Abstract
Populus species are currently being domesticated through intensive time- and resource-dependent programs for utilization in phytoremediation, wood and paper products, and conversion to biofuels. Poplar leaf rust disease can greatly reduce wood volume. Genetic resistance is effective in reducing economic losses but major resistance loci have been race-specific and can be readily defeated by the pathogen. Developing durable disease resistance requires the identification of non-race-specific loci. In the presented study, area under the disease progress curve was calculated from natural infection of Melampsora ×columbiana in three consecutive years. Association analysis was performed using 412 P. trichocarpa clones genotyped with 29,355 SNPs covering 3,543 genes. We found 40 SNPs within 26 unique genes significantly associated (permutated P<0.05) with poplar rust severity. Moreover, two SNPs were repeated in all three years suggesting non-race-specificity and three additional SNPs were differentially expressed in other poplar rust interactions. These five SNPs were found in genes that have orthologs in Arabidopsis with functionality in pathogen induced transcriptome reprogramming, Ca2+/calmodulin and salicylic acid signaling, and tolerance to reactive oxygen species. The additive effect of non-R gene functional variants may constitute high levels of durable poplar leaf rust resistance. Therefore, these findings are of significance for speeding the genetic improvement of this long-lived, economically important organism.
Introduction
Rust fungi cause some of the most important crop and tree diseases worldwide. In Populus species (poplar trees), leaf rust disease is caused by several species of Melampsora. Severe poplar leaf rust infections decrease photosynthetic capacity, reduce biomass, and increase susceptibility to additional pathogens [1]. Reductions in dry weight and fiber volume of P. deltoides x P. balsamifera ‘Northwest’, have been estimated at 57% and 65%, respectively [2]. In North America, hybridization of rust species, M. occidentalis and M. medusae, has produced a new rust pathogen, M. ×columbiana. This hybrid rust has demonstrated high pathogenic diversity. In an initial collection 13 pathotypes (race with unique virulence to specific hosts) were identified. Furthermore, host resistance loci to pathotypes Mxc3 and Mxc4 map to unique chromosomes in poplar and are race-specific [3], [4].
Rust virulence and poplar resistance interact in a classical gene-for-gene model, where host R gene recognition of a pathogen avirulence gene is necessary for resistance [5].The pathogenic diversity of Melampsora is facilitated by obligate sexual reproduction on alternative hosts (Larix species) during winter and migration of wind-dispersed spores. Previous studies of M. medusae populations suggest that inoculum source and genetic composition vary from year to year outside the range of alternative host sympatry [6]. This is also evident in M. ×columbiana populations in the coastal Pacific Northwest, where Larix spp. are not native and rust pathotypes varied across years at single locations [4].
Poplar leaf rust resistance has been extensively studied using the European counterpart, M. larici-populina [7]–[11]. Bi-parental linkage mapping has identified major resistance loci that map to nucleotide binding site – leucine rich repeat (NBS-LRR) R genes on chromosome 19 [7]–[9], [11]. During the mid-1990s, P. trichocarpa × P. deltoides ‘Beaupré’ was bred for rust resistance and then exclusively planted in European plantations. Yet, a rust epidemic occurred after the R gene was defeated by the evolution of virulence factor 7 [1], [12]. In wheat, NBS-LRRs have also conferred race-specific resistance to stem rust; however, association mapping has recently been applied to validate non-R gene non-race-specific resistance loci [13].
Association mapping in plants has the capability to precisely identify a greater number of functional variants which explain smaller proportions of the phenotypic variance than traditional linkage analysis [14]. The development of genomic resources necessary for association analysis in Populus species have been facilitated by its value to wood and paper industries and its potential as a biofuels feedstock. With large unstructured populations and wide phenotypic diversity, wood traits in P. trichocarpa have begun to be studied via association analysis [15]. Here, we report the first multi-year association analysis of poplar leaf rust resistance SNPs in P. trichocarpa.
Methods
Plant Material and Phenotyping
The ramets of 456 genotypes of native black cottonwood were collected from the common garden of British Columbia Ministry of Forest, Lands and Natural Resource Operations (MOFLNRO) at Surrey, BC in March of 2008. MOFLNRO collected these native poplar genotypes from 136 provenances from 44.00 degrees north latitude (Oregon USA) to 59.34 degrees north latitude of (Alaska USA) under the authority of Dr. Alvin Yanchuk, Technical Advisor for the Tree Improvement Branch (Victoria, BC Canada). In June of 2008, four replicates of each genotype were planted in a common garden situated at the University of British Columbia in Vancouver, Canada (49.27 degree north latitude). Replicates were planted in a completely randomized design with 1.5×1.5 meter spacing. In 2008 and 2009, fields were watered daily by rainfall or drip irrigation. No fertility or soil amendments were applied at any time. In 2009, 2010, and 2011 natural infection from Melampsora ×columbiana was scored visually on the basis of pustules present on the leaves. Ratings were taken on a 0–4 scale where (0) = no pustules, (1) = less than five pustules per leaf on less than five leaves, (2) = less than five pustules per leaf on more than five leaves, (3) = more than five pustules per leaf on more than five leaves, (4) = more than five pustules on all leaves. Ratings were taken once a week for 11 consecutive weeks (Julian Day 200–279). Ratings were used to calculated area under the disease curve (AUDPC) using the following equation [16]:
| (1) |
where, Y i is the disease rating at the i th observation, Xi is the time at the i th observation, and n is the total number of observations. Genotypes with missing scores from all four replicates were removed from the study and reduced the population to 412 genotypes. Date of bud set was taken concurrently with disease ratings and varied widely (data not shown). Host age can interact with disease resistance [17] thus, AUDPC scores were adjusted for bud set using ANCOVA in Minitab v16 (Minitab® Statistical Software). Finally, all adjusted AUDPC scores were transformed for normalization using the following equation:
| (2) |
Data normality was tested using Lilliefors (Kolomorov-Smirnov) test in R package “nortest”.
SNP Genotyping
We genotyped a total of 456 clones of the P. trichocarpa population using an Illumina Infinium® genotyping array with a set of 34,131 SNPs in 3,543 candidate genes. The 34K SNP genotyping array we employed was designed to take linkage disequilibrium (LD) into account. SNPs in any given candidate gene represented on the array were chosen to “tag” as many other target SNPs as possible (based on LD calculations), with a SNP density of approximately 1–2 SNPs per candidate gene kb [18]. We eliminated SNPs with: i) minor allele frequency below 0.05, ii) more than 10% missing values, and iii) an Illumina GeneTrain score below 0.5. These three selection criteria reduced the number of SNPs to 29,355. These remaining SNPs were used in all subsequent analyses.
Population Structure
To fit population structure effect, we used a subset of 899 randomly selected SNPs distributed across all 19 chromosomes with complete information (i.e., no missing data) and meeting HWE expectation (tested using “HWChisq” function implemented in “HardyWeinberg” R package [19]. Population fit was done by performing principal component analysis (PCA) in TASSEL [20] and 263 principal components accounting for 90% of the total variance in the SNP data were retained for further determination of their impact. Principal components affecting AUDPC in each year were selected through regression in a stepwise manner using the function “stepwise” implemented in R package “Rcmdr” with “backward” direction and Bayesian information criterion “BIC” as the selection criterion.
Kinship matrix was calculated using the above mentioned 899 SNPs in SPAGeDi [21]. All negative values were set as zero and diagonal elements were set to one [22]. Q matrix and FST were calculated using GENELAND software with a subset of 200 SNPs randomly selected from the 899 used in PCA. Pearson’s product moment correlations for latitude, AUDPC, Q matrix, and PC1 were calculated in R package “Rcmdr”.
Association Analysis
We applied a two-step approach to analyze SNP-AUDPC association [23]. First, a simple linear regression with AUDPC in each year and every SNP was used to pre-select SNPs with the following equation:
| (3) |
where, Y is the observations vector, β is the fixed effects of population mean and SNP effect vector, X is the incidence matrix assigning fixed effects to observations, and e is the residual effect. SNP genotypes were coded as 0, 1, and 2 for common allele homozygote, heterozygote, and rare allele homozygote, respectively. SNPs with significant effect (P<0.001) were included in the second analysis.
Finally, the selected principal components were included in a regression model along the screened SNP individually as follows:
| (4) |
where, Y is vector of measurements, is the population mean, is the SNP effect, the term represents the effect of selected principal components resulting from backward stepwise selection procedure (above), and e is the residual effect. Association analysis was performed in TASSEL [20] employing the GLM procedure. Permutated P value was calculated in TASSEL with 1,000 permutations. The correction for multiple testing was applied at α <0.05. Cumulative effect of SNPs within each year was calculated using method described by Ingvarsson et al. [24]. Pairwise LD plots was calculated using the “LDheatmap” function implemented in the LDheatmap R package [25].
Variance Components and Spatial Analysis
A REML-mixed linear model was used to estimate variance components in SAS and test the effect of clone, year, and clone ×year interaction as follows:
| (5) |
where, Z is vector of measurements (AUDPC) in the ith year, of the jth clone, in the kth ramet, µ is the population mean, Y i = effect of the ith year, Cj = effect of the jth clone, Y i×Cj = effect of the ith year × the jth clone interaction, e is the residual effect. Broad-sense heritability in each year was calculated using methods described in Lynch & Walsh [26].
In each year the mixed linear model implemented in ASReml [27] was used to plot the residuals to their location in the field as follows:
| (6) |
where Y is vector of measurements, β and µ are vectors of fixed (intercept and population) and random (genotypic values) effects assuming U∼N(0, ) and Var(µ) = I, e is vector of residual effects assuming E∼N(0, ) and Var(e) = I where I is identity matrix containing 1′s on diagonal and 0′s at diagonal-off elements, and X and Z are index matrices assigning both fixed and random effects to measurements. Q matrix from GENELAND analysis was used to fit population structure effect.
Results
Disease Analysis
To identify SNPs that confer non-race-specific resistance to M. ×columbiana, we performed association analysis on 412 unrelated P. trichocarpa genotypes from a North American provenance trial ranging from Alaska to Oregon. Poplar leaf rust severity was scored (0–4 worst) on natural infection in a replicated (ramets = 4) common garden experiment where ratings were taken over 11 continuous weeks (Julian days 200 – 279) in each of three consecutive years. Rust severity ranged from zero rust pustules after 11 weeks (complete resistance) to 100% of the leaves covered after four weeks (Julian day 229).
Area under the disease progress curve (AUDPC) was calculated from the disease ratings over time. Previously, AUDPC had the highest broad-sense heritability (H 2 = 0.69) among four other measures of Melampsora resistance in growth chamber assays with artificial inoculations [28]. We estimated broad-sense heritability for AUDPC at H 2 = 0.72, 0.65, and 0.58 for each of the three years, respectively. Analysis of variance indicated that clone and clone × year interaction were significant, while year was not significant (Table 1). Spatial analysis of the experimental plot also demonstrated a change in the pattern of infection across years (Fig. S1).
Table 1. Analysis of variance (ANOVA) testing the effect of clone, year, and clone × year interaction of AUDPC.
| Sources of Variance | Estimate | St. Error | Z value | Pr>Z |
| Clone | 44.6077 | 3.0644 | 14.56 | <0.0001 |
| Year | 27.9939 | 28.0179 | 1 | 0.1589 |
| Clone x Year | 5.1717 | 0.5507 | 9.39 | <0.0001 |
| Residual | 27.2343 | 0.5616 | 48.5 | <0.0001 |
Population Structure and Association Analysis
Analysis of population structure was tested using GENELAND software [29]. An uncorrelated allele frequency model did not detect any population structure while a correlated allele frequency model revealed three sub-populations with weak systemic structure (FST <0.0227) consistent with our previous results [18], [30]. Components of the population structure also displayed strong correlation to AUDPC and latitude (Table 2; Fig. S2). Due to this correlation of phenotype and population structure, the trait-SNP simple model (simple linear regression) produced 941, 1220, and 1093 significant associations at P<1.72×10−6 in each of the three years, respectively (data not shown) and a prodigious inflation of type-1 error (Fig. 1).
Table 2. Correlation coefficients of the population structure, latitude, and AUDPC in each year.
| Latitude | Q1 | Q2 | Q3 | PC1 | AUDPC09 | AUDPC10 | |
| Q1 | −0.37016 | ||||||
| <0.0001a | |||||||
| Q2 | −0.08427 | −0.74541 | |||||
| 0.0876 | <0.0001 | ||||||
| Q3 | 0.65247 | −0.49093 | −0.2148 | ||||
| <0.0001 | <0.0001 | <0.0001 | |||||
| PC1 | −0.62464 | 0.65285 | −0.17031 | −0.73393 | |||
| <0.0001 | <0.0001 | 0.0005 | <0.0001 | ||||
| AUDPC09 | 0.73339 | −0.42216 | 0.00914 | 0.60658 | −0.67059 | ||
| <0.0001 | <0.0001 | 0.8533 | <0.0001 | <0.0001 | |||
| AUDPC10 | 0.67009 | −0.53882 | 0.05547 | 0.71694 | −0.73558 | 0.77123 | |
| <0.0001 | <0.0001 | 0.2613 | <0.0001 | <0.0001 | <0.0001 | ||
| AUDPC11 | 0.73398 | −0.50088 | 0.09273 | 0.61266 | −0.69105 | 0.79764 | 0.8213 |
| <0.0001 | <0.0001 | 0.06 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
P value of Pearson’s correlation coefficient (n = 412).
Q1, Q2, Q3 represent the three sub-populations revealed by GENELAND analysis where pairwise FST were calculated. Q1×Q2 = 0.0118, Q1×Q3 = 0.0226, and Q2×Q3 = 0.02. PC1 represents the first principal component used to correct for population stratification.
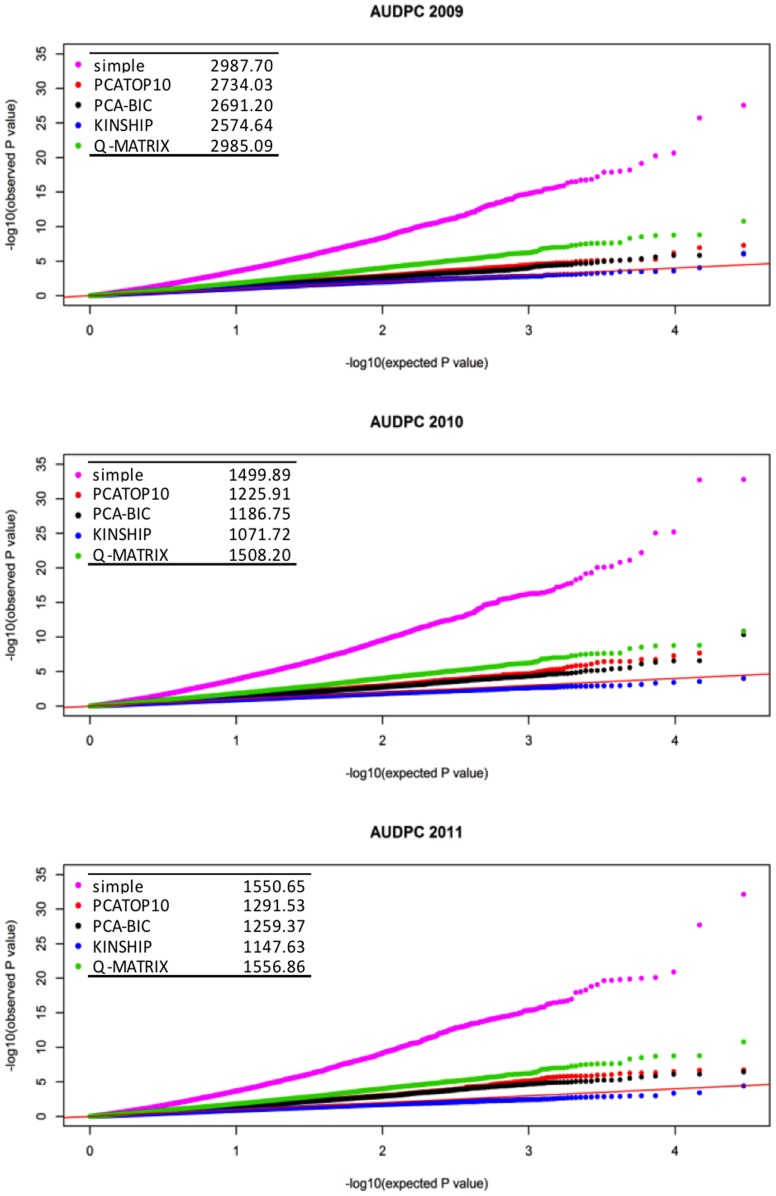
Figure 1. Quantile-quantile plots of expected and observed P values evaluating the type-1 error in a simple model (simple), the top ten principal components (PCA-TOP10), BIC selected PCs (PCA-BIC), the K model (KINSHIP), and the Q model (Q MATRIX) with goodness of fit test using Bayesian Information Criterion for 2009, 2010, and 2011 (top to bottom).
During association analysis, we tested the effects of Q matrix, PCA-based model that uses the first 10 PC’s (PCA-TOP10) [31] PCA-based model with PCs that affect AUDPC selected through a backwards step-wise regression (PCA-BIC), and kinship matrix [32]. In a goodness of fit test, kinship matrix had the lowest Bayesian Information Criterion (BIC) value; however in the 2010 and 2011 quantile-quantile plots (q-q plots), kinship matrix eliminated all of the expected associations. The PCA-BIC displayed the next best model fit without overcorrecting for structure (Fig. 1).
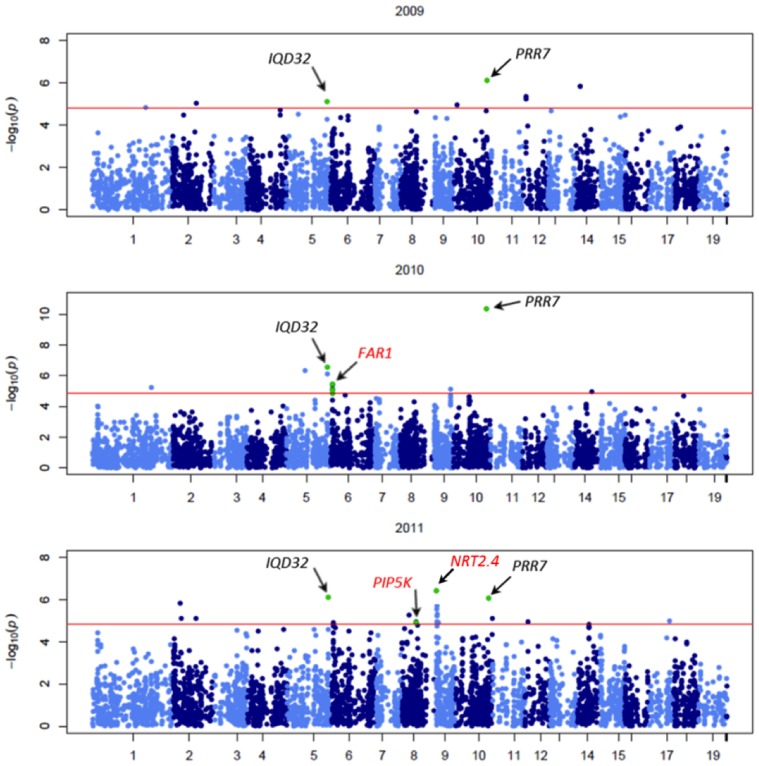
Association analysis was performed with 29,355 SNPs covering 3,543 genes in a linear regression with AUDPC using the PCA-BIC model in each year. In 2009, 2010, and 2011 a total of three, four, and three SNPs were significantly associated at P<1.46×10−6. Bonferroni correction for multiple testing was applied at α = 0.05/29,355 where P = 1.70×10−6. Single SNPs located in PSEUDO RESPONSE REGULATOR7 (PRR7) and IQ-DOMAIN32 (IQD32) were repeated in three and two years respectively. In addition, two SNPs in high linkage disequilibrium (LD) (R2>0.8) located in the intergenic region between NITRATE TRANSPORTER2.1 (NRT2.1) and NITRATE TRANSPORTER2.4 (NRT2.4) were also significant (Table 3).
Table 3. SNPs associated with AUDPC in 2009, 2010, and 2011.
| Year | Scaffold | SNP | Gene Model | SNP location | Arabidopsis Best hit | Annotated Gene | P value | Permutated P value | Marker R2 |
| 2009 | 10 | 19215715 | POPTR_0010s22230 | exon | At5g02810.1 | PRR7 (PSEUDO-RESPONSE REGULATOR 7) | 7.33414E-07 | 0.003 | 0.0289 |
| 14 | 3245282 | POPTR_0014s04070 | intron | At1g19330.1 | unknown protein | 1.45407E-06 | 0.003 | 0.0271 | |
| 14 | 3245414 | POPTR_0014s04070 | intron | At1g19330.1 | unknown protein | 1.45407E-06 | 0.003 | 0.0271 | |
| 12 | 1814218 | POPTR_0012s02170 | intron | At3g49220.1 | pectinesterase family protein | 4.44885E-06 | 0.012 | 0.0250 | |
| 12 | 1814164 | POPTR_0012s02170 | intron | At3g49220.1 | pectinesterase family protein | 5.76602E-06 | 0.018 | 0.0244 | |
| 5 | 23949327 | POPTR_0005s25750 | intergenic | At1g19870.1 | IQD32 (IQ-domain 32) | 7.86988E-06 | 0.025 | 0.0238 | |
| 2 | 13904004 | POPTR_0002s18010 | intergenic | no arabidopsis blast hit | unknown protein | 9.02706E-06 | 0.030 | 0.0236 | |
| 10 | 1844266 | POPTR_0010s01650 | intergenic | At4g15900.1 | PRL1 (PLEIOTROPIC REGULATORY LOCUS 1) | 1.06875E-05 | 0.033 | 0.0253 | |
| 1 | 31118784 | POPTR_0001s32810 | intron | At4g13980.1 | AtHSFA5; DNA binding/transcription factor | 1.40977E-05 | 0.037 | 0.0226 | |
| 2010 | 10 | 19215715 | POPTR_0010s22230 | exon | At5g02810.1 | PRR7 (PSEUDO-RESPONSE REGULATOR 7) | 4.49274E-11 | 0.001 | 0.0476 |
| 5 | 23949327 | POPTR_0005s25750 | intergenic | At1g19870.1 | IQD32 (IQ-domain 32) | 2.71307E-07 | 0.003 | 0.0302 | |
| 5 | 10782555 | POPTR_0005s13780 | intergenic | At2g23760.1 | BLH4 (BEL1-LIKE HOMEODOMAIN 4) | 4.67804E-07 | 0.004 | 0.0292 | |
| 5 | 23952538 | POPTR_0005s25750 | exon | At1g19870.1 | IQD32 (IQ-domain 32) | 7.99477E-07 | 0.004 | 0.0281 | |
| 6 | 1402770 | POPTR_0006s02140 | intergenic | At4g15090.1 | FAR1 (FAR-RED IMPAIRED RESPONSE 1) | 3.6489E-06 | 0.016 | 0.0253 | |
| 6 | 1397889 | POPTR_0006s02140 | 3′-UTR | At4g15090.1 | FAR1 (FAR-RED IMPAIRED RESPONSE 1) | 4.07883E-06 | 0.017 | 0.0250 | |
| 1 | 34721616 | POPTR_0001s36210 | exon | At3g27330.1 | zinc finger (C3HC4-type RING finger) family protein | 6.1898E-06 | 0.024 | 0.0244 | |
| 6 | 1399289 | POPTR_0006s02140 | exon | At4g15090.1 | FAR1 (FAR-RED IMPAIRED RESPONSE 1) | 7.28474E-06 | 0.029 | 0.0238 | |
| 9 | 10970414 | POPTR_0009s13880 | intergenic | At4g02390.1 | APP (ARABIDOPSIS POLY(ADP-RIBOSE) POLYMERASE) | 7.59421E-06 | 0.029 | 0.0239 | |
| 6 | 1402469 | POPTR_0006s02140 | intergenic | At4g15090.1 | FAR1 (FAR-RED IMPAIRED RESPONSE 1) | 8.05648E-06 | 0.031 | 0.0242 | |
| 14 | 10716774 | POPTR_0014s14650 | intron | At5g48560.1 | basic helix-loop-helix (bHLH) family protein | 1.1107E-05 | 0.041 | 0.0230 | |
| 2011 | 9 | 1676227 | POPTR_0009s01420 | intergenic | At1g08090.1 | PtNRT2.1 (NITRATE TRANSPORTER 2.1) | 3.52242E-07 | 0.003 | 0.0342 |
| 5 | 23949327 | POPTR_0005s25750 | intergenic | At1g19870.1 | IQD32 (IQ-domain 32) | 7.88901E-07 | 0.003 | 0.0308 | |
| 10 | 19215715 | POPTR_0010s22230 | exon | At5g02810.1 | PRR7 (PSEUDO-RESPONSE REGULATOR 7) | 8.34729E-07 | 0.003 | 0.0313 | |
| 9 | 1678826 | POPTR_0009s01420 | intergenic | At1g08090.1 | PtNRT2.1 (NITRATE TRANSPORTER 2.1) | 1.9236E-06 | 0.007 | 0.0308 | |
| 9 | 1606213 | POPTR_0009s01330 | exon | At3g45040.1 | phosphatidate cytidylyltransferase family protein | 2.96357E-06 | 0.008 | 0.0279 | |
| 9 | 1857142 | POPTR_0009s01490 | intergenic | At5g60720.1 | unknown protein | 4.83027E-06 | 0.014 | 0.0270 | |
| 8 | 4165833 | POPTR_0008s06920 | intergenic | At5g05610.1 | AL1 (ALFIN-LIKE 1) | 5.71705E-06 | 0.017 | 0.0266 | |
| 9 | 1676590 | POPTR_0009s01420 | 3′-UTR | At1g08090.1 | PtNRT2.1 (NITRATE TRANSPORTER 2.1) | 5.74407E-06 | 0.017 | 0.0266 | |
| 143 | 2955 | POPTR_0143s00200 | exon | At5g60770.1 | PtNRT2.4 (NITRATE TRANSPORTER 2.1) | 5.74407E-06 | 0.017 | 0.0266 | |
| 2 | 13131622 | POPTR_0002s17360 | intergenic | no arabidopsis blast hit | unknown protein | 7.73544E-06 | 0.022 | 0.0259 | |
| 2 | 4627286 | POPTR_0002s06880 | intron | At1g76900.1 | AtTLP1 (TUBBY LIKE PROTEIN 1) | 8.13072E-06 | 0.025 | 0.0259 | |
| 10 | 21451968 | POPTR_0010s26100 | 5′-UTR | At3g54540.1 | AtGCN4; transporter | 8.23563E-06 | 0.025 | 0.0261 | |
| 17 | 12392905 | POPTR_0017s12210 | 3′-UTR | At5g61430.1 | ANAC100 (ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN 100) | 1.02655E-05 | 0.035 | 0.0254 | |
| 9 | 1679212 | POPTR_0009s01420 | intergenic | At1g08090.1 | PtNRT2.1 (NITRATE TRANSPORTER 2.1) | 1.09575E-05 | 0.039 | 0.0252 | |
| 12 | 1811250 | POPTR_0012s02170 | intergenic | At3g49220.1 | pectinesterase family protein | 1.10236E-05 | 0.039 | 0.0253 | |
| 9 | 1679805 | POPTR_0009s01420 | intergenic | At1g08090.1 | PtNRT2.1 (NITRATE TRANSPORTER 2.1) | 1.16257E-05 | 0.040 | 0.0250 | |
| 8 | 8157244 | POPTR_0008s12610 | exon | At5g17350.1 | unknown protein | 1.1642E-05 | 0.040 | 0.0251 | |
| 6 | 1405713 | POPTR_0006s02150 | exon | At3g22170.1 | FHY3 (FAR-RED ELONGATED HYPOCOTYLS 3) | 1.22801E-05 | 0.041 | 0.0250 | |
| 9 | 2563210 | POPTR_0009s01990 | intron | At5g60690.1 | REV (REVOLUTA) | 1.28046E-05 | 0.043 | 0.0249 | |
| 8 | 8261867 | POPTR_0008s12780 | exon | At1g71010.1 | phosphatidylinositol-4-phosphate 5-kinase family protein | 1.31391E-05 | 0.046 | 0.0248 |
Permutated P value of α = 0.05 was used as the threshold for multiple testing corrections after SNP pre-selection.
Bonferroni correction threshold was applied at P = 1.70×10−6 without SNP pre-selection.
Arabidopsis best hit and annotated function is derived from BLAST results of poplar gene models in POPGENIE. R2 value explains the effect of each SNP on the phenotype.
In the final analysis, we used a simple linear regression with AUDPC in each year to preselect SNPs and reduce the constraint of multiple testing correction. Significant SNPs at P<0.001 were selected and re-run in a linear regression with the PCA-BIC model to correct for population structure. In 2009, 2010, and 2011 a total of 9, 11, and 20 SNPs achieved experiment-wide significance at P<1.45×10−5, P<1.15×10−5, and P<1.35×10−5, respectively (Fig. 2; Table 3). Permutated p-value was used as correction for multiple testing at a threshold α <0.05. Individually, these SNPs explain 2.2–4.7% of the phenotypic variance. The cumulative effects of independent SNP associations explain 12.1, 14.2, and 19.6% of the phenotypic variance within each year, respectively (Table 3).
Figure 2. Manhattan plot of the results from association analysis for AUDPC in 2009, 2010, and 2011 (from top to bottom).
The red line represents the P value (P<1.45×10−5, P<1.15×10−5, and P<1.35×10−5 in 2009, 2010, and 2011, respectively) corresponding to permutated P of α = 0.05 as the threshold for multiple testing corrections. SNPs repeated in time are highlighted in green and identified by gene name in black. SNPs within genes showing expression profile changes in response to M. larici-populina are highlighted in green and identified by gene name in red.
Single SNPs located in PRR7 and IQD32 were repeated in all three years (Table 1). In addition, SNPs in FAR-RED IMPAIRED RESPONSE1 (FAR1), a phosphatidylinositol 4-phosphate 5-kinase (POPTR_0008s12780; PIPK5), and NITRATE TRANSPORTER2.4 (NRT2.4) were associated in a single year. In host infection transcriptome analysis, these genes were differentially expressed in incompatible interactions with additional Melampsora species (unpublished data). Arabidopsis orthologs corresponding to genes housing these five SNPs indicate functions in host defense through transcriptome reprogramming, calcium and salicylic acid signaling, and tolerance to reactive oxygen species.
Discussion
In prior descriptions of this population, growth traits and population stratification differentiated in a north to south pattern [15], [18], [30], [33], [34]. This differentiation may be driven by intense selection pressure for adaptation to day-length and physical barriers impeding gene flow [33]. Rust severity was also correlated to latitude. Rust aggressiveness can be reduced in below optimal temperatures (20°C) in both poplar leaf rust and wheat rust interactions [35], [36]. Cooler temperatures at northern latitudes may produce a weaker selection pressure for resistance than in the south, thus creating co-linearity of causal variants with population structure. Co-linearity of allele frequencies via correlation of phenotype and population structure inflates type-1 error in association studies; therefore, it is necessary to correct for the confounding effects of population structure.
Several approaches have been proposed to correct for structure. Yu et al. [22] proposed a unified mixed model which fits both population (Q matrix) and familiar structure (kinship matrix) to precisely eliminate confounding factors. Consideration of a residual polygenic term fitting LD pattern across chromosomes in the model, which is usually fit solely by kinship matrix, has also improved the estimate of the genetic relatedness [37]–[39] especially in complex polygenic traits. Moreover, the kinship matrix itself is considered to fit both confounding factors efficiently [31], [40], [41]. In our study, GENELAND analysis indicated very weak stratification among three sub-populations (FST <0.0227). The use of Q matrix in the association model resulted in decreased fit and inflated type-1 error. Kinship matrix had the lowest BIC value in a goodness of fit test, suggesting that it is the best model to correct for the confounding structure; however the q-q plots indicated that kinship matrix eliminated the expected associations and overcorrected the model (Fig. 1).
Alternatively, Price et al. [42], [43] employed principal component analysis to improve the correction for population stratification and the confounding effects of phenotype – population structure correlation. They proposed using a fixed number of principal components (first 10) or ones selected on the basis of Tracy-Widom statistics [44] when admixture occurred in population regardless of their relationship to phenotype. Methods using a stepwise regression to select a set of SNPs [37], [45] or principal components [23], [46] have been suggested to fit the confounding structure and used as regressors in the final association analysis model. Novembre & Stephens [47] also indicated that inclusion of principal components not correlated with the trait may reduce power. In our study, the inflation of significant associations and the goodness of fit in the PCA-TOP10 model in comparison to the PCA-BIC model further supports this hypothesis (Fig. 1).
We also used SNP pre-selection to reduce the constraint of multiple testing corrections on inflation of false negative associations [23], [48]. We reason that elevating the pre-selection threshold from P<0.05 to P<0.001 would remove erroneous SNPs that would have been selected via the correlation of AUDPC and population stratification and thus increase type-2 error. In 2009, pre-selection at P<0.05 would have selected 10,828 SNPs for AUDPC, where P<0.001 reduced the SNP selection to 3,905 (data not shown). Moreover, SNP associations in FAR1 and PIPK5 were only achieved via SNP pre-selection but correlated to rust resistance through transcriptome analysis during incompatible poplar leaf rust interactions.
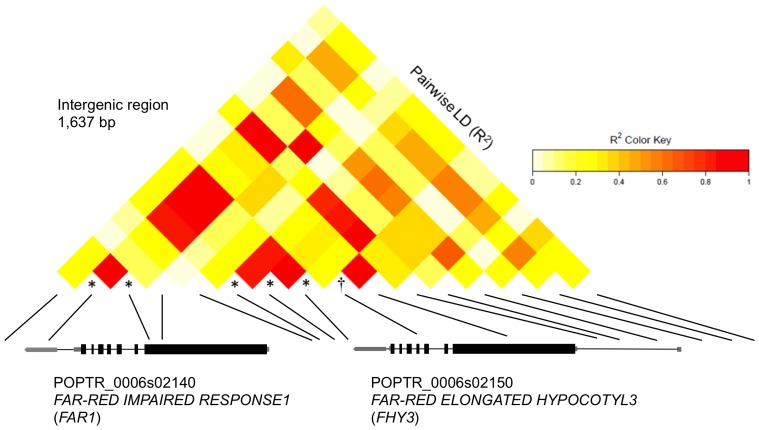
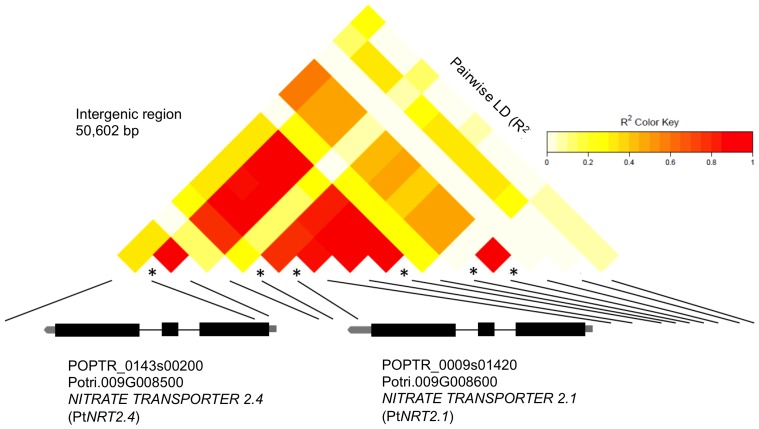
Associated SNPs within PRR7, IQD32, and PIPK5 were in low LD with the adjacent SNPs. Thus, these SNPs may be causative variants or in high LD with the unrepresented causative SNP. Conversely, several SNPs within the neighboring gene pairs; FAR1 and FHY3, and NRT2.4 and NRT2.1, respectively, were in high LD which convolutes the elucidation of the true causative SNP.
Scaffold_10_ 19,215,715 is a non-synonymous polymorphism in the fifth exon of a sequence orthologous to PRR7 (Fig. S3). In Arabidopsis, PRR7 is a gene within a small family of circadian clock gene transcription factors [49]; however, it was not associated with phenological traits in this population (personal communications, Athena McKown). The prr7 loss of function mutants has an ambiguous phenotype, but double and triple mutants accentuate the prr5 and prr9 single mutant phenotypes: arrhythmia with increased hypocotyl elongation, leaf number, and days to flowering [49]. More recently, PRR7 was down-regulated in response to chitooctaose (chitin oligomer; chitin is a component of fungal cell walls). These results would suggest a role of PRR7 transcriptional regulation during host infection [50].
Scaffold_5_23949327 is located 1,826 bp downstream from the 3′UTR of IQD32 (Fig. S4). This gene is orthologous to a calmodulin binding protein that serves as an integral component of Ca2+/calmodulin signaling. In Arabidopsis and rice, IQD gene family members share as many as three calmodulin binding motifs IQ, 1-5-10, and 1-8-14. While IQD gene function has not been well characterized, IQD1 has been shown to function in defense response to herbivory [51].
These single SNPs within PRR7 and IQD32 were associated in all three years. Bi-parental linkage mapping has identified major resistance loci that confer race-specific resistance to M. ×columbiana [4]. In our study, ANOVA and spatial analysis suggest the pathogenicity of the rust population varied across the three years. This is consistent with the reproductive biology of the rust where the non-overlap of poplar-alternative host ranges would affect the genetic composition of the rust population in time. Therefore, we propose that SNP-associations replicated in time and in the diversity of M. ×columbiana across the three years confer non-race-specific resistance.
Numerous signals within FAR1 were also significant in 2010, but not repeated in time (Fig. 3). A homolog of FAR1, FAR-RED ELONGATED HYPOCOTYL3 (FHY3), is a clock gene that indirectly mediates the phytochrome A response, but has additional functions. In Arabidopsis, fhy3 mutants regulate plant architecture and abiotic stress tolerance through suppression of axillary bud outgrowth and repressed leaf growth with decreased tolerance to oxidative stress. Loss of function mutants in far1 and revoluta (REV), a leucine-zipper transcription factor, enhance the fhy3 phenotype [52]. In 2011, scaffold_6_1405713 within FHY3 and scaffold_9_2,563,210 within REV were also significant (Table 3). These signals were not repeated in time; however, FAR1 expression was increased 2-fold 96 hours after inoculation with M. larici-populina (unpublished data).
Figure 3. Pairwise linkage disequilibrium plot of FAR-RED IMPAIRED RESPONSE1 and FAR-RED ELONGATED HYPOCOTYL3 with gene structures.
SNPs significant in 2010 are indicated with an asterisk; SNPs significant in 2011 are indicated with a cross. Scaffold_6_ 1402770 in the intergenic region had the highest significance (P = 3.64×10−6) and explained 2.5% (R2 = 0.025) of the phenotypic variance in 2010.
Likewise, scaffold_8_8261867 in the 12 exon of POPTR_0008s12780; encoding a phosphatidylinositol 4-phosphate 5-kinase (PIP5K), was significant in 2011 (Fig. S5) and has been previously implicated in resistance to Melampsora. In Arabidopsis, lower expression of PIP5Ks leads to accumulation of the raffinose family oligosaccharides that act as osmoprotectants and antioxidants and protect mitochondria and chloroplasts from stress-induced production of reactive oxygen species [53]. In resistant P. trichocarpa × deltoides, this PIP5K gene was shown to have more than a 2-fold decrease in its expression 48 hours after inoculation with M. larici-populina [54].
The SNP at scaffold_143_2955 encodes a non-synonymous mutation in the third exon of NRT2.4 and is in high LD (R2>0.8) with three other associated SNPs in the neighboring NRT2.1 (Fig. 4). These genes are orthologous to the nitrate transporter AtNRT2.1. Nitrate transporters are transmembrane proteins that primarily function in nitrate transport; however, they also function as environmental signal receptors and regulators of biotic and abiotic stress pathways. Recently, it was reported that the mutant nrt2 that lacks the function of both AtNRT2.1 and the adjacent AtNRT2.2 shows decreased susceptibility to Pseudomonas syringae. The decrease in susceptibility is coordinated through an earlier and more robust induction of salicylic acid and up-regulation of defense genes PR1 and PR5 [55]. In poplar, NTR2.4 was down-regulated in incompatible interactions of P. deltoides with M. occidentalis and M. larici-populina (unpublished data). The change in expression levels of genes housing associated SNPs during incompatible poplar leaf rust interactions further implicates a functional role in host defense.
Figure 4. Pairwise linkage disequilibrium plot of NITRATE TRANSPORTER2.1 and NITRATE TRANSPORTER2.4 with gene structures.
SNPs significant are indicated with an asterisk. Scaffold_9_ 1676227 in the intergenic region had the highest significance (P = 3.52×10−7) and largest effect on AUDPC in 2010 (R2 = 0.034). Scaffold_143_2955 was annotated to Potri.009G008500 in JGI Populus trichocarpa genome v3.
In summary, we identified two independent loci that were strongly associated with host defense to M. ×columbiana and through repetition in time confer non-race-specific resistance. Furthermore, three other associated loci have been correlated to poplar leaf rust resistance through transcriptome analysis and may form a functional network with additional genes involved in tolerance to reactive oxygen species. In this long-lived ecologically and economically important tree species, these associations lay the foundation to more efficient breeding of durable disease resistance.
Supporting Information
Spatial distribution of residuals for AUDPC in each year. The scale of residuals ranges from −20 (low disease) to 20 (high disease).
(TIFF)
Population structure estimates and geographical distribution of each sampled tree (n = 412). Colors designate the three sub-populations detected using GENELAND analysis (Q matrix).
(TIFF)
Pairwise linkage disequilibrium plot of PSEUDO-RESPONSE REGULATOR7 and gene structure. Scaffold_10_19215715 is indicated with an asterisk. In 2009, 2010, and 2011 (from left to right) each box plot shows the lower quartile, the median, and the upper quartile values, and the whiskers show the range of the phenotypic variation in the population.
(TIF)
Pairwise linkage disequilibrium plot of IQ-DOMAIN32 and gene structure. Scaffold_5_23949327 is indicated with an asterisk. In 2009, 2010, and 2011 (from left to right) each box plot shows the lower quartile, the median, and the upper quartile values, and the whiskers show the range of the phenotypic variation in the population.
(TIF)
Pairwise linkage disequilibrium plot of PHOSPHATIDYLINOSITOL-4-PHOSPHATE 5-KINASE and gene structure. Scaffold_8_ 8261867 is indicated with an asterisk. In 2011, the box plot shows the lower quartile, the median, and the upper quartile values, and the whiskers show the range of the phenotypic variation in the population.
(TIF)
Acknowledgments
The authors acknowledge Nicolas Feau’s work on RNA-Seq transcriptome analysis of Populus x Melampsora interactions and his review of the manuscript.
Funding Statement
This work was supported by Genome British Columbia (103BIO) Applied Genomics Innovation Program and Genome Canada (168BIO) Large Scale Applied Research Project funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Steenackers J, Steenackers M, Steenackers V, Stevens M (1996) Poplar diseases, consequences on growth and wood quality. Biomass Bioenerg 10: 267–274. [Google Scholar]
- 2. Widin KD, Schipper AL (1981) Effect of Melampsora medusae leaf rust infection on yield of hybrid poplars. Eur J Forest Pathol 11: 438–448. [Google Scholar]
- 3. Newcombe G, Stirling B, Mcdonald S, Bradshaw HD (2000) Melampsora ×columbiana, a natural hybrid of M. medusa and M. occidentalis. Mycol Res 104: 261–274. [Google Scholar]
- 4. Newcombe G, Stirling B, Bradshaw HD (2001) Abundant pathogenic variation in the new hybrid rust Melampsora ×columbiana on hybrid poplar. Phytopathology 91: 981–985. [DOI] [PubMed] [Google Scholar]
- 5. Flor HH (1955) Host-parasite interaction in flax rust - its genetics and other implications. Phytopathology 45: 680–685. [Google Scholar]
- 6. Bourassa M, Bernier L, Hamelin RC (2007) Genetic diversity in poplar leaf rust (Melampsora medusae f. sp. deltoidae) in the zones of host sympatry and allopatry. Phytopathology 97: 603–610. [DOI] [PubMed] [Google Scholar]
- 7. Zhang J, Steenackers M, Storme V, Neyrinck S, Van Montagu M, et al. (2001) Fine mapping and identification of nucleotide binding site/leucine-rich repeat sequences at the MER locus in Populus deltoides ‘S9–2’. Phytopathology 91: 1069–1073. [DOI] [PubMed] [Google Scholar]
- 8. Lescot M, Rombauts S, Zhang J, Aubourg S, Mathé C, et al. (2004) Annotation of a 95-kb Populus deltoides genomic sequence reveals a disease resistance gene cluster and novel class I and class II transposable elements. Theor Appl Genet 109: 10–22. [DOI] [PubMed] [Google Scholar]
- 9. Yin TM, DiFazio SP, Gunter LE, Jawdy SS, Boerjan W, et al. (2004) Genetic and physical mapping of Melampsora rust resistance genes in Populus and characterization of linkage disequilibrium and flanking genomic sequence. New Phytol 164: 95–105. [DOI] [PubMed] [Google Scholar]
- 10. Gérard PR, Husson C, Pinon J, Frey P (2006) Comparison of genetic and virulence diversity of Melampsora larici-populina populations on wild and cultivated poplar and influence of the alternate host. Phytopathology 96: 1027–1036. [DOI] [PubMed] [Google Scholar]
- 11. Bresson A, Jorge V, Dowkiw A, Guerin V, Bourgait I, et al. (2011) Qualitative and quantitative resistances to leaf rust finely mapped within two nucleotide-binding site leucine-rich repeat (NBS-LRR)-rich genomic regions of chromosome 19 in poplar. New Phytol 192: 151–163. [DOI] [PubMed] [Google Scholar]
- 12. Dowkiw A, Voisin E, Bastien C (2010) Potential of Eurasian poplar rust to overcome a major quantitative resistance factor. Plant Pathol 59: 523–534. [Google Scholar]
- 13. Crossa J, Burgueño J, Dreisigacker S, Vargas M, Herrera-Foessel SA, et al. (2007) Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177: 1889–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu C, Gore M, Buckler E, Yu J (2008) Status and prospects of association mapping in plants. The Plant Genome 1: 5–20. [Google Scholar]
- 15. Wegrzyn JL, Eckert AJ, Choi M, Lee JM, Stanton BJ, et al. (2010) Association genetics of traits controlling lignin and cellulose biosynthesis in black cottonwood (Populus trichocarpa, Salicaceae) secondary xylem. New Phytol 188: 515–532. [DOI] [PubMed] [Google Scholar]
- 16. Shaner G, Finney RE (1977) The effect of nitrogen fertilization on the expression of slow mildewing resistance in Knox wheat. Phytopathology 67: 1051–1056. [Google Scholar]
- 17. Rupe JC, Siegel MR, Hartman JR (1982) Influence of environment and plant maturity on gray leaf spot of corn caused by Cercospora zeae-maydis . Phytopathology 72: 1587–1591. [Google Scholar]
- 18.Geraldes A, Difazio SP, Slavov GT, Ranjan P, Muchero W, et al. (2013) A 34K SNP genotyping array for Populus trichocarpa: Design, application to the study of natural populations and transferability to other Populus species. Mol Ecol Resour, in press. [DOI] [PubMed]
- 19. Graffelman J, Morales-Camarena J (2008) Graphical tests for Hardy-Weinberg equilibrium based on the ternary plot. Human Heredity 65: 77–84. [DOI] [PubMed] [Google Scholar]
- 20. Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, et al. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- 21. Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2: 618–620. [Google Scholar]
- 22. Yu J, Pressoir G, Briggs W, Vroh I, Yamasaki M, et al. (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208. [DOI] [PubMed] [Google Scholar]
- 23. Pant SD, Schenkel FS, Verschoor CP, You Q, Kelton DF, et al. (2010) A principal component regression based genome wide analysis approach reveals the presence of a novel QTL on BTA7 for MAP resistance in holstein cattle. Genomics 95: 176–182. [DOI] [PubMed] [Google Scholar]
- 24. Ingvarsson P, Garcia M, Luquez V, Hall D, Jansson S (2008) Nucleotide Polymorphism and Phenotypic Associations Within and Around the phytochrome B2 Locus in European Aspen (Populus tremula, Salicaceae). Genetics 178: 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin J-H, Blay S, McNeney B, Graham J (2006) LDheatmap: An R Function for Graphical Display of Pairwise Linkage Disequilibria Between Single Nucleotide Polymorphisms. Journal of Statistical Software 16: Code Snippet 3.
- 26.Lynch M, Walsh B (1998) Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer Associates, Inc.
- 27.Gilmour AR, Gogel BJ, Cullis BR, Welham SJ, Thompson R (2002) ASReml User Guide Release 1.0: Hemel Hempstead, UK: VSN International Ltd.
- 28. Dowkiw A, Husson C, Frey P, Pinon J, Bastien C (2003) Partial resistance to Melampsora larici-populina leaf rust in hybrid poplars: genetic variability in inoculated excised leaf disk bioassay and relationship with complete resistance. Phytopathology 93: 421–427. [DOI] [PubMed] [Google Scholar]
- 29. Guillot G, Mortier F, Estoup A (2005) Geneland: A program for landscape genetics. Mol Ecol Notes 5: 712–715. [Google Scholar]
- 30. Slavov GT, DiFazio SP, Martin J, Schackwitz W, Muchero W, et al. (2012) Genome resequencing reveals multiscale geographic structure and extensive linkage disequilibrium in the forest tree Populus trichocarpa . New Phytol 196: 713–725. [DOI] [PubMed] [Google Scholar]
- 31. Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, et al. (2007) An arabidopsis example of association mapping in structured samples. PloS Genet 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82: 1420–1425. [Google Scholar]
- 33. Xie C-Y, Ying CC, Yanchuk AD, Holowachuk DL (2009) Ecotypic mode of regional differentiation caused by restricted gene migration: a case in black cottonwood (Populus trichocarpa) along the Pacific Northwest coast. Can J Forest Res 39: 519–526. [Google Scholar]
- 34. Xie C-Y, Carlson MR, Ying CC (2012) Ecotypic mode of regional differentiation of black cottonwood (Populus trichocarpa) due to restricted gene migration: further evidence from a field test on the northern coast of British Columbia. Can J Forest Res 42: 400–405. [Google Scholar]
- 35. Levine MN (1928) Biometric studies of the variation of physiologic forms of Puccinia graminis tritici and the effects of ecological factors on the susceptibility of wheat varieties. Phytopathology 18: 7–123. [Google Scholar]
- 36. Chandrashekar M, Heather WA (1980) Temperature sensitivity of reactions of Populus spp. to races of Melampsora larici-populina . Phytopathology 71: 421–424. [Google Scholar]
- 37. Habier D, Fernando RL, Dekkers J (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karkkainen H, Sillanpaa M (2012) Robustness of Bayesian multilocus association models to cryptic relatedness. Annals of Human Genetics 76: 510–523. [DOI] [PubMed] [Google Scholar]
- 39. Wang M, Jiang N, Jia T, Leach L, Cockram J, et al. (2012) Genome-wide association mapping of agronomic traits in highly structured populations of barley cultivars. Theor Appl Genet 124: 233–246. [DOI] [PubMed] [Google Scholar]
- 40. Astle W, Balding DJ (2009) Population structure and cryptic relatedness in genetic association studies. Stat Sci 24: 451–471. [Google Scholar]
- 41. Stich B, Mohring J, Piepho HP, Heckenberger M, Buckler ES, et al. (2008) Comparison of mixed-model approaches for association mapping. Genetics 174: 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 43. Price AL, Zaitlen NA, Reich D, Patterson N (2010) New approaches to population stratification in genome-wide association studies. Nature Reviews Genetics 11: 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patterson N, Price A, Reich D (2006) Population Structure and Eigenanalysis. PLoS Genet 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Setakis E, Stirnadel H, Balding D (2006) Logistic regression protects against population structure in genetic association studies. Genome Res 16: 290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Q, Wacholder S, Hunter D, Hoover R, Chanock S, et al. (2009) Genetic background comparison using distance-based regression, with applications in population stratification evaluation and adjustment. Genet Epidemiol 33: 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Novembre J, Stephens M (2008) Interpreting principal component analyses of spatial population genetic variation. Nat Genet 40: 646–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Quesada T, Gopal V, Cumbie WP, Eckert AJ, Wegrzyn JL, et al. (2010) Association mapping of quantitative disease resistance in a natural population of loblolly pine (Pinus taeda L.). Genetics 186: 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T (2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46: 686–698. [DOI] [PubMed] [Google Scholar]
- 50. Libault M, Wan J, Czechowski T, Udvardi M, Stacey G (2007) Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant-Microbe In 20: 900–911. [DOI] [PubMed] [Google Scholar]
- 51. Levy M, Wang Q, Kaspi R, Parrella MP, Abel S (2005) Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J 43: 79–96. [DOI] [PubMed] [Google Scholar]
- 52. Stirnberg P, Zhao S, Williamson L, Ward S, Leyser O (2012) FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J 71: 907–920. [DOI] [PubMed] [Google Scholar]
- 53. Vallurua R, Van den Endeb W (2011) Myo-inositol and beyond – Emerging networks under stress. Plant Sci 181: 387–400. [DOI] [PubMed] [Google Scholar]
- 54. Rinaldi C, Kohler A, Frey P, Duchaussoy F, Ningre N, et al. (2007) Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina . Plant Physiol 144: 347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Camañes G, Pastor V, Cerezo M, García-Andrade J, Vicedo B, et al. (2012) A deletion in NRT2.1 attenuates Pseudomonas syringae induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol 158: 1054–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spatial distribution of residuals for AUDPC in each year. The scale of residuals ranges from −20 (low disease) to 20 (high disease).
(TIFF)
Population structure estimates and geographical distribution of each sampled tree (n = 412). Colors designate the three sub-populations detected using GENELAND analysis (Q matrix).
(TIFF)
Pairwise linkage disequilibrium plot of PSEUDO-RESPONSE REGULATOR7 and gene structure. Scaffold_10_19215715 is indicated with an asterisk. In 2009, 2010, and 2011 (from left to right) each box plot shows the lower quartile, the median, and the upper quartile values, and the whiskers show the range of the phenotypic variation in the population.
(TIF)
Pairwise linkage disequilibrium plot of IQ-DOMAIN32 and gene structure. Scaffold_5_23949327 is indicated with an asterisk. In 2009, 2010, and 2011 (from left to right) each box plot shows the lower quartile, the median, and the upper quartile values, and the whiskers show the range of the phenotypic variation in the population.
(TIF)
Pairwise linkage disequilibrium plot of PHOSPHATIDYLINOSITOL-4-PHOSPHATE 5-KINASE and gene structure. Scaffold_8_ 8261867 is indicated with an asterisk. In 2011, the box plot shows the lower quartile, the median, and the upper quartile values, and the whiskers show the range of the phenotypic variation in the population.
(TIF)