Abstract
Monotremes (echidna and platypus) are egg-laying mammals. One of their most unique characteristic is that males have venom/crural glands that are seasonally active. Male platypuses produce venom during the breeding season, delivered via spurs, to aid in competition against other males. Echidnas are not able to erect their spurs, but a milky secretion is produced by the gland during the breeding season. The function and molecular composition of echidna venom is as yet unknown. Hence, we compared the deeply sequenced transcriptome of an in-season echidna crural gland to that of a platypus and searched for putative venom genes to provide clues into the function of echidna venom and the evolutionary history of monotreme venom. We found that the echidna venom gland transcriptome was markedly different from the platypus with no correlation between the top 50 most highly expressed genes. Four peptides found in the venom of the platypus were detected in the echidna transcriptome. However, these genes were not highly expressed in echidna, suggesting that they are the remnants of the evolutionary history of the ancestral venom gland. Gene ontology terms associated with the top 100 most highly expressed genes in echidna, showed functional terms associated with steroidal and fatty acid production, suggesting that echidna “venom” may play a role in scent communication during the breeding season. The loss of the ability to erect the spur and other unknown evolutionary forces acting in the echidna lineage resulted in the gradual decay of venom components and the evolution of a new role for the crural gland.
Introduction
Monotremes are egg-laying mammals and include the platypus (Ornithorhynchus anatinus), the short-beaked echidna (Tachyglossus aculeatus) (shown in Figure 1), and the long-beaked echidnas (Zaglossus sp). It has been long recognised that male platypuses have spurs on their hind legs that are connected to venom glands by venom ducts. The venom glands increase in size during the breeding season [1], [2]. During this period, males exhibit aggressive behaviours towards other males. In the platypus the venom is delivered into its victim through spurring events. The venom contains a complex mixture of peptides that cause intense pain and swelling. The recent sequencing of the platypus genome and the venom transcriptome has allowed us to more comprehensively characterise many of these peptides and determine their evolutionary origins. For instance, we showed that the key component of platypus venom, the defensin-like peptides (DLPs) evolved by gene duplication from the antimicrobial peptide β-defensin genes [3]. The function of DLPs is unknown, although they do not have any antimicrobial characteristics [4]. Other platypus venom components include C-type natriuretic peptides, which is known to cause histamine release [5], and calcium influx [6], as well as hyaluronidase, amide oxidase, protease inhibitor, proteins associated with mammalian stress response pathway and immune molecules, discovered by shot-gun proteomics [7].
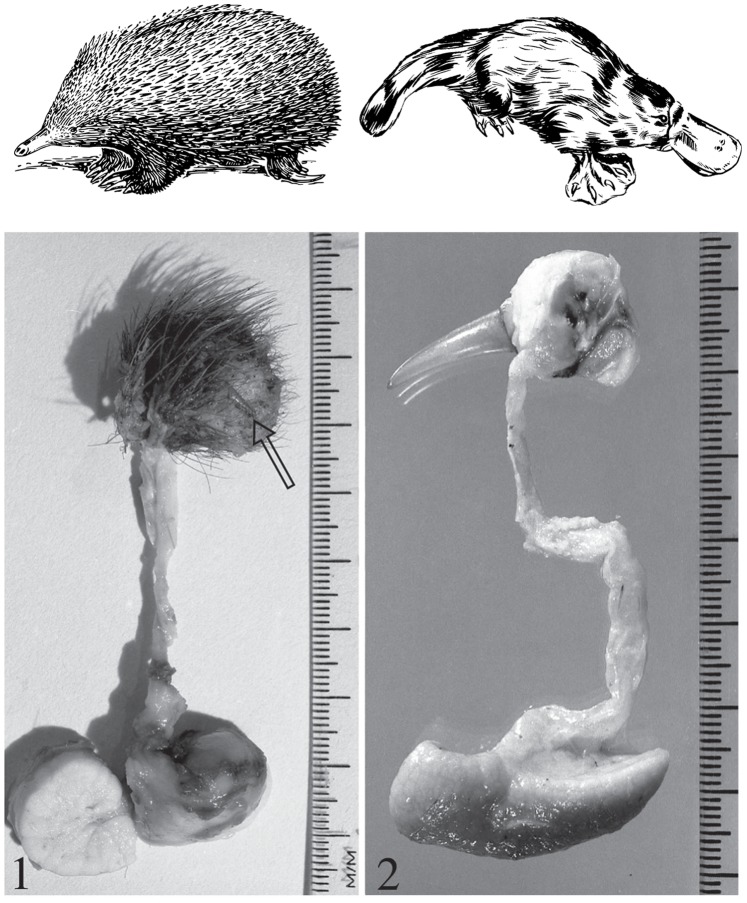
Figure 1. Monotremes and their crural glands.
1) An echidna (image taken from http://faunafemales.wikispaces.com/Echidna) and a dissected crural gland-spur apparatus from an adult male echidna. Photo was taken by William Krause and reprinted with permission from reference 2. Note that the main duct linking the crural gland and spur is shorter in the echidna than in the platypus. The spur (highlighted by an arrow) is not able to be erected in the echidna. 2) A platypus (image taken from http://faunafemales.wikispaces.com/Platypuses) and a dissected gland-spur apparatus taken from an adult male platypus. The original photograph was published by Krause in reference 2 (licence number 3184480567659).
Male echidnas also have spurs on their hind limbs. As in the platypus, the spur is attached to a gland, however, this spur is unable to be erected and used for spurring (Figure 1). The first description of the echidna crural gland [8] described the gland in its inactive phase. However, in 1968 Griffiths [9] noted that the gland increased in size during the breeding season. This gland produced a milky substance which was secreted during the breeding season. However, there have been no accounts of echidna envenomations and it is not known whether the secretions produced by this gland are venomous. Krause recently provided a detailed anatomical description of the echidna crural gland and confirmed cyclic activity of the gland [2]. He showed an increase in secretory granules in the secretory epithelium of the gland during the breeding season. In the platypus, exocytosis of this secretory epithelium was observed. However in the echidna, exocytosis was not observed. This suggests that there may be key differences in the function of the gland between the two species.
There is physiological, molecular and fossil evidence to suggest that the ancestor of platypus and echidnas was venomous. Firstly, both animals possess spurs, apparatuses which were likely developed to pierce the surface of the skin to inject toxic substances. Additionally, using a Bayesian molecular clock dating strategy, we have found that key toxin genes in the platypus evolved through gene duplication prior to the divergence of echidna and platypus lineages [3], which occurred in the early Cretaceous [10]. Monotremes possess a supernumerary bone, the os calcaris, which forms a plate-like bony base of the spur, and multituberculate mammals from the late Cretaceous possess an os calcaris which appears to be homologous to the monotreme structure [11]. Hurum et al argue from this that ancestral mammals were probably venomous, and that the venom gland apparatus of modern monotremes is a plesiomorphic character which has been lost in therian mammals.
To determine whether the echidna crural gland retains its ancestral venomous role, we compared an echidna in-season venom transcriptome with a platypus in-season venom transcriptome. We expected to see high levels of similarity between the venom gene repertoires of the two species. However, we found this not to be the case as platypus venom genes were not highly expressed in the echidna transcriptome.
Methods
Sequence Analysis
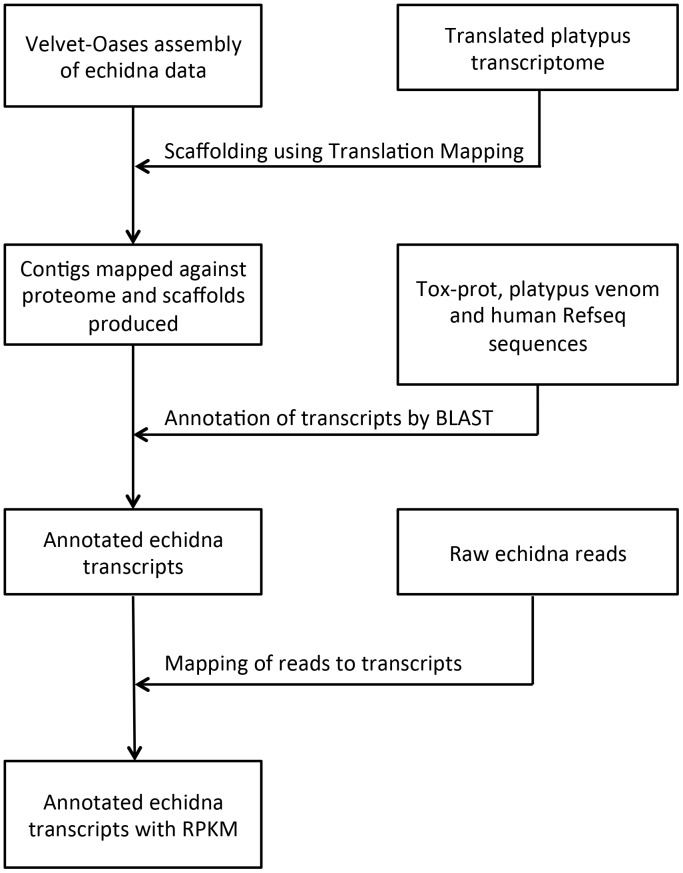
A schematic of the bioinformatics workflow is presented in Figure 2. A Tachyglossus aculeatus venom gland transcriptome was sequenced on an Illumina GAIIx instrument using previously describe methods for sample preparation and sequencing [7]. The venom gland was kindly provided by Frank Grutzner under University of Adelaide Animal Ethics Committee project number S-032-2008. The sequence reads have been deposited under accession number SRP027593 in the SRA database at NCBI. Quality filtered reads were assembled with the Velvet-Oases pipeline (kmer length = 31bp and –ins_length = 260) [12], [13]. To improve the transcriptome assembly, the Scaffolding using Translation Mapping (STM) strategy [14] was used to scaffold Oases contigs using Ensembl predicted proteins, an experimentally derived dataset of platypus venom proteins [5], [7], [15], and all human Refseq proteins as references. Data available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.4qq0v. BLAST [16] searches were performed against a database containing all Tox-Prot proteins [17], additional platypus venom toxins that have not been yet included in the Tox-Prot database [7] and human Refseq sequences using scaffolds derived from STM to identify possible homologies between echidna and other venom toxin proteins.
Figure 2. Schematic of bioinformatics workflow for the assembly, annotation and quantitation of echidna transcripts.
Expression Quantification and Comparison
Expression levels for each scaffold were derived by first mapping the raw sequencing reads to transcript scaffolds using Bowtie [18]. We then generated digital counts for each scaffold by counting reads that map back to transcripts. Multi-mapping reads were discarded. To compare the most highly expressed proteins between echidna and platypus venom, we ranked the expression values (normalized using Reads Per Kilobase per Million (RPKM) [19]) of the echidna transcripts, from highest to lowest, and compared it with similarly ranked list from a platypus transcriptomic dataset described in [20]. Echidna transcripts were annotated with respect to both human and platypus genes. The top 100 genes from both lists were compared and functional enrichment analysis was performed using Ontologizer [21] with orthologous human gene names as input. GoStat2 [22] was used to compare overrepresented ontology terms from highly expressed echidna genes versus the same in platypus. False discovery rate (FDR) [23] corrections were used for all functional term enrichment analyses. Ranked gene lists for comparisons only included genes that map to a homologous protein and did not include unnamed genes. If multiple scaffolds map to the same gene name, the name is only included once in the list.
Results and Discussion
The Oases assembly comprised of 72,549,524 base pairs with an average transcript length of 751 base pairs, a maximum transcript length of 25,037 base pairs and an N50 of 1,474 bases. To improve our assembly we mapped these contigs against the platypus proteome to scaffold contigs aligning to the same protein in the proteome. The platypus proteome was chosen because it is the most closely related species with a sequenced genome. Moreover, it facilitates the identification of the echidna homologs of platypus venom components that have been conserved between the two species. Our reference proteomes contain both Ensembl predicted proteins, derived from the translation of mRNAs of predicted genes in the platypus genome, as well as proteomic data that was generated using shot-gun proteomics [7].
Tox-Prot Comparison
We compared our scaffolds to sequences in a toxin database, Tox-Prot, to identify putative venom proteins based on homology. As venom proteins have typically evolved from body proteins, to distinguish between venom proteins and related body proteins we also searched against the human Refseq sequences. Sequences were considered as possible toxins if they were more similar to known toxins than a homologous protein from human. 30 transcripts from 15 genetic regions matched 12 unique toxin sequences with a lower BLAST e-value than to a human protein (Table 1). Five transcripts of proteins found in platypus venom were identified: CD55, amide oxidase, corticotrophin-releasing factor binding protein, a Kunitz domain-containing protease inhibitor and a cysteine-rich secretory protein (Table 2). Echidna sequences also shared similarity with a subunit stonefish toxin with haemolytic activities [24], snake nerve growth factors, protease inhibitors from snake and sea anemone venom with ion channel inhibitory activity (sea anemone) [25], and myotoxic and neurotoxic snake phospholipase A2s [26], [27].
Table 1. Echidna transcripts that display higher similarity to known toxins than human proteins.
| Toxin name | Transcript locus number | Expression rank (out of 91,300) |
| Stonustoxin subunit alpha (Acc: Q98989) | 726, 11716 | 24,455 |
| Platypus CD55/complement decay accelerating factor (Acc: K4PB89) | 2792, 50224 | 42,570 |
| Platypus amide oxidase (Acc: K4P911) | 8792, 18049 | 71,866 |
| Snake venom nerve growth factor 2 (Tropidechis carinatus) (Acc: Q3HXX7) | 11196 | 36,479 |
| Snake venom nerve growth factor (Naja naja)(Acc: P01140) | 11197 | 20,498 |
| Platypus corticotrophin-releasing factor binding protein (Acc: K4PLU9) | 11716 | 79,938 |
| Platypus Kunitz domain containing protease inhibitor | 16527 | 61,251 |
| Sea anemone Kunitz domain containing protease inhibitor (Acc: Q9TWG0) | 16966 | 41,527 |
| Snake mulgin-3 protease inhibitor (Pseudechis australis) (Acc: Q6ITB9) | 16966, 47678 | 35,771 |
| Platypus cysteine-rich secretory protein (Acc: K4P991) | 40579 | 51,113 |
| Snake venom phospholipase A2 (Echis carinatus)(Acc: P48650) | 43122 | 53,680 |
| Snake venom phospholipase A2 trimucrotoxin (Protobothrops mucrosquamatus ) (Acc: Q90W39) | 49881 | 83,368 |
The name of the known toxin together with the echidna transcript number and ranking of mRNA expression level are shown (a higher ranking indicates higher expression levels relative to other transcripts in the echidna transcriptome).
Table 2. Known platypus venom proteins and their homologs in the echidna transcriptome.
| Platypus toxin | Ancestral activity | Derived activity | Expressed in echidna gland |
| Defensin-likepeptides | Antimicrobial | Unknown | No |
| Nerve growthfactor | Regulation of nerve growth and differentiation; Hyperalgaesia | Possibly hyperalgaesia | No |
| C-type natriureticpeptide-like | Natriuretic, diuretic and vasodilatory functions associatedwith homeostasis and blood pressure control andregulates the growth and differentiation ofcartilaginous growth plate chondrocytes | Relaxant, oedema-producing andmast-cell degranulatingactivities | No |
| Hyaluonidase | Increase tissue permeability by lowering the viscosity of hyaluronan– roles in reproduction | Believed to accelerate the spread oftoxins and hemostatic factorsin toxin | No |
| Amide oxidase | Function in cell differentiation, growth, wound healing,detoxification, cell signaling | May cause platelet aggregation,assist in haemorrhage andcell death | Yes |
| Peptidoglycanrecognitionprotein-1 | Antimicrobial | Unknown | No |
| Serpin | Blood coagulation, complement activation, fibrinolysis,angiogenesis, inflammation, tumour suppression andhormone transport | Possible role in blood coagulationand hypertension | No |
| Kunitz-domaincontaining serineprotease inhibitor | Involved in hemostasis | Possible role in disrupting hemostasis | Yes |
| Nucleobindin | Stimulates autonomic nervous system activity, increasesblood pressure, induces fear in rats | May induce the fear response in victim | No |
| Differentiationfactor-15 | Regulates the inflammatory response and apoptosis | May function to induce pain | No |
| CXC-chemokine | Chemotactic, mediate cell growth and triggers aninflammatory response | Unknown | No |
| Complement decay-acceleration factor | Inhibits complement-mediated lysis | Unknown | Yes |
| L-D-amino-acid-residue isomerase | Unknown | Interconverts the second amino-acidresidue of venom peptides betweenthe L-form and D-form | No |
| Corticotropin-releasingfactor-binding protein | Linked to behavioural and psychological changes andnerve signalling | Suggested to increase submissive behaviourin victim linked to role of venomfor intraspecific competition | Yes |
We further examined the expression levels of these echidna transcripts to gain insights into their putative function. We found that all identified echidna protein-coding transcripts with homology to known toxins were lowly expressed in the transcriptome (Table 2). Pharmacologically characterized venom proteins have been found to be some of the most highly expressed genes in the platypus venom gland transcriptome [7], [20]. Using this logic the low expression of echidna toxin transcripts suggests that venom production is not a key role of this gland.
Analysis of most Highly Expressed Genes
To provide insight into the function of echidna venom, we examine the most highly expressed genes in the echidna venom gland. Quantification of expression levels involved the counting of raw reads that map to assembled scaffolds. The numbers were normalized with respect to the length of the scaffolds. This method allowed us to examine the most highly expressed genes and allowed comparison with similar experiments in platypus. None of the top 50 most highly expressed echidna genes were found in the top 50 of platypus. Similar results were obtained when human protein-coding transcripts instead of platypus ones were used to assemble the contigs. Comparison of the top 200 most abundant transcripts from the two datasets identified three proteins common to both echidna and platypus glands: a mitochondrial phosphate carrier protein (SLC25A3), mesencephalic astrocyte-derived neurotrophic factor (MANF) and a ribosomal protein (RPL9). The most highly expressed genes in the echidna venom gland are shown in Table 2. The most abundant venom components in platypus venom, venom defensin-like peptides and C-type natriuretic peptides [20], were not identified in echidna (Table 3).
Table 3. The most highly expressed proteins in the echidna transcriptome as annotated by platypus Ensembl proteins.
| Transcript reference number | Platypus Ensembl accession | Gene name | Description | RPKM |
| 968 | ENSOANG00000011369 | BCHE | butyrylcholinesterase | 604 |
| 8337 | ENSOANG00000005594 | SLC25A3 | solute carrier family 25 (mitochondrial carrier) | 441 |
| 1723 | ENSOANG00000009799 | CALR | calreticulin | 433 |
| 15082 | ENSOANG00000012964 | EEF1B2 | eukaryotic translation elongation factor 1 beta 2 | 421 |
| 1219 | ENSOANG00000013689 | – | – | 400 |
| 1766 | ENSOANG00000003422 | LTV1 | unknown function | 378 |
| 656 | ENSOANG00000011189 | RPS27A | ribosomal protein S27a | 376 |
| 6048 | ENSOANG00000000439 | – | – | 371 |
| 3603 | ENSOANG00000010996 | PDCD10 | programmed cell death 10 | 367 |
| 871 | ENSOANG00000008043 | – | – | 366 |
Gene Ontology Analysis
We compared gene ontology (GO) enrichment statistics between the expressed genes (RPKM>10) in the echidna and platypus datasets to examine and infer biological differences between the two species by restricting the analysis to the top 200 most highly expressed genes in both datasets. 182 out of the top 200 echidna genes were associated with 1,537 GO annotations that represented 580 unique terms. Compared to the platypus dataset, we detected five significantly enriched GO terms that included adenyl ribonucleotide binding, adenyl nucleotide binding, ATP binding, post-translational modification and plasma membrane (Table 4).
Table 4. Gene ontology (GO) terms that were enriched in the top 200 most highly expressed echidna genes compared to the platypus using GoStat2.
| GOterm | GOname | Echidna genesin group | Adjustedp-value |
| GO:0032559 | adenylribonucleotide binding | chuk; supv3l1; lars; igf1r; helz; tarsl2; tpr; atp2c1; dhx29; wee1;crebbp; smarca1; dhx36; dync1h1; hspa4; smchd1;dync1li1; tcn1; myo10; smarca5; taok1; atp2a2;abl1; ndufa10; trio; atp2b1; ddx50; top2b; afg3l2;hnrnpu; kif5b; ddx6 | 0.0159 |
| GO:0030554 | adenylnucleotide binding | chuk; supv3l1; lars; igf1r; helz; tarsl2; tpr; atp2c1;dhx29; wee1; crebbp; smarca1; dhx36; dync1h1;hspa4; smchd1; dync1li1; tcn1; myo10; smarca5;taok1; atp2a2; abl1; ndufa10; trio; atp2b1; ddx50;top2b; afg3l2; hnrnpu; kif5b; ddx6 | 0.0159 |
| GO:0005524 | ATPbinding | chuk; supv3l1; lars; igf1r; helz; tarsl2; tpr; atp2c1;dhx29; wee1; crebbp; smarca1; dhx36; dync1h1;hspa4; smchd1; dync1li1; tcn1; myo10;smarca5; taok1; atp2a2; abl1; ndufa10; trio;atp2b1; ddx50; top2b; afg3l2; hnrnpu; kif5b; ddx6 | 0.0159 |
| GO:0043687 | post-translationalprotein modification | chuk; mgrn1; cnot4; abl1; usp14; igf1r; trio; ppp2ca;dsp; rb1cc1; twf1; ube2d2; prdx4; march7; wee1;wwp2; crebbp; cul4b; ighm; huwe1; ppp1cc; ppp3ca;ccdc88c; march6; taok1 | 0.0257 |
| GO:0044459 | plasmamembrane part | clcn4; atp2a2; igf1r; slc12a7; mpp1; cltc; dsp;rab5a; cyfip1; enpp3; slc4a4; atp2b1; dtna;apc; calr; cp; cdh1; ighm; tjp1; itgb1; slc7a8; slc25a3 | 0.0843 |
Threshold at adjusted p-value<0.1.
We also performed functional term analyses on the top 200 most highly expressed genes in both echidna and platypus datasets. No gene ontology terms were found to be significant at p<0.05 after multiple testing correction (FDR) in independent tests for each species.
These analyses confirm that the key role of the echidna crural gland is not venom production. The life history of echidnas may point to the likely functional role of this gland. Echidnas are usually solitary, but during the breeding season (June to September) they are believed to use chemical cues to attract mates [1], [28]. Both sexes exude a pungent, musky odour possibly from the cloaca to advertise their reproductive status [29]. Male echidnas also exude a white milky substance from the base of their non-erectable spurs during the breeding season, while females, who lose their spurs before maturity, produce a small amount of solid secretion from the pits previously occupied by the spurs [28]. Several authors have speculated that the primary function of this white exudates acts as a chemical attractant during mating [2], [28].
The recent study by Harris et al. [28] investigated the composition of waxy secretions from the base of the spur using a mass spectrometry strategy to identify non-peptide compounds. The study revealed a variety of large molecules in produced by the tissue around the spur collar including sterols, fatty acids and methyl esters supporting their function in some form of scent communication. Consistent with this finding, stearoyl-CoA desaturase (SCD) – a protein involved in fatty acid biosynthesis was highly expressed in the echidna transcriptome. Other proteins involved in fatty acid metabolism including acyl-CoA dehydrogenase (ACADVL), acetyl-CoA carboxylase beta (ACACB), acyl-CoA synthetase long-chain family member 1 (ACSL1) as well as lecithin-cholesterol acetyltransferase (LCAT), sortilin-related receptor, L(DLR class) Repeats-containing protein (SORL1) and the Niemann-Pick disease protein (NPC1) are also all involved in sterol metabolic processes and expressed in the echidna crural gland transcriptome. Similar sterols and fatty acids are commonly found in scent gland secretions from other animals and can function to prolong the life of a scent mark (sterol esters) and encode complex informational content (long-chain fatty acids) (reviewed in [28]).
The major components of secretory granules in echidna crural glands are proteins [2] and it may be that the spur secretions possess additional, as yet, unknown roles. The most highly expressed gene in the echidna crural gland is most similar to butyrylcholinesterase, a serine hydrolase, whose physiological function in humans is still unclear, although it has been found to be an effective detoxification enzyme [30], [31]. Further research involving functional assays is required to determine the true biological roles of the most highly expressed proteins in echidna spur secretions.
Our results suggest that the primary role of the echidna crural gland is not to produce venom for offensive or defensive purposes. The majority of known platypus toxins were not detected in the transcriptome of the echidna crural gland. 30 echidna scaffolds matched 12 known toxins from a number of venomous species, but these were only lowly expressed in the gland and perhaps provide a remnant of the glands life history. Today the transcriptomes of the echidna and platypus crural glands have distinct expression profiles sharing few similarities in their most highly expressed genes. Should the echidna genome become available a review of the evolutionary remnants of venom genes can be conducted. The echidna gland appears to have evolved to play a role in chemical communication during the breeding season. Although the function of the crural gland has diverged significantly since the last common monotreme ancestor, each lineage continues to use the gland to aid reproduction.
Acknowledgments
We are grateful to ARC Fellow Frank Grutzner from the University of Adelaide for kindly sharing the echidna venom gland material and Dr Shuly Lim for extracting the RNA from the venom gland. Photographs of the echidna and platypus crural gland-spur apparatus were kindly shared by William J Krause from the University of Missouri.
Funding Statement
EW is supported by a University of Queensland Postdoctoral Fellowship. KB is supported by an ARC Future Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Griffiths M (1978) The Biology of the Monotremes. New York: Academic Press, Inc.
- 2. Krause WJ (2010) Morphological and Histochemical Observations on the Crural Gland-Spur Apparatus of the Echidna (Tachyglossus aculeatus) together with Comparative Observations on the Femoral Gland-Spur Apparatus of the Duckbilled Platypus (Ornithorhyncus anatinus) . Cells Tissues Organs 191: 336–354 10.1159/000252802 [DOI] [PubMed] [Google Scholar]
- 3. Whittington CM, Papenfuss AT, Bansal P, Torres AM, Wong ESW, et al. (2008) Defensins and the convergent evolution of platypus and reptile venom genes. Genome Res 18: 986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Whittington CM, Papenfuss AT, Kuchel PW, Belov K (2008) Expression patterns of platypus defensin and related venom genes across a range of tissue types reveal the possibility of broader functions for OvDLPs than previously suspected. Toxicon 52: 559–565 10.1016/j.toxicon.2008.07.002 [DOI] [PubMed] [Google Scholar]
- 5. De Plater GM, Martin RL, Milburn PJ (1998) A C-type natriuretic peptide from the venom of the platypus (Ornithorhynchus anatinus): structure and pharmacology. Comp Biochem Physiol C, Pharmacol Toxicol Endocrinol 120: 99–110. [DOI] [PubMed] [Google Scholar]
- 6. Kita M, Black DS, Ohno O, Yamada K, Kigoshi H, et al. (2009) Duck-billed platypus venom peptides induce Ca2+ influx in neuroblastoma cells. J Am Chem Soc 131: 18038–18039. [DOI] [PubMed] [Google Scholar]
- 7. Wong ESW, Morgenstern D, Mofiz E, Gombert S, Morris KM, et al. (2012) Proteomics and deep sequencing comparison of seasonally active venom glands in the platypus reveals novel venom peptides and distinct expression profiles. Mol Cell Proteomics 11: 1354–1364 10.1074/mcp.M112.017491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacKenzie WC, Owen WJ (1919) The Glandular System in Monotremes and Marsupials: Studies and Observations. Jenkin, Buxton. 79 p.
- 9.Griffiths M (1968) Echidnas. Pergamon Press. 308 p.
- 10. Rowe T, Rich TH, Vickers-Rich P, Springer M, Woodburne MO (2008) The oldest platypus and its bearing on divergence timing of the platypus and echidna clades. PNAS 105: 1238–1242 10.1073/pnas.0706385105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hurum J, Luo Z, Zofia K (2006) Were mammals originally venomous? Acta Palaeontologica Polonica 51: 1–11. [Google Scholar]
- 12.Schulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. Available: http://bioinformatics.oxfordjournals.org/content/early/2012/02/24/bioinformatics.bts094. Accessed 25 February 2013. [DOI] [PMC free article] [PubMed]
- 13. Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Surget-Groba Y, Montoya-Burgos JI (2010) Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res 20: 1432–1440 10.1101/gr.103846.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Torres AM, de Plater GM, Doverskog M, Birinyi-Strachan LC, Nicholson GM, et al. (2000) Defensin-like peptide-2 from platypus venom: member of a class of peptides with a distinct structural fold. Biochem J 348 Pt 3: 649–656. [PMC free article] [PubMed] [Google Scholar]
- 16. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jungo F, Bougueleret L, Xenarios I, Poux S (2012) The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 60: 551–557 10.1016/j.toxicon.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth 5: 621–628 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 20. Whittington CM, Papenfuss AT, Locke DP, Mardis ER, Wilson RK, et al. (2010) Novel venom gene discovery in the platypus. Genome Biol 11: R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bauer S, Grossmann S, Vingron M, Robinson PN (2008) Ontologizer 2.0–a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 24: 1650–1651. [DOI] [PubMed] [Google Scholar]
- 22. Beissbarth T, Speed TP (2004) GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20: 1464–1465 10.1093/bioinformatics/bth088 [DOI] [PubMed] [Google Scholar]
- 23. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 57: 289–300 10.2307/2346101 [DOI] [Google Scholar]
- 24. Chen D, Kini RM, Yuen R, Khoo HE (1997) Haemolytic activity of stonustoxin from stonefish (Synanceja horrida) venom: pore formation and the role of cationic amino acid residues. Biochem J 325 (Pt 3): 685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schweitz H, Bruhn T, Guillemare E, Moinier D, Lancelin JM, et al. (1995) Kalicludines and kaliseptine. Two different classes of sea anemone toxins for voltage sensitive K+ channels. J Biol Chem 270: 25121–25126. [DOI] [PubMed] [Google Scholar]
- 26. Tsai IH, Lu PJ, Wang YM, Ho CL, Liaw LL (1995) Molecular cloning and characterization of a neurotoxic phospholipase A2 from the venom of Taiwan habu (Trimeresurus mucrosquamatus). Biochem J 311 (Pt 3): 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou X, Tan T-C, Valiyaveettil S, Go ML, Kini RM, et al. (2008) Structural characterization of myotoxic ecarpholin S from Echis carinatus venom. Biophys J 95: 3366–3380 10.1529/biophysj.107.117747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris RL, Davies NW, Nicol SC (2012) Chemical Composition of Odorous Secretions in the Tasmanian Short-Beaked Echidna (Tachyglossus aculeatus setosus). Chem Senses. Available: http://chemse.oxfordjournals.org/content/early/2012/08/07/chemse.bjs066. Accessed 27 February 2013. [DOI] [PubMed]
- 29.Augee M, Gooden B, Musser A (1996) Echidna: Extraordinary egg-laying mammal. Collingwood: CSIRO Publishing.
- 30. Masson P, Lockridge O (2010) Butyrylcholinesterase for protection from organophosphorus poisons; catalytic complexities and hysteretic behavior. Arch Biochem Biophys 494: 107 10.1016/j.abb.2009.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chilukuri N, Duysen EG, Parikh K, diTargiani R, Doctor BP, et al. (2009) Adenovirus-transduced human butyrylcholinesterase in mouse blood functions as a bioscavenger of chemical warfare nerve agents. Mol Pharmacol 76: 612–617 10.1124/mol.109.055665 [DOI] [PubMed] [Google Scholar]