Abstract
Derangements in normal cellular homeostasis at the protein level can cause or be the consequence of initiation and progression of pulmonary diseases related to genotype, infection, injury, smoking, toxin exposure, or neoplasm. We discuss one of the fundamental mechanisms of protein homeostasis, the ubiquitin proteasome system (UPS), as it relates to lung disease. The UPS effects selective degradation of ubiquitinated target proteins via ubiquitin ligase activity. Important pathobiological mechanisms relating to the UPS and lung disease have been the focus of research, with inappropriate cellular proteolysis now a validated therapeutic target. We review the contributions of this system in various lung diseases, and discuss the exciting area of UPS-targeting drug development for pulmonary disease.
Keywords: ubiquitin, proteasome, pulmonary disease, drug development, E3 ligase
Introduction
Normal pulmonary physiology can be disrupted by direct contact with the environment, exposure to potentially noxious inhalants, and infection with ensuing inflammatory cell activation. This delicate balance from normal homeostasis is tipped toward inflammatory injury in patients with chronic obstructive pulmonary disease (COPD), cystic fibrosis, acute respiratory distress syndrome (ARDS), acute lung injury (ALI), and pneumonia. Insights into the molecular pathophysiology of these diseases have greatly increased. In this review, we highlight emerging discoveries regarding selective regulation of protein degradation in the lung by the ubiquitin (Ub)–proteasome system (UPS), and how this regulation at the protein level affects critical functions of lung cells with ramifications that can maintain or threaten the vitality of the organism.
Selective Protein Degradation and Cellular Function: E3 Ubiquitin Ligase as a Physiological Switch
Maintenance of any healthy tissue requires stringent quality control at the protein level. All cellular proteins undergo tightly regulated turnover in the cell to prevent improper activity or unnecessary accumulation of dysfunctional proteins. Protein degradation also changes critical cellular protein concentrations in response to chemical signals or for important cellular events such as mitosis. Major protein regulatory systems include the autophagic/lysosomal pathways and the more prevalent UPS (1). Ubiquitin covalently interacts with other proteins whereby a single ubiquitin attaches to one lysine (monoubiquitination) or to multiple lysine residues on the target (multimonoubiquitination), or a ubiquitin chain can be produced at a single lysine residue (polyubiquitination). Ubiquitin contains seven lysine residues, which results in eight different interubiquitin linkage types, in turn leading to binding sites for other ubiquitin molecules and distinct branched ubiquitin chains that determine function. Substrate ubiquitination occurs in an ATP-dependent fashion, through an elaborate enzymatic cascade that adds the ubiquitin protein to, first, a ubiquitin-activating enzyme (E1), and then to a ubiquitin-conjugating enzyme (E2), and last to a specific target protein, a critical event that is catalyzed by a ubiquitin E3 ligase. One or more Ub molecules are thus added to the substrate, with monoubiquitination usually tagging the substrate for endocytic sorting, whereas polyubiquitination tags the substrate for recognition by the proteasome, often resulting in target protein degradation by the 26S proteasome protein complex, which is composed of one 20S and two 19S proteolytic subunits (2) (Figure 1). Overall, this process consumes large amounts of cellular energy, is represented by the largest family of enzymes present in eukaryotes, and accounts for about 5% of the genome. Given the physiological importance of the UPS system to cellular and tissue physiology, failure of the UPS is only rarely seen, but some mutations in proteins identified as UPS E3 ligases result in human familial diseases including Angelman syndrome, Parkinson’s disease, and von Hippel–Lindau (vHL) syndrome (3), and changes in UPS function have been implicated in disorders of the cardiovascular (4), neurological (5), and pulmonary (6) systems.
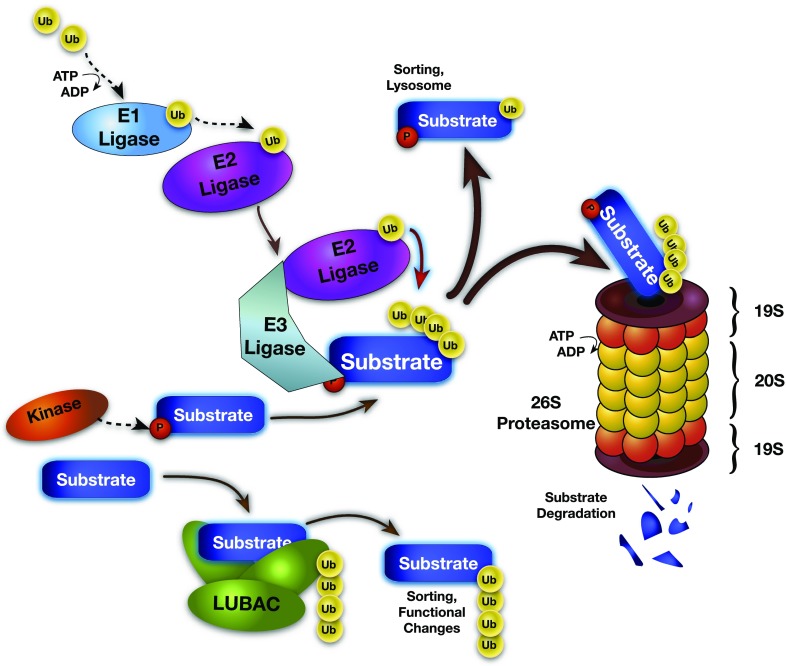
Figure 1.
Schematic of the ubiquitin (Ub)–proteasome system. Protein degradation is a regulated, multistep process. Ubiquitin is loaded onto E1 ligases in an ATP-dependent fashion, and then transferred to E2 ligases. The same E2 ligase can bind many E3 ligases, which in turn can ubiquitinate several target substrate proteins. E3 ligases bind specific substrate proteins based on substrate degron motifs usually consisting of a post-translational modification, such as phosphorylation. Adduction via the K48 residue on Ub tags the substrate for sorting, lysosomal destruction, or proteolytic cleavage and degradation by the 26S ubiquitin proteasome. Linear ubiquitination via the M1 residue by the linear ubiquitination complex (LUBAC) (HOIL-1L, HOIP, and SHARPIN) changes the cellular localization and activity of substrates, including RIP and NEMO for NF-κB signaling. HOIL-1L = heme-oxidized IRP2 (iron-responsive element–binding protein-2) ubiquitin ligase-1; HOIP = HOIL-1L–interacting protein; NEMO = nuclear factor-κB essential modulator; NF-κB = nuclear factor-κB; P = phosphate; RIP = receptor-interacting protein; SHARPIN = SHANK (SH3 and multiple ankyrin repeat domains-2)–associated RH [RBCK1 homology] domain–interacting protein; Ub = ubiqutin.
The system is hierarchical, with one or two E1 enzymes described in mammalian cells, about 40 E2 enzymes, and more than 1,000 E3 ligases described. The targeting, ubiquitination, and degradation of proteins occur in a highly regulated and specific manner, with the E3 ligase usually binding target proteins with some post-translational modification that harbors a specific structural motif, termed a “degron” (7). Two major families of E3 ligases orchestrate ubiquitin addition to target proteins and ultimately determine substantial shifts in cellular behavior. The RING (really interesting new gene) finger and RING-related proteins comprise the largest E3 family, whose members function either independently as monomers or dimers, or in a multisubunit complex to ubiquitinate a broad range of substrates. The HECT (homologous to the E6-AP carboxy terminus) domain–containing proteins are a smaller E3 ligase family whose members are important for regulating many proteins, some of which include the transmembrane surface proteins. There is also a family of “U-box” E3 ligases, which do not have enzymatic activity and mostly bridge catalytically active E2 enzymes, facilitating the transfer of ubiquitin to the target protein. The linear ubiquitination complex (LUBAC) is another multisubunit E3 ligase that polyubiquitinates substrates for sorting and/or degradation by adduction of chains of Ub monomers joined end-to-end (8). LUBAC contains three important components: HOIL-1L (heme-oxidized IRP2 [iron-responsive element–binding protein-2] ubiquitin ligase-1), HOIP (HOIL-1–interacting protein), and SHARPIN (SHANK [SH3 and multiple ankyrin repeat domains-2]–associated RH [RBCK1 homology] domain-3–interacting protein). The E3 ligases are diverse, represented by hundreds of genes in humans, and are highly represented in all eukaryotes (9).
SCF Ligase and the Role of F-Box Proteins
Among RING E3 multisubunit ligases, the Cullin–RING family and anaphase-promoting complex/cyclosome (APC/C) are the best characterized. The largest family of Cullin–RING E3 ligases is the Skp1–Cullin–F-box protein (SCF) family, a canonical multimodule E3 ubiquitination complex often causing target proteasomal degradation. The F-box proteins (FBPs) are interchangeable and confer substrate specificity through recognition of post-translationally modified degron motifs within substrates via target-binding domains, and tether substrate proteins to the Cullin protein of the SCF complex via F-box domains (10). Each FBP causes degradation of a specific set of target proteins by unique degron recognition. Sixty-eight FBPs have been identified in humans, designated FBXL (containing a leucine-rich domain), FBXW (containing a WD-40 domain), or FBXO (containing neither leucine-rich nor WD-40 domains) (11). Overall, only a small fraction of FBP biology is known, and multiple laboratories are describing the diverse and critical activities of FBPs in physiology and disease.
Importance of the Proteasome and Ubiquitin E3 Ligases in Lung Homeostasis and Disease
Oxygen Sensing and Pulmonary Vascular Disease
One example of E3 ligase molecular regulation related to respiration is the oxygen-sensing role of hypoxia-inducible factor (HIF)-1α, a stress response transcriptional activator of chemokines, growth factors, and proteases. In normoxia, HIF-1α is hydroxylated through the action of prolyl hydroxylase domain protein-2 (PHD2), using oxygen as substrate, and the hydroxylated HIF-1α is bound by the vHL protein, which recruits a ubiquitin ligase for HIF-1α polyubiquitination and proteasomal degradation (12). During hypoxia, however, PHD2 activity (and therefore HIF-1α hydroxylation) is decreased, and vHL no longer prevents HIF-1α activation of transcription. Such molecular oxygen sensing is important for normal embryonic development and growth but is a major pathway “hijacked” by cancer cells with limited local oxygen supply to stabilize HIF-1α and augment local tumor growth and metastatic potential (13). In translational biomedical research, the vHL protein and HIF-1α signaling axis was described initially in vHL disease, caused by ineffective vHL; affected patients have polycythemia, pulmonary arterial hypertension, and respiratory insufficiency, attributed to increased HIF-1α signaling.
Host Defense
Immunoproteasome and extracellular alveolar proteasomes.
The proteasome becomes more specialized in the setting of infection or inflammation. For example, tumor necrosis factor (TNF) or IFN release from proinflammatory cells leads to the conversion of 19S elements in the proteasomal machinery to form an “immunoproteasome,” which generates peptides that are trafficked preferentially through antigen-processing machinery and ultimately to the type I major histocompatibility complex to be presented to T lymphocytes that bolster immunity to pathogens (14).
Studies demonstrate the presence of proteasomes outside the cellular environment, such as lung alveolar fluid. Proteasomes in lung fluid are only shown to have the 20S subunit and cleave proteins without requirement for E3 ligases for protein-specific recognition or ubiquitination. It has been proposed that extracellular proteasomes are a consequence of cell lysis and spillage of cellular contents; alternatively, there may be packaging and exocytosis (i.e., active secretion) of intact 20S proteasomes. Regardless of the mechanisms of release, extracellular proteasomes are increased in the setting of acute infection and inflammation, and may play a role in antigen presentation to activate immunity against extracellular microbes the host encounters (15).
Proteasome dysregulation as a microbial pathogenic mechanism
Pathogens can exploit host cell UPS machinery to their advantage. For example, Legionella pneumophila bacteria produce their own FBP, Lpp2082, which is required for infection. This FBP binds and competes the substrate ParvB away from degradation, apparently creating a permissive cellular environment (16). Human adenovirus creates two E3 ligase proteins that cause degradation of the p53 protein, allowing production of viral proteins and genetic material without p53-mediated host cell apoptosis (17). Pseudomonas aeruginosa secretes a toxin, Cif, in vesicles that increases ubiquitination and degradation of cystic fibrosis transmembrane regulator (CFTR) (18), thus making the airway secretions more tenacious. The coronavirus that causes severe acute respiratory syndrome possesses a Ub-like protein that increases pathogenicity; also, proteasome inhibitor pretreatment reduced viral replication and improved survival in mice (19), implicating some role for the UPS in severe acute respiratory syndrome.
Pulmonary Ion Transport and Fluid Balance
Cystic fibrosis.
Cystic fibrosis is due to insufficient CFTR cell surface expression, causing impaired chloride secretion in the airway lumen, with reduced airway surface liquid, conglomeration of proteins, impaired ciliary clearance, and enhanced susceptibility to infection. Cystic fibrosis is most commonly due to CFTR mutation at the position 508 phenylalanine residue (ΔF508); this mutant protein is translated, but intercepted in the endoplasmic reticulum by E3 ligases CHIP and RMA1, ubiquitinated, and degraded by the proteasome before reaching the cell surface (20). C-terminal CFTR deletions are processed normally, but rapidly shuttled to the proteasome for degradation (21), while normal CFTR membrane expression is regulated by E3 ligase C-CBL, mediating ubiquitination and endosomal internalization (22).
Pulmonary edema.
In pulmonary edema, epithelial sodium channel activity regulates apical Na+ entry into the cell, from where it is actively transported out of the cell via the Na-K-ATPase as the critical mechanism for fluid balance in the lungs (23). In addition to its regulation of HIF-1α protein concentrations discussed previously, vHL protein also controls edema clearance during hypoxia, where it mediates degradation of Na-K-ATPase (24). Here, it appears that reactive oxygen species participate in the regulation of the Na-K-ATPase via PKCζ and a member of the LUBAC, HOIL-1L, which leads to impaired lung fluid clearance. Thus, the steady state of both the epithelial sodium channel and Na-K-ATPase are highly regulated by the UPS to critically maintain epithelial function to effect lung fluid balance and normal breathing.
Airway Inflammation
Perhaps the most prominently implicated signal in pulmonary inflammation is the activity of the nuclear factor of κ light polypeptide gene enhancer in B cells, NF-κB (25). When active, this transcription factor master regulator of inflammation leads to expression of cytokines, chemokines, adhesion molecules, matrix metalloproteases, and leukocyte growth factors, among others. The negative regulator of NF-κB is IκB, which usually binds and sequesters NF-κB in the cytosol (26). IκB is degraded by the ubiquitin proteasome via the FBP β-transducin repeat–containing protein (β-Trcp, now designated FBXW1). When IκB is phosphorylated, it is recognized by SCFFBXW1 for ubiquitination and degradation, leaving NF-κB unrestricted to initiate the inflammatory cascade. IκB phosphorylation is in turn regulated by kinases, which are each activated by ligation of receptors, or the activity of protein second messengers, such as the TNF receptor–associated factor (TRAF) proteins.
LUBAC has been described to have an important role in regulating inflammation (27). LUBAC is now known to be part of the TNF receptor signaling complex and participates in signaling processes by end-to-end polyubiquitination of TNF receptor signal modulators RIP1 and NEMO, apparently increasing signal transduction by this particular ubiquitination scheme (28). LUBAC also targets IL-1β, CD40 ligand, and several Toll-like receptors (TLRs). SHARPIN mutant mice develop a proliferative dermatitis, and patients with mutations of HOIL-1L and thus LUBAC deficiency have protracted inflammatory disorders and invasive bacterial infections (29).
Studies indicate that TRAF proteins are targets of the SCFFBXL2 E3 ligase (30). TRAF degradation after overexpression of FBXL2 globally suppresses inflammatory responses in response to endotoxin. Interestingly, another E3 ligase, SCFFBXO3, targets FBXL2 for its degradation; FBXO3 depletion in cells increases FBXL2 and decreases TRAF protein levels, blunting inflammatory cytokine release in vitro. A human FBXO3 polymorphism with a relatively high (∼6%) frequency among individuals of European descent exists, and this mutant FBXO3 decreases FBXL2 ubiquitination; humans with this polymorphism hospitalized with sepsis have lower serum concentrations of inflammatory cytokines (30).
The E3 ligase Itch causes degradation of multiple inflammatory transcription factors and signaling molecules (31), and Itch mutations in mice cause multiorgan inflammatory disease with pulmonary interstitial inflammation (32). A 2010 study of an Amish family with a multiorgan inflammatory–autoimmune syndrome characterized by severe pneumonitis and premature death identified an autosomal recessive mutation in the Itch E3 ligase (33), almost completely recapitulating the disease phenotype initially observed in knockout mice. Miz1 appears to have checkpoint function in the regulation of LPS-induced inflammation where the cytoplasmic Miz1 suppresses LPS- or TNF-induced production of proinflammatory cytokines through inhibition of JNK (c-Jun N-terminal kinase) activation (34). It has been reported that the HECT domain–containing E3 ligase Mule catalyzes TNF-α–induced Miz1 K48-ubiquitination degradation, which is of importance in TNF-α–induced JNK activation and cell death (35). Interestingly, the interaction between Mule and Miz1 occurred TNF-α independently of the pox virus and zinc finger domain of Miz1, which is of potential relevance in the inflammatory pathways during pulmonary infections.
Asthma.
In allergic asthma, an antigen-specific hyperactive helper T-cell type 2 (Th2) immune response causes airway obstruction and hyperresponsiveness. Bronchodilators or immunomodulators including steroids represent the initial therapy, with allergen-specific immunotherapy used to promote immunological tolerance and durable symptom relief. Immune tolerance is mediated, in part, by the Ub E3 ligase Itch, which degrades the phosphorylated JunB, a Th2-specific transcription factor. Itch deficiency causes high JunB levels with increased production of the Th2 cytokines (36). In asthma animal models, Itch knockout mice fail to develop tolerance to ovalbumin antigen–specific immunomodulatory therapy. Likewise, in another ovalbumin-induced asthma study of mice lacking the E3 ligase Cbl-b, immune tolerance was disrupted, although with increased airway neutrophils and Th1 cytokines IL-12 and IFN-γ (37). Itch and Cbl-b therefore each seem to play a role in maintaining immune tolerance in different effector arms of the T-cell system. Another E3 ligase, Midline 1 (MID1), is up-regulated after antigen stimulation with the common asthma allergen house dust mite (38). MID1 targets protein phosphatase-2A (PP2A), an endogenous inhibitor of cytokine signaling that deactivates NF-κB; hence small inhibitory RNA MID1 knockdown suppressed allergic inflammation in vivo in mice and in vitro in human lung cells.
COPD.
With tobacco exposure, some smokers have a sustained inflammatory phenotype, and many develop COPD. UPS activity is dysregulated in this setting, with increased UPS components (39) and impairment of proteasome activity after cigarette smoke administration (40, 41). The epigenetic regulator, histone deacetylase-2 (HDAC2), is degraded by the UPS after cigarette smoke exposure secondary to HDAC2 phosphorylation (42). Although the E3 ligase RING finger LIM domain–binding protein has been shown to target HDAC2 in other systems (43), its role in the lung has not been described; regardless, loss of HDAC2 causes aberrant inflammatory gene transcription and feed-forward inflammation in some smokers, with HDAC2 levels correlating inversely with COPD severity (44).
Acute Respiratory Distress Syndrome
In ALI, many physiological changes involve the UPS (45). TLRs sense pathogen-associated molecular patterns and initiate inflammatory responses. E3 ligase Cbl-b down-regulates TLR signaling, with Cbl-b deficiency potentiating the inflammatory response (46).
IL-33 is a strong inflammatory activator during asthma and ALI through the receptor ST2L. Phosphorylated ST2 is bound and ubiquitinated by SCFFBXL19, causing UPS degradation. Ectopically expressed FBXL19 decreased ST2 and reduced inflammation while improving survival in animal models of ALI (47). In other work, depletion of the proinflammatory FBP FBXO3, which targets the TRAF inhibitor FBXL2 for its disposal, restores FBXL2 protein levels, improves survival, lowers cytokine release, and lessens inflammation histologically in mice in a Pseudomonas and LPS model of ARDS (30).
Lung Disease–associated Myopathy
In severe ARDS and COPD, diaphragmatic and peripheral muscle wasting are common (48). In a mouse ALI model displaying comorbid muscle wasting the E3 ligase muscle RING finger-1 (MuRF1) mediates muscle breakdown as MuRF1 knockdown prevented ALI-associated muscle wasting (49).
Lung Cancer
Many components within the UPS participate in neoplastic processes, including cancer-promoting FBPs β-Trcp (FBXW1) and SKP2 (S-phase kinase-associated protein-2; also known as FBXL1) (50). β-Trcp disinhibits inflammatory NF-κB activity, causing expression of cell-activating cytokines, growth factors, and proteases that augment tumor proliferation and invasion. β-Trcp also targets the β-catenin protein, which mediates cell differentiation through the Wnt signaling pathway. Loss of β-catenin could impair cell differentiation, typical of aggressive malignancies (51). FBXW7, however, is a p53-dependent tumor suppressor that facilitates degradation of oncoproteins (52). FBXL2 destabilizes proteins critical to cell cycle progression, thereby inhibiting lung tumor cell growth (53–55).
In lung cancer, SKP2 is considered a proneoplastic factor that degrades protective p27 and increases tissue invasiveness (56). Studies show that increased SKP2 protein levels in biopsy specimens are associated with increased metastasis. Moreover, decreased p27 in SKP2hi specimens is robustly correlated with shorter survival (57, 58).
Surfactant Metabolism
Pulmonary surfactant is composed of key surfactant-associated proteins and phospholipids, the components of which participate in the innate immune response and maintenance of alveolar stability by lowering surface tension. Mutations of the surfactant protein C (SP-C) gene have been associated with a familial form of usual interstitial pneumonia and pulmonary fibrosis (59, 60). SP-C processing and cell secretion require distinct steps, including ubiquitination by the E3 ligase NEDD4-2 (61, 62); many familial ILD-associated SP-C alleles involve the C terminus of the protein, with ubiquitinated and aggregated SP-C within perinuclear inclusions of one such mutation (63), demonstrating defective trafficking after E3 ligase association.
CTP:phosphocholine cytidylyltransferase (CCTα) is an essential lipogenic enzyme needed for surfactant phospholipid synthesis. CCTα ubiquitination is catalyzed by the SCFFBXL2 E3 ligase complex (64), and FBXL2 depletion stabilizes CCTα levels and stimulates surfactant biosynthesis. Another surfactant enzyme, lysophosphatidylcholine acyltransferase (LPCAT1), is targeted for ubiquitination and degradation by β-Trcp (65). Thus, it is likely that the Ub E3 ligases regulate surfactant components to modulate lung homeostasis.
Pharmacological Targeting of the Proteasome
Bortezomib and Nonselective Proteasome Inhibitors
The first U.S. Food and Drug Administration (FDA)–approved drug that targets the proteasome is bortezomib (Velcade), a reversible 20S proteasome inhibitor. Bortezomib has emerged as an effective agent in the treatment of multiple myeloma, a malignancy previously linked with a dismal prognosis (66, 67). Only one other proteasome inhibitor has been approved by the FDA for use in humans, carfilzomib, as second-line therapy for multiple myeloma and for non-Hodgkin’s lymphoma.
New Proteasome Inhibitors
The success of bortezomib has established the proteasome as a viable target in the current era of drug development, with reports on five “second-generation” proteasome inhibitors that target overlapping aspects of cell signaling in vitro. The preclinical and in vivo targets of these drugs include hematologic malignancies as well as the solid tumors (68). Four of these agents have entered into phase 1 and 2 clinical trials in subjects with solid tumors as well as hematologic malignancies. Neuropathy is a class-wide side effect of proteasomal inhibitors, likely from accumulation of ubiquitin-laden proteins in dorsal root ganglia of patients receiving therapy. This effect could be avoided by targeting factors upstream of the proteasome (Figure 2). The first such compound to be tested in humans is MLN4924, which targets the NEDD8 activating enzyme required for activation of the Cullin–RING proteins (69); this drug globally suppresses ubiquitination through Cullin–RING ligases, which includes all SCF E3 ligase family members and others. In vitro activity of this agent reduces tumor burden in multiple models via tumor cell apoptosis or autophagy. Four phase 1 clinical trials for MLN4924 safety testing are now underway.
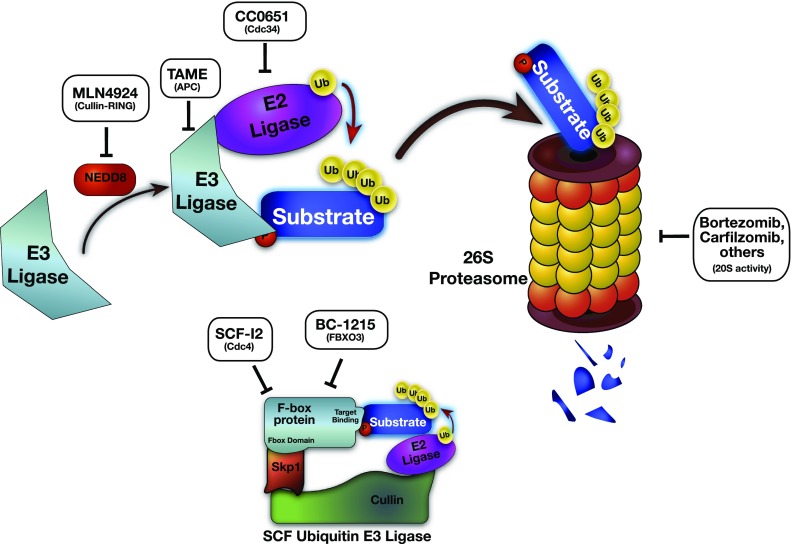
Figure 2.
Drugs targeting the ubiquitin–proteasome system (UPS). Bortezomib and carfilzomib are U.S. Food and Drug Administration–approved proteasome inhibitors that block cleavage of substrate proteins through the 26S proteasome. Upstream inhibitors CC0651 and MLN4924 block ubiquitination by inhibition of the Cullin–RING E3 ligases by inhibition of the Cdc34 E2 ligase and the NEDD8 activator of Cullin–RING E3 ligases, respectively, to temper cell proliferation. TAME blocks APC E3 ligase activity and causes mitotic arrest. The F-box specific inhibitor SCF-I2 blocks the yeast Cdc-4 F-box protein–dependent ubiquitination of substrate proteins (but is not active on human FBPs), and BC-1215 prevents SCFFBXO3-mediated substrate ubiquitination and degradation of the FBXL2 protein, enhancing SCFFBXL2-dependent TRAF degradation, decreasing inflammation. APC = anaphase-promoting complex; FBP = F-box protein; FBXL, FBXO = F-box protein containing a leucine-rich domain, or neither a leucine-rich nor WD-40 domain, respectively; NEDD = neural precursor cell expressed developmentally down-regulated protein; P = phosphate; RING = really interesting new gene; SCF = Skp1–Cullin–F-box protein; TAME = tosyl-l-arginine methyl ester; TRAF = tumor necrosis factor receptor–associated factor; Ub = ubiquitin.
Another newly characterized drug is CC0651, the first small-molecule E2 ligase inhibitor, with high potency and specificity for the E2 ligase Cdc34 (70). This drug suppresses ubiquitination through Cullin–RING E3 ligases that depend on Cdc34. CC0651 and MLN4924 would therefore theoretically have highly overlapping pharmacology, and may have synergistic effects.
The small molecule tosyl-l-arginine methyl ester (TAME) was described as an inhibitor of the APC E3 ligase required for dismantling the spindle assembly checkpoint and completion of mitotic division (71). TAME prevents depletion of cyclin B1, thereby leading to mitotic arrest in metaphase. The next putative selective target of the ubiquitination–proteasome pathways is to target individual subunits of the E3 ligases. Although no drugs with this activity have entered clinical trials, there is significant research in this area.
FBP-specific inhibitors.
The first report of an E3 ligase–targeting drug identified SCF-I2, discovered by small-molecule interrogation of the SCFCdc4 complex by its ability to displace the SCF from its phosphodegron in a yeast system. This molecule antagonized SCFCdc4, but not SCF complexes with related FBPs nor the human ortholog, FBXW7 (72).
Another small molecule inhibits SCFFBXO3 by targeting the FBXO3 C-terminal ApaG domain, a bacteria-like domain found in only two other genes in humans. This compound, BC1215, shows potent blockade of FBXO3 activity with a robust decrease in TRAFs and downstream inflammatory mediators released from endotoxin-stimulated human blood monocytes. BC-1215 effectively reduced inflammation and lung injury in preclinical models of sepsis and ALI (30). This demonstration of a targeted, and specific inhibitor to a single FBP requires further testing, but if successful, has far-reaching implications and may set the stage for a new genus of antiinflammatory drugs.
Conclusions
In summary, the UPS in lung biology is a fundamental area of research and discovery. The activities of many E3 ligases and FBPs remain unknown, and many more discoveries await us in the years to come. However, it is becoming clear that protein processing via the UPS plays a central role in most of the principal types of lung disease (Table 1). Along with these newly described mechanisms of pathobiology come significant advances in our strategies for intervention to expand our arsenal of potential therapies. Although treatment with UPS-targeting medications has thus far been limited to hematologic malignancy, antiinflammatory agents that act on some UPS component may become commonplace in the next decade, especially as more selective compounds with fewer side effects are identified. It is difficult to ascertain how these innovations will impact medicine in the evolution of UPS-targeting therapy; however, it seems that new generations of drugs acting on this important system will become the mainstay of therapy for some diseases.
TABLE 1.
E3 LIGASES AND THEIR TARGETS RELEVANT TO LUNG DISEASE
| Disease/Condition | E3 Ligase/Subunit | Target | Biological Effect |
|---|---|---|---|
| Pulmonary hypertension | vHL | HIF-1α | Prevents HIF-1α transcription of proliferative and invasive/angiogenic genes |
| Legionella pneumonia | Lpp2082* | ParvB | Prevents ParvB degradation and establishes permissive cellular environment for bacteria |
| Adenoviral respiratory infection | E4orf6 and E1B55K† | p53 | Disrupts p53 quality control mechanisms to enhance viral replication |
| Cystic fibrosis | CHIP and RMA1 | CFTR ΔF508 | Premature UPS degradation of CFTR, preventing surface expression |
| C-CBL | CFTR WT | Endosomal internalization and UPS destruction of CFTR | |
| Pulmonary edema | NEDD4-2 | ENaC | Normal ENaC degradation; NEDD4-2−/− mice develop CF phenotype; overexpression impairs fluid clearance |
| vHL | Na-K-ATPase | Decreased Na-K-ATPase activity; impaired sodium and fluid clearance from epithelia and interstitium | |
| HOIL-1L | PKCζ | ||
| Airway inflammation | β-Trcp (FBXW1)‡ | IκB | De-repression of NF-κB and production of multiple proinflammatory cytokines |
| FBXL2‡ | TRAFs 1–6 | Decreased inflammatory signal transduction and decreased NF-κB activity | |
| FBXO3‡ | FBXL2 | Increased TRAF activity and inflammatory signal transduction | |
| Itch | JunB, c-Jun, Notch, PKC, PLCγ, ErbB | Decreased Th2 cytokine production and immunological tolerance; Itch KO results in loss of tolerance; nonfunctional SNP causes lung and multiorgan inflammatory syndrome | |
| Mule | Miz1 | Disinhibits TNF induced inflammatory signaling | |
| Asthma | Cbl-b | Uncharacterized | Decreased Th1 cytokine production and immunological tolerance; Cbl-b KO results in loss of tolerance |
| MID1 | PP2A | Increased NF-κB activity after antigen exposure | |
| COPD | RLIM§ | HDAC2 | Acetylated histones leave chromatin open for transcription of inflammatory genes |
| ALI | Cbl-b | Uncharacterized | Decreased ALI inflammatory response; Cbl-b KO results in increased inflammation and TLR expression |
| FBXL19‡ | ST2 (IL-33R) | Decreased IL-33 signaling and inflammation in ALI and pneumonia | |
| Lung disease–associated myopathy | MuRF1 | Myosin | Skeletal muscle wasting; MuRF1 knockdown in ALI prevents associated muscle wasting |
| Lung cancer | β-Trcp (FBXW1)‡ | IκB | NF-κB derepression with increased cellular activation, proliferation, and invasion |
| β-Catenin | Impaired cell differentiation through Wnt signaling | ||
| SKP2 (FBXL1)‡ | p27, Fox01, p21, and p57 | Loss of tumor suppressor protein activity | |
| FBXW7‡ | Cyclin E1, c-Myc, c-Jun, Notch | Tumor suppression via degradation of oncoproteins | |
| c-CBL | Receptor tyrosine kinases | Reduced proliferation; c-CBL overexpression decreased tumor burden | |
| FBXL2‡ | Cyclin D2, cyclin D3, Aurora B | Reduced proliferation; FBXL2 overexpression decreased tumor burden | |
| Surfactant homeostasis | NEDD4-2 | SP-C | Normal protein sorting and processing; SP-C disease mutants are ubiquitinated but form aggregates in familial ILD |
| FBXL2 | CCTα | Reduced membrane/surfactant phospholipid synthesis | |
| β-Trcp (FBXW1)‡ | LPCAT1 | Impaired surfactant phospholipid remodeling |
Definition of abbreviations: ALI = acute lung injury; C-CBL = C-casitas B-lineage lymphoma E3 ligase; CCT = CTP:phosphocholine cytidylyltransferase; CF = cystic fibrosis; CFTR = cystic fibrosis transmembrane regulator; CHIP = C terminus of Hsc70-interacting protein; COPD = chronic obstructive pulmonary disease; ENaC = epithelial sodium channel; FBXL, FBXW, FBXO = F-box protein containing a leucine-rich domain, a WD-40 domain, or neither a leucine-rich nor WD-40 domain, respectively; HDAC2 = histone deacetylase-2; HIF-1α = hypoxia-inducible factor-1α; HOIL = heme-oxidized IRP2 ubiquitin ligase-1; IκB = inhibitor of NF-κB; IL-33R = IL-33 receptor; ILD = interstitial lung disease; KO = knockout; LPCAT1 = lysophosphatidylcholine acyltransferase-1; Lpp2082 = Legionella pneumophila (strain Paris) F-box protein; MID1 = E3 ubiquitin ligase midline-1; MuRF1 = muscle RING finger-1; NEDD = neural precursor cell expressed developmentally down-regulated protein; NF-κB = nuclear factor-κB; ParvB = parvin B; PKC = protein kinase C; PLC = phospholipase C; RLIM = RING finger LIM domain–binding protein; RMA1 = RING finger protein with membrane anchor-1; SKP2 = S-phase kinase-associated protein-2; SNP = single-nucleotide polymorphism; SP-C = surfactant protein C; Th2 = helper T-cell type 2; TNF = tumor necrosis factor; TRAF = TNF receptor–associated factor; UPS = ubiquitin proteasome system; vHL = von Hippel–Lindau protein; WT = wild type.
Legionella bacteria–derived F-box protein.
Adenovirus-derived E3 ligase.
Part of the SCF (Skp1–Cullin–F-box protein) multisubunit Cullin–RING E3 ligase.
Specific mechanism not fully characterized in lung disease.
Acknowledgments
Acknowledgment
The authors thank Bill Chen and Yutong Zhao for careful review of this manuscript. This material is based on work supported, in part, by the U.S. Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development. This work was supported by a Merit Review Award from the U.S. Department of Veterans Affairs and National Institutes of Health R01 grants HL096376, HL097376, and HL098174 (to R.K.M.) and HL-R37-48129 and HL-PO1-71643 (to J.I.S.). N.M.W. is supported by a research fellowship through the U.S. Department of Veterans Affairs, Veterans Health Administration, VA Pittsburgh Healthcare System.
Footnotes
In order to correct errors in the figures in the original publication of this article, this corrected version was posted on November 8, 2013. In the schematic Figures 1 and 2, E1 and E2 enzymes were originally mislabeled as ligases. The figures and accompanying legends have been corrected.
Originally Published in Press as DOI: 10.1164/rccm.201304-0754PP on May 28, 2013
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 2.Glickman MH, Ciechanover A. The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 3.Jiang YH, Beaudet AL. Human disorders of ubiquitination and proteasomal degradation. Curr Opin Pediatr. 2004;16:419–426. doi: 10.1097/01.mop.0000133634.79661.cd. [DOI] [PubMed] [Google Scholar]
- 4.Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction—Alzheimer’s disease of the heart? N Engl J Med. 2013;368:455–464. doi: 10.1056/NEJMra1106180. [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 6.Bouchecareilh M, Balch WE. Proteostasis: a new therapeutic paradigm for pulmonary disease. Proc Am Thorac Soc. 2011;8:189–195. doi: 10.1513/pats.201008-055MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin–proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 11.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 13.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, Colbert RA. Immunoproteasome assembly: cooperative incorporation of interferon γ (IFN-γ)–inducible subunits. J Exp Med. 1998;187:97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sixt SU, Peters J. Extracellular alveolar proteasome: possible role in lung injury and repair. Proc Am Thorac Soc. 2010;7:91–96. doi: 10.1513/pats.200906-035JS. [DOI] [PubMed] [Google Scholar]
- 16.Lomma M, Dervins-Ravault D, Rolando M, Nora T, Newton HJ, Sansom FM, Sahr T, Gomez-Valero L, Jules M, Hartland EL, et al. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 2010;12:1272–1291. doi: 10.1111/j.1462-5822.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- 17.Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW, Branton PE. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bomberger JM, Ye S, Maceachran DP, Koeppen K, Barnaby RL, O’Toole GA, Stanton BA. A Pseudomonas aeruginosa toxin that hijacks the host ubiquitin proteolytic system. PLoS Pathog. 2011;7:e1001325. doi: 10.1371/journal.ppat.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma XZ, Bartczak A, Zhang J, Khattar R, Chen L, Liu MF, Edwards A, Levy G, McGilvray ID. Proteasome inhibition in vivo promotes survival in a lethal murine model of severe acute respiratory syndrome. J Virol. 2010;84:12419–12428. doi: 10.1128/JVI.01219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 21.Benharouga M, Haardt M, Kartner N, Lukacs GL. COOH-terminal truncations promote proteasome-dependent degradation of mature cystic fibrosis transmembrane conductance regulator from post-Golgi compartments. J Cell Biol. 2001;153:957–970. doi: 10.1083/jcb.153.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye S, Cihil K, Stolz DB, Pilewski JM, Stanton BA, Swiatecka-Urban A. c-Cbl facilitates endocytosis and lysosomal degradation of cystic fibrosis transmembrane conductance regulator in human airway epithelial cells. J Biol Chem. 2010;285:27008–27018. doi: 10.1074/jbc.M110.139881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivera WG, Ridge KM, Sznajder JI. Lung liquid clearance and Na,K-ATPase during acute hyperoxia and recovery in rats. Am J Respir Crit Care Med. 1995;152:1229–1234. doi: 10.1164/ajrccm.152.4.7551375. [DOI] [PubMed] [Google Scholar]
- 24.Zhou G, Dada LA, Chandel NS, Iwai K, Lecuona E, Ciechanover A, Sznajder JI. Hypoxia-mediated Na-K-ATPase degradation requires von Hippel Lindau protein. FASEB J. 2008;22:1335–1342. doi: 10.1096/fj.07-8369com. [DOI] [PubMed] [Google Scholar]
- 25.Li Q, Verma IM. NF-κB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 26.Mercurio F, Manning AM. Multiple signals converging on NF-κB. Curr Opin Cell Biol. 1999;11:226–232. doi: 10.1016/s0955-0674(99)80030-1. [DOI] [PubMed] [Google Scholar]
- 27.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-κB–activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 28.Rieser E, Cordier SM, Walczak H. Linear ubiquitination: a newly discovered regulator of cell signalling. Trends Biochem Sci. 2013;38:94–102. doi: 10.1016/j.tibs.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Boisson B, Laplantine E, Prando C, Giliani S, Israelsson E, Xu Z, Abhyankar A, Israël L, Trevejo-Nunez G, Bogunovic D, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol. 2012;13:1178–1186. doi: 10.1038/ni.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen BB, Coon TA, Glasser JR, McVerry BJ, Zhao J, Zhao Y, Zou C, Ellis B, Sciurba FC, Zhang Y, et al. A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat Immunol. 2013;14:470–479. doi: 10.1038/ni.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melino G, Gallagher E, Aqeilan RI, Knight R, Peschiaroli A, Rossi M, Scialpi F, Malatesta M, Zocchi L, Browne G, et al. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 2008;15:1103–1112. doi: 10.1038/cdd.2008.60. [DOI] [PubMed] [Google Scholar]
- 32.Perry WL, Hustad CM, Swing DA, O’Sullivan TN, Jenkins NA, Copeland NG. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 33.Lohr NJ, Molleston JP, Strauss KA, Torres-Martinez W, Sherman EA, Squires RH, Rider NL, Chikwava KR, Cummings OW, Morton DH, et al. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Hum Genet. 2010;86:447–453. doi: 10.1016/j.ajhg.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Do-Umehara HC, Chen C, Urich D, Zhou L, Qiu J, Jang S, Zander A, Baker MA, Eilers M, Sporn PH, et al. Suppression of inflammation and acute lung injury by Miz1 via repression of C/EBP-δ. Nat Immunol. 2013;14:461–469. doi: 10.1038/ni.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Do H, Tian X, Zhang C, Liu X, Dada LA, Sznajder JI, Liu J. E3 ubiquitin ligase Mule ubiquitinates Miz1 and is required for TNFα-induced JNK activation. Proc Natl Acad Sci USA. 2010;107:13444–13449. doi: 10.1073/pnas.0913690107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venuprasad K, Elly C, Gao M, Salek-Ardakani S, Harada Y, Luo JL, Yang C, Croft M, Inoue K, Karin M, et al. Convergence of Itch-induced ubiquitination with MEKK1-JNK signaling in Th2 tolerance and airway inflammation. J Clin Invest. 2006;116:1117–1126. doi: 10.1172/JCI26858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh SY, Park JU, Zheng T, Kim YK, Wu F, Cho SH, Barber D, Penninger J, Zhu Z. Cbl-b regulates airway mucosal tolerance to aeroallergen. Clin Exp Allergy. 2011;41:434–442. doi: 10.1111/j.1365-2222.2010.03593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collison A, Hatchwell L, Verrills N, Wark PA, de Siqueira AP, Tooze M, Carpenter H, Don AS, Morris JC, Zimmermann N, et al. The E3 ubiquitin ligase midline 1 promotes allergen and rhinovirus-induced asthma by inhibiting protein phosphatase 2A activity. Nat Med. 2013;19:232–237. doi: 10.1038/nm.3049. [DOI] [PubMed] [Google Scholar]
- 39.Carolan BJ, Heguy A, Harvey BG, Leopold PL, Ferris B, Crystal RG. Up-regulation of expression of the ubiquitin carboxyl-terminal hydrolase L1 gene in human airway epithelium of cigarette smokers. Cancer Res. 2006;66:10729–10740. doi: 10.1158/0008-5472.CAN-06-2224. [DOI] [PubMed] [Google Scholar]
- 40.van Rijt SH, Keller IE, John G, Kohse K, Yildirim AO, Eickelberg O, Meiners S. Acute cigarette smoke exposure impairs proteasome function in the lung. Am J Physiol Lung Cell Mol Physiol. 2012;303:L814–L823. doi: 10.1152/ajplung.00128.2012. [DOI] [PubMed] [Google Scholar]
- 41.Min T, Bodas M, Mazur S, Vij N. Critical role of proteostasis-imbalance in pathogenesis of COPD and severe emphysema. J Mol Med (Berl) 2011;89:577–593. doi: 10.1007/s00109-011-0732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adenuga D, Yao H, March TH, Seagrave J, Rahman I. Histone deacetylase 2 is phosphorylated, ubiquitinated, and degraded by cigarette smoke. Am J Respir Cell Mol Biol. 2009;40:464–473. doi: 10.1165/rcmb.2008-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krämer OH. HDAC2: a critical factor in health and disease. Trends Pharmacol Sci. 2009;30:647–655. doi: 10.1016/j.tips.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, Barczyk A, Hayashi S, Adcock IM, Hogg JC, et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 45.Vadász I, Weiss CH, Sznajder JI. Ubiquitination and proteolysis in acute lung injury. Chest. 2012;141:763–771. doi: 10.1378/chest.11-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachmaier K, Toya S, Gao X, Triantafillou T, Garrean S, Park GY, Frey RS, Vogel S, Minshall R, Christman JW, et al. E3 ubiquitin ligase Cblb regulates the acute inflammatory response underlying lung injury. Nat Med. 2007;13:920–926. doi: 10.1038/nm1607. [DOI] [PubMed] [Google Scholar]
- 47.Zhao J, Wei J, Mialki RK, Mallampalli DF, Chen BB, Coon T, Zou C, Mallampalli RK, Zhao Y. F-box protein FBXL19-mediated ubiquitination and degradation of the receptor for IL-33 limits pulmonary inflammation. Nat Immunol. 2012;13:651–658. doi: 10.1038/ni.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Debigaré R, Côté CH, Maltais F. Ubiquitination and proteolysis in limb and respiratory muscles of patients with chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2010;7:84–90. doi: 10.1513/pats.200906-051JS. [DOI] [PubMed] [Google Scholar]
- 49.Files DC, D’Alessio FR, Johnston LF, Kesari P, Aggarwal NR, Garibaldi BT, Mock JR, Simmers JL, DeGorordo A, Murdoch J, et al. A critical role for muscle RING finger-1 in acute lung injury–associated skeletal muscle wasting. Am J Respir Crit Care Med. 2012;185:825–834. doi: 10.1164/rccm.201106-1150OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuchs SY, Spiegelman VS, Kumar KG. The many faces of β-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 52.Tan HL, Regamey N, Brown S, Bush A, Lloyd CM, Davies JC. The Th17 pathway in cystic fibrosis lung disease. Am J Respir Crit Care Med. 2011;184:252–258. doi: 10.1164/rccm.201102-0236OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen BB, Glasser JR, Coon TA, Mallampalli RK. FBXL2 is a ubiquitin E3 ligase subunit that triggers mitotic arrest. Cell Cycle. 2011;10:3487–3494. doi: 10.4161/cc.10.20.17742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen BB, Glasser JR, Coon TA, Mallampalli RK. F-box protein FBXL2 exerts human lung tumor suppressor–like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest. Oncogene. 2012;31:2566–2579. doi: 10.1038/onc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen BB, Glasser JR, Coon TA, Zou C, Miller HL, Fenton M, McDyer JF, Boyiadzis M, Mallampalli RK. F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood. 2012;119:3132–3141. doi: 10.1182/blood-2011-06-358911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hung WC, Tseng WL, Shiea J, Chang HC. Skp2 overexpression increases the expression of MMP-2 and MMP-9 and invasion of lung cancer cells. Cancer Lett. 2010;288:156–161. doi: 10.1016/j.canlet.2009.06.032. [DOI] [PubMed] [Google Scholar]
- 57.Osoegawa A, Yoshino I, Tanaka S, Sugio K, Kameyama T, Yamaguchi M, Maehara Y. Regulation of p27 by S-phase kinase-associated protein 2 is associated with aggressiveness in non–small-cell lung cancer. J Clin Oncol. 2004;22:4165–4173. doi: 10.1200/JCO.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 58.Yokoi S, Yasui K, Mori M, Iizasa T, Fujisawa T, Inazawa J. Amplification and overexpression of SKP2 are associated with metastasis of non–small-cell lung cancers to lymph nodes. Am J Pathol. 2004;165:175–180. doi: 10.1016/S0002-9440(10)63286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas AQ, Lane K, Phillips J, III, Prince M, Markin C, Speer M, Schwartz DA, Gaddipati R, Marney A, Johnson J, et al. Heterozygosity for a surfactant protein C gene mutation associated with usual interstitial pneumonitis and cellular nonspecific interstitial pneumonitis in one kindred. Am J Respir Crit Care Med. 2002;165:1322–1328. doi: 10.1164/rccm.200112-123OC. [DOI] [PubMed] [Google Scholar]
- 60.Nogee LM, Dunbar AE, III, Wert SE, Askin F, Hamvas A, Whitsett JA. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N Engl J Med. 2001;344:573–579. doi: 10.1056/NEJM200102223440805. [DOI] [PubMed] [Google Scholar]
- 61.Conkright JJ, Apsley KS, Martin EP, Ridsdale R, Rice WR, Na CL, Yang B, Weaver TE. Nedd4-2–mediated ubiquitination facilitates processing of surfactant protein-C. Am J Respir Cell Mol Biol. 2010;42:181–189. doi: 10.1165/rcmb.2009-0058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kotorashvili A, Russo SJ, Mulugeta S, Guttentag S, Beers MF. Anterograde transport of surfactant protein C proprotein to distal processing compartments requires PPDY-mediated association with Nedd4 ubiquitin ligases. J Biol Chem. 2009;284:16667–16678. doi: 10.1074/jbc.M109.002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang C, Gale M, Jr, Keller BC, Huang H, Brown MS, Goldstein JL, Ye J. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell. 2005;18:425–434. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 64.Chen BB, Coon TA, Glasser JR, Mallampalli RK. Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis. Mol Cell Biol. 2011;31:1905–1920. doi: 10.1128/MCB.00723-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zou C, Butler PL, Coon TA, Smith RM, Hammen G, Zhao Y, Chen BB, Mallampalli RK. LPS impairs phospholipid synthesis by triggering β-transducin repeat-containing protein (β-TrCP)-mediated polyubiquitination and degradation of the surfactant enzyme acyl-CoA:lysophosphatidylcholine acyltransferase I (LPCAT1) J Biol Chem. 2011;286:2719–2727. doi: 10.1074/jbc.M110.192377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar KGS, Krolewski JJ, Fuchs SY. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor. J Biol Chem. 2004;279:46614–46620. doi: 10.1074/jbc.M407082200. [DOI] [PubMed] [Google Scholar]
- 67.Sonneveld P, Schmidt-Wolf IG, van der Holt B, El Jarari L, Bertsch U, Salwender H, Zweegman S, Vellenga E, Broyl A, Blau IW, et al. Bortezomib induction and maintenance treatment in patients with newly diagnosed multiple myeloma: results of the randomized phase III HOVON-65/GMMG-HD4 trial. J Clin Oncol. 2012;30:2946–2955. doi: 10.1200/JCO.2011.39.6820. [DOI] [PubMed] [Google Scholar]
- 68.Dick LR, Fleming PE. Building on bortezomib: second-generation proteasome inhibitors as anti-cancer therapy. Drug Discov Today. 2010;15:243–249. doi: 10.1016/j.drudis.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 69.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 70.Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 71.Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh D-C, Hathaway N, Dimova N, Cuny GD, King RW. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]