Abstract
The lungworm, Dictyocaulus viviparus, causes parasitic bronchitis in cattle, and is responsible for substantial economic losses in temperate regions of the world. Here, we undertake the first large-scale exploration of available transcriptomic data for this lungworm, examine differences in transcription between different stages/both genders and identify and prioritize essential molecules linked to fundamental metabolic pathways, which could represent novel drug targets. Approximately 3 million expressed sequence tags (ESTs), generated by 454 sequencing from third-stage larvae (L3) as well as adult females and males of D. viviparus, were assembled and annotated. The assembly of these sequences yielded ~61,000 contigs, of which relatively large proportions encoded collagens (4.3%), ubiquitins (2.1%) and serine/threonine protein kinases (1.9%). Subtractive analysis in silico identified 6,928 nucleotide sequences as being uniquely transcribed in L3, and 5,203 and 7,889 transcripts as being exclusive to the adult female and male, respectively. Most peptides predicted from the conceptual translations were nucleoplasmins (L3), serine/threonine protein kinases (female) and major sperm proteins (male). Additional analyses allowed the prediction of three drug target candidates, whose Caenorhabditis elegans homologues were linked to a lethal RNA interference phenotype. This detailed exploration, combined with future transcriptomic sequencing of all developmental stages of D. viviparus, will facilitate future investigations of the molecular biology of this parasitic nematode as well as genomic sequencing. These advances will underpin the discovery of new drug and/or vaccine targets, focused on biotechnological outcomes.
Keywords: Dictyocaulus viviparous, Bovine lungworm, Next-generation sequencing, Bioinformatics, Transcriptome, Ancylostoma-secreted proteins, Drug target prediction
1. Introduction
Parasitic nematodes of livestock are responsible for substantial economic losses due to poor productivity, failure to thrive and deaths (Coles, 2001; Panuska, 2006). Diseases caused by these nematodes cost the meat and livestock industries billions of dollars per annum (Jackson et al., 2007). Lungworms of the genus Dictyocaulus (Strongylida: Dictyocaulidae) cause parasitic bronchitis (= dictyocaulosis) in different ruminant hosts, particularly in young animals (Eysker and van Miltenburg, 1988; David 1997; reviewed by Panuska, 2006). The clinical manifestation of dictyocaulosis varies from mild respiratory signs to emphysema and pneumonia, and can result in death in severely affected animals (Coles, 2001; Schnieder et al., 1991; Panuska, 2006). The life cycle of Dictyocaulus viviparus, the bovine lungworm, is direct (Anderson, 2001). The adult worms live in the bronchi and bronchioles of cattle; the ovoviviparous females produce eggs from which first-stage larvae (L1) hatch within the lungs or the intestinal tract. L1s are excreted in the faeces and develop, under favourable environmental conditions, into third-stage larvae (L3) within ~4–6 days. The cuticle from the second-stage larva (L2) is retained as a sheath around the L3 and protects this stage from adverse environmental conditions. L3s are ingested by grazing cattle, exsheath, penetrate the intestinal wall and migrate to the mesenteric lymph nodes, where they moult to fourth-stage larvae (L4); L4s are then transported to the lungs, where they penetrate the alveoli and develop into adults (within 3–4 weeks). However, larval stages can undergo arrested development in the lungs of the vertebrate host for up to 5 months (von Samson-Himmelstjerna and Schnieder, 1999; Anderson, 2001)..
Traditionally, the control of Dictyocaulus infection and dictyocaulosis has been based on the prophylactic administration of an irradiated larval vaccine (Jarrett et al., 1960) or anthelmintic drugs, such as macrocyclic lactones (Schnieder et al., 1996; Ploeger, 2002). However, the disadvantages of the current (live) vaccination strategies (i.e., instability of the irradiated larvae and inability to confer a sterile immunity and a life-long protection; McKeand, 2000), the unsuccessful attempts to develop a recombinant vaccine and recent reports of emerging anthelmintic resistance (Matthews et al., 2001; Ploeger, 2002; Molento et al., 2006) are driving the search for new intervention targets. The genomic-bioinformatic explorations of fundamental aspects of the molecular biology of D. viviparus could provide a basis for a detailed understanding of mechanisms linked to parasite development, survival, reproduction and interaction/s with the bovine host; such advances in knowledge could represent a platform for the identification of essential genes and/or gene products and the subsequent validation in vitro and in vivo of rationally designed nematocides.
Although knowledge of the genomics and cellular biology of lungworms is limited, recent molecular studies used quantitative real-time PCR to elucidate patterns of transcription of individual genes for different developmental stages of D. viviparus (see Strube et al., 2007a, 2009a, b) and a suppressive-subtractive hybridization approach to explore differential transcription in hypobiosis-induced versus non-induced L3s (Strube et al., 2007b). Another study (Ranganathan et al., 2007) utilised a conventional (Sanger) sequencing approach to determine and analyse ~4,500 expressed sequence tags (ESTs) from adult D. viviparus using a semi-automated bioinformatic pipeline (ESTExplorer; Nagaraj et al., 2007); these authors identified conserved protein domains and linked them to known biological pathways, based on comparative analyses with the free-living nematode Caenorhabditis elegans and/or other organisms, employing data available in public databases. However, in the latter study, differences in transcription among stages and between sexes of D. viviparus were not investigated on a large scale.
Advances in sequencing techniques and computational approaches for the pre-processing, assembly and annotation of sequence data (Morozova and Marra, 2008; Metzker, 2010) are leading to a much better understanding of the transcriptomes of parasitic helminths (e.g., Cantacessi et al., 2010a, b; Wang et al., 2010; Young et al., 2010a, b). In particular, next-generation sequencing technologies (NGS), such as 454-Roche (www.454.com; Margulies et al., 2005) and Illumina-Solexa (www.illumina.com; Bentley et al., 2008), are enhancing our understanding of the molecular processes involved in parasite development, reproduction and interactions with their hosts (see Cantacessi et al., 2010b; Wang et al., 2010). In addition, given that the data files generated by these technologies are often gigabytes (1×109) to terabytes (1×1012) in size, such that many web-interfaces are no longer able to cope with large-scale analyses, we recently developed a semi-automated, custom-built bioinformatic workflow system for the detailed analysis and annotation of NGS data (Cantacessi et al., 2010c). In the present article, through a detailed exploration of available NGS data using this integrated system, we have reviewed and substantially expanded the knowledge of the transcriptome of D. viviparus, and mined for essential genes in each stage/gender in a first effort to predict new drug targets in this parasite.
2. Next-generation technological approaches
Transcriptomic datasets for the L3, adult female and adult male stages of D. viviparus (SRA_XXXXXXX), determined using a massively parallel sequencing approach (see Cantacessi et al., 2010a), were annotated and analysed using a custom-built bioinformatic workflow system (Cantacessi et al., 2010c). FASTA and associated files of sequence quality scores for each dataset were extracted from each SFF-file; sequence adaptors were clipped using the ‘sff_extract’ software (available at http://bioinf.comav.upv.es/sff_extract/index.html). For each stage/sex, sequences were assembled de novo with sequence quality scores using the Contig Assembly Program v.3 (CAP3; Huang and Madan, 1999), employing a minimum sequence overlap length of 40 nucleotides and an identity threshold of 90%. ESTs (and associated sequence quality scores) from all datasets were then combined and assembled using the same parameters as described above. Sequences from adult male, female and L3 of D. viviparus (n = 4,463; www.ncbi.nlm.nih.gov) available from previous studies (Ranganathan et al., 2007; Strube et al., 2007b) were included for comparison. A small number of sequences in the present data (n = 103; i.e., 0.1% of 61,134 contigs) with a perfect match to those available for Bos taurus (GenBank accession numbers T25280-GW425382; e-value cut-off: < 1e-15) were excluded. Contigs and singletons in each of the assemblies were then compared (using BLASTn and BLASTx algorithms; Altschul et al., 1997) with sequences available in public databases, including NCBI (www.ncbi.nlm.nih.gov) and the EMBL-EBI Parasite Genome Blast Server (www.ebi.ac.uk), in order to identify putative homologues (cf. Koonin, 2010) in C. elegans, other nematodes and organisms other than nematodes (e-value cut-off: < 1e-05). WormBase (release WS200; www.wormbase.org) was interrogated extensively for relevant information on C. elegans homologues, including transcriptomic, proteomic, RNAi phenotypic and interactomic data.
Peptides were conceptually translated from contigs and singletons using ESTScan (Iseli et al., 1999). Predicted peptides were classified functionally by InterPro and/or Pfam terms using InterProScan (http://www.ebi.ac.uk/InterProScan/; Hunter et al., 2009) and HMMR (http://hmmer.janelia.org/; Eddy, 1999), respectively. Predicted peptides were then assigned gene ontology (GO) terms (Ashburner et al., 2000) based on their homology to conserved domains and protein families. Peptides were mapped to respective pathways in C. elegans using the KEGG Orthology-Based Annotation System (KOBAS; Wu et al., 2006); conserved metabolic pathways were displayed using the iPath tool (Letunic et al., 2008).
All sequences (i.e., contigs and singletons) in each of the three assemblies were subtracted from one another (in both directions) by in silico-subtraction using a BLASTn algorithm (Altschul et al., 1997), set at a stringent cut-off (e-value cut-off: < 1e-15; see Cantacessi et al., 2010b). Peptides corresponding to transcripts that were unique to a particular dataset were assigned parental (= level 1) InterPro terms and compared, using a BLASTp algorithm (Altschul et al., 1997; e-value cut-off: < 1e-15), with peptides inferred from the assembly of sequences from the combined dataset.
Interaction networks for C. elegans homologues of differentially transcribed molecules were predicted using an established method (Zhong and Sternberg, 2006), and the druggability of C. elegans homologues unique to a particular D. viviparus dataset or common to all datasets was predicted (Cantacessi et al., 2010a). Briefly, InterPro domains of predicted proteins were compared with those linked to known, small molecular drugs, which follow the ‘Lipinski rule of 5’ regarding bioavailability (Lipinski et al., 1997; Hopkins and Groom, 2001). GO terms were mapped to Enzyme Commission (EC) numbers, and a list of enzyme-targeting drugs was compiled based on all data available in the BRENDA database (www.brenda-enzymes.info; [Robertson, 2005; Chang et al., 2009]). Each predicted drug target was selected based on: (i) the presence of homologues linked to gene peturbation phenotypes in C. elegans, Saccharomyces cerevisiae, Drosophila melanogaster and Mus musculus and (ii) their absence from the bovine host. The C. elegans homologues identified were ranked according to the ‘severity’ of non-wildtype RNAi phenotypes (including lethality or sterility at different life cycle stages; see www.wormbase.org; release WS200).
3. Integrated bioinformatic exploration of available transcriptomic datasets, and mining for potential drug targets
3.1. The transcriptomes representing D. viviparus
A total of 2,952,411 ESTs (454 bases ± 86 in length; i.e., average length ± standard deviation) generated by 454 sequencing from L3, and adult female and male D. viviparus were subjected to detailed annotation and analyses using our custom-built workflow system. All previously published ESTs (available via www.ncbi.nlm.nih.gov/; Ranganathan et al., 2007; Strube et al., 2007b) represented ~0.1% of this total number. Following the clipping of adapter sequence residues from all ESTs, only reads of >100 bases (n = 2,943,357; 99.7%) were considered for further analysis. The total numbers of contigs assembled for the three individual datasets are given in Table 1. Briefly, ESTs representing the L3 were assembled into 7,661 contigs and 27,455 singletons, while sequences representing adult female and male were assembled into 9,946 contigs and 28,840 singletons and 9,226 contigs and 28,784 singletons, respectively. The assembly of sequences from all three datasets yielded a total of 6,119 contigs and 55,015 singletons. The mean length (= number of bases) and standard deviation (SD) of assembled contigs included in each of the individual and the combined datasets are given in Table 1. The female dataset had the largest number of sequence clusters with homologues in C. elegans (n = 17,979), in other parasitic nematodes (n = 16,100) and in organisms other than nematodes (n = 19,566) (Table 1).
Table 1.
Summary of available nucleotide sequence data for L3, adult female and adult male of Dictyocaulus viviparus prior to and following in silico-subtraction as well as detailed bioinformatic annotation and analyses.
| L3 | Female | Male | Combined | |
|---|---|---|---|---|
| Expressed sequence tags (ESTs) | ||||
| Number of unassembled ESTs (average length ± SD) | 1,095,063 (539 ± 84) | 955,995 (521 ± 95) | 889,873 (506 ± 99) | 2,943,357 (454 ± 86) |
| Contigs (average length ± range) | 7,661 (461 ± 238) | 9,946 (616 ± 362) | 9,226 (577 ± 307) | 6,119 (214 ± 102) |
| Singletons | 27,455 (317 ± 162) | 28,840 (339 ± 193) | 28,784 (360 ± 181) | 55,015 (202 ± 142) |
| Total | 35,116 | 38,786 | 38,010 | 61,134 (304 ± 141) |
| Containing an Open Reading frame (%) | 31,288 (89) | 34,338 (88.5) | 34,931 (92) | 59,606 (97) |
| Returning Pfam/InterPro results (%) | 12,615 (40) | 16,615 (48.3) | 17,302 (49.5) | 20,974 (37) |
| Gene Ontology (%) | 8,306 (23.6) | 10,514 (27) | 11,822 (31) | 12,151 (21) |
| Number of Biological process terms | 635 | 644 | 656 | 678 |
| Cellular component | 208 | 220 | 214 | 231 |
| Molecular function | 723 | 752 | 736 | 770 |
| With homologues in C. elegans (%) | 13,918 (44) | 17,979 (52) | 17,103 (49) | 18,877 (31.7) |
| other parasitic nematodes (%) | 11,954 (38) | 16,100 (46.8) | 15,928 (45.5) | 14,721 (24.7) |
| other organisms (%) | 14,765 (47) | 19,566 (57) | 18,530 (53) | 17,043 (28) |
| Number of unique C. elegans homologues | 12,641 | 12,853 | 12,258 | 12,938 |
| KOBAS (number of biological pathways predicted) | 193 | 164 | 138 | 245 |
| In silico subtracted datasets | ||||
| Number of EST clusters | 6,928 | 5,203 | 7,889 | |
| Containing an Open Reading frame (%) | 4,902 (70) | 4,947 (95) | 6,003 (76) | |
| Predicted peptides | ||||
| Returning Pfam/InterProScan results (%) | 1,932 (39) | 1,586 (32) | 3,898 (64) | |
| Gene Ontology (%) | 1,246 (25) | 1,027 (20) | 2,724 (45) | |
| Number of Biological process terms | 339 | 291 | 425 | |
| Cellular component | 114 | 106 | 164 | |
| Molecular function | 401 | 366 | 479 | |
| With homologues in C. elegans (%) | 1,608 (33) | 1,816 (37) | 3,890 (64) | |
| other parasitic nematodes (%) | 1,313 (27) | 1,345 (27) | 3,305 (55) | |
| other organisms (%) | 1,901 (39) | 1,888 (38) | 3,895 (65) | |
| KOBAS (number of unique KEGG terms) | 331 | 176 | 131 | |
In total, 59,606 peptides were predicted for all sequences from the combined assembly of the three datasets (Table 1); 20,974 (34%) of them could be mapped to known proteins defined by 1,460 different domains, the most represented being ‘collagen triple helix repeat’ (Pfam code: PF01391; 4.3% of the peptides mapping to a conserved protein motif), ‘ubiquitin’ (PF00240; 2.1%) and ‘serine/threonine protein kinase domain’ (PF00069; 1.9%) (Table 2). GO annotation allowed 12,151 (20.3%) inferred proteins to be assigned to 678 ‘biological process’, 231 ‘cellular component’ and 770 ‘molecular function’ terms (Table 1). The predominant terms were ‘translation’ (GO:0006412; 4.7% of the predicted peptides assigned GO terms), ‘oxidation reduction’ (GO:0055114; 4.3%) and ‘proteolysis’ (GO:0006508; 4.2%) for ‘biological process’; ‘membrane’ (GO:0016020; 8%), ‘nucleus’ (GO:0005634; 7.5%) and ‘intracellular’ (GO:0005622; 7.4%) for ‘cellular component’ and ‘ATP binding’ (GO:0005524; 12%); ‘DNA binding’ (GO:0003677; 66.8%) and ‘protein binding’ (GO:0005515; 5.7%) for ‘molecular function’ (Table 3). Proteins inferred from the combined assembly were predicted to be involved in 245 different biological pathways, defined by 1,889 unique KEGG terms, of which ‘peptidases’, ‘protein kinases’ and ‘chaperones and folding catalysts’ predominated (see Appendix A). Metabolic pathways, defined by KEGG terms inferred from predicted peptides and mapped to the complement of known pathways in C. elegans, are shown in Fig. 1.
Table 2.
Top twenty Pfam protein domains inferred from peptides conceptually translated from individual contigs for Dictyocaulus viviparus (combined dataset for L3, adult female and adult male) and those assigned to predicted peptides unique to each stage following in silico-subtraction.
| Pfam description | Pfam code | Number of predicted peptides |
|---|---|---|
| Combined assembly (1,460)a | ||
| Collagen triple helix repeat | PF01391 | 63 |
| Ubiquitin | PF00240 | 31 |
| Serine/threonine protein kinase domain | PF00069 | 28 |
| Major sperm protein | PF00635 | 16 |
| Ankyrin | PF00023 | 15 |
| WD40 | PF00400 | 15 |
| ATPase, AAA-type | PF00004 | 13 |
| Viral-A type inclusion protein | PF04508 | 13 |
| Septum formation initiator | PF04977 | 13 |
| Kelch repeat type 1 | PF01344 | 12 |
| Leucine zipper, homeobox associated | PF02183 | 11 |
| Major facilitator superfamily | PF07690 | 11 |
| Actin | PF00022 | 10 |
| Ras | PF00071 | 10 |
| RNA recognition motif, RNP-1 | PF00076 | 10 |
| Peptidase C1A | PF00112 | 10 |
| Carboxylesterase | PF00135 | 10 |
| Chaperonin | PF00118 | 9 |
| TGF-beta receptor | PF01064 | 9 |
| IncA | PF04156 | 9 |
| L3 (2,050)a | ||
| Nucleoplasmin | PF03066 | 33 |
| Nop-14-like protein | PF04147 | 29 |
| Microtubule | PF10243 | 26 |
| Mitochondrial substrate, solute carrier | PF00153 | 24 |
| Meiotic nuclear division protein | PF03962 | 19 |
| Nematode cuticle collagen | PF01484 | 17 |
| Myosin tail | PF01576 | 15 |
| Protein synthesis factor | PF00009 | 14 |
| Aminoglycoside phosphotranferase | PF01636 | 14 |
| Peptidase M12A, astacin | PF01400 | 13 |
| Nucleoplasmin | PF03066 | 33 |
| Female (1,741)a | ||
| von Willebrand factor | PF00094 | 77 |
| Serine/threonine protein kinase domain | PF00069 | 25 |
| Transcription factor | PF03153 | 25 |
| IncA | PF04156 | 22 |
| Collagen triple helix repeat | PF01391 | 18 |
| CDC45-like protein | PF02724 | 18 |
| RNA recognition motif, RNP-1 | PF00076 | 16 |
| WD40 repeat, subgroup | PF00400 | 16 |
| Topoisomerase II | PF09770 | 15 |
| Protein of unknown function | PF06156 | 14 |
| Male (2,945)a | ||
| Major sperm protein | PF00635 | 380 |
| SCP-like extracellular | PF00188 | 142 |
| Serine/threonine protein phosphatase | PF00102 | 115 |
| Ryanodine receptor | PF06459 | 81 |
| Neurotransmitter-gated-ion-channel transmembrane domain | PF02932 | 80 |
| Metallophosphoesterase | PF00149 | 72 |
| Nuclear RNA-splicing-associated protein | PF10500 | 63 |
| Aminoglycoside phosphotransferase | PF01636 | 56 |
| RNA polymerase II | PF03985 | 54 |
| ATPase, V0/A0 complex | PF01496 | 51 |
Number of unique Pfam domains assigned to predicted peptides in each dataset
Table 3.
Top five gene ontology (GO) terms, according to the categories ‘biological process’, ‘cellular component’ and ‘molecular function’, assigned to peptides conceptually translated from individual contigs for Dictyocaulus viviparus (combined dataset for L3, adult female and adult male).
| GO description (GO code) | Number of predicted peptides |
|---|---|
| Combined assembly | |
| Biological process (678)a | |
| Translation (GO:0006412) | 572 |
| Oxidation reduction (GO:0055114) | 523 |
| Proteolysis (GO:0006508) | 513 |
| Regulation of transcription, DNA dependent | 512 |
| Metabolic process (GO:0008152) | 503 |
| Cellular component (231)a | |
| Membrane (GO:0016020) | 977 |
| Nucleus (GO:0005634) | 921 |
| Intracellular (GO:0005622) | 903 |
| Integral to membrane (GO:0016021) | 708 |
| Cytoplasm (GO:0005737) | 503 |
| Molecular function (770)a | |
| ATP binding (GO:0005524) | 1,470 |
| DNA binding (GO:0003677) | 812 |
| Protein binding (GO:0005515) | 700 |
| Zinc ion binding (GO:0008270) | 470 |
| Nucleic acid binding (GO:0003676) | 446 |
Total number of unique GO terms assigned to predicted peptides
Fig. 1.
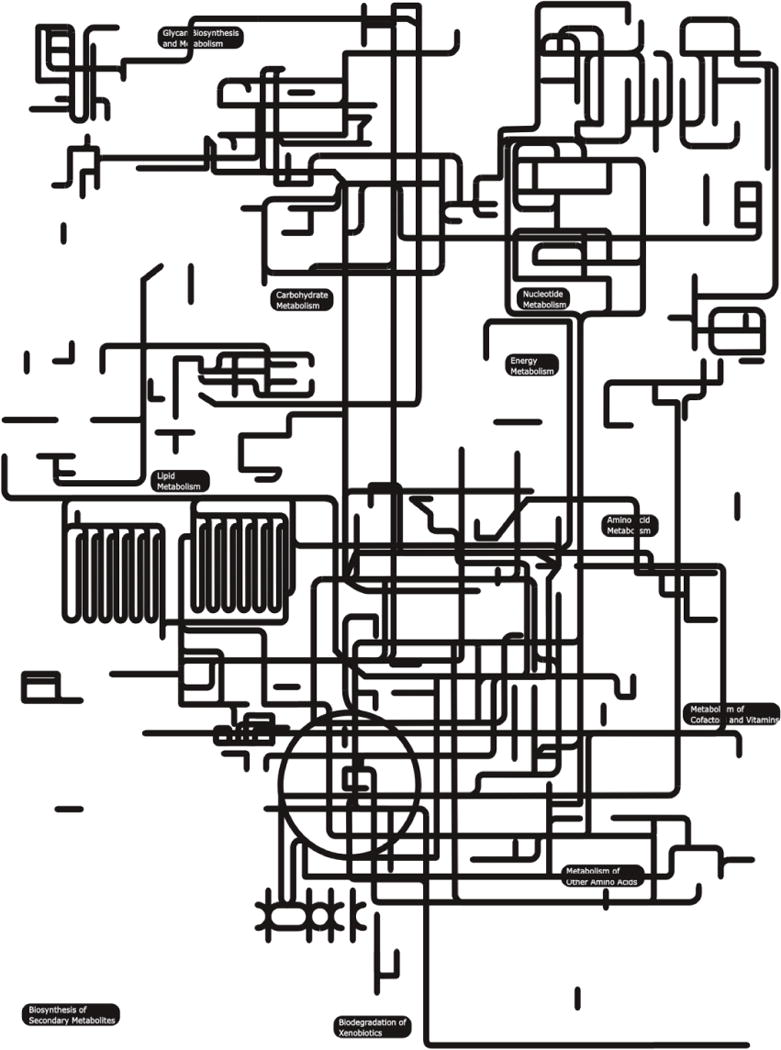
Mapping of peptides inferred from the transcriptome of Dictyocaulus viviparus (combined dataset for L3, adult female and adult male) to known metabolic pathways in Caenorhabditis elegans.
The numbers of peptides predicted from EST clusters in each of the three datasets (i.e., L3, female and male), together with the corresponding number of peptides mapped to known protein domains/motifs and/or assigned GO terms, are reported in Table 1. For both L3 and adult female, the ‘collagen triple helix repeat’ (PF01391; 5.8% and 8.2% of 5,471 and 5,768 protein domains identified in the L3 and female, respectively) and ‘ubiquitin’ (PF00240; 4.5% and 3.7%, respectively) were most commonly detected. Amongst the predicted protein domains identified in the male dataset, ‘major sperm protein’ (PF00635; 43.3% of 5,565 protein domains) and ‘SCP-like extracellular’ (PF00188; 13.2%) were mostly represented (Appendix B). The descriptions of GO terms linked to predicted peptides in each of the three datasets (i.e., L3, female and male), according to the categories ‘biological process’, ‘cellular component’ and ‘molecular function’ are given in Appendix C. For L3, pathway mapping using KOBAS predicted 193 distinct pathways, of which the most represented were ’ribosome’, and ‘receptors and channels’ (see Appendix A). The predicted peptides in the female dataset could be linked to a total of 164 biological pathways, of which ‘receptors and channel’ and ‘protein kinases’ were most commonly represented (Appendix A). For the male, pathway mapping predicted 138 biological pathways, of which ‘glycolysis/gluconeogenesis’ and ‘fructose and mannose metabolism’ were most common (see Appendix A).
3.2. Molecules uniquely transcribed in different stages/sexes, and the inference of drug targets
Subtractive analysis in silico identified 6,928 nucleotide sequences as being uniquely transcribed in the L3 stage, and 5,203 and 7,889 transcripts as exclusive to the female and male dataset, respectively. The specificity of this in silico-subtraction approach has been demonstrated elsewhere (Cantacessi et al., 2010b). Of the 2,050 functional domains inferred for predicted peptides unique to the L3 dataset, ‘nucleoplasmin’ (PF03066; n = 33), and ‘Nop-14-like protein (PF04147; n = 22) were most commonly represented (Table 2). Among the 1,741 functional domains mapped to predicted peptides unique to the female dataset, the ‘von Willebrand factor’ (PF00094; n = 77) and ‘serine/threonine protein kinase’ (PF00069; n = 25) domains had the greatest representation (Table 2). Of the 2,945 protein motifs linked to predicted peptides unique to the male dataset, ‘major sperm protein’ (PF00635; n = 380) and ‘SCP-like extracellular’ (PF00188; n = 142) had the highest representation (Table 2). The descriptions of the most commonly detected GO terms linked to predicted peptides unique to each of the three datasets are given in Table 3. A schematic representation of the biological pathways inferred by KOBAS for predicted peptides unique to each of the three datasets, mapped to the complement of known metabolic pathways in C. elegans, is given in Fig. 2.
Fig. 2.
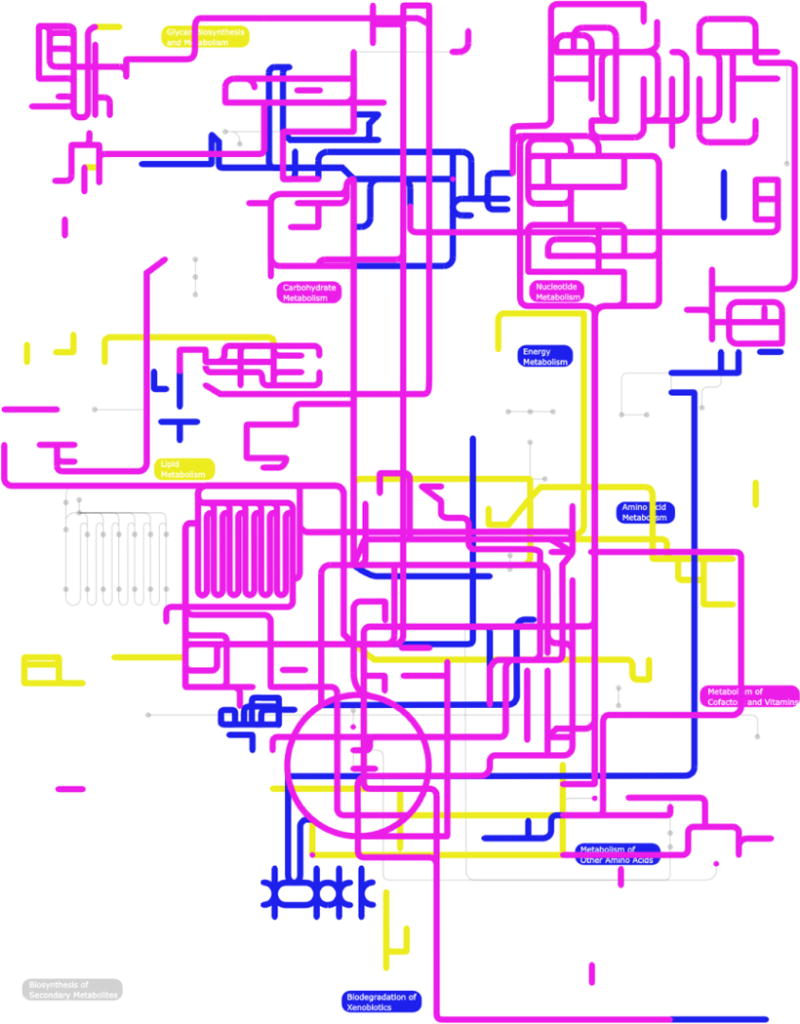
Mapping of predicted peptides unique to either the L3 (yellow), adult female (pink) and adult male (blue) of Dictyocaulus viviparus to known metabolic pathways in Caenorhabditis elegans.
Probabilistic genetic interaction networking predicted 5,196 C. elegans homologues, representing sequence clusters unique to D. viviparus L3, to interact directly with a total of 2,435 other genes (range: 1–271; not shown), including some (e.g., dbl-1; Fig. 3) that are essential to the regulation of body length and size (see www.wormbase.org). The 5,093 C. elegans homologues of contigs and singletons unique to the adult female of D. viviparus were predicted to interact directly with a total of 2,394 other genes (range: 1–260). Amongst them were genes involved in embryonic, larval and vulval development (i.e., glp-1, lin-12, let-92 and mig-2) and behaviour and egg-laying (i.e., goa-1) (Fig. 3; www.wormbase.org). A total number of 7,396 C. elegans homologues of male-unique molecules, were predicted to interact with 2,727 (range: 1–122) other genes, respectively, including some involved in sperm maturation and development (i.e., ima-3 and air-2) (Fig. 3; www.wormbase.org).
Fig. 3.
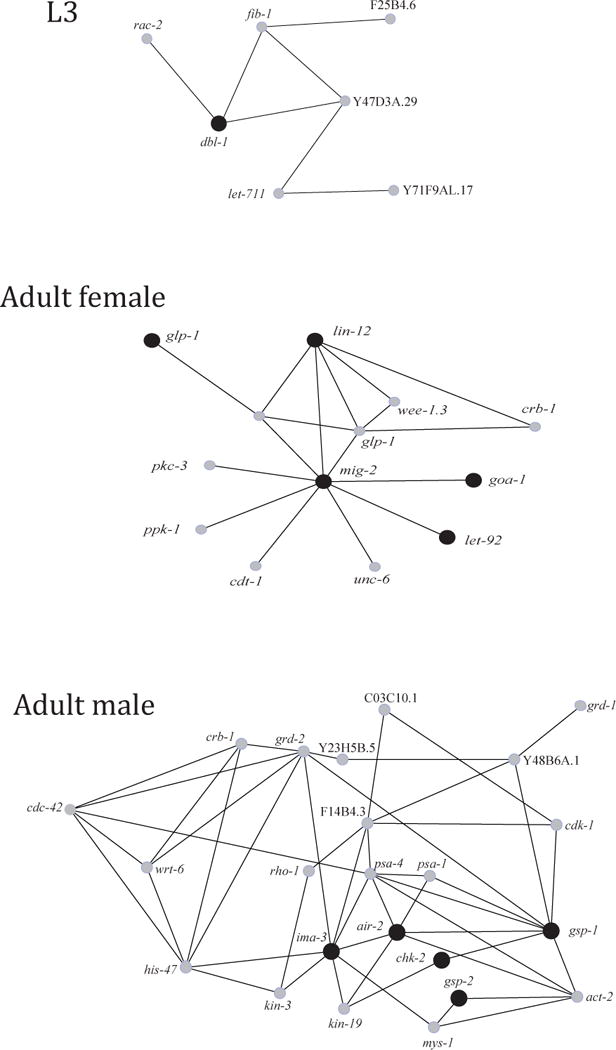
Examples of genetic interaction networks predicted for Caenorhabditis elegans homologues (grey dots) of expressed sequence tags (ESTs; black dots) unique to either L3, adult female and adult male of Dictyocaulus viviparus. Each of the represented C. elegans homologues was predicted to interact with a minimum of 100 other genes. Homologue dbl-1 is linked to regulation of body length and size (L3); glp-1, lin-12, let-92 and mig-2 to embryonic, larval and vulval development and goa-1 to behaviour and egg-laying (adult female); ima-3 and air-2 are associated to sperm maturation and development (adult male).
Of peptides inferred to be unique to L3 (n = 4,902), adult female (4,947) and adult male (6,003) of D. viviparus, 609, 245 and 526 were associated with ‘druggable’ InterPro domains and/or EC numbers, respectively (Table 4). For individual stages/sexes, these ‘druggable’ molecules had significant homologies (e-value cut-off: >1e-05) to ~1,200–2,300 C. elegans homologues linked to RNAi phenotypes, of which ~56–60% were unequivocally linked to lethal or sterile phenotypes (Table 4). Of peptides predicted from the combined assembly of all three sequence datasets (i.e., D. viviparus L3, adult female and adult male), 5,931 were linked to druggable InterPro domains and/or EC numbers (Table 4). These molecules had significant homologies (e-value cut-off: > 1e-05) to ~2,300 C. elegans genes, of which ~73% were linked to lethal or sterile RNAi phenotypes (Table 4). Druggable molecules, linked to a lethal RNAi phenotype, predicted for each of the datasets or common to all three datasets and examples of compounds (from the BRENDA database) are listed in Table 5. For instance, the compounds oligomycin and paclitaxel were inferred to specifically target an homologue represented by gene F09F9.4, whereas tetracyclin and vanadate as well as streptovaricin and tagetotoxin targeted molecules represented by the homologues H27M09.1 and DCR-1, respectively (see Table 5).
Table 4.
Numbers of peptides conceptually translated from individual contigs for Dictyocaulus viviparus (combined dataset for L3, adult female and adult male) and of predicted peptides unique to each stage following in silico-subtraction mapped to druggable InterPro domain and/or Enzyme Commission (EC) number and corresponding homologues in Caenorhabditis elegans, ranked according to the ‘severity’ of known non-wildtype RNA interference (RNAi) phenotypes.
| Descriptions | Datasets | ||||
|---|---|---|---|---|---|
| L3 | Female | Male | Combined | ||
| Numbers of peptides inferred from D. viviparus and linked to: | |||||
| a druggable InterPro domain | 38 | 26 | 163 | 929 | |
| a druggable EC number | 445 | 181 | 299 | 4,919 | |
| both a druggable InterPro domain and EC number | 126 | 38 | 64 | 83 | |
| Totals | 609 | 245 | 526 | 5,931 | |
| Numbers of druggable peptides inferred from D. viviparus with C. elegans homologues linked to RNAi phenotypes: | |||||
| adult lethalitya | 4 | 1 | 5 | 3 | |
| embryonic and/or larval lethality | 456 | 387 | 768 | 635 | |
| sterility | 230 | 233 | 423 | 1,076 | |
| otherb | 537 | 406 | 903 | 622 | |
| Totalsc | 1,227 | 1,027 | 2,099 | 2,336 | |
All C. elegans homologues were linked to multiple non-wildtype RNAi phenotypes.
Other phenotypes (alphabetical): lifespan abnormal (Age), body morphology defect (Bmd), clear (Clr), cytokinesis abnormal (Cyt), dumpy (Dpy), egg laying defective (Egl), general pace of development abnormal early embryo (Emb), asymmetric cell division abnormal early embryo (Emb), pleiotropic defects severe early embryo (Emb), fewer germ cells (Fwr), slow growth (Gro), larval arrest (Lva), pathogen susceptibility increased, protruding vulva (Pvl), exploded through vulva (Rup), sick (Sck), sluggish (Slu), sterile progeny (Ste), uncoordinated (Unc).
Some predicted peptides inferred from D. viviparus (i.e., from 17.7 [female dataset] to 20.1% [male dataset]) had more than one C. elegans homologue.
Table 5.
Caenorhabditis elegans homologues of peptides inferred from contigs unique to each Dictyocaulus viviparus L3, adult female and adult male following in silico-subtraction, and common to all datasets (linked to a ‘lethal’ RNA interference phenotype) and associated with druggable (InterPro) domain and/or Enzyme Commission (EC) number, as well as examples of candidate compounds linked to these domains, predicted using the BRENDA database. Strongylid and non-strongylid nematodes for which homologues are known are also listed.
| C. elegans gene identification code | Gene name | Protein description | Non-wildtype RNAi phenotypesa | References | Presence of homologue/s in strongylid [non-strongylid nematodes]b | Examples of BRENDA compounds | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| L3 | |||||||||||
| WBGene00002845 | LET-711 | NOT1, core subunit of CCR4/NOT complex | Let, Emb, Lvl, Lva, Ste, embryonic defects | Kamath et al. (2003) | Ace[Asu, Bma, Hgl. Mpa, Ovo, Ppa, Ptr. Tmu, Xin] | Neamine; neomycin B; N-ethylmaleimide | |||||
| WBGene00000408 | CDK-7 | cyclin-dependent kinase CDK7 | Let, Emb, Lva, reduced brood size, small | Ceron et al. (2007) | Aca, Hco[Bma, Gpa, Hgl, Mha, Min, Sra, Tsp, Xin] | Guanidine hydrochloride; Iodoacetamide; Mitoxantrone | |||||
| WBGene00004502 | RPT-2 | 26S proteasome regulatory complex, ATPase RPT2 | Let, Ste | Piano et al. (2002) | Aca, Ace, Hco, Oos, Tci[Bma, Gro, Hgl, Mar, Mha, Mja, Ovo, Ppa, Sra, Sst] | Guanidine hydrochloride; Iodoacetamide; Mitoxantrone | |||||
| WBGene00004501 | RPT-1 | 26S proteasome regulatory complex, ATPase RPT1 | Let, Ste | Piano et al. (2002); Kamath et al. (2003) | Ace, Tci[Asu, Bma, Hgl, Mha, Min, Mja, Mpa, Ppa, Sra, Sst, Wba] | Guanidine hydrochloride; Iodoacetamide; Mitoxantrone | |||||
| Female | |||||||||||
| WBGene00003670 | NHR-80 | Hormone receptor | Let, Emb, Ste, thin, embryonic defects | Brock et al. (2006) | [Asu] | Ribostamycin; sparsomycin; streptogramin A | |||||
| Male | |||||||||||
| WBGene00021752 | Y50D7A.2 | RNA polymerase II transcription initiation/nucleotide excision repair factor | Let, Emb, Lva | Eki et al. (2007) | Aca, Nam[Gpa, Hsc, Mja, Ppa, Tsp] | Pyrrole; Pyrrolidine; Tetrahydrofuran | |||||
| WBGene00004189 | PRS-1 | Prolyl-tRNA synthetase region | Let, Lvl, Ste, embryonic defects | Kamath et al. (2003); Rual et al. (2004) | Aca, Hco[Asu, Hgl, Mha, Min, Ppa, Ptr, Sra, Tvu] | Tetracycline; vanadate; viomycin | |||||
| WBGene00021857 | IFFB-1 | Initiation factor five B (eIF5B) | Let, Emb, Ste | Maeda et al. (2001); Sonnichsen et al. (2005) | Aca, Ace[Asu, Gpa, Gro, Hsc, Min, Ppa, Sra, Wba] | Imidodiphosphate; Chloramphenicol; dihydrostreptomycin | |||||
| WBGene00001094 | DRS-1 | Aspartyl-tRNA synthetase | Let, Emb, Lva, Lvl, embryonic defects | Sonnichsen et al. (2005) | Hco[Asu, Dim, Hgl, Mar, Mch, Mja, Ovo, Ppa, Sra, Tsp, Wba] | Chloramphenicol; dihydrostreptomycin; EF-G GTPase inhibitor | |||||
| WBGene00009012 | F21D5.7 | Signal recognition particle protein (SRP54) | Let, Emb, Lva, Lvl, Ste, embryonic defects | Kamath et al. (2003); Rual et al. (2004) | Ace, Hco[Asu, Bma, Hgl, Mch, Ppa, Sra, Sst, Tsp, Tvu] | Guanidine hydrochloride; Iodoacetamide; Mitoxantrone | |||||
| Combined | |||||||||||
| WBGene00017309 | F09F9.4 | Unnamed protein | Let, Emb, Lvl, Lva, | Ceron et al. (2007) | Oligomycin; Paclitaxel; PCMB | ||||||
| Ste, Unc, embryonic defects | |||||||||||
| WBGene00019245 | H27M09.1 | DEAD-box protein | Let, Emb, Lva, Ste | Ceron et al. (2007); Eki et al. (2007) | Hco[Bma, Hgl, Gro, Mar, Mch, Mja, Mpa, Ovo, Ppa, Ptr, Sra, Sst, Tmu, Wba] | Tetracyclin; vanadate; viomycin | |||||
| WBGene00000939 | DCR-1 | dsRNA-specific nuclease Dicer and related ribonucleases | Let, Emb, Lvl, Ste, embryonic defects | Kim et al. (2005); Eki et al. (2007) | [Mch, Min, Ppa, Sst] | Streptovaricin; Tagetitoxin; ureidothiophene | |||||
Abbreviations of RNAi phenotypes (alphabetical): Embryonic lethal (Emb), adult lethal (Let), larval arrest (Lva), larval lethal (Lvl), sterile (Ste), uncoordinated (Unc).
Abbreviations of nematode species (alphabetical): Ancylostoma caninum (Aca), Ancylostoma ceylanicum (Ace), Ascaris suum (Asu), Brugia malayi (Bma), Dirofilaria immitis (Dim), Globodera pallida (Gpa), Globodera rostochiensis (Gro), Haemonchus contortus (Hco), Heterodera glycines (Hgl), Heterodera schachtii (Hsc), Meloidogyne arenaria (Mar), Meloidogyne chitwoodi (Mch), Meloidogyne hapla (Mha), Meloidogyne incognita (Min), Meloidogyne javanica (Mja), Meloidogyne paranaensis (Mpa), Necator americanus (Nam), Onchocerca volvulus (Ovo), Ostertagia ostertagi (Oos), Parastrongyloides trichosuri (Ptr), Pristionchus pacificus (Ppa), Strongyloides ratti (Sra), Strongyloides stercoralis (Sst), Teladorsagia circumcincta (Tci), Trichinella spiralis (Tsp), Trichuris muris (Tmu), Trichuris vulpis (Tvu), Wuchereria bancrofti (Wba), Xiphinema index (Xin).
4. Conclusions – exciting prospects for biological insights, disease intervention and biotechnological outcomes
The present review article substantially enhances our knowledge of the complement of molecules transcribed in different stages and both sexes of D. viviparus. Indeed, contigs assembled from combined sequence data for L3, adult female and male could be mapped with high confidence to ~13,000 unique C. elegans homologues (Table 1), thus increasing the number of newly discovered D. viviparus genes by ~8-fold (cf. Ranganathan et al., 2007). In addition, this result supports those of previous comparative analyses of genetic data sets, showing that strongylid nematodes share at least ~60% of genes with C. elegans (see Blaxter, 1998; Ruvkun and Hobert, 1998; Parkinson et al., 2004). This information has indicated that this free-living nematode provides a surrogate for investigating the function/s of some key genes/gene products in strongylids, further supported by successful genetic complementation studies (Britton and Murray, 2002; Lok, 2009; Hu et al., 2010; Stepek et al., 2010).
Of the ~60,000 peptides predicted from EST clusters assembled from all sequence data for all stages/sexes included (L3, and adult female and male), ~30% could be mapped to known peptides defined according to their conserved protein domains (see section 3.1). Of the 1,460 unique domains identified, the most represented were ‘collagen triple helix repeat’ (PF01391), ‘ubiquitin’ (PF00240) and ‘serine/threonine protein kinase’ (PF00069). Collagens are a large family of structural proteins representing the major component of the extracellular matrix (ECM) of all animals and, in particular, of the external, protective cuticle of nematodes (Page and Winter, 2003). Nematode collagens have been shown to be most similar, although smaller in size, to vertebrate ‘fibril-associated collagens with interrupted triple helices’ collagens (Shaw and Olsen, 1991) and act as molecular bridges required for the organization and stability of ECM (Page and Winter, 2003). Genes encoding collagens have been well characterised in C. elegans, in which they represent almost 1% of all genes (Johnstone, 2000). In this nematode, the biogenesis of the cuticle is known to be dependent upon the function of a group of astacin metalloproteases that are responsible for the cleavage of the C-terminal region of the collagens, thus catalysing the correct formation of ECM (Stepek et al., 2010). In particular, loss of function via gene peturbation of the astacin metalloprotease gene dpy-31 of C. elegans has been shown to result in cuticle defects, abnormal morphology and embryonic lethality, thus indicating that this gene is essential for the formation of the collagenous exoskeleton (Novelli et al., 2004). Recently, the homologues of C. elegans dpy-31 have been isolated and characterised in Brugia malayi, a filarial nematode of humans, and in Haemonchus contortus, a trichostrongylid nematode of small ruminants (Stepek et al., 2010). In the latter study, the expression of the H. contortus homologue in a C. elegans dpy-31 mutant strain resulted in the rescue of the mutant body form, suggesting that the function of DPY-31 is conserved between these two nematodes (Stepek et al., 2010). In H. contortus L3s, astacin metalloproteases are known to mediate the ecdysis from the L2 cuticle through the formation of an anterior refractile ring and removable cap structure (Gamble et al., 1989, 1996). Interestingly, the ‘peptidase M12A, astacin’ (PF01400) was amongst the protein domains most commonly identified in the group of EST clusters unique to the L3 stage of D. viviparus (see section 3). Therefore, considering the results of previous studies, showing that peptidases are essential to the exsheathment process of the D. viviparus L3 in vitro (Silverman and Podger, 1964), it is tempting to speculate a similar role of astacin metalloproteases in the transition from the free-living to the parasitic stage of this nematode in vivo, an hypothesis which requires rigorous testing.
‘Serine/threonine protein kinases’ (PF00069) were also commonly represented in transcriptome of D. viviparus (see section 3). Such PAR-1/MARK serine/threonine protein kinase (STK) subfamily members have been implicated in a number of fundamental pathways of the biology of the cell, such as the establishment and maintenance of cell polarity (i.e., through the activity of the cytoskeleton; Drewes et al., 1998). Particularly in C. elegans and D. melanogaster, a gene (par-1) encoding an STK is known to be required for cytoplasmic partitioning and asymmetric cell division in early embryogenesis (Guo and Kemphues, 1995) and oocyte specification and determination of the embryonic axis, respectively (Cox et al., 2001). An additional role of C. elegans par-1 in the morphogenesis of the vulva has also been implicated, based on the results of a study showing that post-embryonic gene silencing of par-1 causes a ‘protruding vulva’ phenotype, which results from the failure of the two mirror-symmetric halves of the vulva to join into a single organ (Hurd and Kemphues, 2003). Indeed, the in silico-subtraction approach employed in the present study identified the serine/threonine protein kinase’ (PF00069) domain as highly represented amongst the functional domains specific to the adult female of D. viviparus (see section 3). Although the role of STKs in fundamental biological processes of parasitic nematodes is unclear, these results suggest an active involvement into molecular mechanisms linked to the reproductive activity of the adult female. This hypothesis is supported by the localisation of an homologue (i.e., HcSTK) of C. elegans PAR-1 to a cytoplasmic compartment around the nuclei of intestinal and ovarian tissues of the adult stage of H. contortus, a strongylid nematode which is related to D. viviparus (see Nikolaou et al., 2006; Nikolaou and Gasser, 2007).
Other groups of molecules inferred to play fundamental biological roles in reproductive mechanisms in C. elegans were commonly represented in the transcriptome of D. viviparus. For instance, the ‘major sperm protein (MSP)’ domain (PF00635) was commonly identified within peptides inferred from both the combined assembly of data from the L3, adult female and male and specific to the male (cf. section 3). MSPs belong to a large protein family whose members have been recognised to act as bipartite signalling molecules, activating pathways linked to oocyte production and maturation in C. elegans (see Miller et al., 2001, 2003). However, MSPs are known to be predominantly involved in nematode sperm motility through their polymerisation into dense filaments that are required for the movement of the plasma membrane of the amoeboid sperm cells (Roberts and Stewart, 2000; Italiano et al., 2001; Buttery et al., 2003). The pattern of transcription of genes encoding msps throughout different developmental stages has already been investigated in D. viviparus (Strube et al., 2009b). Using quantitative real-time PCR, an up-regulation of genes encoding msps was observed in hypobiotic L5 and adult male of this lungworm (Strube et al., 2009b). Interestingly, in adult males, the authors observed an ~200,000-fold increase in msp transcription levels compared with adult females (Strube et al., 2009b). This difference contrasts the results of other transcriptional analyses of gender-enriched molecules in other species of parasitic nematodes. For instance, a 2-fold increase in msp transcriptional rates were observed in the males of intestinal nematodes of the pig, including Oesophagostomum dentatum and Ascaris suum (see Cottee et al., 2004; Cantacessi et al., 2009a), whereas an ~480-fold increase was detected in males of the filarial nematode B. malayi (see Li et al., 2004). The high transcriptional levels of msps in the adult male of D. viviparus could be indicative of high sperm production or a distinct biological function of this group of molecules in this lungworm, besides the activity in reproductive processes. This question remains to be explored. In addition, a MSP (encoded by a gene designated Dv3–14; Schnieder, 1993) was identified as an immunodominant antigen by immunoblotting using sera from cattle experimentally infected with D. viviparus (see Schnieder, 1992); therefore, given their abundance in the excretory/secretory (ES) products of the adult worm, MSPs (expressed in recombinant form) have been proposed as vaccine candidates (Strube et al., 2009b).
Of the groups of molecules identified herein as being specific to the adult male of D. viviparus, the SCP/Tpx-1/Ag5/PR-1/Sc7 proteins (designated SCP/TAPS; PF00188) are also under investigation for their potential as vaccine targets for other parasitic nematodes (i.e., the human hookworm Necator americanus; Loukas et al., 2006; Bethony et al., 2008; Mendez et al., 2008; Xiao et al., 2008). Such SCP/TAPS are also commonly referred to as Ancylostoma secreted proteins or activation-associated proteins (ASPs), mainly because they were originally discovered in hookworms (Hawdon et al., 1996; Datu et al., 2008; Cantacessi et al., 2009b). Given their abundance in the ES products from serum-activated third-stage larvae (aL3s) of A. caninum and high transcriptional levels in aL3s compared with non-activated L3s, ASPs/asps have been hypothesized to play a major role in the transition from the free-living to the parasitic stage of hookworms (Datu et al., 2008). Other ASP homologues have been characterized in the adult stage of hookworms, where they might play a role in the initiation, establishment and/or maintenance of the host-parasite relationship (Zhan et al., 2003; Cantacessi et al., 2009b; Mulvenna et al., 2009). A male-biased transcription of genes encoding ASPs had been reported for other species of parasitic nematodes, including A. suum, O. dentatum and Trichostrongylus vitrinus (intestinal nematode of small ruminants) (Nisbet and Gasser, 2004; Cottee et al., 2006; Cantacessi et al., 2009a). However, because the sequence datasets analysed here were generated from normalized cDNA libraries, the transcriptional levels of genes encoding ASPs in the adult male of D. viviparus could not be determined. Future work could explore developmentally regulated transcription of these molecules in all developmental stages of D. viviparus, employing, for example, digital gene expression profiling or non-normalized Illumina sequencing (Morrissy et al., 2009, 2010), to assist in understanding their role(s) in the parasite and/or its interplay with the host.
Knowledge of the function of key molecules in a range of organisms (e.g., C. elegans, S. cerevisiae, D. melanogaster and M. musculus) assists in the prediction novel drug targets in parasitic helminths (see Geary et al., 1999; McCarter, 2004; Krasky et al., 2007; Caffrey et al., 2009; Doyle et al., 2010). Such knowledge, combined with the InterPro and/or GO classifications of inferred peptides from the combined assembly of sequence data from L3, adult female and male of D. viviparus as well as peptides unique to each stage/sex, allowed the prediction and prioritisation of molecules considered to be essential in the life cycle of this lungworm and, by inference, other parasitic nematodes (see Table 5). In particular, the C. elegans homologues F09F9.4, H27M09.1 and DCR-1, linked to druggable InterPro domains and/or EC numbers, were associated with an ‘adult lethal’ phenotype for all of the organisms investigated (see section 3.2 and Table 5). Specifically in C. elegans, these molecules are known to be involved in essential molecular processes linked to embryonic and larval development, locomotion and/or reproduction. For instance, the C. elegans gene H27M09.1 encodes an ATP-dependent helicase with a DEAD-box domain (see www.wormbase.org), whose corresponding homologues in B. malayi and Plasmodium falciparum (malaria parasite) have been proposed to represent putative drug targets in these parasites (Tuteja, 2007; Singh et al., 2009). This hypothesis is based on the knowledge that efficacious compounds targeting this group of molecules are available for the treatment of pathogens of humans, such as the Herpes simplex and Hepatitis C viruses (Frick, 2003). Future studies could focus on investigating the transcription levels of the D. viviparus homologue of C. elegans H27M09.1 throughout different life cycle stages and on assessing the specific biochemical activity of the encoded protein (cf. Singh et al., 2009). Insights through such studies could assist in the prediction and optimisation of the chemical structure of this group of molecules via chemoinformatic methods, such as homology modelling and ligand docking (Gasteiger, 2006; Krasky et al., 2007), and might lead to the definition of a target protein structure to assist the design of drugs for in vitro and in vivo testing.
In conclusion, the present study has provided the first large-scale exploration of key transcriptomes of D. viviparus and has predicted, using an integrated bioinformatic approach and knowledge of functional aspects of key genes/gene products in model organisms, molecules unique to and/or essential in different stages of this lungworm. This knowledge, combined with future sequencing efforts using, for instance, Illumina sequencing, should assist in the determination of the genome sequence of D. viviparus, which will constitute a foundation for proteomic and metabolomic studies of this parasite, leading to novel treatment and control strategies as well as major biotechnological outcomes.
Supplementary Material
Acknowledgments
This research was supported by grants from the Australian Research Council, the Australian Academy of Science, the Australian-American Fulbright Commission (RBG) as well as the National Human Genome Research Institute and National Institutes of Health (MM). CC is the grateful recipient of International Postgraduate Research Scholarship from the Australian Government and a fee-remission scholarship through the University of Melbourne as well as the Clunies Ross (2008) and Sue Newton (2009) awards from the School of Veterinary Science of the same university. ARJ is the recipient of a Career Developmental Award (CDA1) from the National Health and Medical Research Council (NHMRC).
Appendix A
Biological pathways assigned to peptides conceptually translated from individual contigs for Dictyocaulus viviparus (combined dataset for L3, adult female and adult male) and to predicted peptides unique to each developmental stage or sex.
Appendix B
Pfam and/or InterPro domains for predicted peptide sequences encoded in either L3, adult female and adult male of Dictyocaulus viviparus.
Appendix C
Gene ontology (GO) terms according to the categories ‘biological process’, ‘cellular component’ and ‘molecular function’ assigned to peptides conceptually translated from individual contigs for Dictyocaulus viviparus representing the combined dataset for L3, adult female and adult male, or individual stages/sexes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RC. Nematode Parasites of Vertebrate: Their Development and Transmission. Wallingford: CABI Publishing; 2001. [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethony JM, Simon G, Diemert DJ, Parenti D, Desrosiers A, Schuck S, et al. Randomized, placebo-controlled, double-blind trial of the Na-ASP-2 hookworm vaccine in unexposed adults. Vaccine. 2008;26:2408–17. doi: 10.1016/j.vaccine.2008.02.049. [DOI] [PubMed] [Google Scholar]
- Blaxter M. Caenorhabditis elegans is a nematode. Science. 1998;282:2041–6. doi: 10.1126/science.282.5396.2041. [DOI] [PubMed] [Google Scholar]
- Britton C, Murray L. A cathepsin L protease essential for Caenorhabditis elegans embryogenesis is functionally conserved in parasitic nematodes. Mol Biochem Parasitol. 2002;122:21–33. doi: 10.1016/s0166-6851(02)00066-x. [DOI] [PubMed] [Google Scholar]
- Brock TJ, Browse J, Watts JL. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006;2:e108. doi: 10.1371/journal.pgen.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery SM, Ekman GC, Seavy M, Stewart M, Roberts TM. Dissection of the Ascaris sperm motility machinery identifies key proteins involved in major sperm protein-based amoeboid locomotion. Mol Biol Cell. 2003;14:5082–8. doi: 10.1091/mbc.E03-04-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey CR, Rohwer A, Oellien F, Marhofer RJ, Braschi S, Oliveira G, et al. A comparative chemogenomics strategy to predict potential drug targets in the metazoan pathogen Schistosoma mansoni. PLoS One. 2009;4:e4413. doi: 10.1371/journal.pone.0004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantacessi C, Zou FC, Hall RS, Zhong W, Jex AR, Campbell BE, et al. Bioinformatic analysis of abundant, gender-enriched transcripts of adult Ascaris suum (Nematoda) using a semi-automated workflow platform. Mol Cell Probes. 2009a;23:205–17. doi: 10.1016/j.mcp.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Cantacessi C, Campbell BE, Visser A, Geldhof P, Nolan MJ, Nisbet AJ, et al. A portrait of the “SCP/TAPS” proteins of eukaryotes–developing a framework for fundamental research and biotechnological outcomes. Biotechnol Adv. 2009b;27:376–88. doi: 10.1016/j.biotechadv.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Cantacessi C, Mitreva M, Jex AR, Young ND, Campbell BE, Hall RS, et al. Massively parallel sequencing and analysis of the Necator americanus transcriptome. PLoS Negl Trop Dis. 2010a;4:e684. doi: 10.1371/journal.pntd.0000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantacessi C, Campbell BE, Young ND, Jex AR, Hall RS, Presidente PJA, et al. Differences in transcription between free-living and CO2-activated third-stage larvae of Haemonchus contortus. BMC Genomics. 2010b;11:266. doi: 10.1186/1471-2164-11-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantacessi C, Jex AR, Hall RS, Young ND, Campbell BE, Joachim A, et al. A practical, bioinformatic workflow system for large datasets generated by next-generation sequencing. 2010c. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceron J, Rual JF, Chandra A, Dupuy D, Vidal M, et al. Large scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev Biol. 2007;7:30. doi: 10.1186/1471-213X-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–92. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles GC. The future of veterinary parasitology. Vet Parasitol. 2001;98:31–9. doi: 10.1016/s0304-4017(01)00421-6. [DOI] [PubMed] [Google Scholar]
- Cottee PA, Nisbet AJ, Boag PR, Larsen M, Gasser RB. Characterization of major sperm protein genes and their expression in Oesophagostomum dentatum (Nematoda: Strongylida) Parasitology. 2004;129:479–90. doi: 10.1017/s003118200400561x. [DOI] [PubMed] [Google Scholar]
- Cottee PA, Nisbet AJ, Abs El-Osta YG, Webster TL, Gasser RB. Construction of gender-enriched cDNA archives for adult Oesophagostomum dentatum by suppressive-subtractive hybridization and a microarray analysis of expressed sequence tags. Parasitology. 2006;132:691–708. doi: 10.1017/S0031182005009728. [DOI] [PubMed] [Google Scholar]
- Cox DN, Lu B, Sun TQ, Williams LT, Jan YN. Drosophila par-1 is required for oocyte differentiation and microtubule organization. Curr Biol. 2001;11:75–87. doi: 10.1016/s0960-9822(01)00027-6. [DOI] [PubMed] [Google Scholar]
- David GP. Survey on lungworm in adult cattle. Vet Rec. 1997;141:343–4. [PubMed] [Google Scholar]
- Datu BJ, Gasser RB, Nagaraj SH, Ong EK, O’Donoghue P, McInnes R, et al. Transcriptional changes in the hookworm, Ancylostoma caninum, during the transition from a free-living to a parasitic larva. PLoS Negl Trop Dis. 2008;2:e130. doi: 10.1371/journal.pntd.0000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MA, Gasser RB, Woodcroft BJ, Hall RS, Ralph SA. Drug target prediction and prioritization: using orthology to predict essentiality in parasite genomes. BMC Genomics. 2010;11:222. doi: 10.1186/1471-2164-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Trinczek B, Illenberger S, Biernat J, Schmitt-Ulms G, Meyer HE, et al. Microtubule associated protein/microtubule affinity-regulating kinase (p110mark). A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J Biol Chem. 1995;270:7679–88. doi: 10.1074/jbc.270.13.7679. [DOI] [PubMed] [Google Scholar]
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–11. [PubMed] [Google Scholar]
- Eki T, Ishihara T, Katsura I, Hanaoka F. A genome-wide survey and systematic RNAi-based characterization of helicase-like genes in Caenorhabditis elegans. DNA Res. 2007;14:183–99. doi: 10.1093/dnares/dsm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eysker M, van Miltenburg L. Epidemiological patterns of gastrointestinal and lung helminth infections in grazing calves in The Netherlands. Vet Parasitol. 1988;29:29–39. doi: 10.1016/0304-4017(88)90005-2. [DOI] [PubMed] [Google Scholar]
- Frick DN. Helicases as antiviral drug targets. Drug News Perspect. 2003;16:355. doi: 10.1358/dnp.2003.16.6.829307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble HR, Lichtenfels JR, Purcell JP. Light and scanning electron microscopy of the ecdysis of Haemonchus contortus infective larvae. J Parasitol. 1989;75:303–7. [PubMed] [Google Scholar]
- Gamble HR, Fetterer RH, Mansfield LS. Developmentally regulated zinc metalloproteinases from third- and fourth-stage larvae of the ovine nematode Haemonchus contortus. J Parasitol. 1996;82:197–202. [PubMed] [Google Scholar]
- Gasteiger J. Chemoinformatics: a new field with a long tradition. Anal Bioanal Chem. 2006;384:57–64. doi: 10.1007/s00216-005-0065-y. [DOI] [PubMed] [Google Scholar]
- Geary TG, Thompson DP, Klein RD. Mechanism-based screening: discovery of the next generation of anthelminthics depends upon more basic research. Int J Parasitol. 1999;29:105–12. doi: 10.1016/s0020-7519(98)00170-2. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–20. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Hawdon JM, Jones BF, Hoffman DR, Hotez PJ. Cloning and characterization of Ancylostoma-secreted protein. A novel protein associated with the transition to parasitism by infective hookworm larvae. J Biol Chem. 1996;271:6672–8. doi: 10.1074/jbc.271.12.6672. [DOI] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Hu M, Lok JB, Ranjit N, Massey HC, Jr, Sternberg PW, et al. Structural and functional characterisation of the fork head transcription factor-encoding gene, Hc-daf-16, from the parasitic nematode Haemonchus contortus (Strongylida) Int J Parasitol. 2010;40:405–15. doi: 10.1016/j.ijpara.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–77. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–15. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd DD, Kemphues KJ. PAR-1 is required for the morphogenesis of the Canorhabditis elegans vulva. Develop Biol. 2003;253:54–65. doi: 10.1006/dbio.2002.0866. [DOI] [PubMed] [Google Scholar]
- Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol. 1999:138–48. [PubMed] [Google Scholar]
- Italiano JE, Jr, Stewart M, Roberts TM. How the assembly dynamics of the nematode major sperm protein generate amoeboid cell motility. Int Rev Cytol. 2001;202:1–34. doi: 10.1016/s0074-7696(01)02002-2. [DOI] [PubMed] [Google Scholar]
- Jackson F, Bartley D, Kenyon F, Sargison N. Dissemination of best practice to producers and advisors. Proc 21st Int Conf WAAVP. 2007:226. [Google Scholar]
- Jarrett WF, Jennings FW, McIntyre WI, Mulligan W, Urquhart GM. Immunological studies on Dictyocaulus viviparus infection. Immunity produced by the administration of irradiated larvae. Immunology. 1960;3:145–51. [PMC free article] [PubMed] [Google Scholar]
- Johnstone IL. The cuticle of the nematode Caenorhabditis elegans. Trends Genetics. 2000;16:21–7. doi: 10.1016/s0168-9525(99)01857-0. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Sohrmann M, Welchman DP, Ziperlen P, Ahringer J, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kim JK, Kaplan JM, Vidal M, Ruvkun G, Gabel HW, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–7. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- Koonin EV. The NCBI Handbook. The National Library of Medicine Press; 2010. The Clusters of Orthologous Groups (COGs) Database: Phylogenetic Classification of Proteins from Complete Genomes. [Google Scholar]
- Krasky A, Rohwer A, Schroeder J, Selzer PM. A combined bioinformatics and chemoinformatics approach for the development of new antiparasitic drugs. Genomics. 2007;89:36–43. doi: 10.1016/j.ygeno.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Letunic I, Yamada T, Kanehisa M, Bork P. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem Sci. 2008;33:101–3. doi: 10.1016/j.tibs.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Li BW, Rush AC, Crosby SD, Warren WC, Williams SA, Mitreva M, et al. Profiling of gender-regulated gene transcripts in the filarial nematode Brugia malayi by cDNA oligonucleotide array analysis. Mol Biochem Parasitol. 2005;143:49–57. doi: 10.1016/j.molbiopara.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Lipinski C, Lombardo F, Dominy B, Feeney P. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Lok JB. Transgenesis in parasitic nematodes: building a better array. Trends Parasitol. 2009;25:345–7. doi: 10.1016/j.pt.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukas A, Bethony J, Brooker S, Hotez P. Hookworm vaccines: past, present, and future. Lancet Infect Dis. 2006;6:733–41. doi: 10.1016/S1473-3099(06)70630-2. [DOI] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–6. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JB, Davidson AJ, Freeman KL, French NP. Immunisation of cattle with recombinant acetylcholinesterase from Dictyocaulus viviparus and with adult worm ES products. Int J Parasitol. 2001;31:307–17. doi: 10.1016/s0020-7519(00)00157-0. [DOI] [PubMed] [Google Scholar]
- McCarter JP. Genomic filtering: an approach to discovering novel antiparasitics. Trends Parasitol. 2004;20:462–8. doi: 10.1016/j.pt.2004.07.008. [DOI] [PubMed] [Google Scholar]
- McKeand JB. Vaccine development and diagnostics of Dictyocaulus viviparus. Parasitology. 2000;120:S17–23. doi: 10.1017/s0031182099005727. [DOI] [PubMed] [Google Scholar]
- Mendez S, D’ Samuel A, Antoine AD, Ahn S, Hotez P. Use of the air pouch model to investigate immune responses to a hookworm vaccine containing the Na-ASP-2 protein in rats. Parasite Immunol. 2008;30:53–6. doi: 10.1111/j.1365-3024.2007.00994.x. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies – the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, et al. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–7. doi: 10.1126/science.1057586. Erratum in: Science 2001;292:639. [DOI] [PubMed] [Google Scholar]
- Miller MA, Ruest PJ, Kosinski M, Hanks SK, Greenstein D. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 2003;17:187–200. doi: 10.1101/gad.1028303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molento MB, Depner RA, Mello MH. Suppressive treatment of abamectin against Dictyocaulus viviparus and the occurrence of resistance in first-grazing-season calves. Vet Parasitol. 2006;141:373–6. doi: 10.1016/j.vetpar.2006.01.061. [DOI] [PubMed] [Google Scholar]
- Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–64. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, et al. Next-generation tag sequencing for cancer gene expression profiling. Genome Res. 2009;19:1825–35. doi: 10.1101/gr.094482.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy S, Zhao Y, Delaney A, Asano J, Dhalla N, et al. Digital gene expression by tag sequencing on the illumina genome analyzer. Curr Protoc Hum Genet. 2010;11:1–36. doi: 10.1002/0471142905.hg1111s65. Unit 11.11. [DOI] [PubMed] [Google Scholar]
- Mulvenna J, Hamilton B, Nagaraj S, Smyth D, Loukas A, Gorman J. Proteomic analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol Cell Proteomics. 2009;8:109–21. doi: 10.1074/mcp.M800206-MCP200. [DOI] [PubMed] [Google Scholar]
- Nagaraj SH, Deshpande N, Gasser RB, Ranganathan S. ESTExplorer: an expressed sequence tag (EST) assembly and annotation platform. Nucleic Acids Res. 2007;35:W143–7. doi: 10.1093/nar/gkm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaou S, Hartman D, Nisbet AJ, Gasser RB. Haemonchus contortus: prokaryotic expression and enzyme activity of recombinant HcSTK, a serine/threonine protein kinase. Exp Parasitol. 2006;113:207–14. doi: 10.1016/j.exppara.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Nikolaou S, Gasser RB. Extending from PARs in Caenorhabditis elegans to homologues in Haemonchus contortus and other parasitic nematodes. Parasitology. 2007;134:461–82. doi: 10.1017/S0031182006001727. [DOI] [PubMed] [Google Scholar]
- Nisbet AJ, Gasser RB. Profiling of gender-specific gene expression for Trichostrongylus vitrinus (Nematoda: Strongylida) by microarray analysis of expressed sequence tag libraries constructed by suppressive-subtractive hybridisation. Int J Parasitol. 2004;34:633–43. doi: 10.1016/j.ijpara.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Novelli J, Ahmed S, Hodgkin J. Gene interactions in Caenorhabditis elegans define DPY-31 as a candidate procollagen C-proteinase and SQT-3/ROL-4 as its predicted major target. Genetics. 2004;168:1259–73. doi: 10.1534/genetics.104.027953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AP, Winter AD. Enzymes involved in the biogenesis of the nematode cuticle. Adv Parasitol. 2003;53:85–148. doi: 10.1016/s0065-308x(03)53003-2. [DOI] [PubMed] [Google Scholar]
- Panuska C. Lungworms of rimunants. Vet Clin North Am Food Anim Pract. 2006;22:583–93. doi: 10.1016/j.cvfa.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Parkinson J, Whitton C, Schmid R, Thomson M, Blaxter M. NEMBASE: a resource for parasitic nematode ESTs. Nucleic Acids Res. 2004;32:D427–30. doi: 10.1093/nar/gkh018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Morton DG, Gunsalus KC, Reinke V, et al. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans Curr Biol. 2002;12:1959–64. doi: 10.1016/s0960-9822(02)01301-5. [DOI] [PubMed] [Google Scholar]
- Ploeger HW. Dictyocaulus viviparus: re-emerging or never been away? Trends Parasitol. 2002;18:329–32. doi: 10.1016/s1471-4922(02)02317-6. [DOI] [PubMed] [Google Scholar]
- Ranganathan S, Nagaraj SH, Hu M, Strube C, Schnieder T, Gasser RB. A transcriptomic analysis of the adult stage of the bovine lungworm, Dictyocaulus viviparus. BMC Genomics. 2007;8:311. doi: 10.1186/1471-2164-8-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Stewart M. Acting like actin. The dynamics of the nematode major sperm protein (msp) cytoskeleton indicate a push-pull mechanism for amoeboid cell motility. J Cell Biol. 2000;149:7–12. doi: 10.1083/jcb.149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JG. Mechanistic basis of enzyme-targeted drugs. Biochemistry. 2005;44:5561–71. doi: 10.1021/bi050247e. [DOI] [PubMed] [Google Scholar]
- Rual JF, van den Heuvel S, Vidal M, Ceron J, Koreth J, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–8. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G, Hobert O. The taxonomy and developmental control in Caenorhabditis elegans. Science. 1998;282:2033–41. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- Schnieder T, Kaup FJ, Drommer W. Morphological investigations on the pathology of Dictyocaulus viviparus infections in cattle. Parasitol Res. 1991;77:260–5. doi: 10.1007/BF00930869. [DOI] [PubMed] [Google Scholar]
- Schnieder T. Dictyocaulus viviparus: isolation and characterization of a recombinant antigen with potential for immunodiagnosis. Int J Parasitol. 1992;22:933–8. doi: 10.1016/0020-7519(92)90050-u. [DOI] [PubMed] [Google Scholar]
- Schnieder T. The diagnostic antigen encoded by gene fragment Dv3–14: a major sperm protein of Dictyocaulus viviparus. Int J Parasitol. 1993;23:383–9. doi: 10.1016/0020-7519(93)90014-p. [DOI] [PubMed] [Google Scholar]
- Schnieder T, Epe C, von Samson-Himmelstjerna G, Kohlmetz C. The development of protective immunity against gastrointestinal nematode and lungworm infections after use of an ivermectin bolus in first-year grazing calves. Vet Parasitol. 1996;64:239–50. doi: 10.1016/0304-4017(95)00896-9. [DOI] [PubMed] [Google Scholar]
- Shaw LM, Olsen BR. FACIT collagens – diverse molecular bridges in extracellular matrices. Trends Biochem Sci. 1991;16:191–4. doi: 10.1016/0968-0004(91)90074-6. [DOI] [PubMed] [Google Scholar]
- Silverman PH, Podger KR. In vitro exsheathment of some nematode infective larvae. Exp Parasitol. 1964;15:314–24. doi: 10.1016/0014-4894(64)90026-8. [DOI] [PubMed] [Google Scholar]
- Singh M, Srivastava KK, Bhattacharya SM. Molecular cloning and characterization of a novel immunoreactive ATPase/RNA helicase in human filarial parasite Brugia malayi. Parasitol Res. 2009;104:753–61. doi: 10.1007/s00436-008-1251-6. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, Cassin E, Hewitson M, Holz C, Khan M, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–9. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Stepek G, McCormack G, Page AP. Collagen processing and cuticle formation is catalysed by the astacin metalloprotease DPY-31 in free-living and parasitic nematodes. Int J Parasitol. 2010;40:533–42. doi: 10.1016/j.ijpara.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Strube C, von Samson-Himmelstjerna G, Schnieder T. Genetic regulation of arrested development in nematodes: are age-1 and daf-gene orthologs present in Dictyocaulus viviparus? Parasitol Res. 2007a;101:1111–5. doi: 10.1007/s00436-007-0594-8. [DOI] [PubMed] [Google Scholar]
- Strube C, Schnieder T, von Samson-Himmelstjerna G. Differential gene expression in hypobiosis-induced and non-induced third-stage larvae of the bovine lungworm Dictyocaulus viviparus. Int J Parasitol. 2007b;37:221–31. doi: 10.1016/j.ijpara.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Strube C, Buschbaum S, von Samson-Himmelstjerna G, Schnieder T. Stage-dependent transcriptional changes and characterization of paramyosin of the bovine lungworm Dictyocaulus viviparus. Parasitol Int. 2009a;58:334–40. doi: 10.1016/j.parint.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Strube C, Buschbaum S, Schnieder T. Molecular characterization and real-time PCR transcriptional analysis of Dictyocaulus viviparus major sperm proteins. Parasitol Res. 2009b;104:543–51. doi: 10.1007/s00436-008-1228-5. [DOI] [PubMed] [Google Scholar]
- Tuteja R. Helicases – feasible antimalarial drug target for Plasmodium falciparum. FEBS J. 2007;274:4699–704. doi: 10.1111/j.1742-4658.2007.06000.x. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G, Schnieder T. Morphology of inhibited larvae of the bovine lungworm Dictyocaulus viviparus. J Helminthol. 1999;73:79–83. doi: 10.1017/s0022149x99000116. [DOI] [PubMed] [Google Scholar]
- Wang Z, Abubucker S, Martin J, Wilson RK, Hawdon J, Mitreva M. Characterizing Ancylostoma caninum transcriptome and exploring nematode parasitic adaptation. BMC Genomics. 2010;11:307. doi: 10.1186/1471-2164-11-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood IB, Amaral NK, Bairden K, Duncan JL, Kassai T, et al. World Association for the Advacement of Veterinary Parasitology (W.A.A.V.P.) second edition of guidelines for evaluating the efficacy of anthelmintics in ruminants. Vet Parasitol. 2005;58:181–213. doi: 10.1016/0304-4017(95)00806-2. [DOI] [PubMed] [Google Scholar]
- Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–4. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Zhan B, Xue J, Goud GN, Loukas A, Liu Y, et al. The evaluation of recombinant hookworm antigens as vaccines in hamsters (Mesocricetus auratus) challenged with human hookworm, Necator americanus. Exp Parasitol. 2008;118:32–40. doi: 10.1016/j.exppara.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Young ND, Hall RS, Jex AR, Cantacessi C, Gasser RB. Elucidating the transcriptome of Fasciola hepatica – a key to fundamental and biotechnological discoveries for a neglected parasite. Biotechnol Adv. 2010a;28:222–31. doi: 10.1016/j.biotechadv.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Young ND, Campbell BE, Hall RS, Jex AR, Cantacessi C, Laha T, et al. Unlocking the transcriptomes of two carcinogenic parasites, Clonorchis sinensis and Opisthorchis viverrini. PLoS Negl Trop Dis. 2010b doi: 10.1371/journal.pntd.0000719. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan B, Liu Y, Badamchian M, Williamson A, Feng J, Loukas A, et al. Molecular characterisation of the Ancylostoma-secreted protein family from the adult stage of Ancylostoma caninum. Int J Parasitol. 2003;33:897–907. doi: 10.1016/s0020-7519(03)00111-5. [DOI] [PubMed] [Google Scholar]
- Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–4. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


