Abstract
MicroRNAs play critical roles in various biological and metabolic processes. The function of miRNAs has been widely studied in model plants such as Arabidopsis and rice. However, the number of identified miRNAs and related miRNA targets in peach (Prunus persica) is limited. To understand further the relationship between miRNAs and their target genes during tissue development in peach, a small RNA library and three degradome libraries were constructed from three tissues for deep sequencing. We identified 117 conserved miRNAs and 186 novel miRNA candidates in peach by deep sequencing and 19 conserved miRNAs and 13 novel miRNAs were further evaluated for their expression by RT-qPCR. The number of gene targets that were identified for 26 conserved miRNA families and 38 novel miRNA candidates, were 172 and 87, respectively. Some of the identified miRNA targets were abundantly represented as conserved miRNA targets in plant. However, some of them were first identified and showed important roles in peach development. Our study provides information concerning the regulatory network of miRNAs in peach and advances our understanding of miRNA functions during tissue development.
Introduction
MicroRNAs (miRNAs) are small, endogenous, non-coding small RNAs that negatively control gene expression by cleaving or inhibiting the translation of target gene transcripts [1], [2]. Increasing evidence indicates that miRNAs play critical roles in nutrient homeostasis, developmental processes, abiotic stresses and pathogen responses [3]–[5]. Plant miRNAs are transcribed by RNA polymerase II to generate primary miRNA (pri-miRNA) transcripts and then cleaved to miRNA precursors (pre-miRNAs), catalysed by a Dicer-like enzyme (DCL1) [6], [7]. The pre-miRNA is further cleaved to a miRNA duplex (miRNA:miRNA*) and one of the strands of this duplex is incorporated into the RNA-induced silencing complex (RISC) [8]. The miRNA* strand is usually degraded, although in some cases it also accumulates at a lower level [9]. The mature miRNA strand guides ARGONAUTE (AGO) to complementary target mRNA resulting in silencing of the target gene [10], [11].
Plant miRNAs have been identified mostly by three strategies: traditional Sanger sequencing, computational prediction and high-throughput sequencing technology. Traditional Sanger sequencing has been used for the identification of conserved miRNAs in Arabidopsis [12], [13], rice [14]–[16], wheat [17], moss [18] and poplar [19]. A computation-based approach can predict miRNAs based on high miRNA conservation, but it cannot be characterized and limited to the discovery of false positive [20]. The application of deep sequencing has greatly facilitated small RNA discovery and increasingly more miRNAs, especially species-specific miRNAs have been identified in Arabidopsis [9], [21], soybean [22], Populus [5], [23], cotton [24], Japanese apricot [25] and Hevea brasiliensis [26].
Peach (Prunus persica L.) is one of the most important fruit crops worldwide and serves as a genome model for fruit trees in the Rosaceae, such as apple, cherry and plum [27]. In 2010, the Genome Sequencing Project of the peach double haploid cultivar ‘Lovell’ was completed, which generated ∼230 Mb genome sequence and 202 assembly scaffolds (http://www.rosaceae.org/node/355). Peach has some unique biological facets not commonly found in model organisms such as a 3–5 year juvenility period before the trees flower and fruit, the formation of fleshy fruit with a hardened endocarp and dormancy triggered by cold weather and/or short photoperiod in the autumn [28]. Recently, some groups have predicted or identified miRNAs and their target genes in peach. Zhang et al. [29] and Gao et al. [30] independently identified miRNAs by computational prediction and experimentally by verifying miRNAs via RT-qPCR and two miRNA target genes were validated using RLM-RACE. Several miRNAs responsive to drought [31] or chilling [32] and miRNAs in different tissues [28] were identified by high-throughput sequencing in peach. However, the majority of miRNA targets in peach were predicted by bioinformatic approaches, and only 80 miRNA target genes were identified by high-throughput degradome library sequencing, which has been developed for the global identification of targets of miRNAs in Arabidopsis [33], [34], rice [35], [36], soybean [37] and grapevine [38] and other plants.
No peach miRNA are annotated in the miRBase (version 19.0). To identify more conserved and peach-specific miRNAs and their target genes and to understand further the mechanism of miRNA-regulated target genes during tissue development in peach, a small RNA library and three degradome libraries were constructed from three different tissues for deep sequencing.
Results
Small RNA Sequencing Analysis
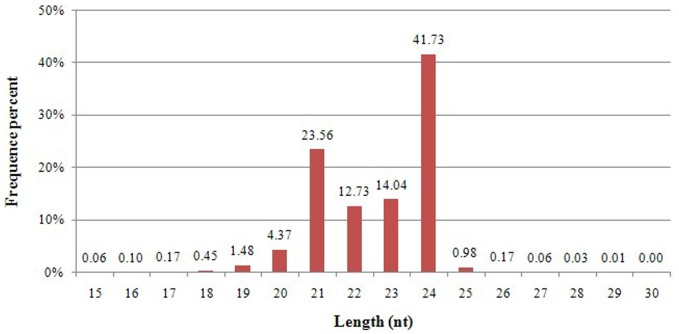
To identify miRNAs involved in tissue development, a small RNA library was constructed from peach from young leaves, young stems and flowers. A total of 14,781,274 high quality reads were obtained from the small RNA library. The small RNA length distribution (15–30 nt) of the library showed that the proportion of 21–24 nt small RNAs reached 92.06%. The 24 nt class was the most abundant (41.73%) group of sRNAs followed by 21 nt sequences (23.56%) (Fig. 1). After removal of adaptor, insert, poly (A) sequences and those reads which were smaller than 18 nt and larger than 30 nt, the remaining 14,693,759 clean reads accounted for 99.41% of the total reads. Clean read sequences were divided into rRNA, tRNA, snRNA, snoRNA, miRNA and unannotated reads, by blasting against Rfam, NCBI and miRBase databases (Table 1). Among the sRNA sequences, unannotated reads accounted for 86.85% of the total (unique, 72.21%), which was the most abundant class.
Figure 1. Sequence length distribution of peach sRNAs.
Table 1. Summary of small RNA sequencing data analysis.
| Type | Unique reads | Count % | Total reads | Count % |
| Clean sequence | 4,259,405 | 100 | 14,693,759 | 100% |
| exon_antisense | 94,683 | 2.22 | 344,177 | 2.34 |
| exon_sense | 132,524 | 3.11 | 532,607 | 3.62 |
| intron_antisense | 92,540 | 2.17 | 294,133 | 2.00 |
| intron_sense | 129,476 | 3.04 | 669,370 | 4.56 |
| rRNA | 63,681 | 1.50 | 1,183,987 | 8.06 |
| tRNA | 6,400 | 0.15 | 154,868 | 1.05 |
| snRNA | 3,026 | 0.07 | 29,905 | 0.20 |
| snoRNA | 666 | 0.02 | 2,064 | 0.01 |
| miRNA | 45,430 | 1.07 | 871,626 | 5.93 |
| unannotated | 3,690,979 | 86.65 | 10,611,022 | 72.21 |
Identification of Conserved miRNAs and Evolutionary Conservation
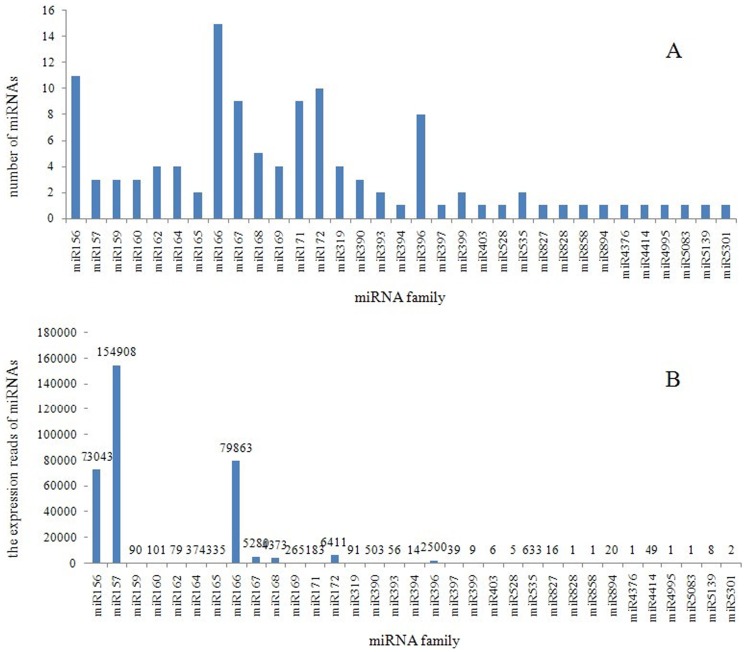
To identify conserved miRNAs, clean reads of the library were blasted against known plant miRNAs from the miRBase 18.0. Only perfectly matched sequences were considered to be conserved miRNAs. Following the BLASTn search and further sequence analysis, 117 conserved miRNAs belonging to 33 families were found to be homologous to known miRNAs from other plant species, which had previously been deposited in the miRBase database. The number of members per miRNA family ranged between one and 15. The miR166, miR156 and miR157 families were the largest, with 15, 11 and 10 members, respectively, whereas 14 miRNA families had only a single member (Fig. 2A). Details of miRNA sequences and their reads are shown in Table S2. The expression of different members of the same family was found to be largely divergent. The abundance of miRNA families also varied drastically: miR157, miR166 and miR156 were most frequently represented in the library, with 154,908, 79,863 and 73,043 reads, whereas miR172, miR167, miR168 and miR396 were moderately abundant in the library with 6,411, 5,280, 4,373 and 2,500 copies. However, the expression of five miRNA families (miR828, miR858, miR4376, miR4495 and miR5083) showed the lowest abundance, with only one read (Fig. 2B).
Figure 2. Member numbers of each identified conserved miRNA family in peach (A).
The expression read of miRNA families in peach (B).
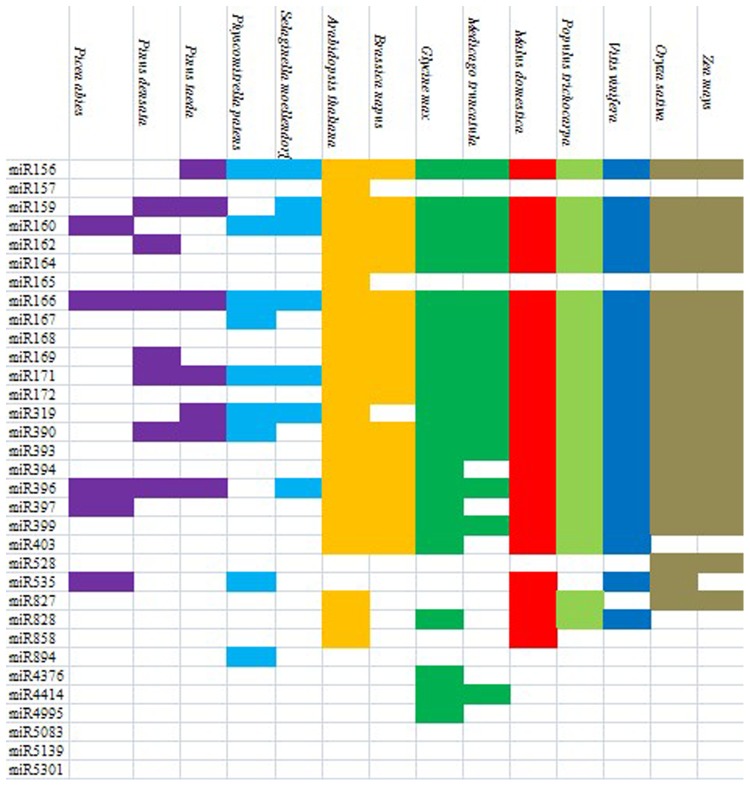
To investigate the evolutionary roles of these conserved miRNAs, extensive comparisons were performed against known miRNAs in 14 other plant species (Fig. 3). Among the miRNA families in peach, miR156, miR159, miR160, miR166, miR171, miR319, miR390 and miR396 showed a high conservation in plants, indicating that these 12 peach miRNA families are ancient. In addition, 11 miRNA families (miR162, miR164, miR167, miR168, miR169, miR172, miR393, miR394, miR397, miR399 and miR827) shared a high conservation in both dicotyledons and monocotyledons. However, 11 miRNA families were less conserved among the 14 plant species. Three families (miR5083, miR5139 and miR5301) had no corresponding homologues. The others (miR157, miR165, miR894, miR4376 and miR4995) had only one homologue among the 14 plant species. Furthermore, miR403 and miR828 was only found in dicots, whereas miR528 was only identified in monocots.
Figure 3. Conserved miRNA families from peach, designated as pmu on the first column, and their homologues in other plant species.
Identification of Peach-specific miRNAs
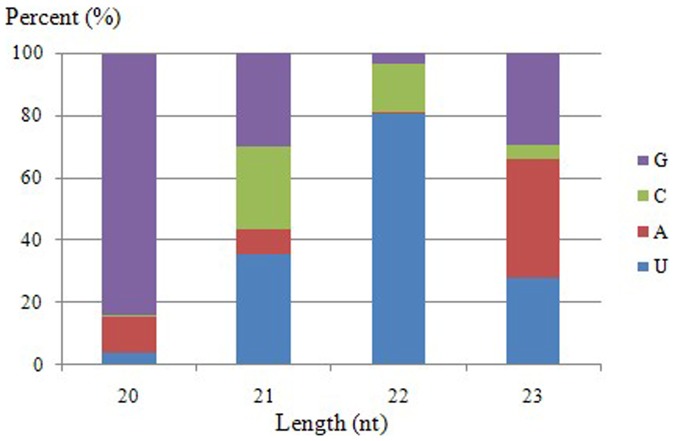
The unannotated small RNAs of 3,690,979 unique reads matching the genome was subjected to secondary structure analysis. After using the criteria suggested by Meyers et al. [39] and the program MIREAP developed by BGI, 186 putative novel miRNAs were identified. The first nucleotide bias analysis showed that uracil was the most frequently used first nucleotide in miRNAs of peach, with 10,801 uracil nucleotides (41.68%). This result was agreement with previously reports that the uracil nucleotide is dominant at the first position of the 5′ end for the majority of miRNAs (Fig. 4). Our sequence analysis showed that the minimum folding free energies (MFEs) of potential novel miRNA precursors ranged from 18.33 to 171.23 kcal/mol with a mean of 53.13 kcal/mol using mFold software (Table S3). Among the miRNA candidates, 46 were identified with complementary miRNA*s. In contrast to the common or evolutionarily conserved miRNAs, the predicted novel miRNAs were often expressed at a very low level, 77.96% of which were sequenced fewer than 50 times. From 186 novel miRNAs candidates, only three miRNAs (miRC57, miRC162 and miRC167) were sequenced more than 1,000 times and 11 miRNAs more than 500 times.
Figure 4. First nucleotide bias in novel miRNA candidates in peach.
miRNA Validation and Quantification by RT-qPCR
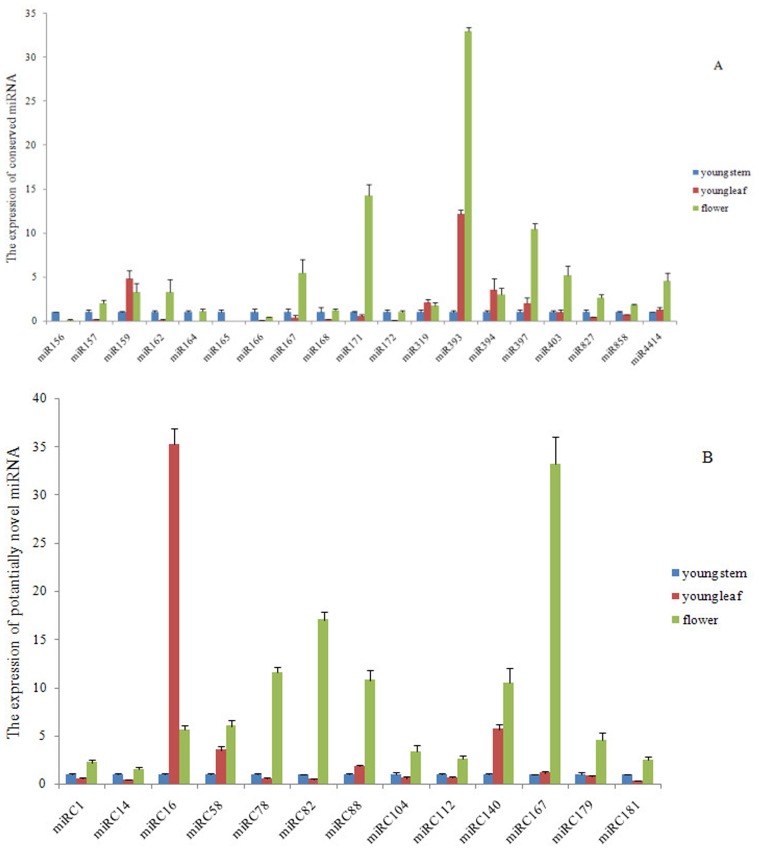
To evaluate the differential expression of the miRNAs in three tissues and to assess their potential roles in regulating the expression of the genes, the expression of 19 conserved miRNAs and 13 novel miRNA candidates was analysed using poly (A) qRT-PCR. These qRT-PCR analyses demonstrated that the expression of miRNAs differed greatly in young leaves, stems and flowers. The expression of 8 conserved (miR166, miR168, miR319, miR394, miR399, miR827, miR894 and miR5139) and six novel miRNAs (miRC1, miRC14, miRC16, miRC112, miRC179 and miRC181) showed no significant change in different tissues (Figs. 5A and 5B). It was notable that the majority of the tested miRNAs were more abundant in flowers than in young leaves and stems, which suggests that these miRNAs might play important roles in peach flower development, whereas miR393 and miRC16 mainly accumulated in young leaves.
Figure 5. Relative expression levels of peach miRNAs in different tissues.
A, Relative expression levels of conserved miRNAs; B, relative expression levels of novel miRNAs.
Summary of the Degradome Library Data
In higher plants, most miRNAs regulate their target genes via cleavage, which normally occurs at the tenth nucleotide of the complementary region between the miRNA and the mRNA [36]. Three degradome libraries, which capture the cleaved mRNAs were constructed from young leaves, stems and flowers in peach. After submitting libraries for parallel analysis of RNA end (PARE) Solexa sequencing [33], [40], a total of 12,295,775, 11,921,348, and 12,004,101 raw reads and 7,974,504, 8,329,611, 8,766,508 unique raw reads were obtained from peach leaf, stem and flower libraries, respectively (Table 2). In total, 5,320,695, 5,004,686 and 5,292,314 reads (referred to as cDNA mapped reads), which perfectly mapped to the peach cDNA database, remained. Finally, 22,058, 2,176 and 22,420 reads, which contained miRNA-mediated cleavage sites, were identified.
Table 2. Data summary of three degradome libraries of peach.
| Type | Leaf library | Stemlibrary | Flowerlibrary |
| Raw Reads | 12,295,775 | 11,921,348 | 12,004,101 |
| Unique Raw Reads | 7,974,504 | 8,329,611 | 8,766,508 |
| cDNA Mapped Reads | 5,320,695 | 5,004,686 | 5,292,314 |
| Total Number of input cDNAs | 34,809 | 28,701 | 28,701 |
| Number of covered cDNAs | 22,058 | 21,767 | 22,420 |
Identification and Classification of Targets for known miRNAs and Novel miRNA Candidates
A total of 259 target mRNAs were identified from three degradome libraries in peach, 172 of which were the target genes for 26 conserved miRNA families and 87 of them for 38 putative novel miRNAs (Table S4). It should be noted that conserved miRNA families on average targeted more gene transcripts compared to novel miRNA candidates. The largest number of targets was shown by miR156, miR172 and miR396, with 25, 21 and 22, respectively. The target genes were classified into five categories (0–4), based on their number. It was shown that 29.30%, 25.90% and 55.43% targets belonged to category 0 from peach flowers, young stems and young leaves, respectively. Target gene categories 1 and 3 had fewer members (Table 3).
Table 3. The percentage of target gene categories in three tissues.
| Category | Flower | Young stem | Young leaf |
| 0 | 29.30% | 25.90% | 55.43% |
| 1 | 5.38% | 0.60% | 0.37% |
| 2 | 25.27% | 30.92% | 24.72% |
| 3 | 1.61% | 2.41% | 0.75% |
| 4 | 38.44% | 40.16% | 18.73% |
Some of the target genes were abundantly represented as conserved miRNA targets, including many transcription factors such as ARF, MYB, NF-Y, GROWTH REGULATORY FACTOR, HD-ZIP, PPR, SBP and the NAD (P)-binding protein-encoding gene and genes encoding ribosomal proteins. In contrast, we found some other miRNA targets such as genes encoding mitochondrial acyl carrier protein, vacuolar ATPase assembly integral membrane protein, proline extensin-like receptor kinase, galactose oxidase/kelch repeat superfamily protein, ankyrin repeat family protein, cell differentiation protein and jasmonate-ZIM-domain protein (Table S4).
Among the 568 miRNA target genes, 197 were present in all three tissues; 123 and 70 targets were only found in peach stems and flowers, respectively, 102 targets were identified in stems and flowers, whereas 76 targets were found in leaves and stems (Table S5).
Discussion
High-throughput Sequencing of Peach Small RNAs
The study of miRNAs as regulators of gene expression has been extensively performed in plants in recent years [41]. The majority of miRNA genes in plants show high conservation and rapid evolution. Deep sequencing technologies allow miRNAs to be identified at an increased depth. Species-specific miRNAs, which should be greater than the number of conserved miRNAs [42] and only 47 peach-specific and 47 known miRNAs or families with distinct expression patterns were identified in peach by deep sequencing [28]; In this study, 117 conserved miRNAs and 186 novel miRNA candidates were identified. We found seven novel lowly-conserved miRNA families (miR528, miR4376, miR4414, miR4995, miR5083, miR5139 and miR5301) and 186 novel miRNA candidates.
miRNA Expression Signatures Associated with Tissue Development
The differential accumulation of miRNAs in different tissues is common [35]. Notably, 12 from 19 conserved miRNAs and 12 from 13 novel miRNA candidates were highly expressed in flowers according to RT-qPCR data. It has been suggested that these miRNAs might play an important role in organ boundary formation or developmental processes. In addition, the expression of miRC16 was much higher in leaves than in flowers and stems, suggesting that it might be associated with leaf development.
Identification of Previously Undiscovered miRNAs Targets
To identify previously undiscovered miRNAs target, three degradome libraries were constructed. The identification of miRNA targets in the libraries was low, regardless of whether they were conserved or novel. It was also noted that miRNAs with a single nucleotide difference within the same family make them functionally distinct because of their interaction with different AGO complexes [43]. Recently, miRNA sequence length variation was also reported to have dramatic effects on miRNA functions [43]–[45]. The targets of conserved and species-specific miRNAs appear to be involved in various processes in plants; many of which were previously uncharacterised. MiR156 and miR157 not only targeted SQUAMOSA promoter-binding protein-like (SBP domain) transcription factors, but also genes encoding proteins associated with energy metabolism, glucose metabolism, redox status and ion transport (Table S4). In this study, miR159 not only targeted MYB transcription factors but also regulated the expression of genes encoding ENTH/VHS family proteins, cytokinin oxidase/dehydrogenase, and transferases. In addition, UDP-XYL synthase was a target of miR164. Thus, the novel peach miRNAs targeted different genes with a wide variety of predicted functions.
Conclusions
In this study, we constructed a small RNA library and three degradome libraries from three tissues for deep sequencing and identified 117 conserved miRNAs and 186 novel miRNA candidates. In addition, we identified 172 targets for conserved miRNA families and 87 for novel miRNAs, using recently developed tools for the global identification of miRNA targets. Peach represents a model for fruit trees in the Rosaceae. We expect to identify more peach-specific miRNAs and their targets and to obtain a better insight into the miRNA networks in peach.
Materials and Methods
Plant Materials
Prunus persica cv. Lovell peach trees were grown in the garden of Nanjing Agricultural University, China. Young emerging leaves, young stems and flower buds were collected from ‘Lovell’ peach trees before the end of flowering. After collection, all samples were immediately frozen in liquid nitrogen and stored at −70°C until use.
Small RNA and Degradome Library Construction for Solexa Sequencing
A low molecular weight (LMW) RNA was isolated from mixed tissues (young leaves, young stems and flower buds) using the CTAB reagent according to the procedure previously described by Wang et al. [46]. The sample was sequenced by the Beijing Genomics Institute (BGI) (Shenzhen, Guangdong Province, China) using high-throughput pyrosequencing technology developed by Solexa. Three total RNAs were extracted from peach: young leaves, young stems and flowers. The degradome cDNA libraries were prepared following the procedures previously described by German et al. [40], [47].
The small RNA library and three degradome libraries sequencing data are available under NCBI-GEO accession no: GSE49579.
Bioinformatic Analysis of Sequencing Data
After removing low quality reads and trimming adaptor sequences, the high-quality small RNA reads ranging from 18 to 30 nucleotides were obtained from the sRNA raw data. Small RNA sequences matching non-coding rRNA, tRNA, snRNA and snoRNA in the Rfam 10 (http://www.sanger.ac.uk/resources/databases/rfam.html) and NCBI Genbank databases were removed. The remaining sequences were searched against the miRBase database v18.0 (http://www.mirbase.org, release 18) with up to two mismatches, to identify conserved miRNAs. The sequences that did not map to any miRNAs in miRBase were analysed for predictions to identify novel miRNAs by the program MIREAP (developed by BGI) with default parameters for mapping the peach genome and obtaining all candidate precursors with hairpin-like structures of novel miRNA candidates [31]. Secondary structures of novel miRNAs were also checked using Mfold 3.2 [48].
The conserved miRNA families identified in peach were compared with known miRNA families in 14 other plant species, including Picea abies, Pinus densata, P. taeda, Physcomitrella patens, Selaginella moellendorffii, Brassica napus, Glycine max, Medicago truncatula, Malus domestica, Populus trichocarpa, Vitis vinifera, Oryza sativa, Zea mays. Peach miRNAs that had corresponding homologues in more than 10 plant species were classified as highly conserved miRNAs. While miRNAs that had corresponding homologues in at most 3 plant species were considered as less conserved miRNAs.
The construction of degradome libraries differed considerably from past efforts [33], [49] and followed the procedure of as Ma et al. [40], [47] with some modifications. (1) Approximately 150 ng of poly (A)+ RNA was used as input RNA and for annealing with biotinylated random primers; (2) Strapavidin capture of RNA fragments was performed via Biotinylated Random Primers; (3) 5′ adaptor ligation was only performed to those RNAs containing 5′-monophosphates; followed by (4) reverse transcription and PCR; (5) libraries were sequenced using the 5′ adapter only, resulting in the sequencing of the first 36 nucleotides of the inserts that represented the 5′ ends of the original RNAs. The purified cDNA library was used for cluster generation on Illumina’s Cluster Station and then sequenced on Illumina GAIIx following the vendor’s instructions for running the instrument. Raw sequencing reads were obtained using Illumina’s Pipeline v1.5 software following sequencing image analysis by the Pipeline Firecrest Module and base-calling by the Pipeline Bustard Module. A Public software package, CleaveLand 3.0 was used for analysing the generated sequencing data.
The peach genome sequences, CDS sequences and gene annotation information were obtained from GDR (www.rosaceae.org).
miRNA Validation and Quantification by RT-qPCR
The LMW RNAs, independently extracted from peach leaves, young stems and flowers were used for RT-qPCR. The cDNA was mixed with 2× SYBR Green Mix (Takara, Japan), together with the miRNA-specific primers and a universal reverse primer (URP) in a final volume of 20 µL. Samples were analyzed in biological triplicate in a 96-well plate. All quantitative PCRs were performed in an ABI 7300 Real-Time PCR System (Bio-Rad) and the PCR conditions were 95°C for 30 s, 40 cycles of: 95°C for 30 s and 60°C for 34 s and a melting curve immediately after amplification. Each reaction was performed in triplicate. The relative miRNA expression was quantified using the comparative ΔΔCT method [50]. All expression profiles were normalised to expression levels in the stem. 5.8S rRNAs [51] was used as an internal control. The primer sequences are shown in Table S1. There three biological replications in every sample.
Additional Information
Accession codes: all of raw sequencing data of this paper have been deposited at GEO under accession GSE49579. P. persica mixed tissues for small RNA sequencing data have been deposited at GEO under accession GSM1202204. P. persica degradome Sequencing data for P. persica are available in the GEO under accession GSM1202205, GSM1202206 and GSM1202207.
Supporting Information
Primer sequences of miRNAs.
(DOC)
Peach homologs of conserved miRNAs.
(XLS)
Detailed information of novel miRNA candidates in peach.
(XLS)
The targets of known miRNA in peach.
(XLS)
Expression of 568 target genes in different tissues.
(XLS)
Funding Statement
The authors gratefully acknowledge the National Science Foundation of China (31101526) and the project of 948 and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) for funding part of this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Joshi T, Yan Z, Libault M, Jeong DH, Park S, et al. (2010) Prediction of novel miRNAs and associated target genes in Glycine max. BMC Bioinformatics 11 Suppl 1S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- 3. Song C, Wang C, Zhang C, Korir NK, Yu H, et al. (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics 11: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science Signaling 301: 336. [DOI] [PubMed] [Google Scholar]
- 5. Lu S, Sun YH, Chiang VL (2008) Stress-responsive microRNAs in Populus. Plant J 55: 131–151. [DOI] [PubMed] [Google Scholar]
- 6. Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, et al. (2008) Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naqvi AR, Sarwat M, Hasan S, Roychodhury N (2012) Biogenesis, functions and fate of plant microRNAs. J Cell Physiol 227: 3163–3168. [DOI] [PubMed] [Google Scholar]
- 8. Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, et al. (2008) High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics 9: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang L, Liu H, Li D, Chen H (2011) Identification and characterization of maize microRNAs involved in the very early stage of seed germination. BMC Genomics 12: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu JK (2008) Reconstituting plant miRNA biogenesis. Proceedings of the National Academy of Sciences 105: 9851–9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 12. Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Science Signaling 16: 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. The Plant Cell Online 14: 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang JF, Zhou H, Chen YQ, Luo QJ, Qu LH (2004) Identification of 20 microRNAs from Oryza sativa . Nucleic Acids Res 32: 1688–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sunkar R, Girke T, Jain PK, Zhu J-K (2005) Cloning and characterization of microRNAs from rice. The Plant Cell Online 17: 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu JK, Sunkar R (2008) Cloning and characterization of microRNAs from rice. Google Patents. [DOI] [PMC free article] [PubMed]
- 17. Yao Y, Guo G, Ni Z, Sunkar R, Du J, et al. (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol 8: R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arazi T, Talmor Neiman M, Stav R, Riese M, Huijser P, et al. (2005) Cloning and characterization of microRNAs from moss. The Plant Journal 43: 837–848. [DOI] [PubMed] [Google Scholar]
- 19. Lu S, Sun YH, Shi R, Clark C, Li L, et al. (2005) Novel and mechanical stress–responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. The Plant Cell Online 17: 2186–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu Q, Liu Y, Zhu A, Wu X, Ye J, et al. (2010) Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics 11: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, et al. (2007) High-Throughput Sequencing of Arabidopsis microRNAs: Evidence for Frequent Birth and Death of MIRNA Genes. PLoS One 2(2): e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kulcheski FR, de Oliveira LF, Molina LG, Almerao MP, Rodrigues FA, et al. (2011) Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li B, Qin Y, Duan H, Yin W, Xia X (2011) Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica . J Exp Bot 62: 3765–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pang M, Woodward AW, Agarwal V, Guan X, Ha M, et al. (2009) Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.). Genome Biol 10: R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao Z, Shi T, Luo X, Zhang Z, Zhuang W, et al. (2012) High-throughput sequencing of small RNAs and analysis of differentially expressed microRNAs associated with pistil development in Japanese apricot. BMC Genomics 13: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gebelin V, Argout X, Engchuan W, Pitollat B, Duan C, et al. (2012) Identification of novel microRNAs in Hevea brasiliensis and computational prediction of their targets. BMC Plant Biol 12: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shulaev V, Korban SS, Sosinski B, Abbott AG, Aldwinckle HS, et al. (2008) Multiple models for Rosaceae genomics. Plant Physiol 147: 985–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu H, Xia R, Zhao B, An YQ, Dardick CD, et al. (2012) Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol 12: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y, Yu M, Yu H, Han J, Song C, et al. (2012) Computational identification of microRNAs in peach expressed sequence tags and validation of their precise sequences by miR-RACE. Mol Biol Rep 39: 1975–1987. [DOI] [PubMed] [Google Scholar]
- 30. Gao Z, Luo X, Shi T, Cai B, Zhang Z, et al. (2012) Identification and validation of potential conserved microRNAs and their targets in peach (Prunus persica). Mol Cells 34: 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eldem V, Celikkol Akcay U, Ozhuner E, Bakir Y, Uranbey S, et al. (2012) Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS One 7: e50298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barakat A, Sriram A, Park J, Zhebentyayeva T, Main D, et al. (2012) Genome wide identification of chilling responsive microRNAs in Prunus persica . BMC Genomics 13: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. German MA, Pillay M, Jeong DH, Hetawal A, Luo S, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946. [DOI] [PubMed] [Google Scholar]
- 35. Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, et al. (2011) Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23: 4185–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou M, Gu L, Li P, Song X, Wei L, et al. (2010) Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. indica). Frontiers in Biology 5: 67–90. [Google Scholar]
- 37. Song QX, Liu YF, Hu XY, Zhang WK, Ma B, et al. (2011) Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, et al.. (2010) Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. The Plant Journal: no-no. [DOI] [PubMed]
- 39. Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, et al. (2008) Criteria for annotation of plant MicroRNAs. Plant Cell 20: 3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. German MA, Luo S, Schroth G, Meyers BC, Green PJ (2009) Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protoc 4: 356–362. [DOI] [PubMed] [Google Scholar]
- 41. An FM, Hsiao SR, Chan MT (2011) Sequencing-based approaches reveal low ambient temperature-responsive and tissue-specific microRNAs in phalaenopsis orchid. PLoS One 6: e18937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lenz D, May P, Walther D (2011) Comparative analysis of miRNAs and their targets across four plant species. BMC Res Notes 4: 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, et al. (2010) 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proceedings of the National Academy of Sciences 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, et al. (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nature structural & molecular biology 17: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu L, Zhou H, Zhang Q, Zhang J, Ni F, et al. (2010) DNA methylation mediated by a microRNA pathway. Mol Cell 38: 465–475. [DOI] [PubMed] [Google Scholar]
- 46. Wang XW, Xiong AS, Yao QH, Zhang Z, Qiao YS (2010) Direct isolation of high-quality low molecular weight RNA of pear peel from the extraction mixture containing nucleic acid. Mol Biotechnol 44: 61–65. [DOI] [PubMed] [Google Scholar]
- 47. Ma Z, Coruh C, Axtell MJ (2010) Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. The Plant Cell Online 22: 1090–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Addo-Quaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25: 130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta] CT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 51. Design RTPCRP (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39: 519–525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences of miRNAs.
(DOC)
Peach homologs of conserved miRNAs.
(XLS)
Detailed information of novel miRNA candidates in peach.
(XLS)
The targets of known miRNA in peach.
(XLS)
Expression of 568 target genes in different tissues.
(XLS)