Abstract
S phase kinase-associated protein 1 (SKP1)–cullin 1 (CUL1)–F-box protein (SCF) ubiquitin ligase complexes use a family of F-box proteins as substrate adaptors to mediate the degradation of a large number of regulatory proteins involved in diverse processes. The dysregulation of SCF complexes and their substrates contributes to multiple pathologies. In the 14 years since the identification and annotation of the F-box protein family, the continued identification and characterization of novel substrates has greatly expanded our knowledge of the regulation of substrate targeting and the roles of F-box proteins in biological processes. Here, we focus on the evolution of our understanding of substrate recruitment by F-box proteins, the dysregulation of substrate recruitment in disease and potential avenues for F-box protein-directed disease therapies.
The irreversibility of ubiquitin-mediated proteolysis makes the ubiquitin–proteasome system the ultimate on–off switch of a cell. It controls many processes, particularly those that must proceed unidirectionally, such as the cell cycle or circadian oscillations. Covalently linked chains of the small protein ubiquitin are generated on substrates through an enzymatic cascade in which ubiquitin is activated by an E1 enzyme, transferred to an E2 ubiquitin-conjugating enzyme and then transferred to a substrate selected by an E3 ubiquitin ligase1. Ubiquitin chains of four or more moieties, linked through either Lys48 or Lys11 of each ubiquitin, direct substrates to the proteasome, which is a large complex of proteases that degrades ubiquitylated proteins2. Multiple monoubiquitins and other, non-Lys63-linked ubiquitin chains have also recently been implicated in protein degradation, and the study of alternative degradation signals is continuing2,3. It is estimated that >80% of proteins undergo ubiquitin-mediated degradation, so the selection of specific substrates by E3 ubiquitin ligases in response to specific stimuli is a crucial factor in cell regulation4. In humans, there are only two E1 enzymes and ~30 E2 enzymes, but there are hundreds of E3 enzymes. Many E3 ligases are modular, based on a core scaffold with interchangeable substrate-targeting subunits, which enable one piece of the core machinery to ubiquitylate many different substrates. The cullin– RING ligase (CRL) family complexes are the archetypes for these modular ubiquitin ligases (FIG. 1), and CRL1 ligases, better known as the S phase kinase-associated protein 1 (SKP1)– cullin 1 (CUL1)–F-box protein (SCF) complexes, are the best characterized5.
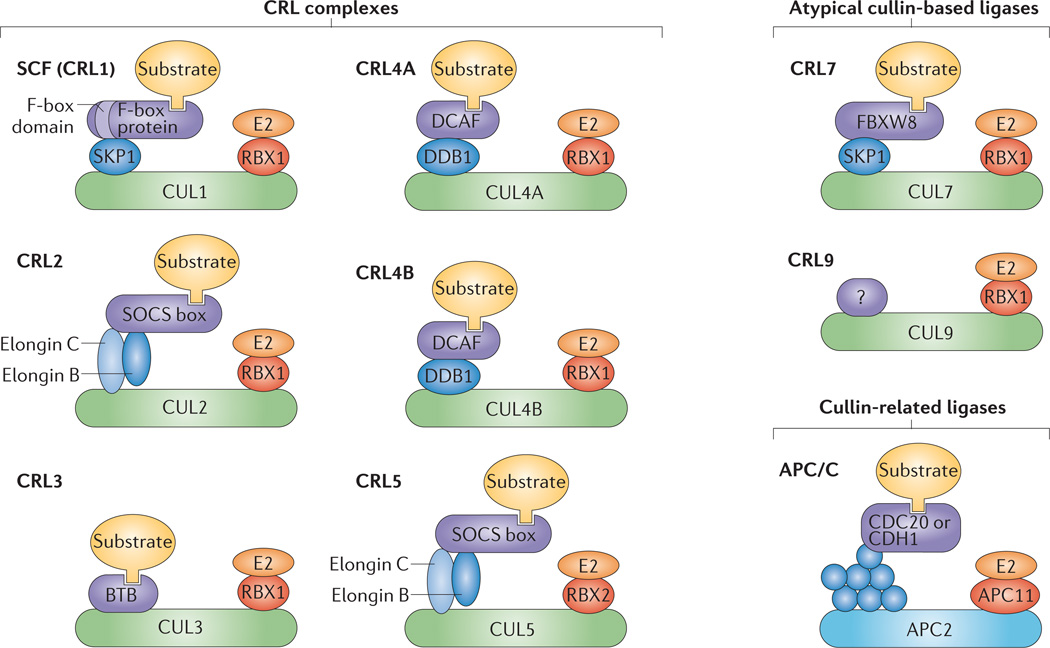
Figure 1. The cullin–RING ligase family.
Cullin (CUL) proteins form the backbones of ubiquitin ligase complexes. CUL7 and CUL9 are atypical cullins owing to their large size and the incorporation of additional homology domains. APC2, the core of the APC/C (anaphase promoting complex; also known as the cyclosome), is distantly related to cullins. Each cullin–RING ligase (CRL) complex is modular, with variable substrate adaptors. CRL1 (better known as S phase kinase-associated protein 1 (SKP1)–CUL1–F-box protein (SCF) complex) uses SKP1 and F-box proteins as substrate adaptors, and CRL2 and CRL5 ligases use elongin B, elongin C and SOCS (suppressor of cytokine signalling) box proteins. The substrate adaptors for CRL3 are BTB (bric-a-brac-tramtrack-broad complex) proteins. CRL4A and CRL4B use DDB1 (DNA damage-binding protein 1) and DCAF (DDB1- and CUL4-associated factor) proteins. CRL7 uses SKP1 and a single F-box protein (F-box and WD40 domain 8 (FBXW8)). The substrate adaptors for CRL9 are not known. The full molecular architecture of the APC/C remains unclear, in part because it contains many more proteins (shown in blue) than the CRL complexes. APC/C uses either CDC20 or CDH1 (CDC20 homologue 1) as a substrate adaptor. The substrate for each complex is shown in yellow. RBX1, RING-box protein 1.
SCF complexes are assembled using the scaffold protein CUL1. CUL1 bridges two essential biochemical functions of the SCF complex. The carboxyl terminus of CUL1 recruits the small RING protein RBX1, which directs the E2 enzyme to the E3 ligase, and the amino terminus of CUL1 binds SKP1 and a variable F-box protein that dictates substrate specificity5. As their name implies, F-box proteins contain the 40-amino-acid F-box domain (first identified in cyclin F (also known as FBXO1)), which binds SKP1 to create a link to CUL1. Sixty-nine human F-box proteins, each targeting multiple substrates, enable the core scaffold to select hundreds of proteins for degradation6,7.
Although the generic SCF complex is considered well-characterized, many questions remain about the functions of specific SCF complexes. One glaring gap in our understanding of the SCF complexes is the number of orphan F-box proteins, for which no substrates are known. Our conceptions of the functions of SCF complexes are based on relatively few unique F-box protein– substrate pairs and the mechanisms controlling these pairings. Therefore, the field is rapidly evolving with the characterization of each orphan F-box protein. This Review highlights key recent advances in our mechanistic understanding of substrate recruitment by F-box proteins and our understanding of the overall biological roles of F-box proteins. An increased understanding of substrate targeting, coupled with knowledge of the roles of F-box proteins in disease, presents new targets for therapeutics, although this area of research remains in its infancy.
The F-box protein families
F-box proteins can be categorized on the basis of the presence of recognizable domains beyond the F-box domain, thereby generating three families6,8,9. In humans, the F-box and WD40 domain (FBXW) family is composed of 12 proteins, and the F-box and Leu-rich repeat (FBXL) family comprises 21 proteins. The remaining 36 F-box proteins were originally categorized as F-box only (FBXO) proteins, but contrary to their name, these F-box proteins often have conserved homology domains that were either not recognized or are not present in a large number of F-box proteins. Therefore, FBXO is more truly an abbreviation for F-box and other domains. At least 21 homology domains have been identified among the FBXO family members, and it is assumed that these homology domains mediate the interactions of F-box proteins with substrates6. So far, this hypothesis has received wide experimental support from studies of cyclin F, FBXO2, FBXO6 and FBXO11 (REFS 10–13).
Substrate recruitment to SCF complexes
SCF complexes are subject to general regulation through multiple mechanisms, including covalent modification with NEDD8 and the binding of various assembly factors (BOX 1). These general regulatory mechanisms are central to the biochemistry of the SCF complexes, but they remain poorly understood and/or controversial. In addition, as global regulators of SCF complexes, these mechanisms do not control specific substrates and biological functions. Overall, the ultimate regulation of the activity of specific SCF complexes occurs at the level of substrate recruitment. In response to stimuli, F-box proteins must rapidly and specifically bind their target proteins in the complex cellular milieu and recruit them to the core SCF scaffold, which exerts ubiquitin ligase activity. Conversely, binding between an F-box protein and its substrate can be perturbed in response to stimuli. Therefore, both F-box protein–substrate interfaces and the F-box proteins themselves are subject to tight regulation.
General regulation of SCF complexes.
Several factors regulate the S phase kinase-associated protein 1 (SKP1)–cullin 1 (CUL1)–F-box protein (SCF) scaffold, and these factors often control all cullin–RING ligase (CRL) complexes. With the exception of CUL7, CRLs are regulated by neddylation, which is the covalent attachment of the small ubiquitin-like protein NEDD8 to the cullin subunit. This modification releases the RING finger subunit on a flexible tether, which positions the E2 enzyme closer to the substrate, thereby potentiating ubiquitylation. The neddylation status of cullins also determines their binding to cullin-associated and neddylation-dissociated 1 (CAND1), which can only bind non-neddylated cullins. CAND1 is an assembly factor that facilitates the rapid exchange of F-box proteins on the SCF scaffold126–128 and other substrate adaptors on the additional CRL scaffolds. Like ubiquitylation, NEDD8 conjugation is accomplished through a cascade of E1, E2 and E3 enzymes. Cullins function as their own E3 ligases for NEDD8 by recruiting a NEDD8-specific E2 (either UBE2M (also known as UBC12) or UBE2F) to the complex. Neddylation is reversed through the action of the COP9 signalosome, a large, multisubunit complex that proteolytically cleaves NEDD8 from cullins. Combined, this system creates a cycle of neddylation and deneddylation that controls the assembly and activity of CRLs. More recently, glomulin was also shown to inhibit CRLs by blocking the access of the E2 enzymes to RING-box protein 1 (RBX1)129. Although the specific phenotypic effects of glomulin loss or inhibition manifest through an SCF complex that contains F-box and WD40 domain 7 (FBXW7), glomulin seems to affect the activity of most CRLs130. It is likely that other general regulators of CRLs will be discovered in the future.
Canonical phosphodegrons
F-box proteins bind substrates in response to various stimuli, and in the canonical model, they bind short, defined degradation motifs (known as degrons)14. For many years, phosphorylation-dependent substrate recruitment was the paradigm for SCF complex function, and the best-characterized F-box proteins bind to phosphodegrons in their substrates. For example, β-transducin repeat-containing protein (βTrCP; which refers to F-box proteins encoded by both FBXW1 and FBXW11) binds the consensus degron Asp-Ser-Gly- Xaa-Xaa-Ser (in which Xaa represents any amino acid and both Ser residues are phosphorylated), and FBXW7 binds the degron Thr-Pro-Pro-Xaa-Ser (in which the Thr and Ser residues are phosphorylated)15,16. Variation from the consensus degrons, such as substitution of Ser and Thr residues or the inclusion of phosphomimicking amino acids, is observed. Simple phosphodegrons can be phosphorylated by a single kinase. Alternatively, degrons with multiple phosphorylation sites can be targeted by multiple kinases and/or use priming phosphorylations, which adds increased complexity and stringency to substrate recognition by F-box proteins. For example, cyclin-dependent kinase 2 (CDK2) and glycogen synthase kinase 3 (GSK3) phosphorylate different residues of the cyclin E degron17, and phosphorylation of the JUN degron by GSK3 requires prior phosphorylation of the degron by an additional kinase18. Degron phosphorylation can also require priming phosphorylations that are near, but not directly at, the F-box protein–substrate interface, such as the priming phosphorylations that are required for β-catenin regulation by βTrCP19.
Finally, phosphodegrons can be used to fine-tune substrate recognition by setting a threshold of kinase activity that creates a direct linkage to a regulatory pathway. The yeast CDK inhibitor Sic1 is degraded to allow S phase entry, and recognition of Sic1 by the Cdc4 F-box protein requires the phosphorylation of at least six of nine specific amino acids of Sic1 by CDKs. These phosphorylations generate multiple suboptimal degrons, the binding affinity of which is sufficient to increase the local concentration of Sic1 associated with SCFCdc4 (an SCF complex containing Cdc4), producing highly specific and efficient binding20–22. CDK activity increases during G1 phase of the cell cycle, eventually reaching a threshold level that is required for the recognition of Sic1 by SCFCdc4, resulting in Sic1 degradation and entry into S phase.
However, as the larger F-box protein family is explored, it is becoming clear that although direct recognition of phosphodegrons is a common mechanism for substrate recruitment, it is not the only mechanism (FIG. 2). In addition, multiple recruitment mechanisms can be combined, which enables more precise regulation of substrate selection by each SCF complex.
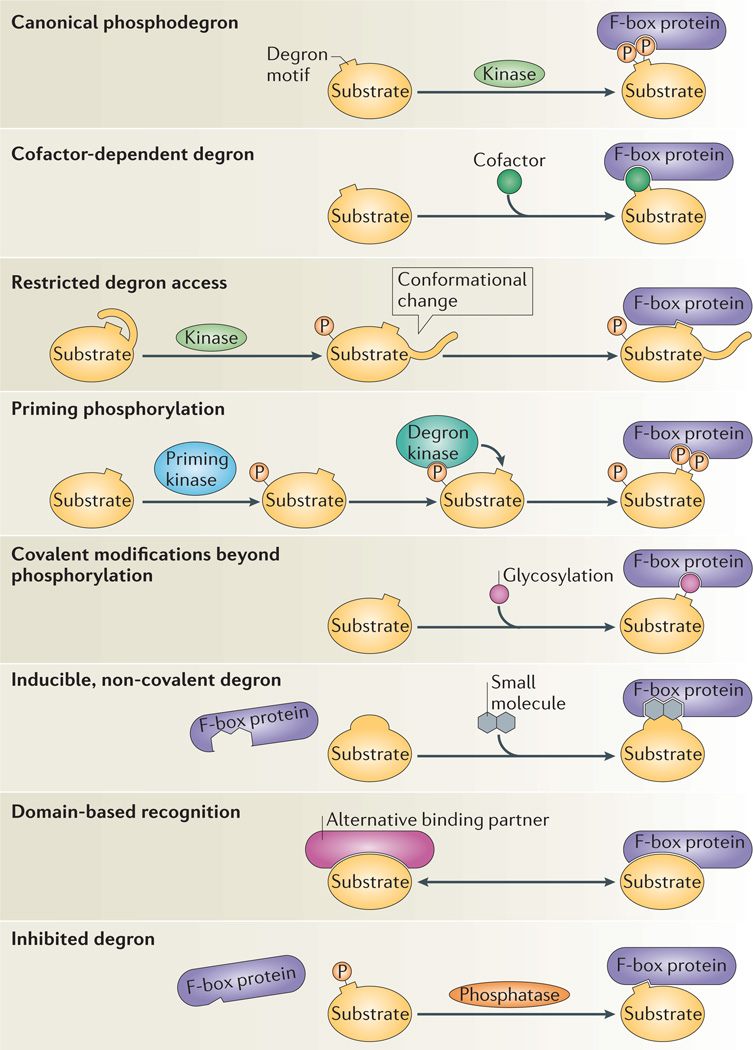
Figure 2. Recognition of substrates by F-box proteins.
F-box proteins recognize their substrates in multiple ways. Often, various recognition mechanisms are combined to impart precise regulation of substrate degradation. Eight different modes of recognition and regulation are shown, as discussed in the main text. These simple concepts of regulation and recognition are often combined in substrates. For example, phosphodegron recognition can be combined with priming phosphorylations, or the recognition of non-modified degrons can be paired with restricted degron access.
Cofactor-dependent substrate recognition
SKP2 (also known as FBXL1)-dependent degradation of the CDK inhibitor p27 presents one permutation of phosphorylation-directed substrate binding. Following the phosphorylation of Thr187 of p27 by a CDK, SKP2 binds p27 leading to its ubiquitylation and degradation. However, when this ubiquitylation reaction is reconstituted in vitro, SCFSKP2 by itself has poor ubiquitylation activity towards p27 (REFS 23,24). Full activity requires CKS1 (CDK regulatory subunit 1), an accessory protein that binds both SKP2 and p27 (REFS 25,26). This model seems to fit the canonical view of phosphorylation-dependent binding of F-box proteins, but the crystal structure of the SKP2–CKS1–p27 phosphopeptide complex indicates otherwise27. CKS1, but not SKP2, forms contacts with phosphorylated Thr187 of p27, so the accessory protein CKS1 is integral to substrate recognition by SKP2. It is currently unclear how many F-box proteins — and belonging to which families — might use accessory proteins and whether all accessory proteins control substrate recognition. In this regard, FBXO4 has been reported to use the chaperone αβ-crystallin as a cofactor for phosphorylation-dependent substrate recognition, but it is not clear whether FBXO4 and/or αβ-crystallin directly recognize the phosphorylated substrate 28,29. Ubiquitylation of the cofactor Met4 in yeast presents a variation on this theme. This transcription factor itself is inactivated by SCFMet30-mediated ubiquitylation and, concomitantly, Met4 facilitates ubiquitylation of several of its cofactors, providing coordinate regulation30.
Unmodified degrons
Degron phosphorylation facilitates the rapid and specific regulation of substrate selection by SCF complexes, but studies of cyclin F show that F-box proteins can also recognize unmodified degrons. Cyclin F does not bind CDKs, but its cyclin-homology domain recognizes an Arg-Xaa-(Ile, Leu) motif in its substrates in a manner similar to the way that other cyclins recognize their substrates for phosphorylation by CDKs10,11. Cyclin F uses this recognition mechanism for all known substrates, including the centrosomal protein CP110 and ribonucleotide reductase subunit M2 (RRM2), which provides evidence of the utility of unmodified degrons10,11. Because the Arg-Xaa-Leu degron is independent of any modification, substrates containing this motif would be degraded constitutively in the absence of additional regulatory mechanisms. Such mechanisms include the restriction of degron access and indirect regulation of substrate recognition through the control of cyclin F localization and stability.
Restricted degron access
Physical access of F-box proteins to substrate degrons can be controlled by the phosphorylation of motifs other than the degron, and such control can be combined with multiple degron types, including non-modified degrons and phosphodegrons. This regulation is exemplified by the ubiquitylation of RRM2 by cyclin F10. Ribonucleotide reductase generates deoxyribonucleotides for DNA synthesis, and in G2 phase of the cell cycle, the enzyme is inactivated by degradation of RRM2. To impart cell cycle-dependent regulation, access to the Arg-Xaa-Ile degron in RRM2 is controlled by CDK-mediated phosphorylation of RRM2. When CDK activity is low, the degron is obscured, but high CDK activity in G2 causes RRM2 phosphorylation at Thr33, which is not part of the degron, enabling recognition of the unmodified Arg-Xaa-Ile degron by cyclin F. Other SCF substrates might also use phosphorylation-regulated degron access, but in the case of phosphodegrons, it is difficult to determine whether these external phosphorylation events are priming events for the degron, control degron access, or both31–33.
Indirect regulation of substrate recognition
SCFcyclin F_ mediated ubiquitylation of RRM2 also depends on the localization of both the substrate and SCF complex. RRM2 is a cytoplasmic protein, and cyclin F is a nuclear protein. Therefore, cyclin F-dependent RRM2 degradation requires nuclear import of RRM2 (REF. 10). Localization also affects the ubiquitylation of other cyclin F substrates, such as CP110. CP110 controls centrosome duplication, and its expression is closely linked to the cell cycle. CP110 localizes exclusively to the centrosome, and its degradation requires a subpopulation of cyclin F at the centrosome11.
Subcellular localization also controls other F-box proteins. FBXL2 and FBXL20 contain Cys-Ala-Ala-Xaa (CAAX) motifs, which undergo isoprenylation, directing the proteins to membranes34,35. CAAX-dependent membrane localization of FBXL20 is required for the degradation of RAB3-interacting molecule 1 (RIM1), and membrane-associated FBXL2 is required for hepatitis C virus (HCV) replication (through unknown substrates). The localization-dependent functions of cyclin F, FBXL2 and FBXL20 highlight the ability of SCF complexes to control substrates in both a spatial and temporal manner.
In fact, the temporal activation of cyclin F is regulated through tight control of its expression levels. Cyclin F levels are modulated throughout the cell cycle by transcription and by degradation involving an unknown ubiquitin ligase10,36. When cyclin F levels increase in G2 phase of the cell cycle, cyclin F substrates are degraded. Regulation of other F-box proteins by degradation has also been reported. The cell cycle-dependent oscillations of SKP2 levels are controlled by the APC/CCDH1 (that is, the anaphase-promoting complex (also known as the cyclosome) containing CDC20 homologue 1), and EMI1 (early mitotic inhibitor 1; also known as FBXO5) is regulated in a cell cycle-dependent manner by SCFβTrCP (REFS 37–40). F-box protein stability might also be controlled through autoubiquitylation mediated by the SCF scaffold, but the specificity of such regulation remains unclear41.
Most examples of F-box protein degradation are linked to the cell cycle, but the regulation of FBXL5 is unique in this regard. FBXL5 mediates the degradation of iron regulatory protein 2 (IRP2) under conditions of high iron and/or oxygen levels, and in turn, IRP2 controls the translation and stability of iron-responsive mRNAs, including those encoding proteins related to iron metabolism and enzymes that are functionally dependent on iron42–44. The key feature of this regulatory system is a haemerythrin domain in FBXL5 that binds two iron ions. In iron-depleted conditions, the haemerythrin domain cannot bind iron, resulting in FBXL5 unfolding and degradation (through an unknown ubiquitin ligase)45,46. The properly folded haemerythrin domain confers stability to FBXL5 in the presence of iron, but it does not affect substrate binding, highlighting the direct regulation of F-box proteins in response to environmental cues. Iron availability might also affect FBXL5–IRP2 binding in a haemerythrin domain-independent manner, but this intriguing mechanism remains to be investigated43. The modification of F-box proteins, whether by iron coordination or covalent modifications, in response to stimuli to increase F-box protein stability and substrate degradation is an emerging theme that is also illustrated by ataxia-telangiectasia mutated (ATM)-mediated phosphorylation of FBXO31 in response to DNA damage47. Covalent modifications of F-box proteins are common, and it is possible that they could have a role beyond F-box protein stability, affecting either substrate recruitment or association with the SCF scaffold48. However, this hypothesis requires further investigation.
Inducible, non-covalent degrons
The regulation of FBXL5 by non-covalent binding of iron underscores that F-box proteins can respond to signals other than covalent posttranslational modifications. Studies of plant responses to the hormones auxin and jasmonate have shown that substrates can be recruited to F-box proteins through degrons that are a combination of protein sequence and non-peptide hormones or small molecules. The receptors for auxin and jasmonate (TIR1 (TRANSPORT INHIBITOR RESPONSE 1) and COI1 (CORONATINEINSENSITIVE 1), respectively) are F-box proteins that target the auxin–indoleacetic acid (AUX–IAA) and JA–ZIM domain (JAZ) families of transcriptional repressors, respectively, for degradation49,50. Binding of the hormones to their receptors fills the gaps between the substrates and the F-box proteins, extending the binding surface and functioning as ‘molecular glue’ to stabilize the interaction. The formation of this protein–hormone hybrid degron is essential for substrate degradation. In addition, both TIR1 and COI1 are similar to FBXL5 in that they incorporate a small molecule into their folded structure. TIR1 incorporates inositol hexakisphosphate, and COI1 incorporates inositol pentakisphosphate. Whether the incorporation of these small molecules has a regulatory role, similar to the incorporation of iron ions in FBXL5, remains to be determined. However, in the case of COI1, inositol pentakisphosphate has a crucial role in degron recognition, helping to coordinate binding to a key carboxyl group in the hormone, and it is likely that inositol hexakisphosphate has a similar role in TIR1 (REF. 49).
Covalent, non-phosphorylation-based degron modifications
F-box proteins also recognize degrons with covalent modifications other than phosphorylation. FBXO2 and FBXO6 bind glycosylated substrates, such as pre-integrin β1 and T cell receptor α-chain, respectively, through F-box-associated (FBA) domains. Based on the presence of FBA domains, FBXO17, FBXO27 and FBXO44 are predicted to also bind glycosylated substrates12,51,52. The recognition of high-mannose oligosaccharides by FBXO2 and FBXO6 is in line with their function in endoplasmic reticulum (ER)-associated degradation (ERAD) of incorrectly folded proteins, as many proteins are glycosylated in the ER during the folding process, but clear functions for other sugarbinding F-box proteins remain unclear. In general, investigating the substrates and specificity of sugarbinding F-box proteins is an underdeveloped area of F-box protein biology, and no substrates have been identified for FBXO17, FBXO27 or FBXO44. However, these F-box proteins are likely to be functionally distinct based on non-overlapping affinities for different glycans and different expression profiles12. Finally, it is unclear whether binding to glycosylated degrons is mutually exclusive with other forms of substrate binding. FBXO6 targets checkpoint kinase 1 (CHK1) for degradation, and although the nature of the CHK1 degron is currently not known, CHK1 has not been reported to be glycosylated53.
Modifications blocking degron recognition
Although post-translational modifications of degrons often direct the binding of F-box proteins to substrates, they can also prevent substrate recognition by the F-box protein. FBXO11 recognizes an unmodified degron in CDT2, and phosphorylation of this degron by a CDK blocks FBXO11 binding54,55. Similarly, FBXL2 binding to p85β is also prevented by phosphorylation56. Although this mechanism has only been reported for two F-box protein– substrate pairs, it highlights an alternative model of degron recognition. Historically, the role of degron phosphorylation in substrate recognition has been viewed through kinase activity. However, phosphatases may facilitate degron recognition, as demonstrated by the regulation of the p85β degron by the protein Tyr phosphatase PTPL1 (REF. 56). Non-phosphorylation-based modifications could also block degron recognition.
Domain-based recognition
Finally, although most of the F-box protein field focuses on the concept of degrons as short stretches of amino acids, several findings indicate that not all F-box proteins recognize such short degrons. The short degron hypothesis has been challenged by reports of F-box protein–substrate interactions that are determined by conserved domain structures. FBXO4, which binds cyclin D1 in a phosphorylation-dependent manner, also binds the telomere protein TRF1 (telomeric repeat-binding factor 1) in a domain-dependent manner. In the FBXO4–TRF1 co-crystal structure, FBXO4 adopts a GTPase-like fold that binds to TRF1 independently of covalent modifications57,58. Only non-telomeric TRF1 is degraded, and this regulation is imparted by the binding of TIN2 (TRF1-interacting nuclear protein 2) to TRF1 at telomeres, which physically blocks the FBXO4-binding site on TRF1.
In addition, FBXL3 recognizes cryptochrome 1 and cryptochrome 2 (CRY1 and CRY2) in a modification-independent manner through extended contacts between its Leu-rich repeats and the surface of the CRY proteins59–62. The C terminus of FBXL3 is also inserted into a conserved pocket in CRY that may be subject to competition with the flavine adenine dinucleotide (FAD) redox cofactor, which would also represent a new way to regulate substrate recruitment. This potentially novel mechanism requires further study. CRY binding to FBXL3 is probably blocked by binding of the transcription factor period (PER), a core oscillatory component of the circadian clock (the levels of which are regulated by βTrCP) that couples CRY oscillations to PER. Other F-box protein–substrate pairs might also use domain or secondary structure recognition instead of degron recognition, but this form of regulation is difficult to prove without crystal structures.
Further diversification and regulation of substrate recruitment
As illustrated above, substrate recruitment by F-box proteins is regulated by multiple mechanisms, at the levels of both the substrate and F-box protein, including restriction of degron access, covalent degron modifications, regulated F-box protein stability and defined F-box protein localization. The diversity of targeting mechanisms for F-box protein substrates is continuing to increase. Methylation-dependent recognition of substrates has recently been reported for a CRL4 ubiquitin ligase through a DCAF (DDB1- and CUL4-associated factor) substrate adaptor63, and the von Hippel–Lindau disease tumour suppressor (VHL), which is a substrate adaptor for CRL2, recognizes hydroxylated Pro residues in substrates64, suggesting that F-box proteins could recognize alternative modifications. In addition, although hormone- and small molecule-regulated substrate binding have only been observed in plants, this mechanism can be functionally reconstituted in mammalian cells, which indicates that similar mechanisms for substrate recognition are possible in higher organisms65. Importantly, small molecule- induced binding of substrates is currently under investigation for the development of drugs that restore, activate or retarget E3 ubiquitin ligase activity in various diseases. The F-box protein–substrate interface is also a highly specific target for the development of therapeutic inhibitors (BOX 2).
F-box protein-targeted therapy.
Rational design of inhibitors
Crystal structures of F-box protein–substrate complexes allow the rational design of inhibitors, as exemplified by the structure of S phase kinase-associated protein 1 (SKP1)–F-box and Leu-rich repeat 3 (FBXL3)–cryptochrome 2 (CRY2)62. Intriguingly, the tail of FBXL3 fits into a pocket in CRY2. In CRY proteins from other organisms, which (unlike mammalian CRY proteins) function as photoreceptors, this conserved pocket binds to flavin adenine dinucleotide (FAD)131, and mammalian CRY proteins also have (extremely weak) binding to FAD. Therefore, FAD analogues could function as competitive inhibitors of FBXL3–CRY2 interactions. A compound that competes for FAD binding, inhibits CRY ubiquitylation and lengthens the circadian cycle was recently identified independently132.
Virtual ligand screening (VLS) has also been used to identify inhibitory compounds in silico based on pockets in structures that are able to bind small molecules, such as the interface between SKP2, cyclin-dependent protein kinase regulatory subunit 1 (CKS1) and p27. This approach facilitated the discovery of compounds that competitively inhibit SKP2-mediated degradation of p27 (REF. 133).
High-throughput screening
High-throughput screens based on ubiquitin ligase activity are also being applied to SCF complexes and can identify inhibitors that target multiple steps in the ubiquitylation process, including kinase inhibitors, inhibitors of SCF complex assembly, allosteric inhibitors and degron inhibitors. One recent screen identified an allosteric inhibitor of yeast Cdc4 that distorts the WD40 domain and prevents substrate binding. This finding validates the concept of allosteric inhibition of F-box proteins134. Another compound disrupts binding between Skp1 and Met30, preventing SCF complex formation135. This compound is surprisingly selective for the Met30 F-box domain and does not inhibit the interaction between Skp1 and other F-box proteins.
De novo protein targeting
Two different strategies can be envisioned for restoring E3 ligase functions: stabilization of the interface between a mutated F-box protein and its substrates or the retargeting of substrates to a different, functional ligase. The first approach draws from the precedent established by auxin and TIR1 (TRANSPORT INHIBITOR RESPONSE 1)50. Provided that the mutated F-box protein is still expressed and only mildly unfolded, it may be possible to use a small molecule as a molecular glue or as an allosteric modifier of the structure of the F-box protein. However, this approach is technically daunting, specific for individual mutants and cannot correct large deletions.
Alternatively, protacs (proteolysis targeting chimaeras) are hetero-bivalent chimeric molecules that recruit an E3 ligase at one end (by mimicking a degron) and bind specific target proteins at the other end, which tethers the ligase to the substrate and results in target degradation136. Both peptide degron-based fusion protacs and small molecule-based protacs have been developed137. Protac technology has several limitations, including the inhibition of the endogenous ligase and nonspecific degradation of non-target proteins.
Finally, the incorporation of F-box proteins into the SCF scaffold is an emerging area of research. SCF complexes can be disassembled in response to specific stimuli. This was shown by the removal of Met30 from SCFMet30 by Cdc48 in response to treatment of yeast with cadmium66. Dimerization of F-box proteins has also been observed, but the in vivo effect of F-box protein dimerization on substrate selection and ubiquitylation remains unclear58,67–70. Whereas dimerization of yeast Cdc4 contributes to substrate ubiquitylation, disrupting the dimerization of FBXW7, the human Cdc4 homologue, does not seem to affect substrate regulation71. The functional relevance of substrate adaptor dimerization is more firmly established for the CRL3 substrate adaptors Speckle-type POZ (SPOP) and Kelch-like ECH-associated protein (KEAP1). In each case, dimerization enables the recognition of two degrons in the substrate and could affect either the avidity of the substrate for the ligase or its positioning within the active ligase complex71.
F-box proteins in disease
As discussed, F-box proteins regulate substrates in diverse biological pathways that control key dimensions of cellular life, including cell growth, cell division, development and differentiation, signalling responses, and cell survival and death (TABLE 1). Therefore, dysregulation of F-box protein-mediated ubiquitylation, which can occur via multiple distinct mechanisms (FIG. 3), has been implicated in many pathologies, including sleep disorders, mood disorders, diabetes, Parkinson’s disease, bacterial infections and viral infections35,72–80. Our knowledge of the involvement of F-box proteins in many of these diseases (and others) will be expanded and cemented in the coming years. Notably, although F-box proteins function in diverse biological pathways, the evolution of the field is closely linked to studies of cell proliferation, so F-box protein function has been viewed largely in the context of cancer biology and the best-characterized F-box proteins, SKP2, FBXW7 and βTrCP (FIG. 4). These studies have provided insights into the ability of F-box proteins to function in a general or context-dependent manner, providing a valuable framework for the study of F-box proteins in other systems and diseases (FIG. 4).
Table 1.
Control of key dimensions of cellular life by F-box proteins
| F-box protein |
Substrate | Function of substrate | Mechanism of degron regulation |
|---|---|---|---|
| Differentiation and development | |||
| βTrCP | β-catenin | WNT signalling | Phosphodegron, priming phosphorylation |
| βTrCP | REST | Neural differentiation, cell cycle | Phosphodegron |
| βTrCP | SNAIL | Epithelial–mesenchymal transition | Phosphodegron, priming phosphorylation |
| FBXL14 | SNAIL | Epithelial–mesenchymal transition | Unknown |
| FBXO11 | BCL6 | B cell development | Unknown |
| FBXO11 | CDT2 | DNA replication, seam cells (in worms) | Blocked by phosphorylation |
| FBXO32 | MyoD | Muscle cell differentiation | Unknown |
| FBXW7 | CEBPα | Adipocyte differentiation | Phosphodegron |
| FBXW7 | SREBP1 | Adipocyte differentiation | Phosphodegron |
| Cell division | |||
| βTrCP | BORA | Mitotic signalling | Phosphodegron |
| βTrCP | EMI1 | APC/C inhibitor | Phosphodegron |
| βTrCP | PLK4 | Centriole duplication | Phosphodegron |
| βTrCP | WEE1 | Kinase, CDK inhibitor | Phosphodegron |
| Cyclin F | CP110 | Centriole duplication | Unmodified, localization dependent |
| Cyclin F | RRM2 | dNTP production | Restricted access |
| FBXW5 | EPS8 | Actin remodelling | Unknown |
| FBXW7 | Cyclin E | Cell cycle | Phosphodegron |
| SKP2 | p27 | CDK inhibitor | Cofactor-dependent |
| Cell death and survival | |||
| βTrCP | BIMEL | Apoptosis | Phosphodegron |
| βTrCP | IκB | Inhibitor of NF-κB signalling | Phosphodegron |
| FBXO7 | cIAP1 | Apoptosis inhibitor | Unknown |
| FBXW7 | MCL1 | Survival factor | Phosphodegron |
| FBXW7 | p100 | Non-canonical NF-κB signalling | Phosphodegron |
| Cell growth | |||
| βTrCP | DEPTOR | Inhibitor of mTOR | Phosphodegron, priming phosphorylation |
| βTrCP | PDCD4 | Inhibits protein synthesis | Phosphodegron |
| FBXO9 | TEL2 and TTI1 | mTOR signalling | Phosphodegron |
| FBXW7 | JUN | Mitogenic signalling | Phosphodegron |
| FBXW7 | MYC | Cell proliferation | Phosphodegron |
| Signalling | |||
| βTrCP | CDC25A | Phosphatase, CDK activator | Phosphodegron |
| βTrCP | PER1 and PER2 | Circadian rhythm | Phosphodegron |
| FBXL2 | p85β | PI3K signalling | Blocked by phosphorylation |
| FBXL3 | CRY1 and CRY2 | Circadian rhythm | Domain recognition, possibly ligand |
| FBXL5 | IRP2 | Iron homeostasis | Unknown |
APC/C, anaphase-promoting complex (also known as the cyclosome); βTrCP, β-transducin repeat-containing protein; BCL6, B cell lymphoma 6; BIMEL, BCL-2-interacting mediator of cell death extra long; BORA, aurora borealis; CDK, cyclin-dependent kinase; CEBPα, CCAAT-enhancer-binding protein-α; cIAP1, cellular inhibitor of apoptosis 1; CP110, centrosomal protein 110; CRY, cryptochrome; DEPTOR, DEP-domain containing mTOR-interacting protein; EMI1, early mitotic inhibitor 1; EPS8, EGFR kinase substrate 8; FBXL, F-box and Leu-rich repeat; FBXO, F-box only; FBXW, F-box and WD40 domain; IκB, inhibitor of κB; IRP2, iron-regulatory protein 2; MCL1, myeloid cell leukaemia sequence 1; mTOR, mammalian target of rapamycin; MyoD, myoblast determination; NF-κB, nuclear factor-κB; PDCD4, programmed cell death 4; PER, period; PLK4, Polo-like kinase 4; REST, RE1-silencing transcription factor; RRM2, ribonucleotide reductase subunit M2; SREBP1, sterol regulatory element-binding protein 1; SKP2, S phase kinase-associated protein 2; TEL2, telomere length regulation protein 2; TTI1, TELO2-interacting protein 1.
Figure 3. Dysregulation of F-box protein-mediated degradation in disease.
Dysregulation of F-box protein-mediated degradation can occur by the overexpression of an F-box protein, deletion of an F-box protein, point mutation of an F-box protein or mutation of substrate degrons. In addition, the expression of microbial proteins can increase or decrease substrate degradation, or lead to the degradation of alternative substrates. Ub, ubiquitylation.
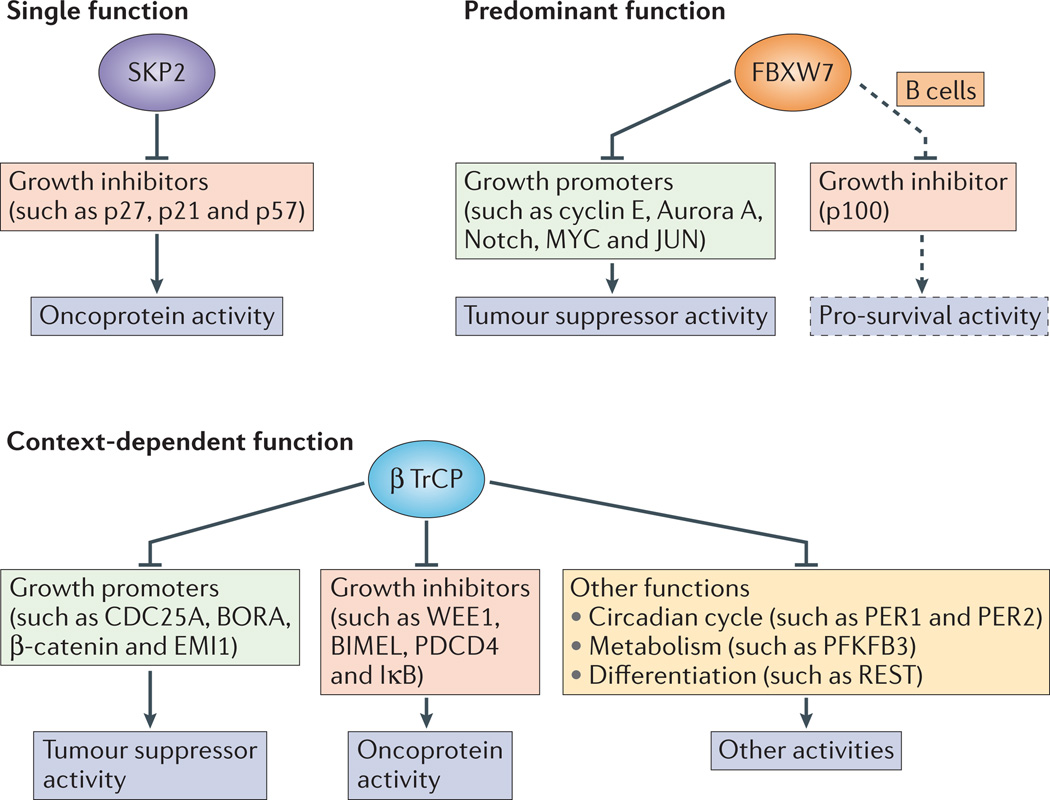
Figure 4. Generalized and context-dependent functions of F-box proteins.
Studies of S phase kinase-associated protein 2 (SKP2), F-box and WD40 domain 7 (FBXW7) and β-transducin repeat-containing protein (βTrCP) show that F-box proteins can have generalized and/or context-dependent functions. As shown in mouse models, SKP2 functions as an oncoprotein that ubiquitylates and hence degrades growth suppressive substrates. Moreover, in mouse models, FBXW7 functions as a tumour suppressor by ubiquitylating growth-promoting substrates, but this role is cell type specific. In B cell lineages, FBXW7 actually has a pro-survival role by mediating the degradation of p100, an inhibitor of nuclear factor-κB (NF-κB) signalling. βTrCP function is highly stimulus and cell type specific, and it has a role in many disparate pathways, including pathways beyond cell growth and proliferation. BORA, aurora borealis; BIMEL, BCL-2-interacting mediator of cell death extra long; EMI1, early mitotic inhibitor 1; IκB, inhibitor of κB; PDCD4, programmed cell death 4; PER, period; PFKFB3, 6-phosphofructo-2-kinase/ fructose-2,6-biphosphatase 3; REST, RE1-silencing transcription factor.
SKP2, FBXW7 and βTrCP in cancer
The misregulated degradation of tumour suppressors or oncoproteins can drive tumorigenesis. Accordingly, F-box proteins can function as oncoproteins when overexpressed (if their substrates are tumour suppressors) or as tumour suppressors (if their substrates are oncoproteins). SKP2 is the archetypal oncogenic F-box protein. It promotes S phase entry through the ubiquitylation and degradation of the CDK inhibitor p27, and mouse models unequivocally confirm the role of the SKP2–p27 axis in tumorigenesis. In the vast majority of tumours, p27 is inactivated by SKP2-mediated degradation, not genetic deletion, and SKP2 overexpression correlates with high tumour grade and poor prognosis in a broad range of cancers16. Although SKP2 functions predominantly in the regulation of p27, it also targets other anti-proliferative substrates, such as p21 and p57, contributing to its function as an oncoprotein7,16.
By contrast, loss-of-function mutations in FBXW7 have been identified in many cancers, which indicates that it might function as a tumour suppressor81. FBXW7 is frequently deleted in tumours, and it is estimated that 6% of cancers have mutations in the FBXW7 gene82,83. FBXW7 mutations are detected most frequently in T cell acute lymphoblastic leukaemia (T-ALL; 31%), but mutations are also found in solid tumours, including cancers of the breast, intestine and bone. Mouse models have confirmed the tumour suppressor function of FBXW7; conditional deletion of Fbxw7 leads to the development of haematological malignancies, such as thymic lymphoma and T-ALL84,85. FBXW7 substrates (including MYC, JUN, cyclin E and Notch) drive proliferation and tumorigenesis in conjunction with other mutations81,86. Fbxw7 deletion causes premature loss of haematopoietic stem cells by promoting cell cycle entry and apoptosis in a p53-dependent manner. p53 loss leads to unchecked cell proliferation and cancer, supporting the idea that FBXW7 and p53 function synergistically to prevent tumorigenesis86. The loss of FBXW7 function also facilitates resistance to certain chemotherapy drugs, such as anti-tubulin therapies. This phenotype might be the result of increased levels of myeloid cell leukaemia sequence 1 (MCL1), an FBXW7 substrate and pro-survival factor that suppresses apoptosis87,88.
Point mutations that dysregulate the SCF-dependent turnover of oncogenic substrates usually affect the F-box protein–substrate interface. Most tumorigenic FBXW7 point mutations are missense mutations in crucial residues of the substrate-binding region, and these mutations interfere with substrate recruitment82,85. Indeed, mutations of the two Arg residues in FBXW7 that directly contact the phosphodegron of the substrate account for 43% of cancer-associated FBXW7 mutations. Other mutations inactivate FBXW7 by interfering with its localization, mutating the F-box domain or resulting in premature termination of translation. The F-box protein–substrate interface can also be affected by mutations in substrates. Degron mutations in oncogenic substrates, including FBXW7 substrates, are common in tumours. For example, the phosphodegron of MYC (Thr58; a GSK3 target site) is frequently mutated in patients with Burkitt’s lymphoma, resulting in MYC stabilization89,90. In addition, mutations that disrupt Notch binding to FBXW7 are found in patients with T-ALL83,91.
As demonstrated by mouse models, the roles of SKP2 and FBXW7 in tumorigenesis provide an idealized framework for the evaluation of F-box proteins and their substrates in disease. However, a closer examination of F-box proteins in cancer shows that their function can be more nuanced. The stimulus- and temporally-regulated control of substrate binding enables an F-box protein to regulate multiple substrates, including substrates with opposing biological roles. Therefore, some F-box proteins can have a marked cell type- and/or contextdependent function (FIG. 4). They can be tumour suppressors or oncoproteins depending on which substrates are dysregulated and/or in which biological compartment they are dysregulated.
For example, although FBXW7 functions as a tumour suppressor in numerous cancers, it can also function in a pro-survival manner in multiple myelomas by mediating the degradation of p100 (also known as nuclear factor-κB2 (NF-κB2)), an inhibitor of NF-κB signalling92. Multiple myeloma cells are ‘addicted’ to NF-κB activity93,94, so degradation of p100 is required for their growth and proliferation. Intriguingly, this function seems to be B cell specific. Whereas FBXW7 mutations have been found in a wide variety of tumours, they have not been detected in tumours from the B cell lineage, including multiple myelomas, which might indicate a tissue-specific pro-survival role of FBXW7 (REF. 92). Notably, the proteasome inhibitor bortezomib is the frontline chemotherapy for multiple myelomas, and although the antitumour effects of bortezomib result from the stabilization of many substrates, p100 stabilization is likely to contribute to the effectiveness of this drug. In tissue culture, the pro-survival role of FBXW7 can also be countered by inhibitors of GSK3, the kinase that phosphorylates the p100 degron, suggesting a new avenue for therapy for B cell malignancies92.
Another F-box protein with context-dependent functions is βTrCP, which has an important role in integrating both positive and negative growth signals throughout the cell cycle. βTrCP mediates the degradation of substrates that promote cell proliferation upon mitogenic stimulation in G1, but it also regulates both positive and negative components of the feedback loops that control the timing and progression of mitosis. βTrCP1 or βTrCP2 is overexpressed in multiple cancers, including colorectal cancer, pancreatic cancer, hepatoblastoma, breast cancer, melanoma and gastrointestinal cancers, which supports an oncogenic function for these proteins. In these settings, βTrCP may target various growth and survival inhibitors, such as BIMEL (an extra-long isoform of BIM), PDCD4 (programmed cell death 4) and IκB (inhibitor of κB) family members16. By contrast, βTrCP substrates with oncogenic properties, such as β-catenin, CDC25A and EMI1, are also overexpressed in cancers95–97. Notably, the contribution of loss of βTrCP function to the overexpression of these proteins is unclear, so although βTrCP regulates oncogenic substrates, it might not function as a bona fide tumour suppressor16.
Context-dependent F-box protein function may also result from the context-dependent functions of substrates. For example, some βTrCP substrates, such as DEPTOR (DEP-domain containing mammalian target of rapamycin (mTOR)-interacting protein), seem to function as both tumour suppressors and oncoproteins. In general, DEPTOR functions as a tumour suppressor by blocking mTOR activity and inhibiting protein synthesis, cell proliferation and cell survival. Following mitogenic stimulation, DEPTOR is targeted for degradation by βTrCP, resulting in mTOR activation and cell proliferation31–33. Accordingly, DEPTOR levels are low in most cancers98. By contrast, DEPTOR levels are increased in many tumours with poor prognosis, which indicates that it might have oncogenic properties. Furthermore, DEPTOR is significantly overexpressed in a subset of multiple myelomas and is required for the survival of these cancer cells. Overexpressed DEPTOR maintains PI3K–AKT activation by disrupting the normal feedback regulation between mTOR complex 1 (mTORC1) and mTORC2. DEPTOR upregulation in multiple myelomas occurs largely as a result of increased transcription; the possibility of disrupted βTrCP-mediated DEPTOR degradation remains to be investigated.
FBXO11, a candidate tumour suppressor
SKP2, FBXW7 and βTrCP are valuable standards for the evaluation of the function of orphan F-box proteins as they are paired with substrates, and they highlight the perils of bestowing a generalized function on F-box proteins on the basis of a limited number of substrates. Recently, FBXO11 was shown to mediate degradation of the oncoprotein BCL6 (B cell lymphoma 6), suggesting that FBXO11 is a tumour suppressor13. BCL6 functions as a transcription factor in B cell development, differentiation and activation, and increased BCL6 expression drives the development of diffuse large B cell lymphoma (DLBCL)99. BCL6 overexpression in DLBCL is caused by multiple mechanisms, including translocation to constitutively active promoters, promoter hypermutation or protein stabilization. Mutations in FBXO11 (deletions or inactivating missense mutations) are seen in ~20% of patients with DLBCL and DLBCL cell lines, corresponding with increased levels and stability of BCL6. These mutations are predominantly located in the CASH repeat domain of FBXO11, the presumptive substratebinding domain, and this is in agreement with the theory that substrate-binding domains are mutational hotspots. Reconstitution of FBXO11 expression in FBXO11-null DLBCL cell lines suppresses tumorigenicity in xenograft models, suggesting that downregulation of BCL6 levels by FBXO11 has an antitumour effect.
However, many questions regarding the FBXO11–BCL6 interaction remain. Similar to the mutation of FBXW7 substrates in cancer, it is possible that mutations in BCL6 that compromise FBXO11 binding could also contribute to high BCL6 levels in DLBCL, so tumour-derived point mutants of BCL6 require further investigation. In addition, the signalling pathways upstream of BCL6 degradation are unknown, although they seem to be separate from B cell receptor signalling and MAPK pathways13. FBXO11 might also have a role in normal B cell development. Notably, although naive B cells express BCL6 mRNA, they do not express BCL6 protein, which indicates that BCL6 expression is post-translationally regulated. Finally, FBXO11 mutations are also observed in cancers of the colon, lung, ovary, head and neck, as well as non-DLBCL lymphomas, which implies that FBXO11 functions as a tumour suppressor in multiple tissues100–103. FBXO11 also targets CDT2 for degradation, but the relevant targets in these other tissues remain unknown54,55. Mouse models investigating FBXO11 and its interactions with BCL6 and other substrates will undoubtedly shed light on its function in normal cells and tumour cells.
Emerging F-box proteins in cancer
Several other F-box proteins regulate the degradation of substrates that are involved in processes fundamentally related to tumorigenesis and tumour progression. For example, defects in centrosome copy number lead to aneuploidy and the loss of genome integrity, as seen in many types of cancer. Moreover, SCF complexes have an emerging role in controlling centrosome numbers, with several F-box proteins regulating the levels of centrosome proteins, including Polo-like kinase 4 (PLK4; which is regulated by βTrCP)104,105, SAS6 (spindle assembly abnormal 6; which is regulated by FBXW5)106 and CP110 (which is regulated by cyclin F)11. In addition to regulating the centrosome, cyclin F controls genome integrity through the degradation of RRM2 (REF. 10), and cyclin F downregulation has been reported in hepatocellular carcinoma107. Finally, FBXO9 mediates the degradation of mTORC1-associated telomere length regulation protein 2 (TEL2) and TELO2-interacting protein 1 (TTI1), removing feedback inhibition of mTORC2 and resulting in an increase in mTOR activity in a manner similar to DEPTOR overexpression108–110. Further characterization of these F-box proteins is required before their true roles in cancer are understood.
F-box proteins beyond cancer
Although F-box proteins have clear roles in cancer, their functions in other pathologies are gradually emerging as research into F-box proteins evolves beyond investigations of cell proliferation. This shift is exemplified by studies of the APC/C, a prominent cell cycle-associated ubiquitin ligase (FIG. 1) that also has roles in post-mitotic cells, such as neurons111. In particular, the regulation of circadian rhythms by βTrCP and FBXL3 has been linked to sleep and mood disorders. βTrCP and FBXL3 control the degradation of PER1, PER2, CRY1 and CRY2, which negatively regulate the activity of the heterodimeric BMAL– CLOCK transcription factor at the core of the circadian system59–61,112,113. A mutation in PER2 has been found in individuals with familial advanced sleep phase syndrome, and this mutation causes increased nuclear export and decreased stability of PER2 (REFS 80,114,115). In addition, inhibitors of casein kinase 1 (CK1), the kinase that phosphorylates the βTrCP degron in PER proteins, have been used to alter circadian cycles in rodents116,117. The involvement of βTrCP in circadian regulation also highlights the context-dependent nature of this multifunctional F-box protein. FBXL3 has not yet been directly linked to a human disorder on the basis of mutation data. However, in addition to exhibiting circadian phenotypes, mice carrying a mutated form of FBXL3 show reduced anxiety- and depression-associated behaviours118, and the FBXL3 substrates CRY1 and CRY2 have been linked to diabetes75,76. The FBXL3–CRY interface, particularly the interaction between the FAD-binding pocket of CRY proteins and the C terminus of FBXL3, is a promising site for pharmacological manipulation62 (BOX 2). As CRY proteins are the only known substrates of FBXL3, it is currently unknown whether FBXL3 has a generalized function in circadian regulation.
Notably, FBXL21, an FBXL3 homologue, also binds CRY proteins, and the interplay of FBXL3 and FBXL21 will be an area of future investigation. It has been reported that FBXL21 is expressed predominantly in the suprachiasmatic nucleus, which is the region in the brain that functions as the master pacemaker in mammals119, but more recent studies have proposed a more universal mechanism, in which cytoplasmic FBXL21 inhibits the degradation of CRY proteins in a manner mediated by nuclear FBXL3 (REFS 120,121). Fbxl21-mutant mice have circadian defects, and FBXL21 has been linked to schizophrenia122. Further research is required to fully integrate FBXL3 and FBXL21 into our understanding of circadian rhythm regulation and circadian-associated pathologies.
F-box proteins are also manipulated by multiple viruses, including HIV and HCV. The HIV viral protein U (Vpu) binds βTrCP and retargets the ubiquitylation activity of SCFβTrCP to CD4 and bone marrow stromal antigen 2 (BST2; also known as tetherin), facilitating the release and dispersion of the virus from the cell72. The hijacking of βTrCP by Vpu might also inhibit the normal functions of βTrCP, such as regulation of the NF-κB pathway78,79. Similarly, the HCV NS5A protein binds FBXL2, and FBXL2 function, including proper localization (see above), is required for HCV replication35. However, the effect of HCV on FBXL2 substrate selection is unclear, as are the substrates required for viral replication. Many other viruses also modulate the activity of host F-box proteins123–125, and several viruses and bacteria encode their own F-box proteins73,74, which indicates that the role of SCF complexes in microbial infections will expand in the future.
Finally, the association of F-box proteins with human disease highlights the importance of identifying more F-box protein substrates. For example, FBXO7 mutations have been identified in a subtype of Parkinson’s disease, but there are few known FBXO7 substrates,with unknown relevance to Parkinson’s disease7, 77. As the F-box field moves beyond cancer biology, the links between F-box proteins and human disease are likely to expand.
Concluding remarks
Fourteen years after the initial annotation of the F-box protein family, our understanding of the biochemical mechanisms of SCF ubiquitin ligases and their biological roles continues to evolve8,9. The old paradigm of phosphorylation-directed substrate recognition by F-box proteins may still be dominant, but it is no longer absolute. The biochemical and biological functions of F-box proteins expand with each new substrate that is described, be it for an F-box protein with established substrates or an orphan F-box protein. The historical linkage between the SCF complex and the cell cycle has driven the investigation of this protein family in cancer, but it is likely that F-box proteins will have important roles in many other pathologies and pathways35,75–77,118. The identification of substrates for F-box proteins is of primary importance for determining the biological roles of each F-box family member, and as our knowledge of SCF biology expands, each SCF complex presents several avenues for the development of new therapies.
Acknowledgements
The authors apologize to their colleagues in the field for omitting their work owing to space constraints. J.R.S. is a Special Fellow of The Leukemia and Lymphoma Society. J.K.P. is supported by a fellowship from the Lymphoma Research Foundation. Work in the Pagano laboratory is supported by grants from the US National Institutes of Health (NIH) (R37CA076584 and R01GM057587). M.P. is an Investigator of the Howard Hughes Medical Institute.
Glossary
- Ubiquitin
A 76-amino-acid protein that can be covalently conjugated to Lys residues in other proteins to specify several protein fates. Polyubiquitin chains can be generated using seven internal Lys residues in ubiquitin. Lys11- or Lys48-linked chains target proteins to the proteasome, whereas other chains, such as Lys63-linked chains, have signalling roles. Monoubiquitylation also has a signalling role.
- RING
Really interesting new gene (RING) proteins coordinate zinc using Cys and His residues in a cross-brace arrangement. RING proteins typically recruit E2 ubiquitin-conjugating enzymes.
- NEDD8
A small ubiquitin-like protein that can be covalently conjugated to other proteins. Cullin proteins are the primary targets for neddylation, which activates the cullin–RING ligase.
- Degrons
Small sections of a substrate that are recognized by a ubiquitin ligase and that are required for substrate degradation. Canonical degrons for F-box proteins are very short, conserved stretches of amino acids.
- β-transducin repeat-containing protein (βTrCP)
Refers to two paralogous F-box proteins, βTrCP1 (FBXW1 (F-box and WD40 domain 1)) and βTrCP2 (FBXW11), that are biochemically indistinguishable. βTrCP recognizes phosphodegrons through WD40 repeats.
- Cyclin-dependent kinase (CDK)
These are drivers of the cell cycle. The activity of these kinases is controlled by the availability of their cognate cyclins. The oscillation of cyclin levels during the cell cycle determines which CDKs are active.
- αβ-crystallin
A chaperone protein that can be induced by heat, but unlike heat shock proteins, it does not re-fold proteins. Instead, αβ-crystallin forms protein aggregates.
- Cyclin-homology domain
This domain normally determines the binding of cyclins to cyclin-dependent kinase substrates. The original cyclins were identified on the basis of their cyclic oscillations, but additional proteins contain a cyclin-homology domain, which has led to their designation as cyclins.
- Ribonucleotide reductase subunit M2 (RRM2)
A subunit of the enzyme that converts ribonucleotides to deoxyribonucleotides for DNA replication. RRM2 is regulated by the cell cycle, whereas the RRM1 subunit is stable throughout the cell cycle.
- Isoprenylation
The transfer of a farnesyl or geranyl–geranyl moiety to a carboxy-terminal Cys residue of a target protein. This modification facilitates protein recruitment to membranes.
- RAB3-interacting molecule 1 (RIM1)
It is expressed near the active zone of the neuronal synapse and interacts with many presynaptic proteins, including RAB3. It controls calcium-evoked neurotransmitter release.
- Auxin
A family of plant hormones required for growth signalling. Indole-3-acetic acid is the most potent auxin. They bind to TRANSPORT INHIBITOR RESPONSE 1, a plant F-box protein, promoting its interaction with auxin– indoleacetic acid transcriptional repressors.
- Jasmonate
A lipid-based plant hormone that binds to CORONATINEINSENSITIVE 1, an F-box protein, and JA–ZIM domain (JAZ), a transcriptional regulator, promoting the degradation of JAZ.
- Checkpoint kinase 1 (CHK1)
A Ser/Thr kinase that is required for checkpoint arrest and DNA repair following DNA damage.
- CDT2
A DDB1- and CUL4-associated factor (DCAF) protein that mediates the degradation of SET domain-containing protein 8, p21 and CDT1, preventing re-replication in S phase.
- Cryptochrome 1 and cryptochrome 2 (CRY1 and CRY2)
Mammalian CRY proteins function as transcriptional repressors within a negative feedback loop of the circadian cycle. CLOCK and BMAL1 activate the transcription of genes encoding period (PER) and CRY proteins, which then negatively regulate transcription by CLOCK–BMAL1. In lower organisms, CRY proteins function as photoreceptors, but this activity is not present in mammals.
- p100
An inhibitor of nuclear factor-κB (NF-κB) signalling and the precursor for p52, which forms an active transcription factor in conjunction with REL family proteins. p100 is part of the non-canonical NF-κB signalling pathway, which largely responds to developmental signals.
- Bortezomib
A proteasome inhibitor approved as frontline therapy for multiple myelomas. Originally, much of its effectiveness was ascribed to inhibition of canonical nuclear factor-κB signalling through stabilization of inhibitor of κB proteins.
- DEPTOR
(DEP-domain containing mTOR-interacting protein). An inhibitor of the mammalian target of rapamycin (mTOR) kinase in the context of both mTOR complex 1 (mTORC1) and mTORC2.
- Mammalian target of rapamycin
(mTOR). A PI3K that is an important regulator of cell growth and metabolism, particularly protein synthesis. mTOR forms two complexes, designated mTOR complex 1 (mTORC1) and mTORC2, which have multiple feedback links.
- Telomere length regulation protein 2 and TELO2-interacting protein 1 (TEL2 and TTI1)
Evolutionarily conserved proteins that interact with all six mammalian PI3K-like protein kinases and control their abundance.
- Familial advanced sleep phase syndrome
An inherited circadian and sleep disorder in which patients have altered circadian rhythms, with early sleep onset and early waking.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Michele Pagano’s homepage: http://pathology.med.nyu.edu/Pagano/The_Michele_Pagano_Laboratory.html
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2. Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328.. An in-depth examination of the current state of our knowledge of ubiquitin as a signal for both proteasomal degradation and other processes.
- 3.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yen HC, et al. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 5.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nature Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 6. Jin J, et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304.. Presents a unified nomenclature for the F-box proteins, details their domain structures and describes their evolutionary relationship. However, certain domains that were initially not detected in FBXO proteins have since been described, such as the haemerythrin domain in FBXL5.
- 7.Skaar JR, et al. SnapShot: F box proteins II. Cell. 2009;137:1358.e1. doi: 10.1016/j.cell.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 8.Cenciarelli C, et al. Identification of a family of human F-box proteins. Curr. Biol. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- 9.Winston JT, et al. A family of mammalian F-box proteins. Curr. Biol. 1999;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- 10.D’Angiolella V, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Angiolella V, et al. SCFCyclin Fcontrols centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glenn KA, et al. Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases. J. Biol. Chem. 2008;283:12717–12729. doi: 10.1074/jbc.M709508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan S, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nature Rev. Mol. Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau AW, et al. The Fbw7 and βTRCP E3 ubiquitin ligases and their roles in tumorigenesis. Front. Biosci. 2012;17:2197–2212. doi: 10.2741/4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nature Rev. Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welcker M, et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell. 2003;12:381–392. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 18.Wei W, et al. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Liu C, et al. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 20.Nash P, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 21.Tang X, et al. Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase. Proc. Natl Acad. Sci. USA. 2012;109:3287–3292. doi: 10.1073/pnas.1116455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koivomagi M, et al. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature. 2011;480:128–131. doi: 10.1038/nature10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutterluty H, et al. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nature Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- 24. Carrano AC, et al. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nature Cell Biol. 1999;1:193–199. doi: 10.1038/12013.. References 23 and 24 describe the SKP2-mediated degradation of p27 and provide insight into the function and clinical relevance of SCF complexes. Subsequent studies have established that SKP2 overexpression and low p27 expression are poor prognostic indicators in patients with various tumours.
- 25.Ganoth D, et al. The cell-cycle regulatory protein Cks1 is required for SCFSkp2-mediated ubiquitinylation of p27. Nature Cell Biol. 2001;3:321–324. doi: 10.1038/35060126. [DOI] [PubMed] [Google Scholar]
- 26.Spruck C, et al. A CDK-independent function of mammalian Cks1: targeting of SCFSkp2 to the CDK inhibitor p27Kip1. Mol. Cell. 2001;7:639–650. doi: 10.1016/s1097-2765(01)00210-6. [DOI] [PubMed] [Google Scholar]
- 27.Hao B, et al. Structural basis of the Cks1-dependent recognition of p27Kip1 by the SCFSkp2 ubiquitin ligase. Mol. Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Lin DI, et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCFFBX4-αB crystallin complex. Mol. Cell. 2006;24:355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.den Engelsman J, et al. The small heat-shock protein αB-crystallin promotes FBX4-dependent ubiquitination. J. Biol. Chem. 2003;278:4699–4704. doi: 10.1074/jbc.M211403200. [DOI] [PubMed] [Google Scholar]
- 30.Ouni I, et al. A transcriptional activator is part of an SCF ubiquitin ligase to control degradation of its cofactors. Mol. Cell. 2010;40:954–964. doi: 10.1016/j.molcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan S, et al. mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol. Cell. 2011;44:317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao D, et al. mTOR drives its own activation via SCFβTrCP-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell. 2011;44:290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, et al. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCFβTrCP E3 ubiquitin ligase and regulates survival and autophagy. Mol. Cell. 2011;44:304–316. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao I, et al. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell. 2007;130:943–957. doi: 10.1016/j.cell.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, et al. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell. 2005;18:425–434. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Bai C, et al. Human cyclin F. EMBO J. 1994;13:6087–6098. doi: 10.1002/j.1460-2075.1994.tb06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bashir T, et al. Control of the SCFSkp2-Cks1 ubiquitin ligase by the APC/CCdh1 ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- 38.Wei W, et al. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- 39.Moshe Y, et al. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc. Natl Acad. Sci. USA. 2004;101:7937–7942. doi: 10.1073/pnas.0402442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margottin-Goguet F, et al. Prophase destruction of Emi1 by the SCFβTrCP/Slimb ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell. 2003;4:813–826. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 41.Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- 42.Vashisht AA, et al. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science. 2009;326:718–721. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salahudeen AA, et al. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science. 2009;326:722–726. doi: 10.1126/science.1176326.. References 42 and 43 establish a new means to regulate F-box protein stability through iron binding. In addition, reference 43 suggests that iron binding might also affect substrate binding, although this remains to be confirmed.
- 44.Sanchez M, et al. Iron regulatory protein-1 and -2: transcriptome-wide definition of binding mRNAs and shaping of the cellular proteome by iron regulatory proteins. Blood. 2011;118:e168–e179. doi: 10.1182/blood-2011-04-343541. [DOI] [PubMed] [Google Scholar]
- 45.Chollangi S, et al. Hemerythrin-like domain within F-box and leucine-rich repeat protein 5 (FBXL5) communicates cellular iron and oxygen availability by distinct mechanisms. J. Biol. Chem. 2012;287:23710–23717. doi: 10.1074/jbc.M112.360404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shu C, et al. The structural basis of iron sensing by the human F-box protein FBXL5. ChemBiochem. 2012;13:788–791. doi: 10.1002/cbic.201200043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santra MK, et al. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bassermann F, et al. NIPA defines an SCF-type mammalian E3 ligase that regulates mitotic entry. Cell. 2005;122:45–57. doi: 10.1016/j.cell.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 49.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731.. References 49 and 50 provide the first demonstration that endogenous small molecules (in this case hormones) can dictate substrate binding to F-box proteins. These studies have important implications not only for other hormone-based recognition systems, but also for the development of drugs targeting the F-box protein-substrate interface.
- 51.Yoshida Y, et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–442. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida Y, et al. Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J. Biol. Chem. 2003;278:43877–43884. doi: 10.1074/jbc.M304157200. [DOI] [PubMed] [Google Scholar]
- 53.Zhang YW, et al. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol. Cell. 2009;35:442–453. doi: 10.1016/j.molcel.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossi M, et al. Regulation of the CRL4 ubiquitin ligase and cell-cycle exit by the SCF ubiquitin ligase. Mol. Cell. 2013;49:1159–1166. doi: 10.1016/j.molcel.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abbas T, et al. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol. Cell. 2013;49:1147–1158. doi: 10.1016/j.molcel.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuchay S, et al. FBXL2- and PTPL1-mediated degrdation of p110-free p85β regulatory subunit controls the PI3K signalling cascade. Nature Cell Biol. 2013 Apr 21; doi: 10.1038/ncb2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng Z, et al. Structural basis of selective ubiquitination of TRF1 by SCFFbx4 . Dev. Cell. 2010;18:214–225. doi: 10.1016/j.devcel.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Hao B. Structural basis of dimerization-dependent ubiquitination by the SCFFbx4 ubiquitin ligase. J. Biol. Chem. 2010;285:13896–13906. doi: 10.1074/jbc.M110.111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 60.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 61.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of Cryptochrome and Period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xing W, et al. SCF ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013;496:64–68. doi: 10.1038/nature11964.. Describes the crystal structure of FBXL3 bound to CRY2 and demonstrates both domain-dependent recognition and the potential for ligand-dependent regulation of recognition.
- 63.Lee JM, et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 64.Kaelin WG. Proline hydroxylation and gene expression. Annu. Rev. Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 65.Nishimura K, et al. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 66.Yen JL, et al. Signal-induced disassembly of the SCF ubiquitin ligase complex by Cdc48/p97. Mol. Cell. 2012;48:288–297. doi: 10.1016/j.molcel.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang X, et al. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki H, et al. Homodimer of two F-box proteins βTrCP1 or βTrCP2 binds to IκBα for signal-dependent ubiquitination. J. Biol. Chem. 2000;275:2877–2884. doi: 10.1074/jbc.275.4.2877. [DOI] [PubMed] [Google Scholar]
- 69.Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hao B, et al. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 71. Zimmerman ES, et al. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010.. Provides an excellent overview of the structures of SCF complexes and other CRLs.
- 72.Sandberg JK, et al. HIV-1 Vpu interference with innate cell-mediated immune mechanisms. Curr. HIV Res. 2012;10:327–333. doi: 10.2174/157016212800792513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L, et al. Interplay between poxviruses and the cellular ubiquitin/ubiquitin-like pathways. FEBS Lett. 2009;583:607–614. doi: 10.1016/j.febslet.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 74.Price CT, et al. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 2009;5:e1000704. doi: 10.1371/journal.ppat.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang EE, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nature Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Di Fonzo A, et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–245. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 78.Akari H, et al. The human immunodeficiency virus type 1 accessory protein Vpu induces apoptosis by suppressing the nuclear factor κB-dependent expression of antiapoptotic factors. J. Exp. Med. 2001;194:1299–1311. doi: 10.1084/jem.194.9.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bour S, et al. The human immunodeficiency virus type 1 Vpu protein inhibits NF-κB activation by interfering with βTrCP-mediated degradation of IκB. J. Biol. Chem. 2001;276:15920–15928. doi: 10.1074/jbc.M010533200. [DOI] [PubMed] [Google Scholar]
- 80.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 81.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nature Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 82.Akhoondi S, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 83.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Onoyama I, et al. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J. Exp. Med. 2007;204:2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thompson BJ, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crusio KM, et al. The ubiquitous nature of cancer: the role of the SCFFbw7 complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Inuzuka H, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wertz IE, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 89.Bahram F, et al. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. [PubMed] [Google Scholar]
- 90.Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt’s lymphoma cells. Mol. Cell. Biol. 2000;20:2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 92. Busino L, et al. Fbxw7α- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nature Cell Biol. 2012;14:375–385. doi: 10.1038/ncb2463.. Shows that FBXW7, an established tumour suppressor in many tissues, is a pro-survival factor in tumours of the B cell lineage and highlights that context-dependent functions of F-box proteins could be exploited through the inhibition of degron kinases, such as GSK3.
- 93.Annunziata CM, et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Keats JJ, et al. Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loffler H, et al. Distinct modes of deregulation of the proto-oncogenic Cdc25A phosphatase in human breast cancer cell lines. Oncogene. 2003;22:8063–8071. doi: 10.1038/sj.onc.1206976. [DOI] [PubMed] [Google Scholar]
- 96.Lehman NL, et al. Oncogenic regulators and substrates of the anaphase promoting complex/cyclosome are frequently overexpressed in malignant tumors. Am. J. Pathol. 2007;170:1793–1805. doi: 10.2353/ajpath.2007.060767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.MacDonald BT, et al. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peterson TR, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol. Rev. 2012;247:172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 100.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 101.Richter J, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nature Genet. 2012;44:1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- 102.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 104.Cunha-Ferreira I, et al. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 105.Guderian G, et al. Plk4 trans-autophosphorylation regulates centriole number by controlling βTrCP-mediated degradation. J. Cell. Sci. 2010;123:2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- 106.Puklowski A, et al. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nature Cell Biol. 2011;13:1004–1009. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- 107.Fu J, et al. Low cyclin F expression in hepatocellular carcinoma associates with poor differentiation and unfavorable prognosis. Cancer Sci. 2013;104:508–515. doi: 10.1111/cas.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kaizuka T, et al. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J. Biol. Chem. 2010;285:20109–20116. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takai H, et al. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 110.Fernandez-Saiz V, et al. SCFFbxo9 and CK2 direct the cellular response to growth factor withdrawal via Tel2/Tti1 degradation and promote survival in multiple myeloma. Nature Cell Biol. 15:72–81. doi: 10.1038/ncb2651. [DOI] [PubMed] [Google Scholar]
- 111.Manchado E, et al. The anaphase-promoting complex/cyclosome (APC/C): cell-cycle-dependent and -independent functions. Biochem. Soc. Trans. 2010;38:65–71. doi: 10.1042/BST0380065. [DOI] [PubMed] [Google Scholar]
- 112.Shirogane T, et al. SCFβ-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 113.Takahashi JS, et al. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nature Rev. Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vanselow K, et al. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shanware NP, et al. Casein kinase 1-dependent phosphorylation of familial advanced sleep phase syndrome-associated residues controls PERIOD 2 stability. J. Biol. Chem. 2012;286:12766–12774. doi: 10.1074/jbc.M111.224014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Walton KM, et al. Selective inhibition of casein kinase 1ε minimally alters circadian clock period. J. Pharmacol. Exp. Ther. 2009;330:430–439. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- 117.Meng QJ, et al. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc. Natl Acad. Sci. USA. 2010;107:15240–15245. doi: 10.1073/pnas.1005101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Keers R, et al. Reduced anxiety and depression-like behaviours in the circadian period mutant mouse afterhours. PLoS ONE. 2012;7:e38263. doi: 10.1371/journal.pone.0038263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dardente H, et al. Implication of the F-Box protein FBXL21 in circadian pacemaker function in mammals. PLoS ONE. 2008;3:e3530. doi: 10.1371/journal.pone.0003530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hirano A, et al. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell. 2013;152:1106–1118. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 121.Yoo SH, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen X, et al. FBXL21 association with schizophrenia in Irish family and case-control samples. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;147B:1231–1237. doi: 10.1002/ajmg.b.30759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baresova P, et al. Kaposi sarcoma-associated herpesvirus vIRF-3 protein binds to F-box of Skp2 protein and acts as a regulator of c-Myc protein function and stability. J. Biol. Chem. 2012;287:16199–16208. doi: 10.1074/jbc.M111.335216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Reviriego-Mendoza MM, Frisque RJ. Interaction and co-localization of JC virus large T antigen and the F-box protein β-transducin-repeat containing protein. Virology. 2011;410:119–128. doi: 10.1016/j.virol.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Isobe T, et al. Adenovirus E1A inhibits SCFFbw7 ubiquitin ligase. J. Biol. Chem. 2009;284:27766–27779. doi: 10.1074/jbc.M109.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu S, et al. CAND1 controls in vivo dynamics of the cullin 1–RING ubiquitin ligase repertoire. Nature Commun. 2013;4:1642. doi: 10.1038/ncomms2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pierce NW, et al. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell. 2013;153:206–215. doi: 10.1016/j.cell.2013.02.024.. An intricate examination of the biochemistry of CAND1 (cullin-associated and neddylation-dissociated 1) and SCF complexes. Demonstrates that CAND1 is an important assembly factor for CRLs.
- 128.Zemla A, et al. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nature Commun. 2013;4:1641. doi: 10.1038/ncomms2628. [DOI] [PubMed] [Google Scholar]
- 129.Duda DM, et al. Structure of a glomulin–RBX1–CUL1 complex: inhibition of a RING E3 ligase through masking of its E2-binding surface. Mol. Cell. 2012;47:371–382. doi: 10.1016/j.molcel.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Tron AE, et al. The glomuvenous malformation protein Glomulin binds Rbx1 and regulates cullin RING ligase-mediated turnover of Fbw7. Mol. Cell. 2012;46:67–78. doi: 10.1016/j.molcel.2012.02.005.. Together, references 129 and 130 introduce a novel player to the regulation of CRLs. Although the phenotypic effects of glomulin are manifested through FBXW7 and its substrates, the biochemical mechanism of glomulin indicates the ability to broadly regulate CRL complexes.
- 131.Chaves I, et al. The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 132. Hirota T, et al. Identification of small molecule activators of cryptochrome. Science. 2012;337:1094–1097. doi: 10.1126/science.1223710.. Although it was discovered before the crystal structure of the FBXL3-CRY complex, the small molecule described in reference 132 probably binds the FAD-binding pocket of CRY, illustrating the feasibility of targeting drugs to the F-box protein-substrate interface.
- 133. Wu L, et al. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem. Biol. 2012;19:1515–1524. doi: 10.1016/j.chembiol.2012.09.015.. This study highlights the use of the F-box protein–substrate interface as a target for new therapies and presents lead compounds for the future development of SKP2 inhibitors for cancer treatment. These lead compounds were identified in silico, on the basis of the SKP2-CKS1-p27 peptide structure.
- 134.Orlicky S, et al. An allosteric inhibitor of substrate recognition by the SCFCdc4 ubiquitin ligase. Nature Biotech. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aghajan M, et al. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nature Biotech. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sakamoto KM, et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. USA. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sakamoto KM, et al. Development of Protacs to target cancer-promoting proteins for ubiquitination and degradation. Mol. Cell Proteom. 2003;2:1350–1358. doi: 10.1074/mcp.T300009-MCP200. [DOI] [PubMed] [Google Scholar]