Abstract
The adaptation of transposable elements inserted within the genome to serve novel functions in a host cell, a process known as molecular domestication, is a widespread phenomenon in nature. Around fifty protein-coding genes in humans have arisen through this mechanism. Functional characterization of these domesticated genes has revealed involvement in a multitude of diverse cellular processes. Some of these functions are related to cellular activities and pathways known to be involved in cancer development. In this mini-review we discuss such roles of domesticated genes that may be aberrantly regulated in human cancer, as well as studies that have identified disrupted expression in tumors. We also describe studies that have provided definitive experimental evidence for transposable element-derived gene products in promoting tumorigenesis.
Keywords: domesticated genes, cancer, transposons, exaptation, Rtl1, LDOC1, LDOC1L
Introduction
Nearly half of the human genome is evolutionarily derived from transposable elements (TEs),1 which are DNA sequences capable of mobilization within the genome of a host organism. These elements are often highly repetitive and can be grouped into two general classes based on their mechanism of transposition.2 Class I TEs, also referred to as retrotransposons, utilize a reverse transcriptase enzyme to generate an RNA intermediate sequence which is used as template to regenerate a copy of the original DNA sequence for integration at a distinct genomic location. This mechanism results in duplication of the transposon. Class I TEs include retrotransposons with long-terminal repeats (LTRs) resembling those of retroviruses, as well as long and short interspersed elements (LINEs and SINEs). Class II TEs, also called DNA transposons, include the TC-1/mariner, hAT, and piggyBac transposon families. They are typically mobilized via a “cut and paste” mechanism wherein the transposon is excised from a donor site by a transposase enzyme and reinserted elsewhere in the genome without duplication. Some members are capable of replicative mobilization through mechanisms not involving RNA.3-5
Following their incorporation into the genome, the vast majority of TEs have been inactivated as a result of accumulating mutations over an evolutionary time scale, preventing them from mobilizing autonomously. Barring some kind of negative selective pressure, inserted TEs can become fixed in the genome of a species and serve as a source for novel genetic loci. In some cases, accumulated mutations have caused neofunctionalization of inserted TEs. This process is termed exaptation (or alternatively molecular domestication or co-option). Positive selective pressure for maintenance of co-opted TEs may be provided by some beneficial cellular function performed by the novel gene product. The process of TE exaptation has contributed significantly to the human genome. Over 10,000 TE-derived genomic regions have been subject to strong purifying selection6 and ~50 protein-coding genes have arisen via this mechanism.1,7 The majority of domesticated genes are functionally uncharacterized; those that have been studied have been found to be involved in a variety of cellular processes, including transcriptional regulation, proliferation, cell cycle progression, and apoptosis.
Aside from functions of their domesticated gene products, there is a growing body of literature describing the involvement of transposable elements in promoting human cancer, a topic which has been the subject of several recent reviews.8-11 The best characterized roles for TEs in tumorigenesis involve active mobilization or genomic rearrangement through recombination events. Although most TE sequences within the human genome have sustained mutations rendering them inactive, an estimated 80–100 elements retain the ability to transpose.12 The activity of these elements is normally repressed in somatic tissues; however, epigenetic modifications, such as those associated with cellular transformation, can lead to their reactivation. Hypomethylation of L1 retrotransposons has been observed in a variety of human cancers, including those of the prostate,13 liver,14 and colon.15 Analyses of somatic TE mobilization have revealed substantial activity in solid tumors with apparent selection for recurrent disruption of genes known to be involved in cancer, suggesting a contributing role for TE-mediated insertional mutagenesis in promoting tumor formation.16,17 Aside from direct disruption via insertion within genes, mobilized TEs can exert position effects on loci near the site of integration by disrupting normal mechanisms of regulated expression.18 In some cases, TE-initiated transcription can proceed through neighboring loci, leading to the generation of chimeric transcripts that may actively promote neoplastic transformation.19 Another mechanism by which TEs have been implicated in human cancer is through the disruption of genomic architecture. This mechanism is independent of transposition, and it involves non-allelic homologous recombination between repetitive regions of TE-derived sequences. Such events can result in deletion, duplication, or translocation of chromosomal regions, promoting genomic instability.20
In addition to the mechanisms already discussed, it has been speculated that gene products expressed from domesticated TEs may play a direct role in human cancer. There is an increasing amount of correlative evidence linking aberrant expression of domesticated gene products to tumor development and/or progression. Validation of putative TE-derived cancer genes in a meaningful in vivo context, however, has been minimal. Recently, we described the identification of Retrotransposon-like 1 (Rtl1) as a gene involved in hepatocellular carcinoma (HCC) development through the use of a Sleeping Beauty transposon forward genetic mutagenesis screen in mice.21 Subsequent in vivo validation of tumor-promoting activity and correlation to expression data from human liver tumors led us to conclude that activation of RTL1 may be a significant driving event in human HCC. In this mini-review, we discuss the functions of various domesticated TE gene products in cellular processes related to cancer, as well as summarize the results of studies correlating expression of this gene class with tumor status. Additionally, we briefly discuss the efforts of our group and another in providing some of the first direct experimental evidence of a functional role for a domesticated TE gene product in promoting cancer.
Gene Products of Co-Opted Transposable Elements have Diverse Roles in Cellular Processes Related to Cancer
Although the majority of domesticated TE genes have not been functionally characterized, those that have are involved in a variety of cellular processes relevant to cancer biology. One explanation for the diversity of cellular functions performed by co-opted genes derived from TEs has to do with the fact that they encode protein domains that are widely utilized by cells, such as those involved in DNA binding, chromatin organization, and transcriptional regulation.22 Genes of this class have been shown to regulate growth factor signaling, cell cycle progression, proliferation, and survival.
One of the clearest examples of the involvement of a domesticated TE in a process related to cancer is that of telomerase reverse transcriptase (TERT). Proposed to have evolved from an ancient L1 retrotransposon insertion,23 TERT is a key component of a system that maintains telomere length in replicating cells. Activation of TERT expression has been observed in the majority of human cancers, where it is associated with replicative immortality, a critical step in neoplastic transformation.24 In addition to its canonical function, roles have been proposed for TERT in Wnt pathway signaling, cellular proliferation, and resistance to apoptosis.
Acquisition of the ability to avoid cell death is a central tenet of the process of cellular transformation and tumorigenesis.24 Involvement of multiple co-opted TE gene products in cell survival has been described. Evolutionarily derived from an LTR retrotransposon of the Ty3/Gypsy family,25 modulator of apoptosis (MOAP1) has been proposed to regulate caspase-dependent apoptosis through interaction with BAX.26 Paternally expressed gene 10 (PEG10) is another Ty3/Gypsy-derived human protein25 with a function related to apoptosis. Through direct physical interaction with the pro-apoptotic E3 ubiquitin ligase seven in absentia homolog 1 (SIAH1), PEG10 was shown to increase cell survival by preventing the induction of apoptosis.27 Also derived from a member of the Ty3/Gypsy family of retrotransposons, leucine zipper, downregulated in cancer 1 (LDOC1) was found to increase cell sensitivity to apoptosis induced by TNF-α.28 This effect is mediated through negative regulation of the anti-apoptotic NF-κB transcriptional network. Taken together, these findings suggest a potential link between Ty3/Gypsy-derived domesticated gene products and regulation of apoptosis in human cells.
The zinc finger, BED-type containing (ZBED) gene family originated through exaptation of DNA transposons of the hAT family.29 This family, which consists of ten members in humans, exemplifies the diverse cellular roles for which TEs have been domesticated. Members have been shown to be involved in transcriptional regulation, growth factor signaling, and proliferation. As an example of potential involvement in cancer, it has been reported that ZBED6 acts as a repressor of insulin-like growth factor 2 (IGF2) through directly binding its DNA sequence and inhibiting transcription.30 A role for IGF2 in promoting human cancer has been well established.31
Progression through the cell cycle is another process that is subject to regulation by domesticated TEs. THAP domain containing, apoptosis associated protein 1 (THAP1), which arose from exaptation of a P element DNA transposon,7 was found to be important for G1/S progression through a mechanism involving the pRB/E2F pathway.32 Centromere protein B (CENPB), derived from a Pogo family DNA transposon,7 is involved in chromatin compaction and centromere assembly during mitosis.33 Both THAP1 and CENPB are derivatives of transposase enzymatic domains from Class II transposons, suggesting a possible broader connection between this type of functional domain and cell cycle progression.
Aberrant Expression of Domesticated Transposable Element Gene Products Observed in Various Types of Human Cancer
Several connections between aberrant expression of co-opted TE gene products and cancer have been described. As discussed above, inappropriate activation of TERT is associated with the majority of human cancers. Additional examples of correlative associations between expression of co-opted gene products and tumor status are discussed below.
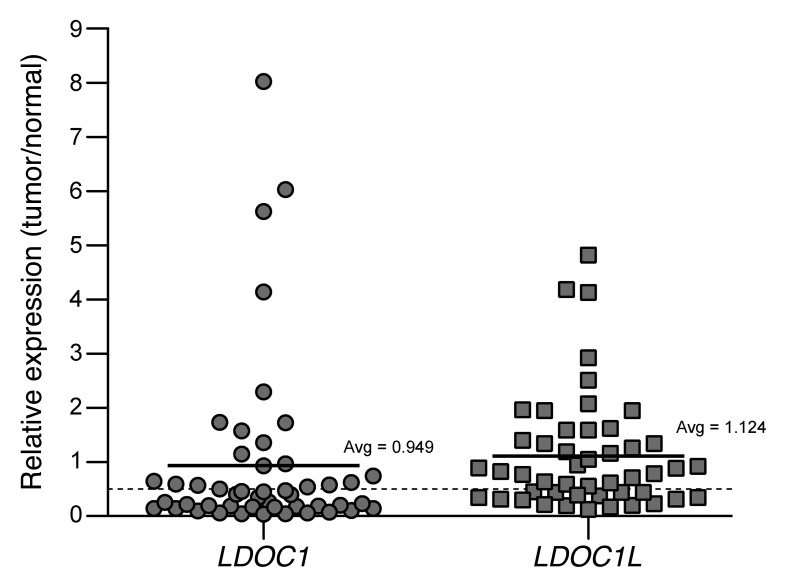
Significantly decreased expression of LDOC1 in tumors relative to normal tissue has been observed in pancreatic,34 gastric,34 prostate,35 and oral squamous cell36 cancer samples, suggesting that it functions as a tumor suppressor. Consistent with this hypothesis, we report here for the first time an observed downregulation of LDOC1 and family member LDOC1-like (LDOC1L) in a significant subset of human hepatocellular carcinoma (HCC). As shown in Figure 1, LDOC1 expression is decreased by greater than 50% in 29 of 48 (60%) HCC samples relative to adjacent tumor-free liver. Similarly, LDOC1L expression is decreased by more than half in 17 of 48 (35%) matched samples. Of the 17 HCC samples with > 50% LDOC1L downregulation, 15 additionally display > 50% decreased LDOC1 levels. These data are based on RNA expression levels in HCC and matched normal liver samples profiled by The Cancer Genome Atlas (TCGA) consortium.
Figure 1. Decreased expression of LDOC1 and LDOC1L in human hepatocellular carcinoma. RNA expression levels of LDOC1 and LDOC1L in human hepatocellular carcinoma (HCC) as reported by TCGA. Plotted values represent the ratio of expression in tumor tissue to that in matched normal liver samples (i.e., a value of one indicates no difference in expression between tumor and normal). The average ratio across all 48 sample pairs is indicated with a solid horizontal line and numerical value. A dashed line is included at 0.5 to distinguish samples with a decrease of more than 50% in tumor relative to normal liver.
PEG10, SETMAR, and GIN1 are additional examples of domesticated genes that are aberrantly expressed in human cancer. Overexpression of PEG10 has been observed in HCC,27 B-cell chronic lymphocytic leukemia,37 gallbladder adenocarcinoma,38 and Wilms’ tumor.39SETMAR, a gene derived from a Tc1/Mariner family DNA transposon, has been found to be highly expressed in acute myeloid and acute lymphoblastic leukemias (AML and ALL), as well as breast cancer cells.40Ty3/Gypsy-derived GIN1 expression has also been detected in a variety of tumor samples.41
Aside from the well-characterized example of TERT, the observed connections between domesticated TE gene products and cancer are almost entirely correlative in nature. Though suggestive of a putative driving role in carcinogenesis, further functional validation of these molecular aberrations in a meaningful in vivo context that accurately recapitulates human tumor formation is necessary before this conclusion can be made definitively.
Direct Evidence of Co-Opted Transposable Element Gene Product Involvement in Cancer
Some studies have been conducted to characterize the roles of TE-derived gene products identified as disrupted in cancer; however, the majority have been performed in cultured cancer cell lines. While this approach can lead to informative results and novel hypotheses, it should be complemented with in vivo functional validation experiments to maximize the utility of any discoveries. Without functional demonstration of a role in tumorigenesis, there is little justification for the pursuit of diagnostics or therapeutics based on a candidate gene. To date, few proposed links between co-opted TE gene products and tumorigenesis have been adequately validated.
Besides TERT, another example of the definitive involvement of domesticated gene products in promoting human cancer involves errors in normal recombination events within lymphoid cells. Recombination activating genes 1 and 2 (RAG1 and RAG2) are evolutionarily derived from ancient insertions of Transib DNA transposons.7 These genes mediate Variable, Diverse, and Joining (V(D)J) recombination in developing lymphocytes, a site-specific recombination process that generates a vast and diverse repertoire of T cell surface receptor and immunoglobulin molecules necessary for a functional immune system. As expected based on the derivation of RAG proteins from Class II TEs, this process involves the generation of double-strand DNA breaks in a way that is mechanistically similar to the “cut” component of “cut and paste” transposition. Normally, genomic modification induced by V(D)J recombination is limited to a defined window within the locus encoding the variable region of B- and T-cell receptors. With very low frequency, however, excised fragments can be reinserted within other regions of the genome, and this RAG-mediated transposition has been associated with lymphoid cancer development.42 An additional mechanism by which RAG proteins may contribute to human tumorigenesis is through the induction of chromosomal translocation events resulting from the utilization of cryptic recombination signal sequences outside of the V(D)J region.43,44
Recently, we described a study characterizing the involvement of Retrotransposon-like 1 (Rtl1), a Ty3/Gypsy family-derived gene, in HCC development.21 We initially identified the Rtl1 locus as a candidate HCC gene based on recurrent detection of its mutation in tumors generated by a Sleeping Beauty (SB) transposon-based forward genetic mutagenesis screen. While not expressed at detectable levels in normal adult mouse liver, we found Rtl1 to be highly overexpressed in SB-induced liver tumors. We also found low to undetectable expression of RTL1 in tumor-free human liver samples, with activated expression detected in ~30% of matched HCCs. Importantly, in vivo overexpression of Rtl1 in the livers of adult mice induced highly penetrant (86%) tumor formation, validating it as a driver of hepatocarcinogenesis. An independent insertional mutagenesis screen using lentiviral vectors also identified recurrent mutations causing overexpression of Rtl1 in mouse HCCs.45 This group also validated in vivo that forced expression of Rtl1 promotes liver tumorigenesis.
Concluding Remarks
It has been proposed that transposable element exaptation during the evolution of species has been driven largely by the utility of inherent TE functional elements in a variety of cellular processes.22,46 Indeed, those domesticated genes that have been functionally characterized are involved in diverse processes including growth factor signaling, cell cycle progression, proliferation, and survival. Given these associations, it is possible that the set of ~50 TE-derived human genes may have a higher propensity for involvement in cancer than other loci. Of the numerous domesticated TE gene products for which a link to human cancer has been identified, very few have been experimentally validated in vivo. Definitive roles for TERT, RAG1/RAG2, and Rtl1 in promoting tumorigenesis have been reported. It is likely that additional co-opted gene products are directly involved in driving cancer development; however, further experimentation is required to substantiate this hypothesis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/26693
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–82. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 3.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–25. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pritham EJ, Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci U S A. 2007;104:1895–900. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenblatt IM, Brink RA. Twin Mutations in Medium Variegated Pericarp Maize. Genetics. 1962;47:489–501. doi: 10.1093/genetics/47.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci U S A. 2007;104:8005–10. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinzelle L, Izsvák Z, Ivics Z. Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell Mol Life Sci. 2009;66:1073–93. doi: 10.1007/s00018-009-8376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ’bout retroelements in cancer. Semin Cancer Biol. 2010;20:200–10. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chénais B. Transposable elements and human cancer: a causal relationship? Biochim Biophys Acta. 2013;1835:28–35. doi: 10.1016/j.bbcan.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Kozeretska IA, Demydov SV, Ostapchenko LI. Mobile genetic elements and cancer. From mutations to gene therapy. Exp Oncol. 2011;33:198–205. [PubMed] [Google Scholar]
- 11.Rodić N, Burns KH. Long interspersed element-1 (LINE-1): passenger or driver in human neoplasms? PLoS Genet. 2013;9:e1003402. doi: 10.1371/journal.pgen.1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Florl AR, Steinhoff C, Müller M, Seifert HH, Hader C, Engers R, Ackermann R, Schulz WA. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer. 2004;91:985–94. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim MJ, White-Cross JA, Shen L, Issa JP, Rashid A. Hypomethylation of long interspersed nuclear element-1 in hepatocellular carcinomas. Mod Pathol. 2009;22:442–9. doi: 10.1038/modpathol.2008.203. [DOI] [PubMed] [Google Scholar]
- 15.Suter CM, Martin DI, Ward RL. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis. 2004;19:95–101. doi: 10.1007/s00384-003-0539-3. [DOI] [PubMed] [Google Scholar]
- 16.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–61. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Cancer Genome Atlas Research Network Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–71. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estécio MR, Gallegos J, Dekmezian M, Lu Y, Liang S, Issa JP. SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters. Mol Cancer Res. 2012;10:1332–42. doi: 10.1158/1541-7786.MCR-12-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruickshanks HA, Tufarelli C. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics. 2009;94:397–406. doi: 10.1016/j.ygeno.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riordan JD, Keng VW, Tschida BR, Scheetz TE, Bell JB, Podetz-Pedersen KM, Moser CD, Copeland NG, Jenkins NA, Roberts LR, et al. Identification of rtl1, a retrotransposon-derived imprinted gene, as a novel driver of hepatocarcinogenesis. PLoS Genet. 2013;9:e1003441. doi: 10.1371/journal.pgen.1003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curcio MJ, Belfort M. The beginning of the end: links between ancient retroelements and modern telomerases. Proc Natl Acad Sci U S A. 2007;104:9107–8. doi: 10.1073/pnas.0703224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Youngson NA, Kocialkowski S, Peel N, Ferguson-Smith AC. A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J Mol Evol. 2005;61:481–90. doi: 10.1007/s00239-004-0332-0. [DOI] [PubMed] [Google Scholar]
- 26.Tan KO, Tan KM, Chan SL, Yee KS, Bevort M, Ang KC, Yu VC. MAP-1, a novel proapoptotic protein containing a BH3-like motif that associates with Bax through its Bcl-2 homology domains. J Biol Chem. 2001;276:2802–7. doi: 10.1074/jbc.M008955200. [DOI] [PubMed] [Google Scholar]
- 27.Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, Nakajima Y, Yamaoka Y, Nakamura Y. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003;63:3043–8. [PubMed] [Google Scholar]
- 28.Nagasaki K, Schem C, von Kaisenberg C, Biallek M, Rösel F, Jonat W, Maass N. Leucine-zipper protein, LDOC1, inhibits NF-kappaB activation and sensitizes pancreatic cancer cells to apoptosis. Int J Cancer. 2003;105:454–8. doi: 10.1002/ijc.11122. [DOI] [PubMed] [Google Scholar]
- 29.Hayward A, Ghazal A, Andersson G, Andersson L, Jern P. ZBED evolution: repeated utilization of DNA transposons as regulators of diverse host functions. PLoS One. 2013;8:e59940. doi: 10.1371/journal.pone.0059940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markljung E, Jiang L, Jaffe JD, Mikkelsen TS, Wallerman O, Larhammar M, Zhang X, Wang L, Saenz-Vash V, Gnirke A, et al. ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 2009;7:e1000256. doi: 10.1371/journal.pbio.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28:20–47. doi: 10.1210/er.2006-0001. [DOI] [PubMed] [Google Scholar]
- 32.Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, Lazar V, Dessen P, Mantovani R, Aguilar L, et al. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood. 2007;109:584–94. doi: 10.1182/blood-2006-03-012013. [DOI] [PubMed] [Google Scholar]
- 33.Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 34.Nagasaki K, Manabe T, Hanzawa H, Maass N, Tsukada T, Yamaguchi K. Identification of a novel gene, LDOC1, down-regulated in cancer cell lines. Cancer Lett. 1999;140:227–34. doi: 10.1016/S0304-3835(99)00087-7. [DOI] [PubMed] [Google Scholar]
- 35.Camões MJ, Paulo P, Ribeiro FR, Barros-Silva JD, Almeida M, Costa VL, Cerveira N, Skotheim RI, Lothe RA, Henrique R, et al. Potential downstream target genes of aberrant ETS transcription factors are differentially affected in Ewing’s sarcoma and prostate carcinoma. PLoS One. 2012;7:e49819. doi: 10.1371/journal.pone.0049819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee CH, Wong TS, Chan JY, Lu SC, Lin P, Cheng AJ, Chen YJ, Chang JS, Hsiao SH, Leu YW, et al. Epigenetic regulation of the X-linked tumour suppressors BEX1 and LDOC1 in oral squamous cell carcinoma. J Pathol. 2013;230:298–309. doi: 10.1002/path.4173. [DOI] [PubMed] [Google Scholar]
- 37.Kainz B, Shehata M, Bilban M, Kienle D, Heintel D, Krömer-Holzinger E, Le T, Kröber A, Heller G, Schwarzinger I, et al. Overexpression of the paternally expressed gene 10 (PEG10) from the imprinted locus on chromosome 7q21 in high-risk B-cell chronic lymphocytic leukemia. Int J Cancer. 2007;121:1984–93. doi: 10.1002/ijc.22929. [DOI] [PubMed] [Google Scholar]
- 38.Liu DC, Yang ZL, Jiang S. Identification of PEG10 and TSG101 as carcinogenesis, progression, and poor-prognosis related biomarkers for gallbladder adenocarcinoma. Pathol Oncol Res. 2011;17:859–66. doi: 10.1007/s12253-011-9394-7. [DOI] [PubMed] [Google Scholar]
- 39.Dekel B, Metsuyanim S, Schmidt-Ott KM, Fridman E, Jacob-Hirsch J, Simon A, Pinthus J, Mor Y, Barasch J, Amariglio N, et al. Multiple imprinted and stemness genes provide a link between normal and tumor progenitor cells of the developing human kidney. Cancer Res. 2006;66:6040–9. doi: 10.1158/0008-5472.CAN-05-4528. [DOI] [PubMed] [Google Scholar]
- 40.Shaheen M, Williamson E, Nickoloff J, Lee SH, Hromas R. Metnase/SETMAR: a domesticated primate transposase that enhances DNA repair, replication, and decatenation. Genetica. 2010;138:559–66. doi: 10.1007/s10709-010-9452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lloréns C, Marín I. A mammalian gene evolved from the integrase domain of an LTR retrotransposon. Mol Biol Evol. 2001;18:1597–600. doi: 10.1093/oxfordjournals.molbev.a003947. [DOI] [PubMed] [Google Scholar]
- 42.Vanura K, Montpellier B, Le T, Spicuglia S, Navarro JM, Cabaud O, Roulland S, Vachez E, Prinz I, Ferrier P, et al. In vivo reinsertion of excised episomes by the V(D)J recombinase: a potential threat to genomic stability. PLoS Biol. 2007;5:e43. doi: 10.1371/journal.pbio.0050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marculescu R, Vanura K, Montpellier B, Roulland S, Le T, Navarro JM, Jäger U, McBlane F, Nadel B. Recombinase, chromosomal translocations and lymphoid neoplasia: targeting mistakes and repair failures. DNA Repair (Amst) 2006;5:1246–58. doi: 10.1016/j.dnarep.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 44.Zhang M, Swanson PC. V(D)J recombinase binding and cleavage of cryptic recombination signal sequences identified from lymphoid malignancies. J Biol Chem. 2008;283:6717–27. doi: 10.1074/jbc.M710301200. [DOI] [PubMed] [Google Scholar]
- 45.Ranzani M, Cesana D, Bartholomae CC, Sanvito F, Pala M, Benedicenti F, Gallina P, Sergi LS, Merella S, Bulfone A, et al. Lentiviral vector-based insertional mutagenesis identifies genes associated with liver cancer. Nat Methods. 2013;10:155–61. doi: 10.1038/nmeth.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]