RNA helicase in Arabidopsis . From left to right are the DEAD-box, DEAH-box and DExD/H-box RNA helicase proteins, respectively. The scale bar represents 0.2, 0.2 and 0.2 substitutions per sequence position, respectively. Sister pairs of paralogous helicase genes were indicated by red shadow, which had very strong bootstrap support (>90%).
(TIF)
Phylogenetic tree analysis of RNA helicase in Oryza sativa . From left to right are the DEAD-box, DEAH-box and DExD/H-box RNA helicase proteins, respectively. The scale bar represents 0.2, 0.2 and 0.2 substitutions per sequence position, respectively. Sister pairs of paralogous helicase genes were indicated by red shadow, which had very strong bootstrap support (>90%).
(TIF)
Phylogenetic tree analysis of RNA helicase in Zea mays . From left to right are the DEAD-box, DEAH-box and DExD/H-box RNA helicase proteins, respectively. The scale bar represents 0.2, 0.2 and 0.2 substitutions per sequence position, respectively. Sister pairs of paralogous helicase genes were indicated by red shadow, which had very strong bootstrap support (>90%).
(TIF)
Phylogenetic tree analysis of RNA helicase in Glycine max . From left to right are the DEAD-box, DEAH-box and DExD/H-box RNA helicase proteins, respectively. The scale bar represents 0.2, 0.2 and 0.2 substitutions per sequence position, respectively. Sister pairs of paralogous helicase genes were indicated by red shadow, which had very strong bootstrap support (>90%).
(TIF)
RNA helicase genes in Arabidopsis, Oryza sativa, Zea mays, Glycine max .
(DOC)
The primers used in the Real-time quantification RT-PCR reactions.
(DOC)
Funding Statement
This work was supported by the National Natural Science Foundation (Grant No. 30970230 and No. 31000121), the Shandong Province Natural Science Foundation (Grant No. ZR2010CQ037), the Open Project Program of the State Key Laboratory of Crop Biology (Grant No. 2013KF07) and the Research Fund for Ph.D in Weifang University (No. 2012BS15) in China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Isono K, Yamamoto H, Satoh K, Kobayashi H (1999) An Arabidopsis cDNA encoding a DNA-binding protein that is highly similar to the DEAH family of RNA-DNA helicase genes. Nucleic Acids Res 27 (18): 3728–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Umate P, Tuteja R, Tueja N (2010) Genome-wide analysis of helicase gene family from rice and Arabidopsis: a comparison with yeast and human. Plant Mol Biol 73 (4–5): 449–465. [DOI] [PubMed] [Google Scholar]
- 3. de la Cruz J, Kressler D, Linder P (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci 24 (5): 192–198. [DOI] [PubMed] [Google Scholar]
- 4. Tanner NK, Linder P (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell 8 (2): 251–262. [DOI] [PubMed] [Google Scholar]
- 5. Tanner NK, Cordin O, Banroques J, Doere M, Linder P (2003) The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell 11 (1): 127–138. [DOI] [PubMed] [Google Scholar]
- 6. Rocak S, Linder P (2004) DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5 (3): 232–241. [DOI] [PubMed] [Google Scholar]
- 7. Lorsch JR (2002) RNA chaperones exist and DEAD box proteins get a life. Cell 109 (7): 797–800. [DOI] [PubMed] [Google Scholar]
- 8. Linder P, Owttrim GW (2009) Plant RNA helicases: linking aberrant and silencing RNA. Trends Plant Sci 14 (6): 344–352. [DOI] [PubMed] [Google Scholar]
- 9. Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444 (2): 139–158. [DOI] [PubMed] [Google Scholar]
- 10. Owttrim GW (2006) RNA helicase and abiotic stress. Nucleic Acids Res 34 (11): 3220–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gong ZZ, Lee H, Xiong LM, Jagendorf A, Stevenson B, et al. (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA 99 (17): 11507–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gong ZZ, Dong CH, Lee H, Zhu JH, Xiong LM, et al. (2005) A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis . Plant Cell 17 (1): 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kant P, Kant S, Gordon M, Shaked R, Barak S (2007) STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-Box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol 145 (3): 814–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Western TL, Cheng YL, Liu J, Chen XM (2002) HUA ENHANCER2, a putative DExH-box RNA helicase, maintains homeotic B and C gene expression in Arabidopsis . Development 129 (7): 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Duby G, Purnelle B, Boutry M (2000) Tobacco VDL gene encodes a plastid DEAD box RNA helicase and is involved in chloroplast differentiaton and plant morphogenesis. Plant Cell 12 (11): 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacobsen SE, Running MP, Meyerowitz EM (1999) Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126 (23): 5231–5243. [DOI] [PubMed] [Google Scholar]
- 17. Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana . Curr Biol 12 (17): 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gendra E, Moreno A, Albà MM, Pages M (2004) International of the plant glycine-rich RNA-binding protein MA16 with a novel nuclear DEAD box RNA helicase protein from Zea mays . The Plant J 38 (6): 875–886. [DOI] [PubMed] [Google Scholar]
- 19. Inagaki S, Suzuki T, Ohto MA, Urawa H, Horiuchi T, et al. (2006) Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell 18 (4): 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kobayashi K, Otegui MS, Krishnakumar S, Mindrinos M, Zambryski P (2007) INCREASED SIZE EXCLUSION LIMIT2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell 19 (6): 1885–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li D, Liu H, Zhang H, Wang X, Song F (2008) OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J Exp Bot 59 (8): 2133–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Köhler D, Schmidt-Gattung S, Binder S (2010) The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana . Plant Mol Biol 72 (4–5): 459–467. [DOI] [PubMed] [Google Scholar]
- 23. Liu M, Shi DQ, Yuan L, Liu J, Yang WC (2010) SLOW WALKER3, encoding a putative DEAD-box RNA helicase, is essential for female gametogenesis in Arabidopsis . J Integr Plant Biol 52 (9): 817–828. [DOI] [PubMed] [Google Scholar]
- 24. Xu RR, Qi SD, Lu LT, Chen CT, Wu CA, et al. (2011) A DExD/H box RNA helicase is important for potassium deprivation responses and tolerance in Arabidopsis thaliana . FEBS J 278(13): 2296–2306. [DOI] [PubMed] [Google Scholar]
- 25. Li X, Gao X, Wei Y, Deng L, Ouyang Y, et al. (2011) Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-Box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell 23(4): 1416–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He J, Duan Y, Hua D, Fan G, Wang L, et al. (2012) DExH box RNA helicase–mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 24 (5): 1815–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aubourg S, Kreis M, Lecharny A (1999) The DEAD box RNA helicase family in Arabidopsis thaliana . Nucleic Acids Res 27(2): 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iost I, Dreyfus M (2006) DEAD-box RNA helicases in Escherichia coli . Nucleic Acids Res 34 (15): 4189–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perutka J, Wang W, Goerlitz D, Lambowitz AM (2004) Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol 336 (2): 421–439. [DOI] [PubMed] [Google Scholar]
- 30. Kosarev P, Mayer KF, Hardtke CS (2002) Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol 3(4): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, et al. (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16 (5): 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, et al. (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11 (4): 192–198. [DOI] [PubMed] [Google Scholar]
- 33. Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, et al. (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5 (3): 479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gupta M, Qiu X, Wang L, Xie W, Zhang C, et al. (2008) KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol Genet Genomics 280 (5): 437–452. [DOI] [PubMed] [Google Scholar]
- 35. Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, et al. (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465(1–2): 30–44. [DOI] [PubMed] [Google Scholar]
- 36. Gao LL, Xue HW (2011) Global analysis of expression profiles of rice receptor-like kinase genes. Mol Plant 5 (1): 143–153. [DOI] [PubMed] [Google Scholar]
- 37. Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17 (6): 369–381. [DOI] [PubMed] [Google Scholar]
- 38. Hu X, Cheng X, Jiang H, Zhu S, Cheng B, et al. (2010) Genome-wide analysis of cyclins in maize (Zea mays). Genet Mol Res 9 (3): 1490–1503. [DOI] [PubMed] [Google Scholar]
- 39. Feng J, Fu XQ, Wang TT, Tao YS, Gao YJ, et al. (2011) Genome-Wide analysis of MuDR-related transposable elements insertion population in maize. Acta Agron Sin 37 (5): 772–777. [Google Scholar]
- 40. Gómez-Anduro G, Ceniceros-Ojeda EA, Casados-Vázquez LE, Bencivenni C, Sierra-Beltrán A, et al. (2011) Genome-wide analysis of the beta-glucosidase gene family in maize (Zea mays L. var B73) Plant Mol Biol 77. (1–2): 159–183. [DOI] [PubMed] [Google Scholar]
- 41. Xing H, Pudake RN, Guo G, Xing G, Hu Z, et al. (2011) Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12: 178–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chu ZX, Ma Q, Lin YX, Tang XL, Zhou YQ, et al. (2011) Genome-wide identification, classification, and analysis of two-component signal system genes in maize. Genet Mol Res 10 (4): 3316–3330. [DOI] [PubMed] [Google Scholar]
- 43. Silverman E, Edwalds-Gilbert G, Lin RJ (2003) DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene 312: 1–16. [DOI] [PubMed] [Google Scholar]
- 44. Fuller-Pace FV (2006) DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res 34: 4206–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kemp C, Imler JL (2009) Antiviral immunity in drosophila. Curr Opin Immunol 21: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sahni A, Wang N, Alexis JD (2010) UAP56 is an important regulator of protein synthesis and growth in cardiomyocytes. Biochem Biophys Res Commun 393: 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, et al. (2007) Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol 178: 6444–6455. [DOI] [PubMed] [Google Scholar]
- 48. Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, et al. (2007) The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35: D883–D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poole RL (2007) The TAIR database. Methods Mol Biol 406: 179–212. [DOI] [PubMed] [Google Scholar]
- 50. The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- 51. International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- 52. Palmer LE, Rabinowicz PD, O’Shaughnessy AL, Balija VS, Nascimento LU, et al. (2003) Maize genome sequencing by methylation filtration. Science 302(5653): 2115–2117. [DOI] [PubMed] [Google Scholar]
- 53. Whitelaw CA, Barbazuk WB, Pertea G, Chan AP, Cheung F, et al. (2003) Enrichment of gene-coding sequences in maize by genome filtration. Science 302(5653): 2118–2220. [DOI] [PubMed] [Google Scholar]
- 54. Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- 55. Brown JW, Simpson CG (1998) Splice site selection in plant premRNA splicing. Annual Review of Plant Physiology and Plant Molecular Biology 49: 77–95. [DOI] [PubMed] [Google Scholar]
- 56. Lorković ZJ, Wieczorek Kirk DA, Lambermon MH, Filipowicz W (2000) Pre-mRNA splicing in higher plants. Trends Plant Sci 5(4): 160–167. [DOI] [PubMed] [Google Scholar]
- 57. Reddy ASN (2001) Nuclear pre-mRNA splicing in plants. Critical Reviews in Plant Sciences 20: 523–571. [Google Scholar]
- 58. Li J, Li X, Guo L, Lu F, Feng X, et al. (2006) A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J Exp Bot 57(6): 1263–1273. [DOI] [PubMed] [Google Scholar]
- 59. Lee JH, Kim SH, Kim JJ, Ahn JH (2012) Alternative splicing and expression analysis of High expression of osmotically responsive genes1 (HOS1) in Arabidopsis . BMB Rep 45(9): 515–520. [DOI] [PubMed] [Google Scholar]
- 60. Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336. [DOI] [PubMed] [Google Scholar]
- 61. Graveley BR (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17: 100–107. [DOI] [PubMed] [Google Scholar]
- 62. Severing EI, van Dijk AD, van Ham RC (2011) Assessing the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana using proteomics data. BMC Plant Biol 11: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lu T, Lu G, Fan D, Zhu C, Li W, et al. (2010) Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res 20: 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, et al. (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana . Genome Res 20: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yoo HH, Kwon C, Chung IK (2010) An Arabidopsis splicing RNP variant STEP1 regulates telomere length homeostasis by restricting access of nuclease and telomerase. Mol Cells 30: 279–283. [DOI] [PubMed] [Google Scholar]
- 66. Li YZ, Wu BJ, Yu YL, Yang GD, Wu CA, et al. (2011) Genome-wide analysis of the RING finger gene family in apple. Mol Genet Genomics 286: 81–94. [DOI] [PubMed] [Google Scholar]
- 67. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 68. Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, et al. (2002) The pfam protein families database. Nucleic Acids Res 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain. Nucleic Acids Res 40: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci. 23: 403–405. [DOI] [PubMed] [Google Scholar]
- 71. Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, et al. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31: 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10): 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree analysis of RNA helicase in Arabidopsis . From left to right are the DEAD-box, DEAH-box and DExD/H-box RNA helicase proteins, respectively. The scale bar represents 0.2, 0.2 and 0.2 substitutions per sequence position, respectively. Sister pairs of paralogous helicase genes were indicated by red shadow, which had very strong bootstrap support (>90%).
(TIF)
Phylogenetic tree analysis of RNA helicase in Oryza sativa . From left to right are the DEAD-box, DEAH-box and DExD/H-box RNA helicase proteins, respectively. The scale bar represents 0.2, 0.2 and 0.2 substitutions per sequence position, respectively. Sister pairs of paralogous helicase genes were indicated by red shadow, which had very strong bootstrap support (>90%).
(TIF)
Phylogenetic tree analysis of RNA helicase in Zea mays . From left to right are the DEAD-box, DEAH-box and DExD/H-box RNA helicase proteins, respectively. The scale bar represents 0.2, 0.2 and 0.2 substitutions per sequence position, respectively. Sister pairs of paralogous helicase genes were indicated by red shadow, which had very strong bootstrap support (>90%).
(TIF)
Phylogenetic tree analysis of RNA helicase in Glycine max . From left to right are the DEAD-box, DEAH-box and DExD/H-box RNA helicase proteins, respectively. The scale bar represents 0.2, 0.2 and 0.2 substitutions per sequence position, respectively. Sister pairs of paralogous helicase genes were indicated by red shadow, which had very strong bootstrap support (>90%).
(TIF)
RNA helicase genes in Arabidopsis, Oryza sativa, Zea mays, Glycine max .
(DOC)
The primers used in the Real-time quantification RT-PCR reactions.
(DOC)