Abstract
Quinone oxidoreductases are a class of membrane enzymes that catalyse the oxidation or reduction of membrane-bound quinols/quinones. The conversion of quinone/quinol by these enzymes is difficult to study due to the hydrophobic nature of the enzymes and their substrates. We describe some biochemical properties of quinones and quinone oxidoreductases and then look in more detail at two model membranes that can be used to study quinone oxidoreductases in a native-like membrane environment with their native lipophylic quinone substrates. The results obtained with these model membranes are compared to classical enzyme assays that use water-soluble quinone analogues.
Keywords: ubiquinone, co-enzyme Q, oxidative phosphorylation, solid-supported bilayer lipid membrane, protein-film voltammetry, enzyme kinetics
Introduction
Biological cell membranes are an essential component of all living cells. They are ~5 nm thick two-dimensional fluids composed of different lipids, carbohydrates and proteins which form a characteristic bilayer structure and control the import and export of vital ions and nutrients to the cell via highly regulated mechanisms. The specialised structure of the bilayer enables it to act as a spatially constrained ‘reaction vessel’ in which many essential biochemical processes such as photosynthesis and oxidative phosphorylation occur within. The lipid composition can modulate enzyme activity and the function of some membrane enzymes are dependent on not just the chemical properties of the membrane but also on the physical properties such as lateral tension, membrane curvature, hydrophobic matching and electrostatic effects[1].
An important class of membrane proteins are redox enzymes which are largely involved in energetic processes such as photosynthesis and oxidative phosphorylation. Quinone oxidoreductases are a subclass of redox enzymes which are essential in all electron transport chains. This class of enzymes is active in the oxidation or reduction of quinones, which are hydrophobic electron carriers which mediate electron transfer between these enzyme complexes. Characterising quinone oxidoreductase activity is difficult due to the hydrophobic nature of the enzymes and substrates, but this has recently been addressed by assaying activity using biomimetic membrane models. In this mini-review we will first describe some biochemical properties of quinones and quinone oxidoreductases, followed by some model membranes used to study enzyme activity.
Function and Characterisation of Quinones in the Lipid Bilayer
Besides mediating electron transfer between enzyme complexes, many more roles have recently been discovered for quinones. They have been found to protect against lipid peroxidation in the membrane, regenerate vitamin E, prevent DNA damage, control low density lipoprotein peroxidation and regulate the permeability of transition pores in mitochondria and physiochemical properties of the membranes itself[2]. There is also increasing use of quinones for treatment of diseases, including myopathies, cardiovascular diseases and age related degenerative diseases.
The most widely studied class of quinones is ubiquinone, or coenzyme Q, which has a long isoprenoid chain with a benzoquinone ring. Other quinones which occur in nature include menaquinone, plastoquinone and a number of quinone derivatives such as plumbagin, phylloquinone and juglone. An important property of quinones is their hydrophobicity which restricts them to within the bilayer, but also enables them to diffuse freely within it. Despite the fundamental importance of quinones in biology there is still controversy about the localisation and orientation within the membrane. Many different theoretical and experimental studies have been carried out to try and resolve some of these issues. It is generally accepted that the hydrophobic polyisoprene chain which anchors the quinone in the bilayer is located in the hydrophobic core of the membrane parallel to the membrane surface[3], although there is still debate as to whether the tail is in a linear or bent conformation. The position of the polar headgroup is less well agreed upon. It is reported that they are localised at the midplane of the bilayer, but also that they reside at the tail-headgroup interface of the phospholipids and, finally, that their position oscillates within the membrane[4]. Furthermore, the position of the ring in the bilayer has been shown to depend on its oxidation state, with the more polar reduced quinol situated closer to the phospholipid headgroups than the oxidised quinone form[5]. The lateral diffusion of quinones within phospholipid bilayers has been studied using a range of techniques and the diffusion constant is around 10−7 cm2/s as determined by fluorescence quenching[6] and about an order of magnitude lower as determined by chronocoulometry[7] and fluorescence recovery after photobleaching (FRAP) measurements. Further eluciation of the physical properties of quinones in the bilayer is required to fully understand the properties quinone oxidoreductases.
Biochemistry of Quinone Oxidoreductases
As described, quinones are involved in a broad range of cellular processes, the main one being their role as diffusible electron carriers in respiratory chains. In branched bacterial respiratory pathways they serve as a general pool which can couple input of electrons from a wide variety of substrates with a variety of terminal reductases which are dependent on specific growth conditions[8, 9]. Quinone oxidoreductases occur in all respiratory pathways and include the alternative oxidase in plants[10], complex I in mitochondria[11] and ubiquinol oxidases in Escherichia coli[12]. Despite the ubiquity of these enzymes and their fundamental importance in bioenergetic pathways there are still many unanswered questions regarding the location and specificity of quinone binding sites, concentration of protein-bound and freely-diffusible quinone in the bilayer, quinone specificity and kinetic properties. One major difficulty in resolving these questions is that it is challenging to assay quinone oxidoreductases under native-like conditions as they are membrane associated proteins which couple the oxidation or reduction of a quinone in the bilayer with the reduction or oxidation of another soluble substrate.
One model protein for the study of quinone oxidoreductases is cytochrome bo3 (cbo3) from the aerobic respiratory chain in E. coli. cbo3 belongs to the haem-copper oxidase superfamily and closely resembles cytochrome c oxidase from eukaryotes[13]. cbo3 couples the oxidation of ubiquinol to ubiquinone to the reduction of oxygen to water and also pumps protons across the periplasmic membrane. It contains two ubiquinol binding sites, a low affinity site, QL, which is in equilibrium with the quinone pool in the membrane and mutagenesis studies have shown it to reside in the extrinsic domain of subunit II close to the head groups of the lipid bilayer. cbo3 also has a high affinity site, QH, which contains a much more tightly bound quinone, which does not exchange with the quinone pool during turn-over and is buried deep within the lipid bilayer in subunit I close to the binuclear centre. Despite the difference in electron input to the enzyme, the main catalytic cycle steps at the binuclear centre are the same as that of cytochrome c oxidase.
As discussed above, it is problematic to characterise the activity of quinone oxidoreductases using native substrates with the enzyme in its native, membrane associated state. Many studies thus investigate the kinetics using hydrophilic quinone analogues, rather than the hydrophobic natural substrates due to ease of handling in biochemical assays. A complication arises when exploring the kinetic data as the partitioning of the quinone analogues between the aqueous bulk phase and the membrane or enzyme micelle influences the observed kinetics[14]. Consequently, there are many published KM values for cbo3 which vary considerably (Table 1). This is largely due to the different quinone substrates and enzyme preparations used in the assays. Furthermore, these quinone analogues are not necessarily truly representative of the biological processes. Short chain analogues have been shown to localise in different positions and orientations within a membrane compared to long-chain quinones[15] and have higher diffusion constants.
Table 1.
Reported KM values for cytochrome bo3 (cbo3) with different water-soluble ubiquinol/ubiquinone analogues.
| Quinone Substrate | Reported KM |
|---|---|
| ubiquinone-1 | 10 μM [16]; 18-45 μM [13]a; 36 μM [17]; 40-625 μM [18]b; 48 μM [19]; 60.7 μM [20]; 75 μM [21]; 103 μM [22]; 172 μM [23] |
| ubiquinone-2 | 14 μM [22]; 15 μM [17]; 18.5 μM [20]; 65 μM [23] |
| ubiquinone-6 | 50 μM [18] |
| menadiol | 38.4 μM [19] |
| dimethoxybenzoquinol | 152 μM [20]; 166 μM [22] |
| duroquinol | 129 μM [24]; 170-200 μM [25] |
KM shown to be dependent on detergent used for purification;
ibid for the phospholipid used to reconstitute purified cbo3 complex.
To overcome some of the problems identified above, model membrane systems have been used to characterise the activity of quinone oxidoreductases. In the next section, we will briefly describe some of these model systems.
Model membranes used to study quinone oxidoreductases
The earliest and simplest artificial membrane model is the black lipid membrane (BLM) developed in the late 1960s[26]. BLMs are formed by forming a thin phospholipid film across a small aperture in a septum separating two aqueous reservoirs. This model has the advantage that it has large aqueous spaces either side of the bilayer mimicking a cellular membrane but they are very fragile and sensitive to mechanical shocks and low resistivity to electric fields. The stability of a lipid bilayer can be significantly enhanced by forming it on a solid support such as mica or glass[27]. This solid-supported bilayer lipid membrane (sBLM) can be characterised using a wide range of surface sensitive techniques such as atomic force microscopy (AFM), electrochemical impedance spectroscopy (EIS), total internal reflection TIRF, FT-IR, surface plasmon resonance (SPR), quartz crystal microbalance with dissipation (QCM-D) and X-ray and neutron scattering[28]. The behaviour of a sBLM is influenced by the surface it is formed on. For studies with integral membrane proteins, the surface substrate can have a significant effect on the protein structure and function such as retarding diffusion within the membrane or denaturation of the protein due to adsorption of extrinsic domains to the supporting substrate. These effects need to be taken into account when using these systems. A variety of approaches have been developed to prevent or minimise the interaction of membrane proteins with the surface substrate. These include using soft hydrophilic polymers to coat the substrate surface[28] or the use of tether molecules to anchor the bilayer to the surface support and provide a space between the lower leaflet of the bilayer and the solid surface. A full review of model membrane systems is beyond the scope of this mini-review and we refer to one of the following reviews[29-31], although many more excellent reviews are available.
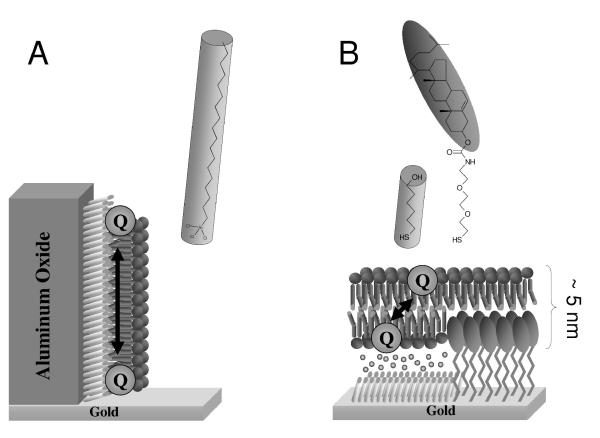
Two model membrane systems have been used to study quinone oxidoreductases, which will briefly be described here. The first model membrane system is the so-called hybrid bilayer, which consist of an alkylated support with a single lipid monolayer on top. The group of Bourdillon [32] have prepared such a hybrid bilayer on porous aluminum oxide to study a peripheral membrane enzyme, pyruvate oxidase (Figure 1A). The second model membrane is a so-called tethered lipid bilayer membrane (tBLM) in which a lipid bilayer is tethered to the solid support (gold) via synthetic tether lipids (Figure 1B). These tether molecules generally contain three distinct parts, a surface reactive group which attaches to the surface substrate, a spacer group and a membrane anchor which interacts with the membrane. The overall quality and properties of the tethered membrane, especially in relation to its electrical properties is dependent on many parameters, like roughness of the underlying substrate surface, grafting density of the tether molecules and their chemical composition[31]. We have used a tBLM on gold to study the ubiquinol oxidase, cbo3[33, 34]. We have made particular progress using the EO3-cholesterol tether molecule. With this system the concentration of the EO3-cholesterol tether on the electrode surface is diluted by mixing it with short chain spacer thiols such as 6-mercaptohexanol[35]. The use of a mixed self-assembled monolayer (SAM) results in phase separated domains in the nanoscale range which allows large transmembrane proteins such as cbo3 to be incorporated into the tBLM. tBLMs have also been formed using native membrane extracts in which bilayers are formed from extracted cell membranes rather than purified lipids and proteins[34, 36-38]. These methods are of particular interest in characterising protein activity as the membrane proteins are entirely retained in their native environment throughout the experimental procedure.
Figure 1.
Model membrane systems used to study quinone oxidoreductases. (A) A hybrid bilayer on alumin oxide modified with trichloro(octadecyl)silane (OTS). This system has been used to study a peripheral membrane enzyme, pyruvate-ubiquinone oxidoreductase[32]. (B) A tethered bilayer lipid membrane (tBLM) formed on a mixed self-assembled monolayer of EO3-Cholesterol and 6-mercaptohexanol. This system has been used to study a ubiquinol oxidase, cytochrome bo3[33, 34].
Electrochemical Assay of Quinone Oxidoreductase Activity using Model Membrane Systems
Electrochemical methods are now widely used to probe the activity of various redox proteins and enzymes[39, 40]. In the overall majority of cases this method, also known as protein-film voltammetry, has been used to characterise soluble redox proteins and enzymes. In the few cases that complete quinone oxidoreductases are studied (e.g., ref. [41, 42]), detergent-solubilised proteins samples are commonly used, which are not reconstituted into a membrane. Furthermore, only the oxidation/reduction reaction coupled to the quinone/quinol conversion is investigated and not the quinone/quinol conversion itself. Even if the quinone/quinol conversion were probed, the enzyme kinetics within the lipid bilayer are likely to differ from those in a detergent solution. Membrane enzymes catalyse reactions in a highly organised specific environment in which the electron carriers (quinones) move laterally within the bilayer and the diffusion of the substrate and reaction product in the bilayer must be considered together with catalysis to fully characterise the system[43, 44].
A novel approach to assay membrane associated enzymes with native like hydrophobic quinones was described by the group of Bourdillon[32]. This approach used microporous alumina on gold electrodes to form hybrid bilayers (see above) with different concentrations of ubiquinone-8 incorporated and a peripheral membrane enzyme, pyruvate oxidase from E. coli. In this architecture there is no water soluble mediator present and all catalytic results occur through coupling to the electrode via the quinone pool. With this method they provided the first value of a KM for electron exchange reaction in two dimensions. The determined KM of 1.8 ± 0.7 pmol/cm2 is necessarily expressed as a 2-dimensional concentration and is very close to the physiological concentration of ubiquinone-8 in the E. coli membrane (2-3 pmol/cm2). As this model membrane is based on hybrid bilayers, it probably will be unsuitable for integral transmembrane proteins as the alkylated surface does not allow space of the membrane proteins to incorporate.
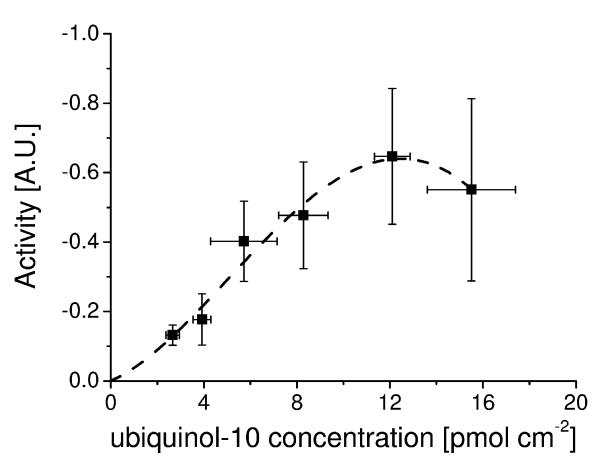
A related approach has been used by us in which tBLMs have been created to study the reaction of ubiquinone-10 in bilayers with an ubiquinol oxidase from E. coli, cytochrome bo3. Using this model membrane the apparent KM for cbo3 with oxygen (1.1 ± 0.4 μM) was determined and also the turnover number which were both shown to agree with previously published values[34]. The concentration of ubiquinone-10 within the bilayer was varied and cbo3 activity determined at saturating oxygen levels (Figure 2). It was shown that the cbo3 activity varies with ubiquinone concentration but not with a typical Michaelis-Menten behaviour. The observed quinone behaviour in this assay is at odds with previous experiments which have characterised cbo3 activity with short chain hydrophilic quinone analogues. We speculate that this difference in activity may be due to the different localisation of hydrophobic (long-chain) quinones in the lipid bilayers compared to the water-soluble quinone analogues. We note that complex I has been shown to have different binding sites for quinones[45] and a similar situation, although unlikely, cannot be ruled out in cbo3 as well. Similar to the case of pyruvate oxidase, the KM for cbo3 for ubiquinol-10 seems to lie in the same of order of magnitude as the physiological ubiquinol-10 concentration. Finally, at very high ubiquinol-10 concentration a drop in enzyme activity might be present. However, at this ubiquinol concentration a large experimental error is observed, which might be due to the destabilisation of the membrane under these conditions.
Figure 2.
The enzyme activity of cytochrome bo3 from E. coli as a function of ubqiuinol-10 concentration in the membrane. Data determined using a tethered bilayer lipid membrane system as explained in the text. The dotted line is guide for the eye to highlight possible cooperative behaviour as described in the text. Details of the experiment are described in [34].
Future Directions
Some progress has been made in using model biomimetic membranes to study quinone oxidoreductases and the first results have now been published. However, thus far, relatively few model membranes have been used to investigate the activity of transmembrane redox proteins and only two enzymes have been investigated with the native (or native-like) hydrophobic quinone substrates in the membrane. The group of Bourdillon has in fact extended their model membrane (Figure 1A) into a tethered lipid bilayer system and have shown that total mitochondrial membrane extract can be used to form the tethered membrane[37]. Regretfully, to date they have not published any results on enzyme kinetics of the mitochondrial quinone enzymes.
More studies will also need to be done to establish what influence the hydrophobic environment and the 2-dimentional substrate diffusion has on the enzyme kinetics and other properties. Some work on the ‘two dimension’ question has already been reported. In the hybrid bilayer system (Figure 1A), the lateral diffusion of the quinol/quinone forms an integral part of the system and needs to be modelled when analysing the electrochemical data[7, 32]. In the planar tethered bilayer system (Figure 1B) we have recently shown that quinone diffusion still influences the observed enzyme kinetics in spite of the ultra-thin membrane layer[46]. Impedance spectroscopy indicated that it takes the quinone 0.05-1 seconds to diffuse from the electrode surface to cbo3 and we hypothesised that this time represents the perpendicular diffusion time of quinone across the membrane (i.e., the “flip” time).
Acknowledgments
Financial support by the BBSRC (BB/D008131/1) is gratefully acknowledged.
Footnotes
This document is the Accepted Manuscript version of a Published Work that appeared in the final form in the Biochemistry Society Transactions, copyright Biochemical Society after peer review and technical editing by the publisher. To access the final edited and published work see http://dx.doi.org/ 10.1042/BST0370707. http://dx.doi.org/ 10.1042/BST0370707.
References
- 1.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 2.Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta-Biomembr. 2004;1660:171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Hauss T, Dante S, Haines TH, Dencher NA. Localization of coenzyme Q(10) in the center of a deuterated lipid membrane by neutron diffraction. Biochim. Biophys. Acta-Bioenerg. 2005;1710:57–62. doi: 10.1016/j.bbabio.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Samori B, Lenaz G, Battino M, Marconi G, Domini I. On coenzyme-Q orientation in membranes - a linear dichroism study of ubiquinones in a model bilayer. J. Membrane Biol. 1992;128:193–203. doi: 10.1007/BF00231812. [DOI] [PubMed] [Google Scholar]
- 5.Ausili A, Torrecillas A, Aranda F, de Godos A, Sanchez-Bautista S, Corbalan-Garcia S, Gomez-Fernandez JC. Redox state of coenzyme Q(10) determines its membrane localization. J. Phys. Chem. B. 2008;112:12696–12702. doi: 10.1021/jp802215s. [DOI] [PubMed] [Google Scholar]
- 6.Blackwell MF, Gounaris K, Zara SJ, Barber J. A method for estimating lateral diffusion-coefficients in membranes from steady-state fluorescence quenching studies. Biophys. J. 1987;51:735–744. doi: 10.1016/S0006-3495(87)83400-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchal D, Boireau W, Laval JM, Moiroux J, Bourdillon C. Electrochemical measurement of lateral diffusion coefficients of ubiquinones and plastoquinones of various isoprenoid chain lengths incorporated in model bilayers. Biophys. J. 1998;74:1937–1948. doi: 10.1016/S0006-3495(98)77902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soballe B, Poole RK. Microbial ubiquinones: Multiple roles in respiration, gene regulation and oxidative stress management. Microbiology-Sgm. 1999;145:1817–1830. doi: 10.1099/13500872-145-8-1817. [DOI] [PubMed] [Google Scholar]
- 9.Poole RK, Cook GM. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Academic Press Ltd; London: 2000. pp. 165–224. ( Advances in microbial physiology, vol 43). [DOI] [PubMed] [Google Scholar]
- 10.Berthold DA, Andersson ME, Nordlund P. New insight into the structure and function of the alternative oxidase. Biochim. Biophys. Acta-Bioenerg. 2000;1460:241–254. doi: 10.1016/s0005-2728(00)00149-3. [DOI] [PubMed] [Google Scholar]
- 11.Brandt U. Energy converting NADH : Quinone oxidoreductase (complex i) Annu. Rev. Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 12.Tsubaki M, Hori H, Mogi T. Probing molecular structure of dioxygen reduction site of bacterial quinol oxidases through ligand binding to the redox metal centers. J Inorg. Biochem. 2000;82:19–25. doi: 10.1016/s0162-0134(00)00140-9. [DOI] [PubMed] [Google Scholar]
- 13.Abramson J, Riistama S, Larsson G, Jasaitis A, Svensson-Ek M, Laakkonen L, Puustinen A, Iwata S, Wikstrom M. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nature Struct. Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- 14.Fato R, Castelluccio C, Palmer G, Lenaz G. A simple method for the determination of the kinetic constants of membrane enzymes utilizing hydrophobic substrates - ubiquinol cytochrome-c reductase. Biochim. Biophys. Acta. 1988;932:216–222. doi: 10.1016/0005-2728(88)90158-2. [DOI] [PubMed] [Google Scholar]
- 15.Roche Y, Peretti P, Bernard S. Influence of the chain length of ubiquinones on their interaction with DPPC in mixed monolayers. Biochim. Biophys. Acta-Biomembr. 2006;1758:468–478. doi: 10.1016/j.bbamem.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Matsushita K, Patel L, Kaback HR. Purification and reconstitution of the cytochrome o-type oxidase from Escherichia coli. Methods In Enzymology. 1986;126:113–122. doi: 10.1016/s0076-6879(86)26013-9. [DOI] [PubMed] [Google Scholar]
- 17.Sato-Watanabe M, Mogi T, Sakamoto K, Miyoshi H, Anraku Y. Isolation and characterizations of quinone analogue-resistant mutants of bo-type ubiquinol oxidase from escherichia coli. Biochemistry. 1998;37:12744–12752. doi: 10.1021/bi981184l. [DOI] [PubMed] [Google Scholar]
- 18.Kita K, Konishi K, Anraku Y. Terminal oxidases of escherichia-coli aerobic respiratory-chain .1. Purification and properties of cytochrome-b562-o complex from cells in the early exponential phase of aerobic growth. J. Biol. Chem. 1984;259:3368–3374. [PubMed] [Google Scholar]
- 19.Kita K, Konishi K, Anraku Y. Purification and properties of 2 terminal oxidase complexes of Escherichia-coli aerobic respiratory-chain. Methods In Enzymology. 1986;126:94–113. doi: 10.1016/s0076-6879(86)26012-7. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto K, Miyoshi H, Ohshima M, Kuwabara K, Kano K, Akagi T, Mogi T, Iwamura H. Role of the isoprenyl tail of ubiquinone in reaction with respiratory enzymes: Studies with bovine heart mitochondrial complex I and Escherichia coli bo-type ubiquinol oxidase. Biochemistry. 1998;37:15106–15113. doi: 10.1021/bi981193u. [DOI] [PubMed] [Google Scholar]
- 21.Sato-Watanabe M, Mogi T, Miyoshi H, Iwamura H, Matsushita K, Adachi O, Anraku Y. Structure-function studies on the ubiquinol oxidation site of the cytochrome bo complex from Escherichia-coli using p-benzoquinones and substituted phenols. J. Biol. Chem. 1994;269:28899–28907. [PubMed] [Google Scholar]
- 22.Sakamoto K, Miyoshi H, Takegami K, Mogi T, Anraku Y, Iwamura H. Probing substrate binding site of the Escherichia coli quinol oxidases using synthetic ubiquinol analogues. J. Biol. Chem. 1996;271:29897–29902. doi: 10.1074/jbc.271.47.29897. [DOI] [PubMed] [Google Scholar]
- 23.Musser SM, Stowell MHB, Lee HK, Rumbley JN, Chan SI. Uncompetitive substrate inhibition and noncompetitive inhibition by 5-nundecyl-6-hydroxy-4,7-dioxobenzothiazole (UHDBT) and 2-n-nonyl-4-hydroxyquinoline-n-oxide (NQNO) is observed for the cytochrome bo(3) complex: Implications for a Q(H2)-loop proton translocation mechanism. Biochemistry. 1997;36:894–902. doi: 10.1021/bi961723r. [DOI] [PubMed] [Google Scholar]
- 24.Moody AJ, Mitchell R, Jeal AE, Rich PR. Comparison of the ligand-binding properties of native and copper-less cytochromes bo from escherichia coli. Biochem. J. 1997;324:743–752. doi: 10.1042/bj3240743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prutsch A, Lohaus C, Green B, Meyer HE, Lubben M. Multiple posttranslational modifications at distinct sites contribute heterogeneity of the lipoprotein cytochrome bo(3) Biochemistry. 2000;39:6554–6563. doi: 10.1021/bi992193c. [DOI] [PubMed] [Google Scholar]
- 26.Mueller P, Rudin DO, Tien HT, Wescott WC. Reconstitution of cell membrane structure in vitro and its transformation into an excitable system. Nature. 1962;194:979. doi: 10.1038/194979a0. &. [DOI] [PubMed] [Google Scholar]
- 27.Castellana ET, Cremer PS. Solid supported lipid bilayers: From biophysical studies to sensor design. Surface Science Reports. 2006;61:429–444. doi: 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M, Sackmann E. Polymer-supported membranes as models of the cell surface. Nature. 2005;437:656–663. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]
- 29.Boxer SG. Molecular transport and organization in supported lipid membranes. Curr. Opin. Chem. Biol. 2000;4:704–709. doi: 10.1016/s1367-5931(00)00139-3. [DOI] [PubMed] [Google Scholar]
- 30.Janshoff A, Steinem C. Transport across artificial membranes - an analytical perspective. Anal. Bioanal. Chem. 2006;385:433–451. doi: 10.1007/s00216-006-0305-9. [DOI] [PubMed] [Google Scholar]
- 31.Koper I. Insulating tethered bilayer lipid membranes to study membrane proteins. Mol. Biosyst. 2007;3:651–657. doi: 10.1039/b707168j. [DOI] [PubMed] [Google Scholar]
- 32.Marchal D, Pantigny J, Laval JM, Moiroux J, Bourdillon C. Rate constants in two dimensions of electron transfer between pyruvate oxidase, a membrane enzyme, and ubiquinone (coenzyme Q(8)), its water-insoluble electron carrier. Biochemistry. 2001;40:1248–1256. doi: 10.1021/bi002325y. [DOI] [PubMed] [Google Scholar]
- 33.Jeuken LJC, Connell SD, Henderson PJF, Gennis RB, Evans SD, Bushby RJ. Redox enzymes in tethered membranes. J. Am. Chem. Soc. 2006;128:1711–1716. doi: 10.1021/ja056972u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss SA, Bushby RJ, Evans SD, Henderson PJF, Jeuken LJC. Characterization of cytochrome bo3 activity in a native-like surface-tethered membrane. Biochem. J. 2009;417:555–560. doi: 10.1042/BJ20081345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeuken LJC, Daskalakis NN, Han XJ, Sheikh K, Erbe A, Bushby RJ, Evans SD. Phase separation in mixed self-assembled monolayers and its effect on biomimetic membranes. Sens. Actuator B-Chem. 2007;124:501–509. [Google Scholar]
- 36.Dodd CE, Johnson BRG, Jeuken LJC, Bugg TDH, Bushby RJ, Evans SD. Native E. coli inner membrane incorporation in solid supported lipid bilayer membranes. Biointerphases. 2008;3:FA59–FA67. doi: 10.1116/1.2896113. [DOI] [PubMed] [Google Scholar]
- 37.Elie-Caille C, Fliniaux O, Pantigny J, Maziere JC, Bourdillon C. Self-assembly of solid-supported membranes using a triggered fusion of phospholipid-enriched proteoliposomes prepared from the inner mitochondrial membrane. Langmuir. 2005;21:4661–4668. doi: 10.1021/la046973k. [DOI] [PubMed] [Google Scholar]
- 38.Jeuken LJC, Connell SD, Nurnabi M, O’Reilly J, Henderson PJF, Evans SD, Bushby RJ. Direct electrochemical interaction between a modified gold electrode and a bacterial membrane extract. Langmuir. 2005;21:1481–1488. doi: 10.1021/la047732f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leger C, Bertrand P. Direct electrochemistry of redox enzymes as a tool for mechanistic studies. Chem. Rev. 2008;108:2379–2438. doi: 10.1021/cr0680742. [DOI] [PubMed] [Google Scholar]
- 40.Armstrong FA. Recent developments in dynamic electrochemical studies of adsorbed enzymes and their active sites. Curr. Opin. Chem. Biol. 2005;9:110–117. doi: 10.1016/j.cbpa.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Christenson A, Gustavsson T, Gorton L, Hagerhall C. Direct and mediated electron transfer between intact succinate : quinone oxidoreductase from Bacillus subtilis and a surface modified gold electrode reveals redox state-dependent conformational changes. Biochim. Biophys. Acta-Bioenerg. 2008;1777:1203–1210. doi: 10.1016/j.bbabio.2008.05.450. [DOI] [PubMed] [Google Scholar]
- 42.Elliott SJ, Hoke KR, Heffron K, Palak M, Rothery RA, Weiner JH, Armstrong FA. Voltammetric studies of the catalytic mechanism of the respiratory nitrate reductase from Escherichia coli: How nitrate reduction and inhibition depend on the oxidation state of the active site. Biochemistry. 2004;43:799–807. doi: 10.1021/bi035869j. [DOI] [PubMed] [Google Scholar]
- 43.Melo E, Martins J. Kinetics of bimolecular reactions in model bilayers and biological membranes. A critical review. Biophys. Chem. 2006;123:77–94. doi: 10.1016/j.bpc.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Gelb MH, Jain MK, Hanel AM, Berg OG. Interfacial enzymology of glycerolipid hydrolases - lessons from secreted phospholipases a(2) Annu. Rev. Biochem. 1995;64:653–688. doi: 10.1146/annurev.bi.64.070195.003253. [DOI] [PubMed] [Google Scholar]
- 45.Helfenbaum L, Ngo A, Ghelli A, Linnane AW, Esposti MD. Proton pumping of mitochondrial complex I: Differential activation by analogs of ubiquinone. J. Bioener. Biomem. 1997;29:71–80. doi: 10.1023/a:1022415906999. [DOI] [PubMed] [Google Scholar]
- 46.Jeuken LJC, Weiss SA, Henderson PJF, Evans SD, Bushby RJ. Impedance spectroscopy of bacterial membranes: Coenzyme-Q diffusion in a finite diffusion layer. Anal. Chem. 2008;80:9084–9090. doi: 10.1021/ac8015856. [DOI] [PMC free article] [PubMed] [Google Scholar]