Abstract
Though operationally defined as cis-regulatory elements, enhancers can also communicate with promoters on a separate homolog in trans, a mechanism that has been suggested to account for the ability of certain alleles of the same gene to complement one another in a process otherwise known as transvection. This homolog-pairing dependent process is facilitated in Drosophila by chromatin-associated pairing proteins, many of which remain unknown and their mechanism of action uncharacterized. Here we have tested the role of the gypsy chromatin insulator in facilitating pairing and communication between enhancers and promoters in trans using a transgenic eGFP reporter system engineered to allow for targeted deletions in the vestigial Boundary Enhancer (vgBE) and the hsp70 minimal promoter, along with one or two flanking gypsy elements. We found a modest 2.5-3x increase in eGFP reporter levels from homozygotes carrying an intact copy of the reporter on each homolog compared to unpaired hemizygotes, although this behavior was independent of gypsy. However, detectable levels of GFP protein along the DV wing boundary in trans-heterozygotes lacking a single enhancer and promoter was only observed in the presence of two flanking gypsy elements. Our results demonstrate that gypsy can stimulate enhancer-promoter communication in trans throughout the genome in a context-dependent manner, likely through modulation of local chromatin dynamics once pairing has been established by other elements and highlights chromatin structure as the master regulator of this phenomenon.
Introduction
Unlocking the mechanism by which gene regulatory elements (enhancers, promoters and other transcription factor binding sites) coordinate gene expression in a precise spatio-temporal manner is critical to understanding how eukaryotic genomes function in vivo. Chromatin structure plays a central role in this process, where nucleosome position and density, chromatin insulators, histone modifications and their associated proteins function to modulate the properties of these regulatory elements. Most of the effort in understanding this interplay has focused on their behavior in cis—that is, interactions occurring along the same chromosome. Indeed, enhancers are defined as cis-regulatory elements that function to stimulate gene expression when located either distal or proximal to their cognate promoters, and can function over large distances [1-4]. This is achieved through physical association between the enhancer and the promoter, mediated by a number of regulatory proteins, general transcription factors, RNA Pol II and chromatin binding proteins that result in the formation of chromatin loop structures that are critical for transcriptional activation [5-9].
However, the ability of enhancers to also act in trans (i.e., on a separate DNA molecule) on promoters has been observed both in vitro and in vivo [7,10-14] where such behavior has been suggested to account for a number of homolog pairing-dependent phenotypes, such as in the phenomenon of transvection (see 15-19 for review). The term transvection was first coined by E.B. Lewis in 1954 to describe the ability of certain Drosophila alleles of Ultrabithorax (Ubx) to complement one another, leading to partial rescue of the mutant phenotype [20]. Importantly, this rescue failed when the locus on either homolog was relocated to a new position on the chromosome, suggesting that somatic pairing between homologous chromosomes is essential for transvection. This type of intragenic complementation has been reported almost exclusively in Drosophila, with transvection effects observed at a number of loci: yellow (y) [21,22], decapentaplegic (dpp) [23], eyes absent (eya) [24], white (w) [25], Gpdh [26], hedgehog (hh) [27], wingless (wg) [28], engrailed (en) [29], pointed (pnt) [30], cubitus interrruptus (ci) [31], sex combs reduced (scr) [32], brown (bw) [33,34] and vestigial (vg) [35]. Recent studies have concluded that transvection is pervasive throughout the entire Drosophila genome [11,36], a finding not surprising given that homologs remain paired in somatic nuclei during interphase in flies [37]. In other eukaryotes, homologs in somatic tissue do not remain in close synapse throughout interphase, yet quite a few cases of transvection and other pairing dependent phenomenon have been reported in model systems other than Drosophila, including yeast [38], plants [39] and mammals [40-42], suggesting that eukaryotes possess evolutionarily conserved mechanisms that allow homologous chromosomes to communicate in trans.
Given the need for physical associations between enhancers and promoters to generate a sustained transcriptional output, homolog pairing in trans can facilitate contacts between a functional enhancer located on one homolog and a functional promoter located on the other, increasing the frequency of collisions between the two elements and thus the probability that a stable ternary complex is established. A number of proteins have been shown to be required for homolog pairing in Drosophila meiosis, including the multi-subunit cohesin complex (SMC1, SMC3, SCC1/RAD21, SCC3), although it does not appear to be required for somatic paring [43]. In mammals, this complex has been shown to be involved in stable cis looping contacts between enhancers and promoters, suggesting it may play a more direct role in gene regulation [44-46]. Only a handful of Drosophila genes, mainly involved in mitotic functions, cell cycle control and chromatin organization, including Topoisomerase II (Top2) have been shown to promote somatic pairing [43,47], although other chromatin binding proteins, such as Zeste and members of the Polycomb Group Complex, have been shown to be required for transvection in a number of cases, including those involving communication between enhancers and promoters in trans [25,32,48-52].
In theory, however, any DNA element and its associated proteins that can mediate stable long-range contacts between distant genomic sites could potentially function to stabilize homolog pairing to facilitate enhancer-promoter interactions in trans. Chromatin insulators are well-suited for this task, given their ability to mediate long-distance contacts along the chromatin fiber in vivo. These DNA elements were first identified in Drosophila based on their ability to block enhancer-promoter communication and heterochromatin spreading along the chromatin fiber in transgenic assays. Such properties are conferred by insulator-binding proteins, seven of which have been characterized in Drosophila, including Su(Hw), CP190, BEAF-32, Mod(mdg4)67.2, dCTCF, GAF and Zw5, with mammals containing only a CTCF ortholog highly divergent in amino acid similarity to its Drosophila counterpart [53]. The gypsy insulator located within the 5' LTR of the gypsy retrotransposon is perhaps the most well characterized insulator, whose enhancer and heterochromatin blocking properties are conferred by Su(Hw), CP190 and Mod(mdg4)67.2. However, in addition to gypsy there are thousands of endogenous insulator sites located throughout the genome, where combinatorial binding of insulator proteins to each of these sites suggests a complex landscape whose functional consequences remain poorly understood [54-56]. Recent work has shown that these elements help mediate long-range contacts between enhancers, promoters and other insulator sites in order to direct transcriptional outputs, maintain regions of histone modifications, and establish gene regulatory and physical domains [2,57-60].
Though most of these interactions are thought to occur in cis, potential interactions in trans are not out of the question, even for long range contacts between non-homologous sites, analogous to the behavior observed for olfactory receptor choice by the H enhancer and various olfactory gene promoters located on different chromosomes in mice [61]. Interestingly, gypsy insulators have been previously implicated in transvection, both directly and indirectly, at the yellow locus [21,62]. Additionally, the bx 34e allele, used by E.B. Lewis in his original description of the phenomenon [20], results from a gypsy retrotransposon insertion between the bx enhancer and the promoter of Ubx and subsequent loss of enhancer-promoter communication in cis due to the enhancer blocking property of the gypsy insulator. Furthermore, reduction of Su(Hw) has been shown to reduce somatic pairing by ~30% in embryos [63], suggesting that insulators might contribute to pairing dependent enhancer-promoter communication in trans.
Here we have used a reporter construct designed to elucidate the role of the gypsy insulator in transvection. We engineered our system with the vestigial (vg) boundary enhancer (vgBE) and a minimal hsp70 promoter to drive eGFP expression, flanked by a gypsy insulator upstream and downstream (2-insulator), upstream only (1-insulator) or no gypsy insulator (0-insulator). Using the Cre/loxP and Flp/FRT system to delete the promoter or enhancer, respectively, “promoterless” and “enhancerless” flies were created and crossed to measure transvection effects. Quantitative fluorescent microscopy and qPCR of wing discs from 3rd instar larvae reveal a pairing dependence in the non-deletion constructs that is independent of gypsy, whereas transvection was only visually observed along the DV wing boundary in lines containing two flanking insulators. Interestingly, the vgBE alone can drive expression of a large pool of eGFP transcripts in cis in the absence of a functional promoter, the majority of which are not translated into protein. Taken collectively, our results demonstrate that the gypsy insulator can contribute to transvection in a dose-dependent manner, likely through modulation of local chromatin dynamics once other chromatin elements have established homolog pairing.
Materials and Methods
Fly Stocks & Husbandry
Flies were cultured on standard cornmeal-agar media and maintained at 25°C. Flippase (FLP) (y 1 w 1118 P{ry[+t7.2]=hsFLP}1; DrMio/TM3, ry * Sb 1), Cre recombinase (y 1 w 67c23; nocSco/CyO, P{w +mC =Crew}DH1) and su(Hw) mutant flies (w 1118 ; PBac{RB}Su(Hw)e04061/TM6B, Tb 1) were obtained from the Bloomington Stock Center. Microinjection to generate 2-insulator (yw; P{Ins-vgBE-eGFP-Ins, w +mC}), 1-insulator (yw; P{Ins-vgBE-eGFP, w +mC}) and 0-insulator (yw; P{vgBE-eGFP, w +mC}) transgenics was performed by Genetivision (Houston, TX). The w; nocSco/CyO; MKRS, Sb1/TM6B, Tb 1 double balancer line was a gift from Bruce McKee.
Construct Design and Plasmid Generation
The reporter construct consisted of 5 core elements: the vestigial boundary enhancer (vgBE), loxP and FRT sites, 5xUAS/hsp-70 minimal promoter, and an eGFP coding sequence. Additional gypsy insulator sequences were present as required to generate either the 1- or 2-insulator construct (Figure 1A). The pGREEN pelican plasmid, consisting of an eGFP reporter with a 5' MCS flanked by two gypsy insulators, served as the vector backbone for these constructs [64]. First, the 5xUAS/hsp-70 minimal promoter was amplified from the pUAST vector using specific primers designed to insert NheI and XhoI cut sites and a single loxP site oriented in the same direction on both sides of the promoter. The PCR product was digested with NheI and XhoI and cloned into the pGREEN pelican vector digested with NheI and XhoI to obtain the pGP-hsp70 plasmid. The wing and haltere disc vestigial Boundary Enhancer (vgBE) present in the second intron of the vg gene [65] was amplified from yw gDNA with primers engineered with BbvCI and BamHI sites, digested and cloned into pCR 2.1-λ- FRT that was available in the lab. This plasmid consists of λ DNA (~800bp) and FRT sequences in the same orientation flanking the multiple cloning site. The cloned vgBE enhancer and its flanking FRT sites were digested as a KpnI-SacII fragment and cloned into pGP-hsp70 to obtain the 2-insulator construct Ins-VgBE-eGFP-Ins. To obtain the 1-insulator construct (Ins-VgBE-eGFP), the 3' insulator downstream of eGFP was deleted from this vector by restriction digestion with SpeI and Eco47III. The sticky ends generated by SpeI digestion were end-filled using Pfu DNA polymerase and blunt ends were ligated. To generate the 0-insulator construct (VgBE-eGFP), both insulators were deleted from the pGP-hsp70 plasmid, generating a KpnI-λ-vgBE-eGFP-SacII cassette that was reinserted into the insulator-less pGP-hsp70 plasmid. All constructs were microinjected into y 1 w 67c23 flies and individual lines established by w + selection.
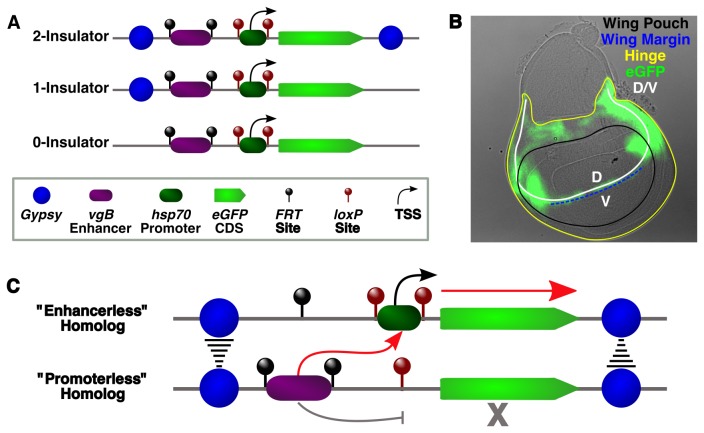
Figure 1. Schematic of the reporter system used in this study.
All constructs contained the vgBE enhancer flanked by FRT sites, a minimal hsp70 promoter flanked by loxP sites, an eGFP coding sequence, and either zero, one or two gypsy insulators (A). Differential Interference Contrast (DIC)-GFP overlay of a 3rd instar wing disc showing the expression pattern of the vgBE along the dorsal-ventral (D/V) boundary, particularly within the hinge and wing margin (B). Communication in trans between a functional enhancer (“promoterless”) on one homolog and a functional promoter (“enhancerless”) on the other homolog might be facilitated by trans interactions between flanking gypsy insulators, leading to expression only from the “enhancerless” homolog (C).
Insertion Mapping & Inverse PCR
Individual lines were mapped using both classical and molecular genetics. A single male homozygous for the transgene (w +) from each line was crossed to w; nocSco/CyO; MKRS, Sb1/TM6B, Tb 1 virgins. Progeny males carrying w + , CyO and TM6B were then crossed to yw virgins and the resulting offspring scored to determine how w + segregated with respect to the dominantly-marked CyO and TM6B balancers. Inverse PCR to identify the precise insertion position in the genome was carried out as described (Berkley Drosophila Genome Project; http://www.fruitfly.org/about/methods/inverse.pcr.html). Genomic DNA was extracted from adult flies with DNAzol (Invitrogen), ~1μg digested with HinPI, Sau3AI or MspI in a 30 μl reaction volume for 3 hrs at 37°C and ligated (T4 DNA ligase, NEB) overnight at 4°C in a 400 μl reaction volume. DNA was EtOH precipitated and washed followed by resuspension in 10 μl H2O and amplified with both the Pry1/Pry4 and PwhtI/Plac1 primer sets. Samples producing a single strong band with minimum background were PCR purified and sequenced with either the Sp1 or Spep1 primer. Sequences were mapped to the latest version of the Drosophila genome using the BLAST algorithm at FlyBase.
Inducing Artificial Mutations in the Enhancer and Promoter
To delete the promoter, transgenic males carrying an intact construct were crossed with virgin females carrying Cre recombinase. Individual progeny males (w +) were then crossed to yw virgins and gDNA extracted from adult progeny using DNAzol (Invitrogen), followed by PCR to screen for promoter removal. A similar crossing scheme was used to remove the vgBE enhancer, with transgenic males carrying an intact construct crossed with virgins carrying flippase under control of the hsp-70 promoter. Larvae were subjected to daily heat shock by submerging vials in a 37°C water bath for 1 hr. Individual males (w +) were then crossed to yw virgins and gDNA extracted from adult progeny to screen for enhancer removal by PCR.
Genotype Nomenclature
For each line, a total of seven genotypes were analyzed: (1) the intact construct containing enhancer and promoter homozygote (P+E+/P+E+); (2) the intact construct containing enhancer and promoter hemizygote (P+E+/+); (3) deleted “promoterless” homozygote (P-/P-); (4) deleted “promoterless” hemizygote (P-/+); (5) deleted vgBE “enhancerless” homozygote (E-/E-); (6) deleted vgBE “enhancerless” hemizygote (E-/+) and (7) Trans-heterozygote (transvection) (P-/E-), derived from crossing “promoterless” and “enhancerless” homozygotes. All hemizygotes were obtained by crossing the homozygote to yw or y 2 wct 6 ; su(Hw)e04061/TM6B, Tb1.
Immunostaining and Microscopy
Immunostaining was performed as described [66]. Wing imaginal discs were dissected from the late third instar larvae in SFX-Insect media (Hyclone) and fixed with 500µl Fixation Buffer (4% PFA/0.5%Triton/1xPBS) for 30 min at RT with rotation. The discs were washed 3X with Block-Permeabilization solution (1% BSA/0.5% Triton/1X PBS) for 10 min each, and then incubated in the same solution for 1 hr at RT with rotation. Discs were then incubated with α-GFP (Invitrogen) diluted 1:350 in wash solution (1% BSA/0.1% Triton/1X PBS) for 1 hr at RT. After washing 3X for 10 min each, discs were incubated with α-rabbit IgG-Texas Red secondary antibody (Jackson ImmunoResearch) at 1:500 for 1 hr at RT, washed 3X with PBST for 10 min each, counterstained with DAPI and rinsed with H2O. Discs were then oriented on coverslips containing Poly-L lysine and mounted in Vectashield.
Images were obtained on a Lecia DM6000B widefield epifluorescent microscope equipped with a Hamamatsu ORCA-ER CCD camera and a HC PL FLUOTAR 20x/.50NA objective. Simple PCI (v6.6) was used for acquisition of raw images, which were processed using AutoQuant's 3D Deconvolution Algorithm utilizing an adaptive (blind) PSF implemented into Lecia Deblur (v2.3.2) software. All seven genotypes for each line were processed and imaged at the same time using identical immunostaining, microscope, camera and software settings. Image level normalization, minimum/maximum correction and false coloring were performed using ImageJ (v1.47n).
RNA Isolation & cDNA Synthesis
Total RNA was isolated from wing imaginal discs dissected from late third instar larva. 10-15 discs were dissected in SFX media and homogenized in 300 µl TRIzol (Invitrogen) by vortexing for 30 sec. 60 µl chloroform was added and vortexed for 15 s, centrifuged at 12,000g for 10 min at 4°C and the upper aqueous layer precipitated with 150μl isopropanol. Samples were incubated at RT for 10 min and centrifuged at 12,000g for 10 min at 4°C. RNA pellets were washed with 80% EtOH and resuspended in 8.5 μl nuclease-free H2O. Genomic DNA was removed by DNAse treatment (TURBO DNA-Free, Ambion/Life Technologies) by incubating at 37°C for 20 min. Concentrations and purity were determined using a NanoDrop spectrophotometer, and 500ng of RNA was used for cDNA synthesis using either the iScriptTM cDNA synthesis kit containing a blend of oligo dT and random hexamers or the iScriptTM Select cDNA synthesis kit with oligo dT primers only (su(Hw)e04061 mutant analysis) (BioRad) for 1 hr at 42°C.
Real Time PCR & Data Normalization
qPCR runs were performed on a BioRad iQ5 cycler using iQ SYBR Green Supermix (BioRad) using 1 μl of cDNA and primers specific for eGFP and Rp49. Both primer sets displayed 99-101% efficiencies. Three biological replicates for each genotype and 3 technical replicates were used and the relative expression was calculated by comparing eGFP Ct values to Rp49 Ct values following the ΔCt method. For each line, the relative expression for the intact homozygote genotype (P+E+/P+E+) was taken to be 1 and the other genotypes normalized accordingly. To derive the final data, the normalized values from all available lines were averaged. Error bars represent standard error of the mean (S.E.M.). Primer sequences are available upon request.
Results
A P-element eGFP Reporter System Engineered to Induce Artificial Mutations in Enhancers and Promoters Shows Insertion Bias To Endogenous Insulators
The somatic pairing dependence required for transvection ultimately derives from the ability of enhancers to act on promoters in close proximity in trans when preferential interactions with promoters in cis are lost. This feature is supported by the fact that a majority of classical complementing mutant alleles disrupt either the enhancer or the promoter [15]. Our P-element reporter constructs were designed with site-specific recombination sites flanking both these elements in order to selectively delete either the enhancer or the promoter. We chose the vestigial boundary enhancer (vgBE), which drives expression in a small stripe of cells at the dorsal/ventral boundary in developing wing and haltere imaginal discs [65] and flanked it with FRT sites. The minimal hsp70 promoter was flanked by loxP sites and followed by the eGFP reporter. Two other derivatives of this construct were created by adding either a single gypsy insulator upstream of the vgBE or two gypsy insulators flanking the entire construct (Figure 1A).
Following microinjection, we recovered 11 independent insertions for the 2-insulator construct, 18 insertions for the 1-insulator construct and 42 insertions for the 0-insulator construct. Analysis of homozygous individuals carrying two copies of the construct from all eleven 2-insulator lines displayed strong eGFP expression in the D/V compartment boundary of the wing discs, particularly along the wing margin within the wing pouch and the hinge region (Figure 1B), whereas only nine 1-insulator line homozygotes and seven 0-insulator line homozygotes displayed detectable eGFP levels, likely the result of position effects [67]. Mapping of these lines revealed a noticeable bias towards the 5' end of genes, transposable elements and other transposon “hotspots”, similar to observations in previous reports [68-70]. Although no correlation with chromatin states/domains were observed [71], nearly all insertions outside of transposons were within 1 Kb of an endogenous protein-bound insulator, particularly those bound by GAF and/or CP190 regardless of whether a gypsy insulator was present in the construct (Table 1-3).
Table 1. Chromosome and genomic coordinates for 2-insulator lines.
| LINE | CHRM. | GENOMIC COORD. | INSERTION TYPE | HOTSPOTb | CHROMATIN TYPEd | INSULATOR PROTEINS |
|---|---|---|---|---|---|---|
| PGP-13B | 3 | 3L: 7,127,316 | Promoter (melted) | * | Blue | CP190, BEAF, Mod, CTCF, GAF, Su(Hw) |
| PGP-20 | 2 | -c | Transposon (Invader) | - | - | - |
| PGP-22A | 3 | 3L: 13,221,600 | Promoter (caps) | * | Blue | CP190, BEAF, Mod, CTCF, GAF |
| PGP-23A | 3 | -c | Transposon (F) | - | - | - |
| PGP-25A | 3 | 3R: 22,360,900 | Promoter (CG6490) | * | Blue/Red | CP190, Mod, GAF |
| PGP-28a | 2 | 2L: 12,018,983 | Exon (CG6734) | - | Yellow | - |
| PGP-33Aa | 2 | 2L: 7,576,480 | Promoter (RapGAP1) | * | Red | CP190, Mod, GAF, SuHw |
| PGP-39Aa | 2 | 2L: 8,989,200 | Promoter (rost) | - | Red | - |
| PGP-50Aa | 2 | 2L: 9,758,467 | 5' UTR (zf30C) | * | Black | CP190, BEAF, Mod, CTCF, GAF |
| PGP-104A1a | 2 | 2L: 20,163,757 | Transposon (Invader) | - | - | - |
| PGP-146C2 | 3 | 3R: 8,326,193 | Intergenic | - | Black | - |
a Used for Su(Hw) mutant analysis.
b Transposon hotspot if more than 5 insertions for other P-element or Pbacs were found at this position in FlyBase.
c Not enough flanking sequence recovered to accurately predict insertion position.
d From: Filion et al (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212-224.
Table 2. Chromosome and genomic coordinates for 1-insulator lines.
| LINE | CHRM. | GENOMIC COORD. | INSERTION TYPE | HOTSPOTa | CHROMATIN TYPEc | INSULATOR PROTEINS |
|---|---|---|---|---|---|---|
| gi-ve 2 | 3 | 3L: 5,177,560 | Intron (shep) | * | Red | GAF |
| gi-ve 2A | 3 | 3R: 24,816,510 | Intergenic | * | Black | GAF |
| gi-ve 9A | 2 | -b | Transposon (gypsy) | - | - | - |
| gi-ve 20 | 3 | -b | Transposon (Diver) | - | - | - |
| gi-ve 29 | 3 | 3R: 7,392,923 | Promoter (sea/fabp) | * | Red | CTCF, GAF, Su(Hw) |
| gi-ve 31 | 2 | 2R: 6,762,150 | Promoter (CG30015) | * | Red/Yellow | CP190, BEAF, CTCF, GAF |
| gi-ve 40 | 3 | 3L: 13,470,406 | Promoter (stv) | * | Black | GAF |
| gi-ve 45 | 3 | -b | Transposon (Doc) | - | - | - |
| gi-ve 7D | X | X: 13,072,760 | Intron (CG34411) | - | Black | BEAF, CP190, CTCF |
a Transposon hotspot if more than 5 insertions for other P-element or Pbacs were found at this position in FlyBase.
b Not enough flanking sequence recovered to accurately predict insertion position.
c From: Filion et al (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212-224.
Table 3. Chromosome and genomic coordinates for 0-insulator lines.
| LINE | CHRM. | GENOMIC COORD. | INSERTION TYPE | HOTSPOTa | CHROMATIN TYPEb | INSULATOR PROTEINS |
|---|---|---|---|---|---|---|
| PNG-10 | X | X: 19,671,651 | Exon (vfl) | - | Blue | - |
| PNG-11 | X | X: 17,793,173 | Promoter (CG32495) | - | Yellow | CP190, BEAF, CTCF, Mod |
| PNG-20 | 2 | 2R: 17,557,761 | 5' UTR (NC2alpha) | - | Yellow | CP190, BEAF |
| PNG-44 | 3 | 3L: 6,957,768 | Promoter (sgl) | * | Yellow | CP190, BEAF, GAF |
| PNG-46 | 3 | 3R: 24,953,620 | Transposon (Opus) | - | - | - |
| PNG-50 | 3 | 3R: 16,886,068 | Promoter (Mvl) | * | Yellow | CP190, BEAF, CTCF, Mod, GAF |
| PNG-1C | 2 | 2L: 9,616,828 | 5' UTR (GlcAT-S) | * | Red | CP190, BEAF, CTCF, Mod, GAF |
a Transposon hotspot if more than 5 insertions for other P-element or Pbacs were found at this position using FlyBase.
b From: Filion et al (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212-224.
To test the gypsy insulator's contribution to transvection, we generated “enhancerless” flies lacking the vgBE and “promoterless” flies lacking the hsp70 promoter for each transgenic line by crossing with flies expressing either Flp or Cre recombinase. Trans-heterozygous progeny for each line were then generated by simply crossing “enhancerless” and “promoterless” homozygotes, allowing for a direct readout of transvection effects. We hypothesized that loss of communication between the vgBE and the hsp70 promoter in cis due to promoter removal could be restored in trans by stable interactions between gypsy components, inducing eGFP expression from the “enhancerless” homolog along the D/V boundary of the wing disc (Figure 1B & 1C).
Homolog Pairing Increases Reporter Levels Independently of gypsy
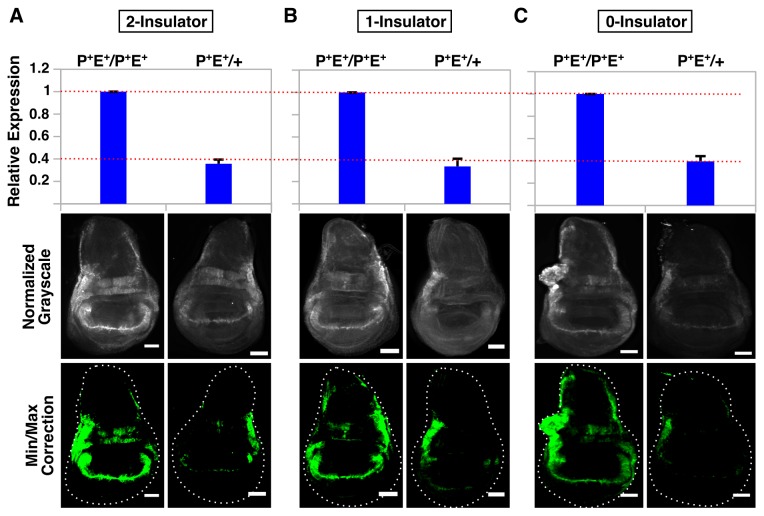
Previous studies in wing imaginal disc have suggested that pairing synapsis between homologous chromosomes can stimulate enhanced transcription of Ubx from both homologs [83]. We began by examining reporter levels in homozygous and hemizygous flies carrying an intact (i.e., non-deleted enhancer/promoter) construct. Immunostaining of wing discs revealed a significant decrease in the amount of GFP protein in hemizygous larvae carrying the construct on only one homolog compared to homozygous larvae carrying a copy on each homolog, a pattern independent of gypsy presence (Figure 2A-2C). In terms of dosage, the expression level of GFP in homozygotes would be expected to be twice the amount observed in hemizygotes, particularly if levels were influenced solely by enhancer-promoter communication in cis. Using qPCR, we measured the levels of GFP expression and found that transcript levels were reduced 2.5-3X in hemizygotes compared to homozygotes, suggesting that pairing in trans can stimulate increased transcription (Figure 2A-2C) in agreement with previous findings [83]. However, this behavior was not significantly influenced by the gypsy insulator, as the 2.5x, 3x, and 2.8x reduction observed for the 0-, 1- and 2-insulator hemizygotes, respectively, rules out any synergistic effect that would be expected if this element contributed to pairing.
Figure 2. Pairing can stimulate eGFP transcription more than two-fold independently of gypsy presence.
QPCR analysis (top graph) and immunostaining (bottom panels) of wing discs from intact homozygotes (P+E+/P+E+) and intact hemizygotes (P+E+/+) for 2-insulator (A), 1-insulator (B) and 0-insulator (C) lines. For microscopy, images were normalized to P+E+/P+E+ for each respective line and minimum/maximum level corrections were applied equally to both genotypes using ImageJ and false-colored green. Error bars represent standard error of the mean (S.E.M) and scalebars are 50 μm.
The vgBE Can Drive Reporter Expression In The Absence of a Functional Promoter
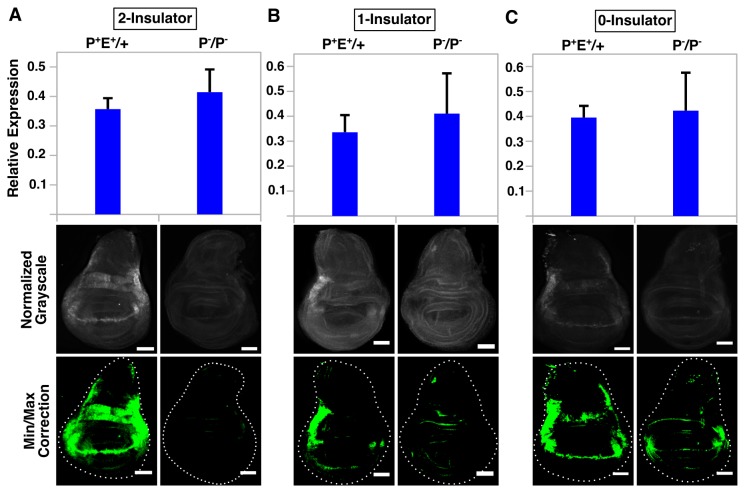
Next, we tested whether gypsy might influence enhancer-promoter communication in trans by removing either the vgBE or the hsp70 minimal promoter to generate “enhancerless” and “promoterless” lines (Figure S1A) and combining them to create trans-heterozygous individuals. However, qPCR revealed a large amount of GFP transcript present in promoterless lines, with levels in the promoterless homozygote (P-/P-) equal or greater to the levels observed in the non-deleted hemizygote (P+E+/+). This pattern of expression was consistent in every single line examined, independent of gypsy insulator presence (Figure 3A-3C), suggesting that the vgBE can drive expression in the absence of a functional promoter. We suspected that perhaps vgBE's proximity to the coding region of GFP (~183 bp after hsp70 promoter deletion) might explain this result, as any RNA Pol II recruited to the vgBE and able to find a suitable transcription start site (TSS) might be able to generate a transcript. We found two TSSs in potential promoters within the vgBE using a neural network prediction algorithm (minimum promoter score=0.8) [72]; however, using qPCR primers designed to only measure mRNAs arising from these TSSs, we could not detect a sufficient amount of transcripts to fully account for the total pool of GFP mRNA (Figure S1B). It is worth noting that our GFP qPCR primers are located at the 3' end of the transcript (within 130 bp of the stop codon), which would fail to distinguish whether other cryptic TSSs located downstream of our vgBE qPCR primers might be utilized and therefore contribute to the pool of transcripts as well.
Figure 3. The vgBE can drive eGFP reporter expression in the absence of a functional promoter but this does not correlate with GFP protein levels.
QPCR analysis (top graph) and immunostaining (bottom panels) of wing discs from intact hemizygotes (P+E+/+) and “promoterless” homozygotes (P-/P-) for 2-insulator (A), 1-insulator (B) and 0-insulator (C) lines. For microscopy, images were normalized to P+E+/+ for each respective line and minimum/maximum level corrections were applied equally to both genotypes using ImageJ and false-colored green. Error bars represent standard error of the mean (S.E.M) and scalebars are 50 μm.
Interestingly, GFP transcript levels as measured by qPCR did not correlate with the amount of GFP protein. Analysis of wing discs stained with α-GFP revealed only a small amount of signal, barely above background levels, or no signal at all along the DV boundary within the wing pouch and hinge in promoterless homozygotes (P-/P-). Non-deleted hemizygotes (P+E+/+), on the other hand, displayed strong signal along the boundary (Figure 3A-3C). This pattern was consistent in all lines examined and independent of gypsy insulator presence. Background levels of expression in other parts of the wing disc were similar between the two genotypes, ruling out the possibility that misexpression by other regulatory elements might contribute to the large amount of GFP transcript in promoterless individuals. Although we have not measured mRNA or protein stability directly in these lines, it should be noted that cDNA generated using two different priming methods (a mix of random hexamers and oligo dT primers or oligo dT primers only) gave identical results (Figure S1C), suggesting that our results are not due to primer bias during cDNA synthesis and that the majority of transcripts being measured were polyadenylated. Taken collectively, our results suggest that although the vgBE can drive reporter expression in the absence of a functional promoter, many of these transcripts do not give rise to functional protein.
Two gypsy Insulators Can Facilitate Enhancer-Promoter Communication in Trans
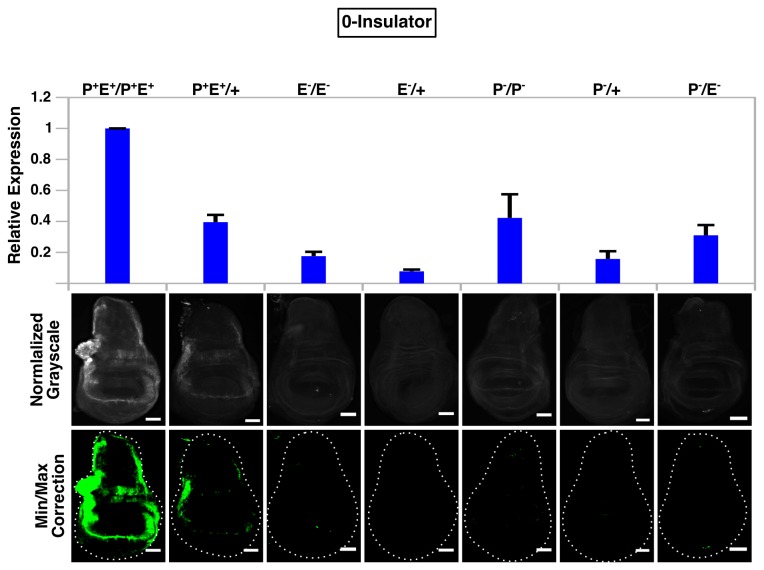
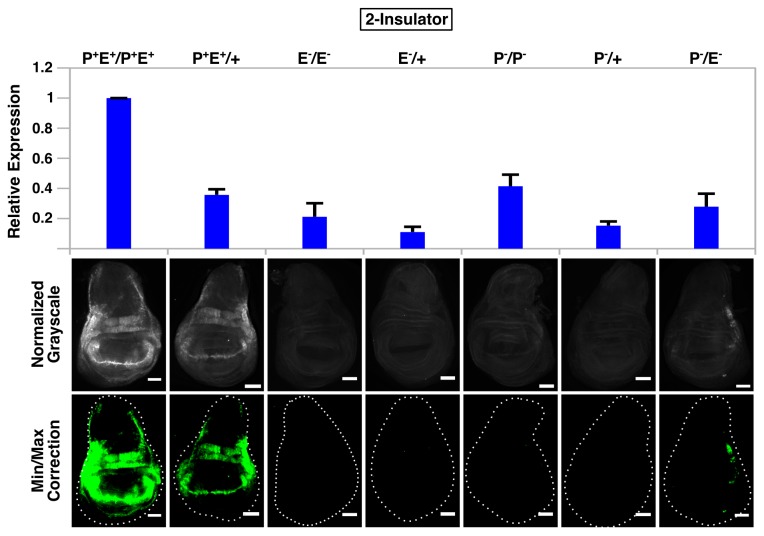
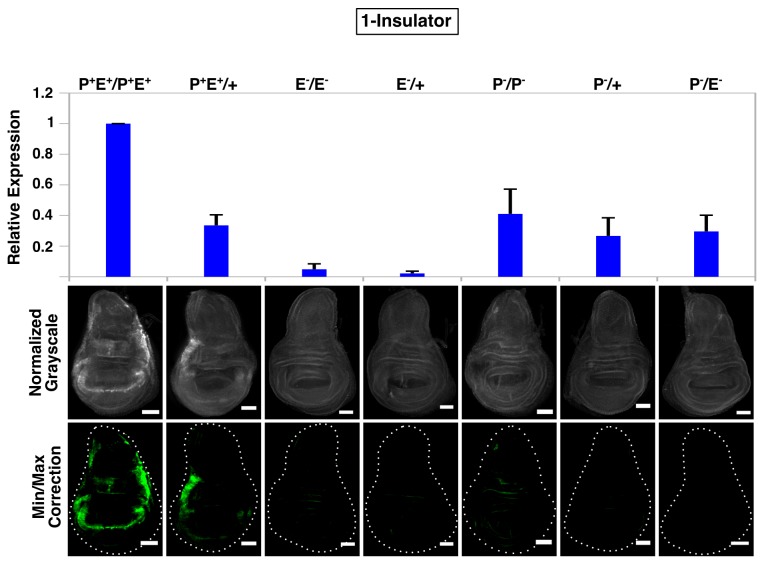
We next determined whether gypsy could promote enhancer-promoter communication in trans by staining wing discs with α-GFP from larvae containing a single functional enhancer on one homolog and a single functional promoter on the other. GFP signal was barely above background or undetectable along the wing margin and hinge region in 0-insulator and 1-insulator trans-heterozygotes (P-/E-) (Figure 4 and Figure 5). However, moderate levels of GFP were observed in 2-insulator trans-heterozygotes, with most of the expression concentrated in the hinge region as staining along the wing margin was considerably weaker and variegated (Figure S2). No signal was observed for either the promoterless (P-/+) or enhancerless (E-/+) hemizygote, and although a small amount of protein could be detected in the promoterless homozygote (P-/P-), the GFP signal was much stronger in the trans-heterozygote (P-/E-) (Figure 6). Taken collectively, these data suggest that the presence of two flanking gypsy insulators is sufficient to support enhancer-promoter communication in trans. Strangely, however, our qPCR results did not agree with our image analysis—we were unable to detect the additional transcripts that should have been present in the 2-insulator trans-heterozygote. Instead, the 2-insulator, 1-insulator and 0-insulator lines all displayed the same behavior: the trans-heterozygote expression level was always the sum of the promoterless (P-/+) and enhancerless (E-/+) hemizygote expression levels, a finding that would be expected if homologs were acting independently of one another (i.e., in cis) and transvection was absent. However, given the caveats of qPCR analysis of our system described earlier (Figure 3), we believe that our immunostaining and subsequent image analysis is the most accurate reflection of whether trans-interactions are occurring or not and therefore conclude that two gypsy insulators are sufficient to mediate transvection.
Figure 4. Absence of gypsy fails to promote enhancer-promoter communication in trans.
QPCR analysis (top graph) and immunostaining (bottom panels) of wing discs from all seven 0-insulator genotypes. For microscopy, all images were normalized to P+E+/P+E+ and minimum/maximum level corrections were applied equally to all genotypes using ImageJ and false-colored green. Error bars represent standard error of the mean (S.E.M) and scalebars are 50 μm.
Figure 6. Flanking gypsy insulators promote enhancer-promoter communication in trans.
QPCR analysis (top graph) and immunostaining (bottom panels) of wing discs from all seven 2-insulator genotypes. For microscopy, all images were normalized to P+E+/P+E+ and minimum/maximum level corrections were applied equally to all genotypes using ImageJ and false-colored green. Error bars represent standard error of the mean (S.E.M) and scalebars are 50 μm.
Figure 5. A single gypsy fails to promote enhancer-promoter communication in trans.
QPCR analysis (top graph) and immunostaining (bottom panels) of wing discs from all seven 1-insulator genotypes. For microscopy, all images were normalized to P+E+/P+E+ and minimum/maximum level corrections were applied equally to all genotypes using ImageJ and false-colored green. Error bars represent standard error of the mean (S.E.M) and scalebars are 50 μm.
Mutations in Su(Hw) Reduce Reporter Levels in a Pairing-Independent manner
Finally, to confirm that the gypsy insulator plays a direct role in mediating transvection, we combined a subset of our 2-insulator lines into a genetically null su(Hw)e04061 mutant background and measured GFP reporter levels by both immunostaining and qPCR. Su(Hw) was the first protein identified shown to be critical for the insulator properties of gypsy and has been shown to be required for somatic pairing in embryos [63,73-75], suggesting that Su(Hw) and other insulator proteins distributed throughout the genome might contribute to transvection in a global manner. If the reduction in transcription levels observed in mutants is attributable to pairing influences, then we would expect that only homozygous individuals carrying the reporter on each homolog would be affected, as hemizygous individuals lacking a suitable pairing region on the opposite homolog should not be affected by loss of such pairing. Immunostaining and qPCR analysis of mutant wing discs revealed a significant decrease in reporter expression for both homozygotes and hemizygotes (Figure S3A). Since transcription does not appear to be globally perturbed in a su(Hw)e04061 background (Figure S3B), it is likely that the reduced reporter expression we observe is due to position effects resulting from the failure of the flanking gypsy insulators to prevent repressive chromatin from spreading into the reporter locus in cis due to loss of Su(Hw) [67], rather than a reduction or loss of homolog pairing.
Discussion
In this work we have utilized a transgenic reporter engineered to induce artificial mutations in the vgBE enhancer and hsp70 promoter in order to test the role of the gypsy insulator in the phenomenon of transvection. We find that although pairing does appear to modestly increase the amount of eGFP transcript arising from intact constructs containing functional regulatory elements in cis on each homolog, this behavior is not dependent on the gypsy insulator. However, flanking gypsy insulators are required for transvection when enhancerless and promoterless homologs were combined in trans. Interestingly, we also found that the vgBE can drive transcription of the reporter in the absence of a functional promoter. From these findings, we conclude that gypsy can support interactions in trans in a dose-dependent manner, although we cannot rule out that other insulator binding sites may contribute to pairing in cooperation with other factors at specific genomic locations, depending on the local chromatin landscape.
Recent reports have utilized a reporter scheme analogous to ours, whose main advantage lies in the fact that transvection can be tested in a tightly controlled manner using fluorophores that allow for single-cell analysis of enhancer-promoter communication in trans, as opposed to phenotypic analysis of whole animals [10,36]. However, unlike those reports, which took advantage of the φC31 integration system to test only a handful of characterized integration sites in the genome [76], we utilized a P-element transformation method to integrate our reporters into a number of different regions of the genome, particularly repetitive sequences, transposon hotspots and the 5' end of genes [68-70] in order to provide a more global analysis of transvection effects.
Image analysis strongly supports the conclusion that the 2-insulator construct can support transvection, as moderate amounts of GFP along the DV boundary, primarily in the hinge region, was readily observable at levels much higher than either promoterless (P-/+) or enhancerless (E-/+) hemizygote, which displayed no signal whatsoever. Perhaps most tellingly, the amount of signal in the promoterless homozygotes (P-/P-) was much lower than the trans-heterozygote, suggesting that pairing a functional enhancer with a functional promoter in trans can positively stimulate transcription above background levels if aided by two gypsy insulators. However, using qPCR we were unable to detect the expected increase in transcripts in the 2-insulator trans-heterozygous lines as compared to either the 1- or 0-insulator lines. We have shown that transcript levels, as measured by qPCR, do not accurately reflect the amount of protein present in the promoterless genotypes—in all lines, regardless of insulator presence, transcript levels were consistently and reproducibly equal to or higher in all promoterless homozygotes (P-/P-) as compared to non-deleted hemizygotes (P+E+/+), despite weak levels of GFP protein staining for P-/P- and high levels for P+E+/+. The most likely explanation for this observation is that the vgBE enhancer in the promoterless transgene can activate transcription using cryptic promoters that lead to a pool of eGFP transcripts that remain untranslated or that are translated into non-functional protein. Therefore, we argue that our immunostaining and image analysis are the most appropriate metric by which to evaluate gypsy insulator function in transvection.
Such spurious transcripts might also explain why we do not observe an increase in the relative number of total transcripts in the 2-insulator transvection lines as compared to the 1- and 0-insulator lines. Rather, perhaps it is simply a shift in the relative ratio of the different types of messages. For simplicity, one could imagine two types of GFP transcripts that differ in their 5' ends: one that gives rise to a functional GFP protein and the other that gives rise to a non-functional protein. In this case, the functional transcript would only arise from the homolog containing the intact promoter (which would only be possible in trans) whereas the non-functional transcript would arise from the promoterless homolog in cis. For 1- and 0-insulator trans-heterozygotes, most eGFP transcripts would be the non-functional message driven in cis due to the failure of the vgBE to stably communicate with the functional promoter in trans. However, in 2-insulator trans-heterozygous individuals, this proportion would be reversed, with most eGFP transcripts being functional as a result of stable communication in trans between the vgBE and functional promoter due to the influence of the flanking gypsy insulators. Note that this argument is only valid if we assume that the total output of the vgBE is equal regardless of whether cis or trans interactions predominate and only if it is acting in either conformation at a given point in time, not both simultaneously. This idea is supported by the fact that although regulatory elements prefer to act in cis [22,77-80], competition between promoters for a single enhancer ultimately dictates whether the enhancer functions primarily in cis or in trans within the same cell [10].
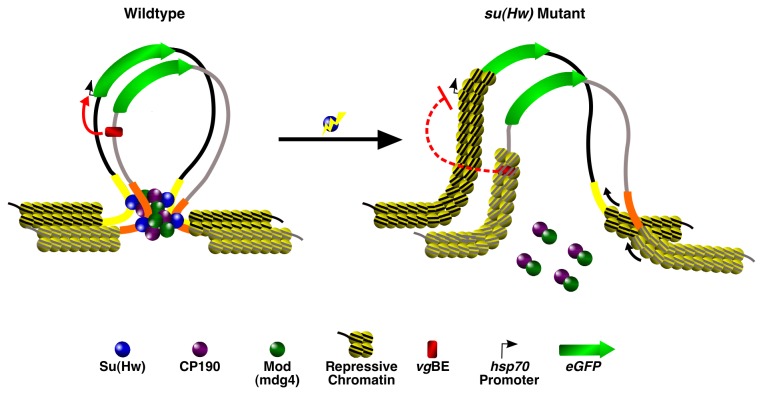
Finally, why do two flanking gypsy insulators, but not a single upstream gypsy insulator, support transvection? One might assume that if gypsy contributes to homolog pairing, then a single insulator located just upstream of the enhancer and promoter would still be capable of ensuring that those two elements remain in close proximity in trans. However, we argue that the most critical determinant is chromatin structure itself—it is widely accepted as a key regulator of transcription in cis, so the same principles would also apply in trans. Even if pairing were to bring enhancers and promoters in close proximity, the underlying chromatin must still be permissible in order for transcription to occur. Insulators were originally identified based on their ability to buffer the effects of surrounding chromatin influences (i.e., position effects) on transgene expression [81,82], and regardless of the mechanism by which insulators accomplish this task (chromatin looping, etc.) it is likely that the transvection we observe is due to the flanking insulators establishing a permissive chromatin environment favorable for transcription. The single gypsy insulator, on the other hand would not be able to establish the same environment and therefore even if pairing were established by other elements, transcription would still be unlikely to occur given the lack of a suitable chromatin landscape (Figure 7). Our su(Hw) mutant data supports this hypothesis, as significant reductions in GFP expression were observed in both homozygotes and hemizygotes, highlighting the importance of chromatin structure on transgene expression regardless of pairing influences. Our findings, along with a number of other studies linking chromatin proteins to transvection [25,32,48-52] and the failure of other studies to observe transvection except when their reporters were located in defined PhiC31 genomic sites that are highly permissible to transcription [36,67], suggests that chromatin itself is the master regulator of this phenomenon.
Figure 7. A model for gypsy insulator function in transvection.
Looping in cis between flanking insulators in a wildtype background prevents silencing of regulatory elements located on opposite homologs. Mutations in su(Hw) disrupt cis looping contacts and alter the local chromatin landscape to a more repressive state, preventing stable communication between regulatory elements even if pairing between homologs is maintained.
Supporting Information
Confirmation of vgBE and hsp70 promoter deletion for a single representative 2-insulator, 1-insulator and 0-insulator line. Schematic (right) shows position of PCR primers (red arrowheads) and the size of each respective element (A). Schematic showing potential position of cryptic transcription start sites (TSSs) in the vgBE (red triangles), the distance between the vgBE and eGFP start codon following promoter removal and the position of test primers (tan and blue arrows) used for qPCR. Graphs showing transcript levels based on these primers for a single “promoterless” (P-/P-) representative from 2-insulator, 1-insulator and 0-insulator lines are shown (B). qPCR analysis of eGFP transcript levels reverse-transcribed using either a mixture of random hexamers+oligo dT primers or oligo dT primers alone for cDNA synthesis for the indicated 2-insulator genotypes (C). All error bars represent the standard error of the mean (S.E.M.).
(TIFF)
2-insulator trans-heterozygote (P-/E- ) GFP protein is enriched primarily in the hinge with a variegated pattern of weak expression in the wing margin. Shown is a collection of 8 different wing discs from a representative 2-insulator P-/E- line (grayscale images) with magnified panels below showing a closeup of the yellow boxed region false-colored in green. All grayscale levels were normalized equally, while min/max corrections to magnified panels were performed separately for each disc using ImageJ. Scalebars in grayscale panels are 50 μm and 10 μm in magnified panels.
(TIFF)
Mutations in su(Hw) decrease reporter level expression in a pairing-independent manner. QPCR analysis (top graph) and immunostaining (bottom panels) of wing discs from all seven 2-insulator genotypes in a TM6B-balanced or su(Hw)e04061 mutant background. For microscopy, all grayscale images were normalized to the balanced P+E+/P+E+, while minimum/maximum level corrections were applied equally to both balanced and su(Hw)e04061 backgrounds based on each individual reporter genotype. Thus, each of the 14 genotypes irrespective of genetic background are directly comparable in the normalized grayscale panels, while only a single genotype (such as P-/P-) is directly comparable between backgrounds in the minimum/maximum corrected panels (A). qPCR analysis of SUMO, Actin and JNK (bsk) expression levels in a balanced or su(Hw)e04061 background (B). Error bars represent standard error of the mean (S.E.M) and scalebars are 50 μm.
(TIFF)
Acknowledgments
We thank Hyuck-Joon Kang for his assistance during the cloning of the p-VgEGFP plasmid and Mark Remec for assistance in mapping. T.S. and R.R. are recipients of Graduate Research Fellowships from the National Science Foundation.
Funding Statement
The authors thank the BCMB Department, the College of Arts and Sciences and Office of Research at the University of Tennessee for support in addition to US Public Health Service Award GM78132-2 from the NIH and MCB-0616081 from NSF to ML. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bulger M, Groudine M (2010) Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol 339: 250-257. doi: 10.1016/j.ydbio.2009.11.035. PubMed: 20025863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krivega I, Dean A (2012) Enhancer and promoter interactions-long distance calls. Curr Opin Genet Dev 22: 79-85. doi: 10.1016/j.gde.2011.11.001. PubMed: 22169023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smallwood A, Ren B (2013) Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol 25: 387-394. doi: 10.1016/j.ceb.2013.02.005. PubMed: 23465541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Szutorisz H, Dillon N, Tora L (2005) The role of enhancers as centres for general transcription factor recruitment. Trends Biochem Sci 30: 593-599. doi: 10.1016/j.tibs.2005.08.006. PubMed: 16126390. [DOI] [PubMed] [Google Scholar]
- 5. Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA et al. (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430-435. doi: 10.1038/nature09380. PubMed: 20720539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M et al. (2009) Transcription factors mediate long-range enhancer-promoter interactions. Proc Natl Acad Sci U S A 106: 20222-20227. doi: 10.1073/pnas.0902454106. PubMed: 19923429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ronshaugen M, Levine M (2004) Visualization of trans-homolog enhancer-promoter interactions at the Abd-B Hox locus in the Drosophila embryo. Dev Cell 7: 925-932. doi: 10.1016/j.devcel.2004.11.001. PubMed: 15572134. [DOI] [PubMed] [Google Scholar]
- 8. Sanyal A, Lajoie BR, Jain G, Dekker J (2012) The long-range interaction landscape of gene promoters. Nature 489: 109-113. doi: 10.1038/nature11279. PubMed: 22955621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su W, Porter S, Kustu S, Echols H (1990) DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci U S A 87: 5504-5508. doi: 10.1073/pnas.87.14.5504. PubMed: 2164685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bateman JR, Johnson JE, Locke MN (2012) Comparing enhancer action in cis and in trans. Genetics 191: 1143-1155. doi: 10.1534/genetics.112.140954. PubMed: 22649083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen JL, Huisinga KL, Viering MM, Ou SA, Wu CT et al. (2002) Enhancer action in trans is permitted throughout the Drosophila genome. Proc Natl Acad Sci U S A 99: 3723-3728. doi: 10.1073/pnas.062447999. PubMed: 11904429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunaway M, Dröge P (1989) Transactivation of the Xenopus rRNA gene promoter by its enhancer. Nature 341: 657-659. doi: 10.1038/341657a0. PubMed: 2797192. [DOI] [PubMed] [Google Scholar]
- 13. Müeller-Storm HP, Sogo JM, Schaffner W (1989) An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell 58: 767-777. doi: 10.1016/0092-8674(89)90110-4. PubMed: 2548735. [DOI] [PubMed] [Google Scholar]
- 14. Wedel A, Weiss DS, Popham D, Dröge P, Kustu S (1990) A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science 248: 486-490. doi: 10.1126/science.1970441. PubMed: 1970441. [DOI] [PubMed] [Google Scholar]
- 15. Duncan IW (2002) Transvection effects in Drosophila. Annu Rev Genet 36: 521-556. doi: 10.1146/annurev.genet.36.060402.100441. PubMed: 12429702. [DOI] [PubMed] [Google Scholar]
- 16. Kennison JA, Southworth JW (2002) Transvection in Drosophila. Adv Genet 46: 399-420. doi: 10.1016/S0065-2660(02)46014-2. PubMed: 11931232. [DOI] [PubMed] [Google Scholar]
- 17. Müller HP, Schaffner W (1990) Transcriptional enhancers can act in trans. Trends Genet 6: 300-304. doi: 10.1016/0168-9525(90)90236-Y. PubMed: 2238088. [DOI] [PubMed] [Google Scholar]
- 18. Tartof KD, Henikoff S (1991) Trans-sensing effects from Drosophila to humans. Cell 65: 201-203. doi: 10.1016/0092-8674(91)90153-P. PubMed: 2015622. [DOI] [PubMed] [Google Scholar]
- 19. Wu CT, Morris JR (1999) Transvection and other homology effects. Curr Opin Genet Dev 9: 237-246. doi: 10.1016/S0959-437X(99)80035-5. PubMed: 10322135. [DOI] [PubMed] [Google Scholar]
- 20. Lewis EB (1954) The Theory and Application of a New Method of Detecting Chromosomal Rearrangements in Drosophila melanogaster. Am Nat 88: 225-239. doi: 10.1086/281833. [DOI] [Google Scholar]
- 21. Geyer PK, Green MM, Corces VG (1990) Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. EMBO J 9: 2247-2256. PubMed: 2162766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morris JR, Chen J, Filandrinos ST, Dunn RC, Fisk R et al. (1999) An analysis of transvection at the yellow locus of Drosophila melanogaster. Genetics 151: 633-651. PubMed: 9927457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gelbart WM (1982) Synapsis-dependent allelic complementation at the decapentaplegic gene complex in Drosophila melanogaster. Proc Natl Acad Sci U S A 79: 2636-2640. doi: 10.1073/pnas.79.8.2636. PubMed: 6806814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zimmerman JE, Bui QT, Liu H, Bonini NM (2000) Molecular genetic analysis of Drosophila eyes absent mutants reveals an eye enhancer element. Genetics 154: 237-246. PubMed: 10628984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Babu P, Bhat S (1980) Effect of zeste on white complementation. Basic Life Sci 16: 35-40. PubMed: 6779800. [DOI] [PubMed] [Google Scholar]
- 26. Gibson JB, Reed DS, Bartoszewski S, Wilks AV (1999) Structural changes in the promoter region mediate transvection at the sn-glycerol-3-phosphate dehydrogenase gene of Drosophila melanogaster. Biochem Genet 37: 301-315. doi: 10.1023/A:1018707226595. PubMed: 10626038. [DOI] [PubMed] [Google Scholar]
- 27. Lee JJ, von Kessler DP, Parks S, Beachy PA (1992) Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell 71: 33-50. doi: 10.1016/0092-8674(92)90264-D. PubMed: 1394430. [DOI] [PubMed] [Google Scholar]
- 28. Buratovich MA, Phillips RG, Whittle JR (1997) Genetic relationships between the mutations spade and Sternopleural and the wingless gene in Drosophila development. Dev Biol 185: 244-260. doi: 10.1006/dbio.1997.8562. PubMed: 9187086. [DOI] [PubMed] [Google Scholar]
- 29. Condie JM, Brower DL (1989) Allelic interactions at the engrailed locus of Drosophila: engrailed protein expression in imaginal discs. Dev Biol 135: 31-42. doi: 10.1016/0012-1606(89)90155-3. PubMed: 2504633. [DOI] [PubMed] [Google Scholar]
- 30. Scholz H, Deatrick J, Klaes A, Klämbt C (1993) Genetic dissection of pointed, a Drosophila gene encoding two ETS-related proteins. Genetics 135: 455-468. PubMed: 8244007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Locke J, Tartof KD (1994) Molecular analysis of cubitus interruptus (ci) mutations suggests an explanation for the unusual ci position effects. Mol Gen Genet 243: 234-243. PubMed: 8177220. [DOI] [PubMed] [Google Scholar]
- 32. Pattatucci AM, Kaufman TC (1991) The homeotic gene Sex combs reduced of Drosophila melanogaster is differentially regulated in the embryonic and imaginal stages of development. Genetics 129: 443-461. PubMed: 1683847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dreesen TD, Henikoff S, Loughney K (1991) A pairing-sensitive element that mediates trans-inactivation is associated with the Drosophila brown gene. Genes Dev 5: 331-340. doi: 10.1101/gad.5.3.331. PubMed: 1848201. [DOI] [PubMed] [Google Scholar]
- 34. Henikoff S, Dreesen TD (1989) Trans-inactivation of the Drosophila brown gene: evidence for transcriptional repression and somatic pairing dependence. Proc Natl Acad Sci U S A 86: 6704-6708. doi: 10.1073/pnas.86.17.6704. PubMed: 2505257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williams JA, Bell JB, Carroll SB (1991) Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes Dev 5: 2481-2495. doi: 10.1101/gad.5.12b.2481. PubMed: 1752439. [DOI] [PubMed] [Google Scholar]
- 36. Mellert DJ, Truman JW (2012) Transvection is common throughout the Drosophila genome. Genetics 191: 1129-1141. doi: 10.1534/genetics.112.140475. PubMed: 22649078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McKee BD (2004) Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta 1677: 165-180. doi: 10.1016/j.bbaexp.2003.11.017. PubMed: 15020057. [DOI] [PubMed] [Google Scholar]
- 38. Aramayo R, Metzenberg RL (1996) Meiotic transvection in fungi. Cell 86: 103-113. doi: 10.1016/S0092-8674(00)80081-1. PubMed: 8689677. [DOI] [PubMed] [Google Scholar]
- 39. Matzke M, Mette MF, Jakowitsch J, Kanno T, Moscone EA et al. (2001) A test for transvection in plants: DNA pairing may lead to trans-activation or silencing of complex heteroalleles in tobacco. Genetics 158: 451-461. PubMed: 11333252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu H, Huang J, Wang J, Jiang S, Bailey AS et al. (2008) Transvection mediated by the translocated cyclin D1 locus in mantle cell lymphoma. J Exp Med 205: 1843-1858. doi: 10.1084/jem.20072102. PubMed: 18625744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rassoulzadegan M, Magliano M, Cuzin F (2002) Transvection effects involving DNA methylation during meiosis in the mouse. EMBO J 21: 440-450. doi: 10.1093/emboj/21.3.440. PubMed: 11823436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sandhu KS, Shi C, Sjölinder M, Zhao Z, Göndör A et al. (2009) Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev 23: 2598-2603. doi: 10.1101/gad.552109. PubMed: 19933149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Joyce EF, Williams BR, Xie T, Wu CT (2012) Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLOS Genet 8: e1002667 PubMed: 22589731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dorsett D (2011) Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 21: 199-206. doi: 10.1016/j.gde.2011.01.018. PubMed: 21324671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T et al. (2009) Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410-413. PubMed: 19458616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S et al. (2009) Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLOS Genet 5: e1000739 PubMed: 19956766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Williams BR, Bateman JR, Novikov ND, Wu CT (2007) Disruption of topoisomerase II perturbs pairing in drosophila cell culture. Genetics 177: 31-46. doi: 10.1534/genetics.107.076356. PubMed: 17890361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jack JW, Judd BH (1979) Allelic pairing and gene regulation: A model for the zeste-white interaction in Drosophila melanogaster. Proc Natl Acad Sci U S A 76: 1368-1372. doi: 10.1073/pnas.76.3.1368. PubMed: 16592632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leiserson WM, Bonini NM, Benzer S (1994) Transvection at the eyes absent gene of Drosophila. Genetics 138: 1171-1179. PubMed: 7896099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pirrotta V, Steller H, Bozzetti MP (1985) Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J 4: 3501-3508. PubMed: 3004963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sipos L, Mihály J, Karch F, Schedl P, Gausz J et al. (1998) Transvection in the Drosophila Abd-B domain: extensive upstream sequences are involved in anchoring distant cis-regulatory regions to the promoter. Genetics 149: 1031-1050. PubMed: 9611211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu CT, Jones RS, Lasko PF, Gelbart WM (1989) Homeosis and the interaction of zeste and white in Drosophila. Mol Gen Genet 218: 559-564. doi: 10.1007/BF00332424. PubMed: 2511424. [DOI] [PubMed] [Google Scholar]
- 53. Schoborg TA, Labrador M (2010) The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific. J Mol Evol 70: 74-84. doi: 10.1007/s00239-009-9310-x. PubMed: 20024537. [DOI] [PubMed] [Google Scholar]
- 54. Bushey AM, Ramos E, Corces VG (2009) Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23: 1338-1350. doi: 10.1101/gad.1798209. PubMed: 19443682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K et al. (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24-32. PubMed: 19056695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA et al. (2010) A comprehensive map of insulator elements for the Drosophila genome. PLOS Genet 6: e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Handoko L, Xu H, Li G, Ngan CY, Chew E et al. (2011) CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43: 630-638. doi: 10.1038/ng.857. PubMed: 21685913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hou C, Li L, Qin ZS, Corces VG (2012) Gene Density, Transcription, and Insulators Contribute to the Partition of the Drosophila Genome into Physical Domains. Mol Cell 48: 471-484. doi: 10.1016/j.molcel.2012.08.031. PubMed: 23041285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B et al. (2012) Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell 148: 458-472. doi: 10.1016/j.cell.2012.01.010. PubMed: 22265598. [DOI] [PubMed] [Google Scholar]
- 60. Yang J, Corces VG (2012) Insulators, long-range interactions, and genome function. Curr Opin Genet Dev 22: 86-92. doi: 10.1016/j.gde.2011.12.007. PubMed: 22265227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J et al. (2006) Interchromosomal interactions and olfactory receptor choice. Cell 126: 403-413. doi: 10.1016/j.cell.2006.06.035. PubMed: 16873069. [DOI] [PubMed] [Google Scholar]
- 62. Georgiev PG, Corces VG (1995) The su(Hw) protein bound to gypsy sequences in one chromosome can repress enhancer-promoter interactions in the paired gene located in the other homolog. Proc Natl Acad Sci U S A 92: 5184-5188. doi: 10.1073/pnas.92.11.5184. PubMed: 7761470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fritsch C, Ploeger G, Arndt-Jovin DJ (2006) Drosophila under the lens: imaging from chromosomes to whole embryos. Chromosome Res 14: 451-464. doi: 10.1007/s10577-006-1068-z. PubMed: 16821139. [DOI] [PubMed] [Google Scholar]
- 64. Barolo S, Carver LA, Posakony JW (2000) GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques 29: 726, 11056799. [DOI] [PubMed] [Google Scholar]
- 65. Williams JA, Paddock SW, Vorwerk K, Carroll SB (1994) Organization of wing formation and induction of a wing-patterning gene at the dorsal/ventral compartment boundary. Nature 368: 299-305. doi: 10.1038/368299a0. PubMed: 8127364. [DOI] [PubMed] [Google Scholar]
- 66. Saint Phalle B (2003) Immunostaining of Whole-Mount Imaginal Discs. Drosophila Cytogenetics Protocols. pp. 373-387. [Google Scholar]
- 67. Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N (2008) Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40: 476-483. doi: 10.1038/ng.101. PubMed: 18311141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bellen HJ, Levis RW, Liao G, He Y, Carlson JW et al. (2004) The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761-781. doi: 10.1534/genetics.104.026427. PubMed: 15238527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Oh SW, Kingsley T, Shin HH, Zheng Z, Chen HW et al. (2003) A P-element insertion screen identified mutations in 455 novel essential genes in Drosophila. Genetics 163: 195-201. PubMed: 12586707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T et al. (1999) The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153: 135-177. PubMed: 10471706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J et al. (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212-224. doi: 10.1016/j.cell.2010.09.009. PubMed: 20888037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Reese MG (2001) Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26: 51-56. doi: 10.1016/S0097-8485(01)00099-7. PubMed: 11765852. [DOI] [PubMed] [Google Scholar]
- 73. Modolell J, Bender W, Meselson M (1983) Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A 80: 1678-1682. doi: 10.1073/pnas.80.6.1678. PubMed: 6300868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Parkhurst SM, Corces VG (1986) Interactions among the gypsy transposable element and the yellow and the suppressor of hairy-wing loci in Drosophila melanogaster. Mol Cell Biol 6: 47-53. PubMed: 3023836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Parkhurst SM, Harrison DA, Remington MP, Spana C, Kelley RL et al. (1988) The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev 2: 1205-1215. doi: 10.1101/gad.2.10.1205. PubMed: 2462523. [DOI] [PubMed] [Google Scholar]
- 76. Groth AC, Fish M, Nusse R, Calos MP (2004) Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775-1782. doi: 10.1534/genetics.166.4.1775. PubMed: 15126397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Casares F, Bender W, Merriam J, Sánchez-Herrero E (1997) Interactions of Drosophila Ultrabithorax regulatory regions with native and foreign promoters. Genetics 145: 123-137. PubMed: 9017395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Geyer PK, Corces VG (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 1865-1873. doi: 10.1101/gad.6.10.1865. PubMed: 1327958. [DOI] [PubMed] [Google Scholar]
- 79. Gohl D, Müller M, Pirrotta V, Affolter M, Schedl P (2008) Enhancer blocking and transvection at the Drosophila apterous locus. Genetics 178: 127-143. doi: 10.1534/genetics.107.077768. PubMed: 18202363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Martínez-Laborda A, González-Reyes A, Morata G (1992) Trans regulation in the Ultrabithorax gene of Drosophila: alterations in the promoter enhance transvection. EMBO J 11: 3645-3652. PubMed: 1396564. PubMed: 1396564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kellum R, Schedl P (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell 64: 941 - 950. doi: 10.1016/0092-8674(91)90318-S. PubMed: 1848159. [DOI] [PubMed] [Google Scholar]
- 82. Udvardy A, Maine E, Schedl P (1985) The 87A7 chromomere : Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol 185: 341-358. doi: 10.1016/0022-2836(85)90408-5. PubMed: 2997449. [DOI] [PubMed] [Google Scholar]
- 83. Goldsborough AS, Kornberg TB (1996) Reduction of transcription by homologue asynapsis in Drosophila imaginal discs. Nature 381: 807-810. doi: 10.1038/381807a0. PubMed: 8657287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confirmation of vgBE and hsp70 promoter deletion for a single representative 2-insulator, 1-insulator and 0-insulator line. Schematic (right) shows position of PCR primers (red arrowheads) and the size of each respective element (A). Schematic showing potential position of cryptic transcription start sites (TSSs) in the vgBE (red triangles), the distance between the vgBE and eGFP start codon following promoter removal and the position of test primers (tan and blue arrows) used for qPCR. Graphs showing transcript levels based on these primers for a single “promoterless” (P-/P-) representative from 2-insulator, 1-insulator and 0-insulator lines are shown (B). qPCR analysis of eGFP transcript levels reverse-transcribed using either a mixture of random hexamers+oligo dT primers or oligo dT primers alone for cDNA synthesis for the indicated 2-insulator genotypes (C). All error bars represent the standard error of the mean (S.E.M.).
(TIFF)
2-insulator trans-heterozygote (P-/E- ) GFP protein is enriched primarily in the hinge with a variegated pattern of weak expression in the wing margin. Shown is a collection of 8 different wing discs from a representative 2-insulator P-/E- line (grayscale images) with magnified panels below showing a closeup of the yellow boxed region false-colored in green. All grayscale levels were normalized equally, while min/max corrections to magnified panels were performed separately for each disc using ImageJ. Scalebars in grayscale panels are 50 μm and 10 μm in magnified panels.
(TIFF)
Mutations in su(Hw) decrease reporter level expression in a pairing-independent manner. QPCR analysis (top graph) and immunostaining (bottom panels) of wing discs from all seven 2-insulator genotypes in a TM6B-balanced or su(Hw)e04061 mutant background. For microscopy, all grayscale images were normalized to the balanced P+E+/P+E+, while minimum/maximum level corrections were applied equally to both balanced and su(Hw)e04061 backgrounds based on each individual reporter genotype. Thus, each of the 14 genotypes irrespective of genetic background are directly comparable in the normalized grayscale panels, while only a single genotype (such as P-/P-) is directly comparable between backgrounds in the minimum/maximum corrected panels (A). qPCR analysis of SUMO, Actin and JNK (bsk) expression levels in a balanced or su(Hw)e04061 background (B). Error bars represent standard error of the mean (S.E.M) and scalebars are 50 μm.
(TIFF)