Circadian clocks provide competitive advantage in an environment that is heavily influenced by the rotation of the Earth1,2 by driving daily rhythms in behavior, physiology and metabolism in bacteria, fungi, plants and animals3,4. Circadian clocks comprise transcription-translation feedback loops, which are entrained by environmental signals such as light and temperature to adjust the phase of rhythms to match the local environment3. Production of sugar from photosynthesis is a key metabolic output of the circadian clock in plants2,5. Here we show that these rhythmic endogenous sugar signals can entrain circadian rhythms in Arabidopsis by regulating circadian clock gene expression early in the photoperiod to define a ‘metabolic dawn’. By inhibiting photosynthesis we demonstrate that endogenous oscillations of sugars provide metabolic feedback to the circadian oscillator through the morning-expressed PSEUDO RESPONSE REGULATOR 7 (PRR7) and identify that prr7 mutants are insensitive to the effects of sugar on circadian period. Thus, photosynthesis has a profound effect on the entrainment and maintenance of robust circadian rhythms in Arabidopsis, demonstrating a critical role for metabolism in regulation of the circadian clock.
In plants, energy is derived from photosynthesis in chloroplasts by fixing CO2 into sugar in a light-dependent manner. Net C assimilation and starch metabolism are under circadian regulation2,6–8 as are transcripts associated with chlorophyll biosynthesis and photosynthetic apparatus, peaking ~ 4 h after dawn5. In Arabidopsis seedlings, addition of sucrose to the growth media shortens circadian period in continuous light9 and can sustain circadian rhythms in continuous dark10. Since exogenous sugars can influence the circadian oscillator, we sought to investigate whether endogenous sugars derived from photosynthesis are part of the circadian network in plants.
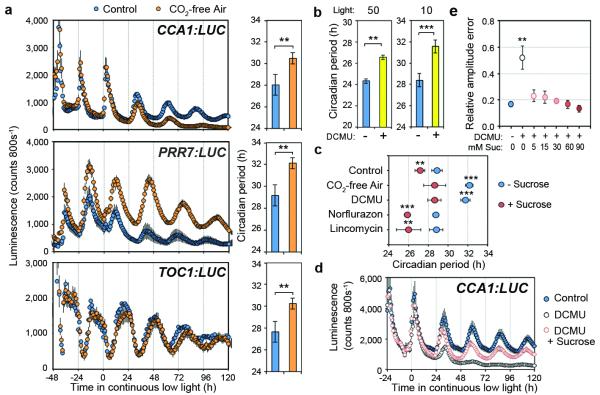
To investigate whether photosynthesis can influence the core circadian clock in Arabidopsis, we inhibited photosynthesis by growing seedlings in CO2-free air or in media containing 3-(3,4-Dichlorophenyl)-1,1-dimethylurea (DCMU), an inhibitor of Photosystem II and monitored circadian rhythms of transcriptional LUCIFERASE (LUC) reporters for core clock gene promoters. We performed these experiments in continuous low light (10 μmol m−2 s−1) because we observed that sucrose shortened circadian period strongly in low light, compared to subtle effects in higher light (50 μmol m−2 s−1) (Extended Data Fig. 2a, b). CO2-depletion (Fig. 1a) or DCMU treatment (Extended Data Fig. 2c, d) lengthened period of clock reporters by an average of 2.9 h and 2.5 h, respectively, compared to controls. Either treatment increased activity of PRR7 promoter:LUC (PRR7:LUC) and reduced activity of CIRCADIAN CLOCK ASSOCIATED1 (CCA1):LUC, which damped towards arrhythmia (Fig. 1a and Extended Data Fig. 2b). In contrast to exogenous sucrose, the effects of DCMU were similar in high or low light (Fig. 1b, Extended Data Fig. 2b). Exogenous sucrose is likely ineffective in altering period in higher light because the response is already saturated by higher endogenous sugars produced from photosynthesis, whereas complete inhibition of photosynthesis will be effective in either light condition.
Figure 1. Photosynthetic sugars influence the circadian clock in Arabidopsis.
a, LUC reporter rhythms (mean ± s.e.m.) and period estimates in seedlings grown in ambient or CO2-free air in 10 μmol m−2 s−1 light, n =4. b, Period estimates of PRR7:LUC in continuous light in the presence or absence of DCMU (mean ± s.d.) n = 4. c, Period estimates of PRR7:LUC rhythms in continuous low light treated with inhibitors in the presence or absence of exogenous sucrose, (mean ± s.d.) n = 4. d, CCA1:LUC rhythms (mean ± s.e.m.) and e, relative amplitude error of CCA1:LUC rhythms in seedlings treated with DCMU in the presence or absence of exogenous sucrose, (mean ± s.d.) n = 4. ** P < 0.01 *** P < 0.001 by t-test.
In light-dark cycles there are robust endogenous rhythms of soluble sugars, which peak ~4-8 h after dawn6,11 (Extended Data Fig. 3a). Inhibition of photosynthesis by either CO2-depletion or DCMU treatment reduced endogenous sugar concentrations (Extended Data Fig. 3b, c). To test whether the effects of inhibition of photosynthesis on the circadian oscillator were due to reduced sugar production, we re-supplied exogenous sucrose to CO2-depleted or DCMU-treated seedlings. Period lengthening by either treatment was suppressed by addition of exogenous sucrose (Fig. 1c). The effects of DCMU treatment on CCA1:LUC rhythms were reversed by addition of as little as 5 mM (0.15 % w/v) exogenous sucrose to the growth media (Fig. 1d, e, Extended Data Fig. 4a). We also tested the effect of norflurazon or lincomycin, which both trigger retrograde signalling from the chloroplast to the nucleus12. Neither treatment lengthened circadian period of PRR7:LUC (Fig. 1c) or inhibited CCA1:LUC activity (Extended Data Fig. 4b) in the presence or absence of exogenous sucrose. Furthermore, we did not find evidence that photosynthesis might affect clock function through mechanisms associated with reactive oxygen species (ROS) production (Extended Data Fig. 5), consistent with a recent report13.
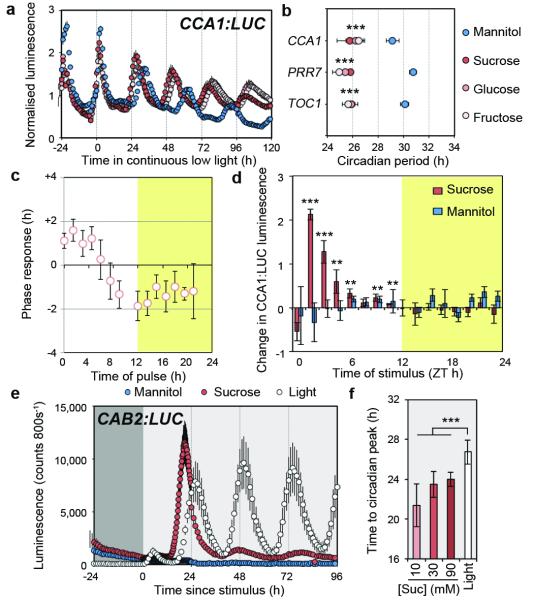
Since our data suggest that the effects of photosynthesis on the circadian clock are mediated by sugars, we investigated the role for sugars in circadian function in more detail. We first tested whether the effects of exogenous sucrose represent a general response to sugar. Circadian period of CCA1:LUC, PRR7:LUC and TIMING OF CAB1 (TOC1):LUC were on average 4.2 h shorter in seedlings grown in media containing 90 mM sucrose (3% w/v), glucose or fructose compared to mannitol-treated controls in continuous low light (Fig. 2a, b). Similarly, exogenous sucrose, glucose or fructose, but not mannitol or a non-metabolisable glucose analog 3-O-methyl glucose, were able to sustain circadian rhythms in continuous dark (Extended Data Fig. 6a). These data suggest that the effects of exogenous sucrose on circadian rhythms represent a general response to metabolically active sugars.
Figure 2. Metabolically active sugar is a zeitgeber which acts differently to light.
a, CCA1:LUC rhythms (mean ± s.e.m) and b, period estimates in seedlings grown in continuous low light with the indicated sugar n =4. c, Phase response of CCA1:LUC rhythms to pulses of sucrose in continuous low light, (mean ± s.d.) n = 8. d, Change in normalised CCA1:LUC activity 3 h after sucrose or mannitol treatment in continuous low light, (mean ± s.d.) n = 4. e, CAB2:LUC rhythms in dark-adapted seedlings treated with sucrose or mannitol in continuous dark or transferred to continuous light, (mean ± s.e.m.) n = 4. f, Time to first circadian peak of CAB2:LUC after treatment with sucrose or light as in e, (mean ± s.d.) n = 4. ** P < 0.01 *** P < 0.001 by t-test.
Oscillations of circadian reporters are absent or very low in continuous dark10 (Extended Data Fig. 6b). Exogenous sucrose can reinitiate circadian oscillations of the clock output reporter CHOROPHYLL A/B BINDING PROTEIN2 (CAB2):LUC in dark-adapted seedlings and the phase is set to the time of sucrose addition after 72 h (subjective dawn) or 60 h (subjective dusk) in continuous dark10. We observed the same behaviour for reporters of the core circadian oscillator and confirmed that exogenous sucrose led to increased CCA1 transcripts in dark-adapted seedlings (Extended Data Fig. 7). The phase-setting of the clock indicates that sucrose is not simply amplifying damped rhythms in dark-adapted seedlings through increased availability of ATP and suggests a role for sugar in entrainment. To directly test whether sugars act in entrainment, we determined a phase-response curve (PRC) for exogenous sucrose, which assesses the ability of a stimulus to alter circadian phase across a circadian cycle14. In continuous low light, the phase of CCA1:LUC and TOC1:LUC peak activity were shifted by pulses of exogenous sucrose inducing phase advances up to 2 h around dawn and phase delays around dusk (Fig. 2c, Extended Data Fig. 8). We observed subtle differences between reporters, similar to phase-setting by temperature15.The phase shifts were not due to effects on circadian period or an osmotic signal (Extended Data Fig. 8c). These data are consistent with metabolically active sugar acting as a Type 1 zeitgeber participating in circadian entrainment14.
A key feature of entrainment is variation, or ‘gating’, of the response to the zeitgeber in a time-dependent manner3. Sucrose application during the first subjective day of continuous low light significantly induced CCA1:LUC activity during the day, but had little effect during the subjective night (Fig. 2d). This effect was most pronounced before midday (ZT6). These data demonstrate that input of sugar to CCA1:LUC activity is gated to be most responsive to sucrose availability early in the light period.
We next compared responses of the Arabidopsis circadian system to sucrose and light because light can act as a strong, Type 0 zeitgeber3,14 and drives sugar production from photosynthesis. In dark-adapted seedlings, there was a similar transient increase in CAB2:LUC peaking ~5 h after treatment with light or sucrose (Fig. 2e). By contrast, the first circadian peak of CAB2:LUC occurred 26.9 h after onset of light compared to 22.8 h after sucrose addition, indicating a 4.1 h advanced phase set by sucrose compared to light (Fig. 2e, f). The difference in phase setting was not concentration-dependent for sucrose, or quantity-dependent for light within the range tested (Fig. 2f, Extended Data Fig. 8d). When photosynthetic sugar production was inhibited in the light by DCMU, the phase set by light was delayed by a further 2.5 to 3.5 h (Extended Data Fig. 8e). These data demonstrate that these two zeitgebers both act in discrete (non-parametric) entrainment. The difference in phase might be due to period effects, but could also indicate distinct phase-setting. The phase difference coincides with the delay between dawn and highest endogenous sugar concentrations (Extended Data Fig.3). We propose that a concentration threshold of photosynthetically-derived sugar provides input to the central oscillator acting as a ‘metabolic dawn’ that contributes to entrainment of the Arabidopsis circadian clock (Extended Data Fig. 1).
Having established that sugars derived from photosynthesis contribute to circadian entrainment in Arabidopsis, we next investigated how this might occur. The increase in PRR7:LUC in DCMU-treated or CO2-depleted seedlings (Fig. 1a, Extended Data Fig. 2) suggested that photosynthesis regulates PRR7 abundance. We measured transcript levels of morning-expressed circadian clock genes in shoots of control and DCMU-treated seedlings (Fig. 3). PRR7 transcript levels were 3.7- to 8.2-fold higher in DCMU-treated seedlings than controls between ZT10 and ZT16 and this difference was suppressed when sucrose was added to media. PRR5 transcripts were only 1.6- to 2.9-fold higher in DCMU-treated seedlings around dawn, and PRR9 transcript levels were unaffected. CCA1 and LHY transcripts were 3.0- to 8.4-fold lower at around dawn, following the increase in PRR7, in DCMU-treated seedlings compared to controls. These data are consistent with the LUC reporter data (Extended Data Fig. 2) and suggest that the effect of photosynthesis is most pronounced on PRR7.
Figure 3. Photosynthetic sugar represses PRR7 late in the photoperiod.
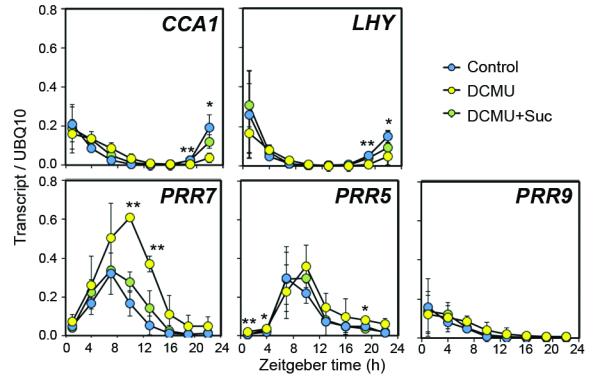
Leaf transcript levels relative in seedlings treated with DCMU in the presence or absence of exogenous sucrose in light-dark conditions, (mean ± s.d.) n = 3. * P < 0.05 ** P < 0.01 by t-test.
These data led us to hypothesise that photosynthetic input to the circadian oscillator might act through PRR7, a transcriptional repressor that acts on the CCA1 promoter during the night16. We first tested the short-term effect of exogenous sucrose on PRR7 promoter activity. In contrast to CCA1:LUC (Fig. 2d), PRR7:LUC activity was significantly repressed during the day and subjective night, but this was most pronounced during the morning (Extended Data Fig. 9a). We tested whether induction of CCA1 depends on PRR7. CCA1:LUC induction was significantly attenuated in prr7-11 mutants compared to wild type (Extended Data Fig. 9b). These data are consistent with sugars activating CCA1 through repression of PRR7. Next we examined whether PRR7 contributes to circadian period adjustment by sucrose. Exogenous sucrose shortened the period of circadian rhythms of CCA1:LUC by 2.7 h in wild-type whereas rhythms in prr7-11 mutants were not shortened by exogenous sucrose (Fig. 4a). Similarly, the period of circadian rhythms of delayed fluorescence17 was also shortened by exogenous sucrose in the wild type, but not in prr7-11 (Extended Data Fig. 9c). To assess whether there is also a role for PRR7 in circadian entrainment by sugars, we determined a PRC for prr7-11 to pulses of exogenous sucrose. In contrast to the wild type (Fig. 2c), sucrose did not induce phase advances in prr7-11 mutants (Extended Data Fig. 9d). Since SENSITIVE TO FREEZING 6 (SFR6), a subunit of the mediator complex, contributes to period adjustment by sucrose by an unknown mechanism9, we determined whether other previously identified pathways participate in the regulation of the circadian oscillator by sugar. We measured rhythms in a range of circadian, sugar insensitive and light signalling mutants. With the exception of cca1-11, all of the tested mutants had significantly shorter circadian period in the presence of sucrose compared to control media (Fig. 4b, Extended Data Fig. 10). Together, these data indicate a specific role for PRR7, acting through CCA1, in regulation of the circadian clock by photosynthetically-derived sugars, and that this might occur through a novel signaling pathway.
Figure 4. PRR7 contributes to circadian sugar signalling.
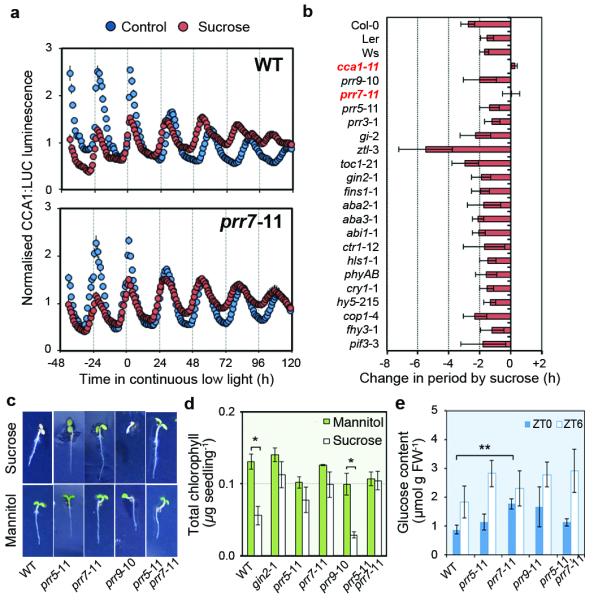
a, CCA1:LUC rhythms in wild-type and prr7-11 seedlings in continuous low light in the presence or absence of exogenous sucrose, (mean ± s.e.m.) n = 4. b, Change in period of CCA1:LUC rhythms in circadian, sugar and light signalling mutants, CAB2:LUC (Ws and cca1-11) or CCR2:LUC (toc1-21) grown in the presence of sucrose compared to control media in continuous low light (mean ± s.d.) n = 8. c, Seedlings germinated on 180 mM sucrose or mannitol. d, Total chlorophyll content of seedlings germinated on sucrose or mannitol, (mean ± s.d.) n = 3. d, Glucose content of seedlings, (mean ± s.d.) n = 3. * P < 0.05 ** P < 0.01 by t-test.
Our findings led us to test whether PRR7 might be more widely involved in sugar signalling. When germinated on media containing 180 mM (6% w/v) sucrose, prr7-11 mutants were resistant to repression of chlorophyll accumulation (Fig 4c, d), similar to that observed in glucose insensitive2/hexokinase1 (gin2-1) mutants18. In addition, prr7-11 contained elevated endogenous sugar concentrations around dawn (Fig. 4e) suggesting a role for PRR7 in regulating endogenous sugar accumulation. This is consistent with previous reports of involvement of PRR proteins in regulating chlorophyll biosynthesis and primary metabolism19–21.
Altered feeding cycles can influence phase of peripheral clocks in animals22,23. Similarly, it was previously suggested that a shoot-derived photosynthate might regulate a simplified circadian oscillator in Arabidopsis roots24. Photosynthesis contributes to entrainment by an unknown mechanism in the green algae, Chlamydomonas reinhardtii25. From analysis of the effects of altered photosynthates on free-running circadian rhythms and examining the role of PRR7, we have demonstrated that photosynthetically-derived sugars act to provide metabolic feedback that entrains the Arabidopsis circadian clock in shoots. We propose that following light-activation of PRR7 at dawn, accumulation of endogenous sugars from photosynthesis repress the PRR7 promoter, leading to de-repression of CCA1. Thus, PRR7 expression is coordinately modulated by light and photosynthesis, permitting PRR7 to act as a transcriptional repressor in circadian sugar signaling (Extended Data Fig. 1). This defines a novel metabolic feedback loop that contributes to circadian entrainment in plants.
Methods
Plant materials and growth methods
CCA1:LUC, TOC1:LUC, PRR7:LUC and CCR2:LUC are in Col-0 ecotype, GI:LUC, PRR9:LUC and CAB2:LUC are in Ws ecotype. CCA1:LUC was introduced into Ler and prr5-11, prr7-11, prr9-1026, prr3-127, gigantea (gi-2)28, zeitlupe (ztl-3)29, gin2-118, fructose insensitive 1 (fins1-1)30, abscisic acid deficient (aba2-1/gin1)31, aba3-1/gin532, abscisic acid insensitive 1 (abi1-1)33, constitutive triple response 1 (ctr1-12/gin4)34, hookless 1 (hls1-1)35, phytochrome A (phyA-201 phyB-5)36, cryptochrome 1 (cry1-1)37, long hypocotyl 5 (hy5-215)38, constitutive photomorphogenic 1 (cop1-4)39, far-red elongated hypocotyl 3 (fhy3-1)40, phytochrome interacting factor 3 (pif3-3)41, spindly (spy-3)42 by crossing. Ler/CCA1:LUC and gin2-1/CCA1:LUC were backcrossed to Ler or CCA1:LUC, respectively, two times. The gin2-1(Col-0) line was used for sugar-insensitivity experiments to allow direct comparison to prr mutants and Col-0. Surface-sterilised seeds were sown on half-strength Murashige & Skoog media (1/2 MS), pH 5.7 without sucrose and solidified with 0.8% (w/v) Bacto agar. After sowing, seeds were kept at 4°C in darkness for 2 d, then grown in 12 h light-12 h dark cycles under 50 μmol m−2 s−1 cool fluorescent white light at constant 19°C.
Photon counting experiments
Clusters of 5-10 seedlings were grown in 1/2 MS agar media and entrained in light-dark cycles (50 μmol m−2 s−1 light). For LUC measurement, seedlings were dosed twice with 1-2 mM D-luciferin between 12 and 48 h before commencing photon counting. Seedlings were released into continuous light after 7-11 d in light-dark cycles. Luminescence was detected for 800 s at each time point with an HRPCS4 (Photek) or a LB985 Nightshade (Berthold) camera. Delayed fluorescence17 was measured for 5 s in a LB985 Nightshade camera. During photon counting, light was supplied from red (660 nm) and blue (470 nm) LEDs at 50 μmol m−2 s−1 during light-dark cycles and either 50 μmol m−2 s−1 (continuous light) or 10 μmol m−2 s−1 (continuous low light). Where indicated, data was normalised to average counts across the experiment for each replicate. All period and relative amplitude error estimates where performed on rhythms between 24-120 h in continuous conditions on non-normalised data using Fast-Fourier Transformed Non-Linear Least Squares (FFT-NLLS) analysis, implemented in Biological Rhythms Analysis Software Suite (BRASS) (http://millar.bio.ed.ac.uk/PEBrown/BRASS/BrassPage.htm). All n values represent biological replicates, and all data are representative of independently repeated experiments. Tests were justified by determining minimum difference of means with a power of 0.9. Two-sided statistical tests, including assessment of normal distribution and equal variance, were performed in Excel.
Gating of short-term responses of LUC reporters to sugars at 1.5 h intervals for 24 h was performed in 8 d old seedlings from ZT0 in continuous low light. Signal was normalised to average signal across a time-course of several days. The change in normalised LUC reporter activity in seedlings before and after 3 h exposure to 90 mM sugars was subtracted from the change in normalised LUC reporter activity in untreated seedlings.
PRC experiments were performed in 8 d old seedlings from ZT0 in continuous low light. Seedlings growing on 1 μm nylon mesh on 1/2 MS were transferred to 1/2 MS containing 90 mM sugars for 3 h at 1.5 h intervals. Phase was determined based on the time of the circadian peaks following sugar pulses and the PRC was determined relative to phase in control seedlings after accounting for period differences as described14.
Treatments
Sugars were added to media for a final concentration of 90 mM (3% w/v sucrose) unless indicated otherwise. Chemical treatments were added to media at the following concentrations: 20 μM DCMU, 5 μM norflurazon, 220 μg ml−1 lincomycin. Seedlings were transferred to treatments 48-60 h before release into continuous conditions. CO2-free air was produced by pumping ambient air through self-indicating soda lime, a 0.45 μm filter and autoclaved deionized water into a sealed growth plate with an outlet. CO2 concentration of the air from the outlet was confirmed at < 1ppm using an infra-red gas analyser (ADC 255-MK3). For experiments with dark-adapted seedlings, sugars were added with a micropipette as ~ 0.1 vol of the growth media to give ~30 mM final concentration. For gating and PRC experiments, seedlings were transferred to media containing 90 mM sugars. Treatments in the dark were performed under dim green light.
Quantitative real-time PCR
Ten d old seedlings growing in light-dark cycles (50 μmol m−2 s−1) were transferred to treatments at dusk and leaf tissue was collected at 3 h intervals between 37 and 58 h later. Total RNA was extracted from three biological replicates of frozen leaf tissue using RNeasy Plant Mini Kit (Qiagen) and RNase-free DNase on-column treatment (Qiagen). cDNA was synthesised from 1 μg RNA with RevertAid First Strand cDNA Synthesis Kit (Fermentas) using oligo-dT primer. Technical replicates of gene specific products were amplified in 10 μL reactions using Rotor-Gene SYBR Green PCR Kit (Qiagen) on a Rotor-Gene 6000 real-time PCR machine fitted with a Rotor-Disc 100 (Qiagen). Primers were UBQ10-F 5′ GGCCTTGTATAATCCCTGATGAATAAG 3′ UBQ10-R 5′ AAAGAGATAACAGGAACGGAAACATAGT 3′ CCA1-F 5′ GATGATGTTGAGGCGGATG 3′ CCA1-R 5′ TGGTGTTAACTGAGCTGTGAAG 3′ LHY-F 5′ ACGAAACAGGTAAGTGGCGACATT 3′ LHY-R 5′ TGGGAACATCTTGAACCGCGTT 3′ PRR9-F 5′ CCACAGTAACGAATCAGAAGCAA 3′ PRR9-R 5′ TTGTCCAGCAATCCCCTCA 3′ PRR7-F 5′ GGAAACTTGGCGGATGAAAA 3′ PRR7-R 5′ CGAGGGCGTTGTTCTGCT 3′ PRR5-F 5′ CCGAATGAAGCGAAAGGACA 3′ PRR5-R 5′ GGATTGGACTTGACGAACG 3′. Relative transcript levels were determined by incorporating PCR efficiencies as described43.
Sugar and chlorophyll measurements
For soluble sugar measurements, 50-100 mg frozen tissue was extracted twice in 80% (v/v) ethanol and used immediately to determine sugar concentrations with a Sucrose/Fructose/D-Glucose Assay Kit (Megazyme). For chorophyll measurements, fresh tissue was extracted in methanol and concentrations determined as described44.
Supplementary Material
Extended Data Figure 1. A model for entrainment of the Arabidopsis circadian clock by photosynthetic sugars. From dawn, light activates PRR7 and drives photosynthesis. The concentrations of simple sugars produced from photosynthesis accumulate within the plant during the day (red dashed line), peaking around 4-8 h after dawn. High endogenous sugar concentrations lead to suppression of the PRR7 promoter, contributing to the phase of PRR7 rhythms. PRR7 is a transcriptional repressor of the circadian clock component CCA1. Thus, rhythms of endogenous sugars derived from photosynthesis contribute to circadian entrainment through PRR7. We propose that the timing of these events represent a ‘metabolic dawn’. Dawn is a time-dependent gradient of light intensity, whereas ‘metabolic dawn’ represents a gradient of increasing metabolite concentration. The metabolic gradient lags behind that of light and contributes to setting of the circadian clock. In the model, previously established relationships are shown by black connectors, novel relationships proposed in this work are shown by orange connectors.
Extended Data Figure 10. Effect of exogenous sucrose on circadian period in circadian, sugar and light signalling mutants. LUC reporter rhythms in circadian, sugar and light signalling mutants in continuous low light in media with or without exogenous sucrose (mean ± s.e.m.) n = 4. Reporter is CCA1:LUC in all lines except for Ws, cca1-11 (CAB2:LUC) and toc1-21 (CCR2:LUC). Period estimates are shown in blue (control) and red (sucrose) for each line (mean ± s.d.) n = 8.
Extended Data Figure 2. Effects of exogenous sucrose and inhibition of photosynthesis on circadian rhythms. a, b, Period estimates for rhythms of promoter LUC reporters in a, continuous low light or b, continuous light grown in media with or without sucrose added, (mean ± s.d.) n = 4. c, d, Promoter LUC rhythms (means ± s.e.m.) and relative amplitude error versus period plots for seedlings in media with or without DCMU in c, continuous low light or d, continuous light, n = 4.* P < 0.05 ** P < 0.01 *** P < 0.001 by t-test.
Extended Data Figure 3. Rhythms of endogenous sugars peak in the morning and are reduced by inhibition of photosynthesis. a, Leaf sucrose and glucose concentrations in 10 d old seedlings growing in a light-dark cycle, (mean ± s.d.) n = 3. b, Glucose, fructose and sucrose concentrations 4 h after subjective dawn in 13 d old seedlings grown in CO2-free air or ambient air in continuous low light for 5 d (mean ± s.d.) n = 3. c, Glucose concentration in 10 d old seedlings growing in a light-dark cycle 12-36 h after transfer to DCMU or control media at dusk (mean ± s.d.) n = 3. * P < 0.05 ** P < 0.01 by t-test compared to ZT0 in a and compared to control conditions in b and c.
Extended Data Figure 4. Effects of DCMU, norflurazon or lincomycin in CCA1:LUC rhythms in the presence or absence of exogenous sucrose. a, CCA1:LUC rhythms in continuous low light for seedlings transferred to media containing DCMU in the presence of the indicated exogenous sucrose concentrations compared to control media, (mean ± s.e.m.) n = 4. b, CCA1:LUC rhythms in continuous light for seedlings transferred to media containing DCMU, norflurazon or lincomycin in and absence (left) or presence (right) of exogenous sucrose, (mean ± s.e.m.) n = 4.
Extended Data Figure 5. Altering ROS production does not influence circadian rhythms. a, CAB2:LUC rhythms in seedlings transferred to continuous light and treated with 1 mM glutathione or 5 mM ascorbate. The short-period mutant toc1-1 and long-period mutant ztl-1 were included as positive controls, (means ± s.d.) n = 2-3. b, Relative amplitude error versus period plot for leaf movement rhythms in wild-type and NADPH oxidase rbohD,F mutants45 in continuous light (means ± s.e.m.). c, Promoter LUC rhythms and relative amplitude error versus period plots for seedlings in continuous light or continuous low light treated with 10 μM DPI or 0.1% (v/v) DMSO at 0 h, (mean ± s.e.m.) n = 4.
Extended Data Figure 6. Metabolically active sugars sustain circadian rhythms in darkness. a, CCA1:LUC rhythms in continuous dark in seedlings grown in media containing the indicated sugars or controls (mean ± s.e.m.) n = 4. b, Promoter LUC rhythms (mean ± s.e.m, n = 4) and relative amplitude error versus period plots (n = 4-8) for seedlings in continuous dark in media with or without sucrose added. Note that rhythms could not be detected in seedlings grown without sucrose for morning expressed CCA1:LUC or PRR9:LUC rhythms, but could be detected for evening expressed GI:LUC and TOC1:LUC, despite the low amplitude.
Extended Data Figure 7. Exogenous sugar can set circadian phase in dark-adapted seedlings. a, Time to first circadian peak of promoter LUC reporters in seedlings treated with sucrose after 72 h (subjective dawn, CT0) or 84 h (subjective dusk, CT12) in continuous dark (mean ± s.d.) n = 4. b, Promoter LUC rhythms of seedlings after sucrose or mannitol treament as in a, (mean ± s.e.m.) n = 4. c, CCA1 transcript level relative to UBQ10 in seedlings treated with sucrose or mannitol after 72 h in continuous dark (mean ± s.d) n = 3. ** P < 0.01 *** P < 0.001 by t-test.
Extended Data Figure 8. Phase-setting by sugar and light. a, Change in period of CCA1:LUC after pulses of sucrose compared to control seedlings in continuous low light (mean ± s.d.) n = 8. b, Phase response of TOC1:LUC to pulses of sucrose for seedlings in continuous low light (mean ± s.d.) n = 8. c, Phase response of CCA1:LUC to pulses of sucrose (reproduced data from Fig. 2c) or mannitol (mean ± s.d.) n = 8. d, LUC reporter rhythms (mean ± s.e.m.), time to circadian peak (mean ± s.d.) and period estimates (mean ± s.d.) in seedlings grown in continuous darkness for 72 h then transferred to continuous light or continuous low light, n = 4. e, CCA1:LUC rhythms (mean ± s.e.m.) and time to circadian peak in seedlings following transfer to, continuous light or continuous low light in control media or in the presence of DCMU or DCMU with sucrose after 72 h in continuous dark, n = 4. * P < 0.05 *** P < 0.001 by t-test.
Extended Data Figure 9. Regulation of the circadian clock by sugar requires PRR7. a, Change in PRR7:LUC luminescence after 3 h treatment with sucrose relative to control (mean ± s.d.), n = 4. Data were normalised across the time-series and change relative to untreated plants was plotted. b, Change in CCA1:LUC luminescence in wild type and prr7-11 after 3 h treatment with sucrose relative to control (mean ± s.d.), n = 8. Data were normalised across the time-series and change relative to untreated plants of the appropriate genotype was plotted. c, Period estimates of rhythms of delayed fluorescence in wild type and mutant seedlings in continuous low light in media with or without exogenous sucrose, (mean ± s.d.) n = 4. d, Phase response of CCA1:LUC to pulses of sucrose in prr7-11 seedlings in continuous low light (mean ± s.d.) n = 8. Compare to sucrose PRC for CCA1:LUC in wild type seedlings in Fig. 2c. * P < 0.05 ** P < 0.01 *** P < 0.001 by t-test compared to control in a and c and compared to wild type in b.
Acknowledgments
This work was supported by BBSRC grant BB/H006826/1. We thank J. O’Neill, J. Davies and J. Hibberd for comments on the manuscript.
Footnotes
Author information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks : an experimental assessment in Cyanobacteria. Curr. Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 2.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–3. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 3.Harmer SL. The circadian system in higher plants. Ann. Rev. Plant Biol. 2009;60:357–77. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 4.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–56. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 5.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–13. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y, Gehan JP, Sharkey TD. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005;138:2280–2291. doi: 10.1104/pp.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graf A, Schlereth A, Stitt M, Smith AM. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl Acad. Sci. USA. 2010;107:9458–9463. doi: 10.1073/pnas.0914299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noordally ZB, et al. Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science. 2013;339:1316–1319. doi: 10.1126/science.1230397. [DOI] [PubMed] [Google Scholar]
- 9.Knight H, Thomson AJW, McWatters HG. Sensitive to freezing6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol. 2008;148:293–303. doi: 10.1104/pp.108.123901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalchau N, et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl Acad. Sci. USA. 2011;108:5104–9. doi: 10.1073/pnas.1015452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bläsing OE, et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koussevitzky S, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–9. [PubMed] [Google Scholar]
- 13.Lai AG, et al. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl Acad. Sci. USA. 2012;109:17129–34. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson CH. Phase Response Curves: What can they tell us about circadian clocks? Circadian Clocks from Cell to Human. 1992:209–249. [Google Scholar]
- 15.Michael TP, Salome P. a, McClung CR. Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc. Natl Acad. Sci. USA. 2003;100:6878–83. doi: 10.1073/pnas.1131995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamichi N, et al. PSEUDO-RESPONSE REGULATORS 9,7 and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould PD, et al. Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J. 2009;58:893–901. doi: 10.1111/j.1365-313X.2009.03819.x. [DOI] [PubMed] [Google Scholar]
- 18.Moore B, et al. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light , and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- 19.Kato T, et al. Mutants of circadian-associated PRR genes display a novel and visible phenotype as to light responses during de-etiolation of Arabidopsis thaliana seedlings. Biosci., Biotech. and Biochem. 2007;71:834–839. doi: 10.1271/bbb.60642. [DOI] [PubMed] [Google Scholar]
- 20.Fukushima A, et al. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl Acad. Sci. USA. 2009;106:7251–7256. doi: 10.1073/pnas.0900952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamichi N, et al. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl Acad. Sci. USA a. 2012;109:17123–8. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Devel. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stokkan K. a, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–3. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 24.James AB, et al. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science. 2008;322:1832–5. doi: 10.1126/science.1161403. [DOI] [PubMed] [Google Scholar]
- 25.Johnson CH, Kondo T, Hastings JW. Action spectrum for resetting the circadian phototaxis rhythm in the CW15 strain of Chlamydomonas. Plant Physiol. 1991;97:1122–9. doi: 10.1104/pp.97.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 26.Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005;46:686–98. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 27.Para A, et al. PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell. 2007;19:3462–3473. doi: 10.1105/tpc.107.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park DH. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 29.Jarillo JA, et al. An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature. 2001;410:487–90. doi: 10.1038/35068589. [DOI] [PubMed] [Google Scholar]
- 30.Cho Y-H, Yoo S-D. Signaling role of fructose mediated by FINS1/FBP in Arabidopsis thaliana. PLoS Genetics. 2011;7:e1001263. doi: 10.1371/journal.pgen.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González-Guzmán M, et al. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell. 2002;14:1833–1846. doi: 10.1105/tpc.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–83. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung J, et al. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–52. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- 34.Gibson SI, Laby RJ, Kim D. The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem. Biophys. Res. Comm. 2001;280:196–203. doi: 10.1006/bbrc.2000.4062. [DOI] [PubMed] [Google Scholar]
- 35.Lehman a, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 1996;85:183–94. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- 36.Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and Phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 38.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng XW, et al. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 40.Hudson ME, Lisch DR, Quail PH. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 2003;34:453–71. doi: 10.1046/j.1365-313x.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- 41.Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–67. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 42.Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl Acad. Sci. USA. 1996;93:9292–6. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talke IN, Hanikenne M, Krämer U. Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol. 2006;142:148–67. doi: 10.1104/pp.105.076232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porra RJ, Thompson WA, Kreidemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents : verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta. 1989;975:384–394. [Google Scholar]
- 45.Kwak JM, et al. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003;22:2623–33. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Figure 1. A model for entrainment of the Arabidopsis circadian clock by photosynthetic sugars. From dawn, light activates PRR7 and drives photosynthesis. The concentrations of simple sugars produced from photosynthesis accumulate within the plant during the day (red dashed line), peaking around 4-8 h after dawn. High endogenous sugar concentrations lead to suppression of the PRR7 promoter, contributing to the phase of PRR7 rhythms. PRR7 is a transcriptional repressor of the circadian clock component CCA1. Thus, rhythms of endogenous sugars derived from photosynthesis contribute to circadian entrainment through PRR7. We propose that the timing of these events represent a ‘metabolic dawn’. Dawn is a time-dependent gradient of light intensity, whereas ‘metabolic dawn’ represents a gradient of increasing metabolite concentration. The metabolic gradient lags behind that of light and contributes to setting of the circadian clock. In the model, previously established relationships are shown by black connectors, novel relationships proposed in this work are shown by orange connectors.
Extended Data Figure 10. Effect of exogenous sucrose on circadian period in circadian, sugar and light signalling mutants. LUC reporter rhythms in circadian, sugar and light signalling mutants in continuous low light in media with or without exogenous sucrose (mean ± s.e.m.) n = 4. Reporter is CCA1:LUC in all lines except for Ws, cca1-11 (CAB2:LUC) and toc1-21 (CCR2:LUC). Period estimates are shown in blue (control) and red (sucrose) for each line (mean ± s.d.) n = 8.
Extended Data Figure 2. Effects of exogenous sucrose and inhibition of photosynthesis on circadian rhythms. a, b, Period estimates for rhythms of promoter LUC reporters in a, continuous low light or b, continuous light grown in media with or without sucrose added, (mean ± s.d.) n = 4. c, d, Promoter LUC rhythms (means ± s.e.m.) and relative amplitude error versus period plots for seedlings in media with or without DCMU in c, continuous low light or d, continuous light, n = 4.* P < 0.05 ** P < 0.01 *** P < 0.001 by t-test.
Extended Data Figure 3. Rhythms of endogenous sugars peak in the morning and are reduced by inhibition of photosynthesis. a, Leaf sucrose and glucose concentrations in 10 d old seedlings growing in a light-dark cycle, (mean ± s.d.) n = 3. b, Glucose, fructose and sucrose concentrations 4 h after subjective dawn in 13 d old seedlings grown in CO2-free air or ambient air in continuous low light for 5 d (mean ± s.d.) n = 3. c, Glucose concentration in 10 d old seedlings growing in a light-dark cycle 12-36 h after transfer to DCMU or control media at dusk (mean ± s.d.) n = 3. * P < 0.05 ** P < 0.01 by t-test compared to ZT0 in a and compared to control conditions in b and c.
Extended Data Figure 4. Effects of DCMU, norflurazon or lincomycin in CCA1:LUC rhythms in the presence or absence of exogenous sucrose. a, CCA1:LUC rhythms in continuous low light for seedlings transferred to media containing DCMU in the presence of the indicated exogenous sucrose concentrations compared to control media, (mean ± s.e.m.) n = 4. b, CCA1:LUC rhythms in continuous light for seedlings transferred to media containing DCMU, norflurazon or lincomycin in and absence (left) or presence (right) of exogenous sucrose, (mean ± s.e.m.) n = 4.
Extended Data Figure 5. Altering ROS production does not influence circadian rhythms. a, CAB2:LUC rhythms in seedlings transferred to continuous light and treated with 1 mM glutathione or 5 mM ascorbate. The short-period mutant toc1-1 and long-period mutant ztl-1 were included as positive controls, (means ± s.d.) n = 2-3. b, Relative amplitude error versus period plot for leaf movement rhythms in wild-type and NADPH oxidase rbohD,F mutants45 in continuous light (means ± s.e.m.). c, Promoter LUC rhythms and relative amplitude error versus period plots for seedlings in continuous light or continuous low light treated with 10 μM DPI or 0.1% (v/v) DMSO at 0 h, (mean ± s.e.m.) n = 4.
Extended Data Figure 6. Metabolically active sugars sustain circadian rhythms in darkness. a, CCA1:LUC rhythms in continuous dark in seedlings grown in media containing the indicated sugars or controls (mean ± s.e.m.) n = 4. b, Promoter LUC rhythms (mean ± s.e.m, n = 4) and relative amplitude error versus period plots (n = 4-8) for seedlings in continuous dark in media with or without sucrose added. Note that rhythms could not be detected in seedlings grown without sucrose for morning expressed CCA1:LUC or PRR9:LUC rhythms, but could be detected for evening expressed GI:LUC and TOC1:LUC, despite the low amplitude.
Extended Data Figure 7. Exogenous sugar can set circadian phase in dark-adapted seedlings. a, Time to first circadian peak of promoter LUC reporters in seedlings treated with sucrose after 72 h (subjective dawn, CT0) or 84 h (subjective dusk, CT12) in continuous dark (mean ± s.d.) n = 4. b, Promoter LUC rhythms of seedlings after sucrose or mannitol treament as in a, (mean ± s.e.m.) n = 4. c, CCA1 transcript level relative to UBQ10 in seedlings treated with sucrose or mannitol after 72 h in continuous dark (mean ± s.d) n = 3. ** P < 0.01 *** P < 0.001 by t-test.
Extended Data Figure 8. Phase-setting by sugar and light. a, Change in period of CCA1:LUC after pulses of sucrose compared to control seedlings in continuous low light (mean ± s.d.) n = 8. b, Phase response of TOC1:LUC to pulses of sucrose for seedlings in continuous low light (mean ± s.d.) n = 8. c, Phase response of CCA1:LUC to pulses of sucrose (reproduced data from Fig. 2c) or mannitol (mean ± s.d.) n = 8. d, LUC reporter rhythms (mean ± s.e.m.), time to circadian peak (mean ± s.d.) and period estimates (mean ± s.d.) in seedlings grown in continuous darkness for 72 h then transferred to continuous light or continuous low light, n = 4. e, CCA1:LUC rhythms (mean ± s.e.m.) and time to circadian peak in seedlings following transfer to, continuous light or continuous low light in control media or in the presence of DCMU or DCMU with sucrose after 72 h in continuous dark, n = 4. * P < 0.05 *** P < 0.001 by t-test.
Extended Data Figure 9. Regulation of the circadian clock by sugar requires PRR7. a, Change in PRR7:LUC luminescence after 3 h treatment with sucrose relative to control (mean ± s.d.), n = 4. Data were normalised across the time-series and change relative to untreated plants was plotted. b, Change in CCA1:LUC luminescence in wild type and prr7-11 after 3 h treatment with sucrose relative to control (mean ± s.d.), n = 8. Data were normalised across the time-series and change relative to untreated plants of the appropriate genotype was plotted. c, Period estimates of rhythms of delayed fluorescence in wild type and mutant seedlings in continuous low light in media with or without exogenous sucrose, (mean ± s.d.) n = 4. d, Phase response of CCA1:LUC to pulses of sucrose in prr7-11 seedlings in continuous low light (mean ± s.d.) n = 8. Compare to sucrose PRC for CCA1:LUC in wild type seedlings in Fig. 2c. * P < 0.05 ** P < 0.01 *** P < 0.001 by t-test compared to control in a and c and compared to wild type in b.