Abstract
Compelling evidence indicates that two autosomal recessive Parkinson’s disease genes, PINK1 (PARK6) and Parkin (PARK2), co-operate to mediate the autophagic clearance of damaged mitochondria (mitophagy). Mutations in the F-box domain containing protein Fbxo7 (PARK15) also cause early onset autosomal recessive Parkinson’s disease by an unknown mechanism. Here we show that Fbxo7 participates in mitochondrial maintenance through direct interaction with PINK1 and Parkin and plays a role in Parkin-mediated mitophagy. Cells with reduced Fbxo7 expression show deficiencies in Parkin mitochondrial translocation, ubiquitination of mitofusin 1 and mitophagy. In Drosophila, ectopic overexpression of Fbxo7 rescued loss of Parkin supporting a functional relationship between the two proteins. Parkinson’s disease-causing mutations in Fbxo7 interfere with this process, emphasising the importance of mitochondrial dysfunction in Parkinson’s disease pathogenesis.
Keywords: Fbxo7, Parkin, PINK1, mitofusin 1, mitophagy, Drosophila, Parkinson’s disease
Parkinson’s disease is the second most common neurodegenerative disorder. Mutations in several genes have been linked to familial forms of Parkinson’s disease, and studying their function has been valuable in highlighting pathways that may be important in the pathogenesis of familial and sporadic forms of the disease. Two of these, PINK1 (PTEN-induced putative kinase 1; PARK6), a mitochondrial kinase, and Parkin (PARK2), an E3-ubiquitin ligase, act in a common pathway to control mitochondrial turnover. It has been shown that upon treatment of cells with a mitochondrial uncoupler, carbonyl cyanide 3-chlorophenylhydrazone (CCCP), Parkin is recruited to depolarised mitochondria which enables their selective autophagic clearance (mitophagy)1. This recruitment requires the kinase activity of PINK12-4, which is cleaved from the mitochondria under basal conditions, but whose full-length form accumulates on depolarised mitochondria under CCCP treatment5-7. Parkin recruitment to mitochondria leads to ubiquitination of several targets including mitochondrial fusion proteins, mitofusins (Mfns)8-13, which promotes mitochondrial fragmentation prior to engulfment by the autophagosome14.
Mutations in the gene encoding F-box only protein 7 (Fbxo7) have been identified in a number of families with a severe form of autosomal recessive early onset Parkinson’s disease similar to that caused by mutations in PINK1 or Parkin15-17. F-box domain-containing proteins target substrates to SCF-type (Skp1-Cul1-F-box) E3-ubiquitin ligase complexes, usually by recruiting the substrate through a protein interaction domain and assembling a functional E3 ligase by associating with the adaptor protein Skp1 through an F-box domain18. Fbxo7 has a C-terminal proline rich substrate recruiting domain and also an N-terminal ubiquitin-like (Ubl) domain which is present in the major isoform of Fbxo7 but is spliced out in a second isoform (Fig. 1a). Fbxo7 has both SCF-dependent and SCF-independent activities19-22; however, little is understood of its function in Parkinson’s disease pathogenesis23. Here we demonstrate that Fbxo7 acts in a common pathway with Parkin and PINK1 to induce mitophagy in response to mitochondrial depolarisation and that disease-associated mutations in Fbxo7 interfere with this pathway. We also show that Fbxo7 expression rescues the phenotype of Drosophila parkin mutants, confirming that the genes function in a common mechanism in vivo.
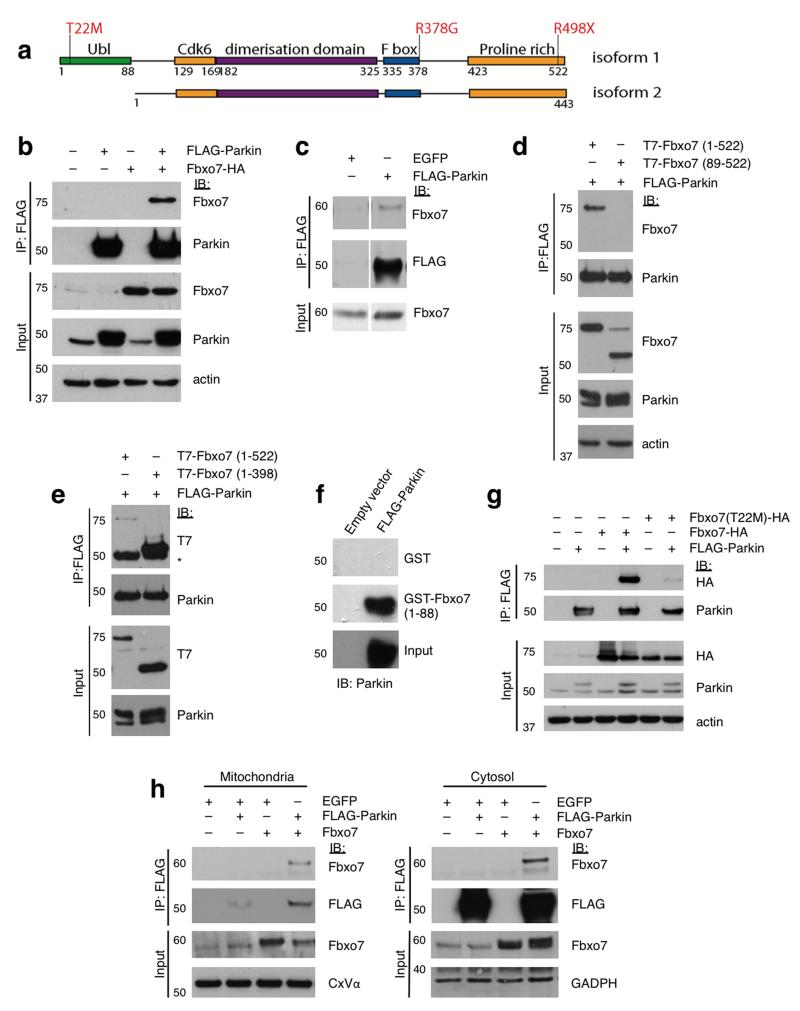
Figure 1. The amino-terminal Ubl domain of Fbxo7 interacts directly with Parkin.
a, Schematic diagram of Fbxo7 isoforms 1 and 2, showing the location of functional domains and the Parkinson’s disease-associated mutations. b, Co-immunoprecipitation of Fbxo7-HA and FLAG-Parkin in whole cell lysates from U2OS cells overexpressing both proteins. c, Co-immunoprecipitation of endogenous Fbxo7 with FLAG-Parkin in HEK293T cells transfected with FLAG-Parkin or a control protein (EGFP). d, FLAG-immunoprecipitation was repeated in U2OS cells transfected with FLAG-Parkin and either full length (1-522) T7-Fbxo7 or an N-terminal truncation lacking the Ubl domain (89-522). e, as with (d) using lysates from U2OS cells expressing FLAG-Parkin and T7-Fbxo7, either full length (1-522) or a C-terminal deletion of the proline rich region (1-398). * indicates IgG heavy chain. f, In vitro translated (IVT) FLAG-Parkin was incubated with bacterially expressed glutathione-S-transferase (GST) or GST fused to the Fbxo7 Ubl domain (1-88) immobilised on glutathione beads. Bead-bound proteins and inputs were analysed by immunoblotting with anti-Parkin antibodies. g, The disease-causing mutation T22M interferes with Fbxo7’s interaction with Parkin. Co-immunoprecipitation was performed as with (b) using lysates from U2OS cells expressing FLAG-Parkin and wild-type or T22M Fbxo7-HA. h, Co-immunoprecipitation of FLAG-Parkin and Fbxo7 in the mitochondrial and cytosolic fractions of HEK293T cells overexpressing both proteins. All representative western blots were performed at least three times. Full-length blots are presented in Supplementary Figure S9.
Results
Fbxo7 interacts directly with Parkin
Given the clinical overlap between patients with Fbxo7 and Parkin mutations, we investigated whether these two proteins might be functionally related. First, we tested whether they physically interact. In whole cell lysates extracted from U2OS osteosarcoma cells expressing Fbxo7-HA and FLAG-Parkin or in HEK293 cells expressing FLAG-Parkin, both exogenously expressed and endogenous Fbxo7 were detected in complex with Parkin after immunoprecipitation (Fig. 1b,c). Performing the reverse experiment, immunoprecipitating overexpressed untagged Fbxo7 and immunoblotting for FLAG-Parkin, confirmed their interaction (Supplementary Fig. S1a).
To identify the interacting region, T7-tagged Fbxo7 mutants lacking specific domains were tested using co-immunoprecipitation assays. The N-terminal 88 amino acids of Fbxo7 which comprise the Ubl domain appeared essential for an interaction (Fig. 1d), and this was supported by the finding that FLAG-Parkin interacted with isoform 1 of Fbxo7 but not with isoform 2, a naturally occurring splice variant of Fbxo7 which lacks the Ubl domain (Supplementary Fig. S1b). Conversely, the removal of the C-terminal proline rich region of Fbxo7 did not affect the interaction with Parkin (Fig. 1e). In vitro pull-down assays were also performed using a GST-Fbxo7 (1-88) construct which was found to be sufficient for binding, confirming that the Ubl domain of Fbxo7 directly mediates Parkin interaction (Fig. 1f). The Parkinson’s disease-associated mutation T22M lies within this domain (Fig. 1a), and this mutation significantly reduced Fbxo7 binding to Parkin by 88% ± 3.5% (Fig. 1g and Supplementary Fig. S1d). The R378G mutation caused a non-significant reduction in Parkin binding, and the R498X mutation had no effect (Supplementary Fig. S1c,d). Immunoprecipitating FLAG-Parkin from cytosolic and mitochondrial fractions revealed that although both proteins localise predominantly to the cytosol, the interaction occurred to a greater extent in the mitochondrial fraction (Fig. 1h).
Fbxo7 participates in Parkin recruitment to mitochondria
As Parkin has been shown to translocate to mitochondria upon treatment with the mitochondrial uncoupler CCCP1-4, we investigated whether Fbxo7 also relocates to depolarised mitochondria. Over a time-course of CCCP treatment, endogenous Fbxo7 levels were found to decrease steadily in the cytosolic fraction and increase concurrently in the mitochondrial fractions of from HEK293T cells (Fig. 2a) and SH-SY5Y cells (Supplementary Fig. S2a). Immunoprecipitating FLAG-Parkin complexes from the mitochondrial fraction of SH-SY5Y cells revealed an increase in Fbxo7 protein levels in complex with FLAG-Parkin after CCCP treatment, consistent with an increase in the association of both proteins with the mitochondria following depolarisation (Fig. 2b).
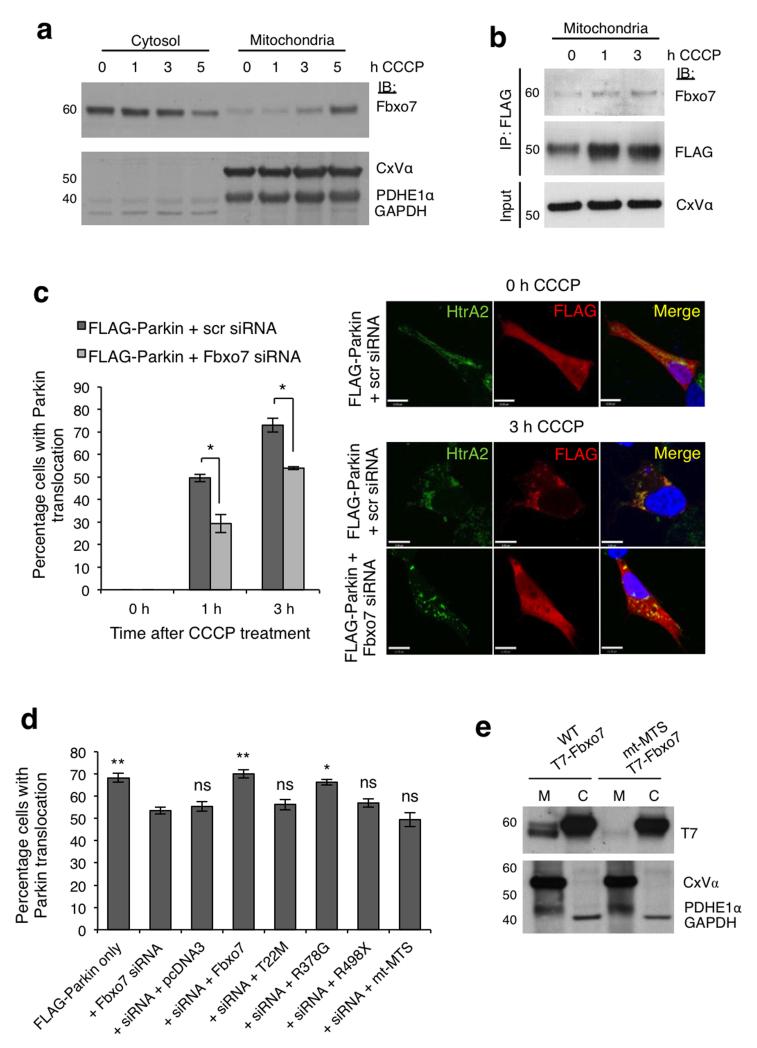
Figure 2. Fbxo7 participates in CCCP-induced accumulation of Parkin at the mitochondria.
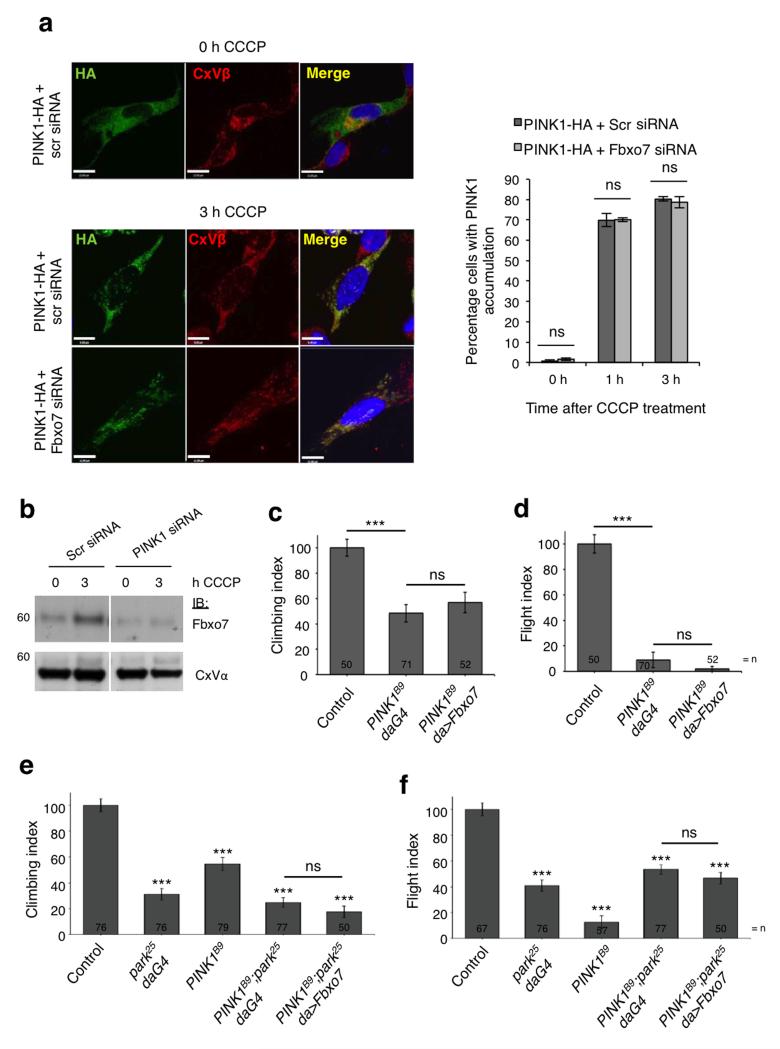
a, Fbxo7 relocates from the cytosolic to the mitochondrial fractions of HEK293T cells treated with CCCP (10 μ M). b, Fbxo7 levels are increased in FLAG-Parkin complexes immunoprecipitated from the mitochondrial fraction of HEK293T cells transfected with FLAG-Parkin and untagged Fbxo7 following 1 or 3 h treatment with CCCP (10 μM). c, Parkin localisation at the mitochondria was assessed by immunocytochemistry in SH-SY5Y cells transfected with FLAG-Parkin plus scrambled (scr) or Fbxo7 siRNA, following 1 or 3 h treatment with CCCP (10 μM). Cells were scored visually for the co-localisation of FLAG-Parkin with HtrA2, a mitochondrial marker. Representative images are displayed for cells transfected as indicated, following 0 or 3 h CCCP treatment. For corresponding images at 0 h and 1 h treatment, see Supplementary Figure S2c. Scale bar, 10 μm. d, Loss of FLAG-Parkin translocation upon Fbxo7 silencing is rescued by WT and R378G Fbxo7, but not by T22M Fbxo7, R498X Fbxo7, or by Fbxo7 in which the mitochondrial targeting sequence is mutated (mt-MTS). For c, and d, histograms indicate the percentage of cells in which Parkin localised to the mitochondria. Data are presented as mean of three experiments ± S.E.M. * p < 0.05, ** p < 0.01 compared to cells transfected with FLAG-Parkin plus Fbxo7 siRNA. e, T7-tagged WT Fbxo7 localises to both cytosolic (C) and mitochondrial (M) fractions of transfected HEK293T cells, but MTS mutant (mt-MTS) Fbxo7 localises only to the cytosolic fraction. All representative western blots were performed at least three times. Full-length blots are presented in Supplementary Figure S9.
We next examined whether Fbxo7 regulates Parkin translocation in SH-SY5Y cells co-transfected with FLAG-Parkin and either non-targeting, scrambled (Scr) siRNA or Fbxo7 siRNA, which substantially attenuates Fbxo7 expression (Supplementary Fig. S2b). At 1 and 3 h CCCP treatment, silencing Fbxo7 significantly reduced the percentage of cells in which Parkin localised to the mitochondria compared to control (Fig. 2c, Supplementary Fig. S2c, d), as scored by eye by an unbiased observer (see online methods). These data were independently confirmed by analysing the images using Pearson’s correlation co-efficient (Rr) (Supplementary Fig. S2e). The reduction in Parkin translocation upon Fbxo7 silencing was rescued by overexpression of WT or R378G mutant Fbxo7 but not by T22M or R498X mutant Fbxo7 (Fig. 2d), suggesting that in addition to binding Parkin through its N-terminal Ubl domain, Fbxo7 requires its C-terminal proline rich region to recruit Parkin to the mitochondria.
Although Parkin is known to localise to depolarised mitochondria it is not predicted to contain a mitochondrial targeting sequence (MTS), raising the possibility that this translocation could depend on its interaction with a mitochondrially-targeted protein. In silico analysis of the primary sequence of the Fbxo7 protein suggested that it may possess such an MTS at the N-terminus of isoform 1, and this prediction depended on two critical residues (Arg-2 and Arg-6). To investigate the relevance of this putative MTS, we produced both a T7-tagged and an untagged Fbxo7 construct in which both residues are mutated (hereafter designated mt-MTS), and found that while a portion of WT T7-Fbxo7 was found to localise to the mitochondrial fraction of SH-SY5Y cells, mt-MTS T7-Fbxo7 did not (Fig. 2e). Furthermore, CCCP-induced mitochondrial translocation of FLAG-Parkin was not rescued by expression of mt-MTS Fbxo7 following silencing of endogenous Fbxo7 (Fig. 2d), indicating that this MTS is necessary for Fbxo7’s effect on Parkin relocation. None of the three Parkinson’s disease-associated mutations affected the mitochondrial localisation of Fbxo7 (Supplementary Fig. S2f). Overall, these data indicate that Fbxo7 facilitates Parkin translocation to the mitochondria in response to depolarisation.
Overexpression of Fbxo7 rescues parkin mutant flies
To assess a functional relationship between Fbxo7 and Parkin in vivo we used a well-established Drosophila model, which has proven useful in uncovering the neurodegenerative and mitochondrial phenotypes in PINK1/parkin mutants and genetic dissection of this pathway24-27. Drosophila parkin mutants recapitulate several features of the disease phenotype including characteristic locomotor deficits in flight and climbing ability as well as dopaminergic neurodegeneration. The profound mitochondrial disruption evident in these mutants also leads to flight muscle degeneration and male sterility. Initial studies of transgenic lines bearing human Fbxo7 showed that ectopic expression of Fbxo7 caused no apparent phenotypes in a wild type background (Supplementary Fig. S3a-d). Notably, expression of Fbxo7 significantly rescued the parkin phenotypes, including locomotor defects, dopaminergic neuron loss, muscle degeneration, and mitochondrial disruption (Fig. 3a-f). In contrast, the expression of neither the pathogenic mutants T22M, R378G and R498X, nor mutants lacking a functional MTS (mt-MTS or isoform 2) provided any rescue of flight or climbing ability (Fig. 3g,h). These results show that human Fbxo7 isoform 1 can functionally substitute for loss of Parkin in vivo and that this rescue is abolished by pathogenic mutations, suggesting they share a common role in mitochondrial biology.
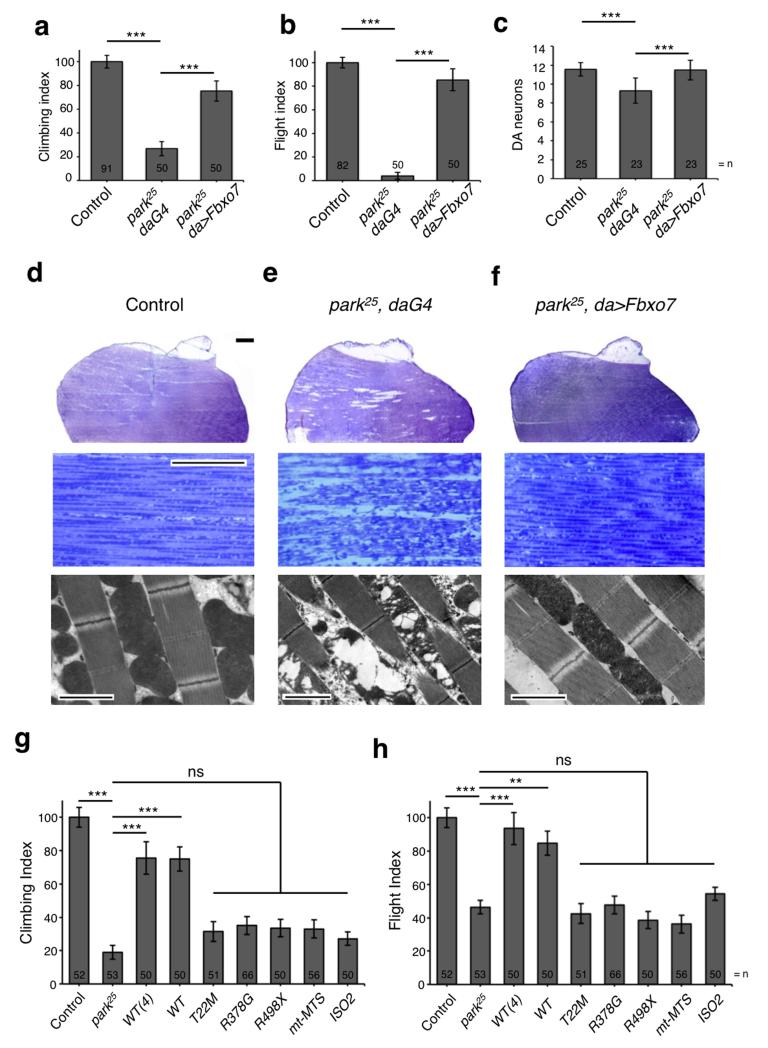
Figure 3. Expression of Fbxo7 rescues parkin mutant phenotypes.
a-b, Overexpression of Fbxo7 suppresses (a) climbing and (b) flight defects of parkin mutants. c, Overexpression of Fbxo7 also suppresses dopaminergic neurodegeneration in the PPL1 cluster of parkin mutants. d-f, (top and middle panels) Toluidine blue stained sections of adult thorax and (bottom panels) TEM images of muscle show that Fbxo7 overexpression suppresses muscle degeneration and mitochondrial disruption in parkin mutants. Toluidine blue scale bars show 200μm (top) and 20μm (middle). TEM scale bars show 2μm. Images are representative of three animals per genotype. g-h, Overexpression of Fbxo7 pathogenic mutants, mt-MTS or isoform 2 by da-GAL4 fails to rescue (g) climbing and (h) flight deficits in parkin mutants. Control genotype is park25/+; da-GAL4/+. Histograms indicate mean ± S.E.M. Significance was determined by one-way ANOVA with Bonferroni correction (*** p < 0.001, ** p < 0.01). For climbing and flight assays at least 50 flies were assessed.
We next sought to determine whether Drosophila encodes an Fbxo7 homologue that may be required for Parkin function. Previous studies have suggested Drosophila nutcracker (ntc) is an Fbxo7 orthologue with some conserved functions28,29. Although the F-box domain of ntc shares 59% identity and 65% similarity with Fbxo7, their overall sequence homology is low (28% identity, 45% similarity). In addition, ntc entirely lacks the proline rich region of Fbxo7, with which some substrates interact. If ntc functions with Parkin in Drosophila, similar loss-of-function phenotypes would be expected. While ntc mutant males are sterile28 they exhibit no overt disruption of the musculature or mitochondria, no loss of flight ability, or dopaminergic neurodegeneration (data not shown), hence do not phenocopy parkin mutants. We also found that expression of Fbxo7 was unable to rescue male sterility in these mutants (data not shown), indicating that ntc is not likely a functional homologue of Fbxo7.
Fbxo7 functionally interacts with PINK1
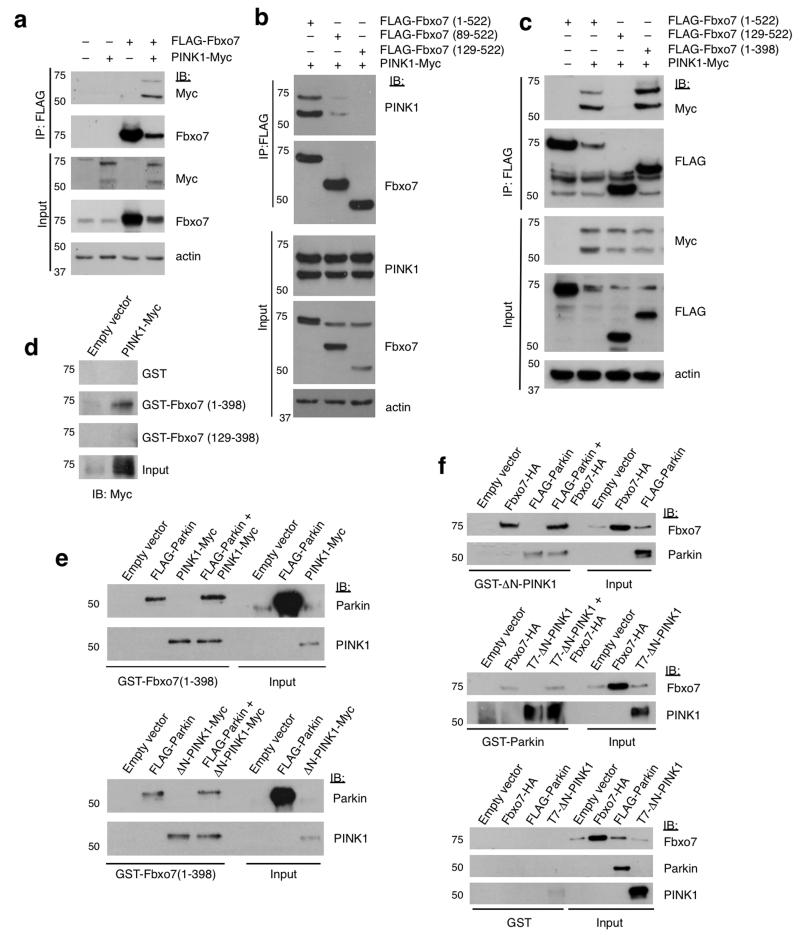
Parkin functions in a common pathway with PINK126,27,30,31, with Parkin translocation to the mitochondria following CCCP preceded by the accumulation of full-length PINK1 on the outer mitochondrial membrane (OMM). To investigate whether Fbxo7 also interacts with PINK1, U2OS cells were transfected with FLAG-Fbxo7 and PINK1-Myc and complexes were immunoprecipitated with FLAG beads. Both full-length and cleaved forms of PINK1 were found to co-immunoprecipitate with FLAG-Fbxo7 (Fig. 4a), and this was also true for the reciprocal immunoprecipitation (Supplementary Fig. S4a). This interaction was unaffected by Fbxo7 pathogenic mutations (Supplementary Fig. S4b). Immunoprecipitations using N- or C-terminal deletion mutants of Fbxo7 showed this interaction also mapped to the N-terminus of Fbxo7 (Fig. 4b,c). However, unlike Parkin, PINK1 retained partial binding to mutant Fbxo7 lacking the Ubl domain (89-522); a further truncation to amino acid 129 ablated Fbxo7 interaction with PINK1 (Fig. 4b). An in vitro binding assay using GST-Fbxo7 (1-398) or GST-Fbxo7 (129-398) with in vitro translated (IVT) PINK1-Myc verified that the PINK1 interaction required the first 129 amino acids and was direct (Fig. 4d). In support of these data, we found that PINK1 retained partial binding to Fbxo7 isoform 2, which contains amino acids 92-522 of isoform 1 (Supplementary Fig. S4c). These data suggest that PINK1 binding is mediated by sequences within amino acids 92-129 of Fbxo7 and raise the possibility that the interactions of Fbxo7, Parkin and PINK1 may be mutually exclusive or form ternary complexes. Fbxo7 was found to interact with both full-length PINK1 and an N-terminal truncation (ΔN-PINK1) in vitro (Fig. 4e), indicating that the first 103 amino acids of PINK1 are dispensable for the interaction. In vitro pull downs of GST-Fbxo7 with either full-length or ΔN-PINK1, Parkin or a combination of the two revealed that neither Parkin nor PINK1 had any significant effect on the interaction of Fbxo7 with the other protein (Fig. 4e). Performing the reciprocal pull down with GST-Parkin or GST-ΔN-PINK1 confirmed these findings (Fig. 4f). These data suggest that Fbxo7, Parkin, and PINK1 interact in a non-competitive, non-synergistic manner.
Figure 4. PINK1 interacts directly with the amino-terminus of Fbxo7.
a, Co-immunoprecipitation of PINK1-Myc and FLAG-Fbxo7 in whole cell lysates from U2OS cells overexpressing both proteins. b, Co-immunoprecipitation of PINK1-Myc with full length and two N-terminal deletions of FLAG-Fbxo7. PINK1-Myc is detected at low levels in complex with FLAG-Fbxo7(89-522) but not with FLAG-Fbxo7(129-522). c, as with (b) using lysates from U2OS cells expressing PINK1-Myc, and FLAG-Fbxo7 containing either N- or C-terminal truncations. d, In vitro pull down experiments were performed using in vitro translated (IVT) PINK1-Myc and either GST or GST fusions of Fbxo7 containing (1-398) or (129-398) immobilised on glutathione beads. e, Competitive binding assays using immobilised GST-Fbxo7(1-398) incubated with IVT FLAG-Parkin and/or full length (top panel) or N-terminally truncated (bottom panel) PINK1-Myc. Input and bead-bound proteins were analysed by immunoblotting as indicated. f, as with (e) immobilised GST-ΔN-PINK1 (top panel), GST-Parkin (middle panel) and immobilised GST alone (bottom panel) were incubated with combinations of IVT FLAG-Parkin, T7-ΔN-PINK1 and Fbxo7-HA as indicated. Input and bead-bound proteins were analysed by immunoblotting with the indicated antibodies. All western blots were performed a minimum of three times. Full-length blots are presented in Supplementary Figure S9.
Since Fbxo7 and PINK1 interact, we sought to determine whether Fbxo7 was also required for PINK1 stabilisation at the mitochondria after CCCP treatment. Consistent with previous reports3,4, PINK1-HA accumulates on mitochondria following CCCP treatment. However, cells co-transfected with PINK1-HA and Fbxo7 siRNA showed no significant effect on PINK1 accumulation compared to controls (Fig. 5a, Supplementary Figs. S5a and S5b), indicating that Fbxo7 is not required for PINK1 stabilisation. This was confirmed by immunoblot of both endogenous and overexpressed PINK1 (Supplementary Fig. S5c,d). To determine whether Fbxo7 instead functions downstream of PINK1 in the pathway, CCCP-induced relocation of endogenous Fbxo7 to the mitochondria was assessed by immunoblotting in cells transfected with scrambled or PINK1 siRNA, or in cells overexpressing PINK1-HA. Fbxo7 relocation was substantially reduced in cells transfected with PINK1 siRNA (Fig. 5b) and increased in cells overexpressing PINK1-HA (Supplementary Fig. S5e), indicating that PINK1 is required for its recruitment to the mitochondria.
Figure 5. Functional interaction of Fbxo7 with PINK1.
a, PINK1 localisation at the mitochondria was assessed by immunocytochemistry in SH-SY5Y cells transfected with PINK1-HA plus scrambled (scr) or Fbxo7 siRNA following 1 or 3 h treatment with CCCP (10 μM). Cells were scored visually for the co-localisation of PINK1-HA with complex V β subunit (CxVβ), a mitochondrial marker. Histograms indicate the percentage of cells in which PINK1-HA accumulated at the mitochondria. Data are presented as mean ± S.E.M., * p < 0.05. Representative images are displayed for cells transfected as indicated, following 0 or 3 h CCCP treatment. For corresponding images at 0 h and 1 h treatment, see Supplementary Figure 5a. Scale bar, 10 μm. b, Fbxo7 accumulation in the mitochondrial fraction following treatment with CCCP (10 μM) is impaired in SH-SY5Y cells transfected with PINK1 siRNA compared to scrambled siRNA (scr). Full-length blots are presented in Supplementary Figure S9. c-f, Overexpression of Fbxo7 does not rescue (c, e) climbing or (d, f) flight defects in PINK1 male mutants (PINK1B9) or PINK1:parkin double mutants (PINK1B9;park25, daG4). Control genotype is (c, d) PINK1B9/+;da-GAL4/+ and (e, f) da-GAL4/+. Histograms indicate mean ± S.E.M. Significance was determined by one-way ANOVA with Bonferroni correction (*** p < 0.001). For climbing and flight assays at least 50 flies were assessed.
To further investigate the functional relationship between Fbxo7 and PINK1 in vivo we again turned to Drosophila models in which PINK1 mutants phenocopy parkin mutants26,27. Fbxo7 expression was not able to rescue the PINK1 locomotor deficits, neuron loss, muscle degeneration or mitochondrial disruption (Fig. 5c,d, and Supplementary Fig. S6a,b), suggesting that Fbxo7 and PINK1 may lie in alternate pathways. However, using a PINK1-overexpression phenotypic assay previously used to study PINK1 genetic interactions7,32, we found that Fbxo7 does genetically interact with PINK1. While expression of Fbxo7 alone causes no phenotype, when co-expressed with PINK1 it substantially enhances the PINK1 overexpression ‘rough eye’ phenotype (Supplementary Fig. S6c).
These genetic interaction studies are consistent with Fbxo7 having a positive influence on the PINK1/Parkin pathway, but the inability of Fbxo7 to rescue PINK1 mutants suggests that its function may be dependent on PINK1 activity. To test this hypothesis directly, we assessed the ability of Fbxo7 expression to suppress phenotypes in PINK1:parkin double mutants. In contrast to the parkin mutants Fbxo7 was unable to rescue climbing and flight defects in the PINK1:parkin double mutant (Fig. 5e,f). Thus, even though overexpression of human Fbxo7 can suppress parkin mutants, Fbxo7 requires PINK1 activity for its protective function consistent with the data showing Fbxo7 requires PINK1 for its translocation to the mitochondria.
Fbxo7 participates in Mfn ubiquitination
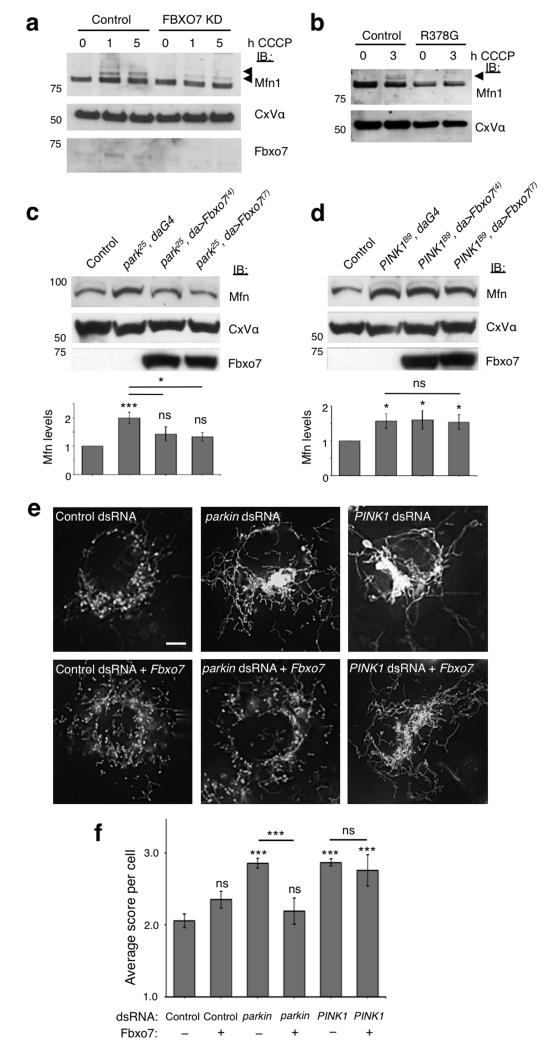
Recent studies have demonstrated that amongst a number of OMM proteins, Mfns are substrates for Parkin-mediated ubiquitination and degradation in response to CCCP treatment8-10,14. We therefore used ubiquitination of Mfn1 as an additional readout for activation of the Parkin-mediated mitophagy pathway. Treatment with CCCP results in the appearance of ubiquitinated forms of Mfns, as described previously8,10,12,14. Immunoblotting of mitochondrial fractions revealed that ubiquitination of Mfn1 was significantly reduced in stable Fbxo7 knockdown SH-SY5Y cells compared to control cells (Fig. 6a and Supplementary Fig. S7a; for level of knockdown see Supplementary Fig. S7b). Mfn1 ubiquitination was rescued by overexpression of WT Fbxo7 in the knockdown cells, confirming that this effect is caused by loss of Fbxo7 (Supplementary Fig. S7c). Moreover, expression of WT Fbxo7 rescued the loss of Mfn1 ubiquitination caused by knockdown of Parkin, but not PINK1 (Supplementary Fig. S7d). Mfn1 ubiquitination was additionally found to be reduced in patient fibroblasts homozygous for the R378G mutation (Fig. 6b), indicating that this mutation interferes with Fbxo7’s ability to facilitate Parkin-mediated Mfn1 ubiquitination. Basal Mfn protein levels frequently appeared reduced in Fbxo7 deficient cells compared to controls. This occurred despite a clear reduction in CCCP-induced ubiquitination which has previously been shown to be Parkin-dependent8-10,14, pointing to a possible additional effect of Fbxo7 on Mfn1 levels. Further experiments would be necessary in order to fully investigate this phenotype.
Figure 6. Fbxo7 promotes Mfn1 ubiquitination and restores Mfn levels and mitochondrial morphology in Parkin but not PINK1 deficient cells.
a-b, Ubiquitination of Mfn1 following treatment with CCCP (10 μM) is reduced in the mitochondrial fraction of both (a) SH-SY5Y cells stably expressing Fbxo7 shRNA (Fbxo7 KD) compared to an empty vector control line and (b) patient fibroblasts with homozygous R378G mutation compared to fibroblasts from healthy controls. Arrows indicate ubiquitinated Mfn1. c-d, Fbxo7 expression restores elevated Mfn steady state levels in (c) parkin but not (d) PINK1 mutant Drosophila. Histograms show mean ± S.E.M. of densitometry analysis of Mfn immunoblots above, normalised to Complex V α (CxV α). Control genotype is (c) park25/+;da-GAL4/+ and (d) PINK1B9/+;da-GAL4/+. e, Mitochondria in control Drosophila S2R+ cells stained with MitoTracker Red show a heterogeneous morphology, with a mixture of tubules and fragmented mitochondria. RNAi knockdown of parkin or PINK1 causes excessive fusion and elongated mitochondria compared to control dsRNA (C. elegans gene ZK686.3). Expression of Fbxo7 restores parkin but not PINK1 knockdown phenotype to wild type appearance. Scale bar shows 5 μm. f, Quantification of mitochondrial morphology in dsRNA treated cells. Score system; 1=fragmented, 2=wild type, 3=tubular, 4=hyper-fused (clumped). Histograms indicate mean ± S.E.M. Significance was determined by two-tailed Student t-tests (*** p < 0.001, * p < 0.05). All western blots were performed a minimum of three times and images are representative of 100 cells scored per condition. Full-length blots are presented in Supplementary Figure S9.
In Drosophila loss of parkin or PINK1 leads to an accumulation of unmodified Mfn and mitochondrial hyperfusion8. In agreement with the previous results, expression of Fbxo7 significantly restored the steady state level of Mfn in parkin mutants but was unable to do so in PINK1 mutants (Fig. 6c,d). Similar results were observed with RNAi knockdown of parkin and PINK1 in cultured Drosophila cells (Supplementary Fig. S7e). Consistent with these findings Fbxo7 could prevent mitochondrial hyperfusion in parkin but not PINK1 knockdown cells (Fig. 6e,f). Altogether these results provide further substantial support in vitro and in vivo that, similar to Parkin, Fbxo7 expression can regulate Mfn levels but is unable to do this in the absence of PINK1.
Fbxo7 plays a role in CCCP-induced mitophagy
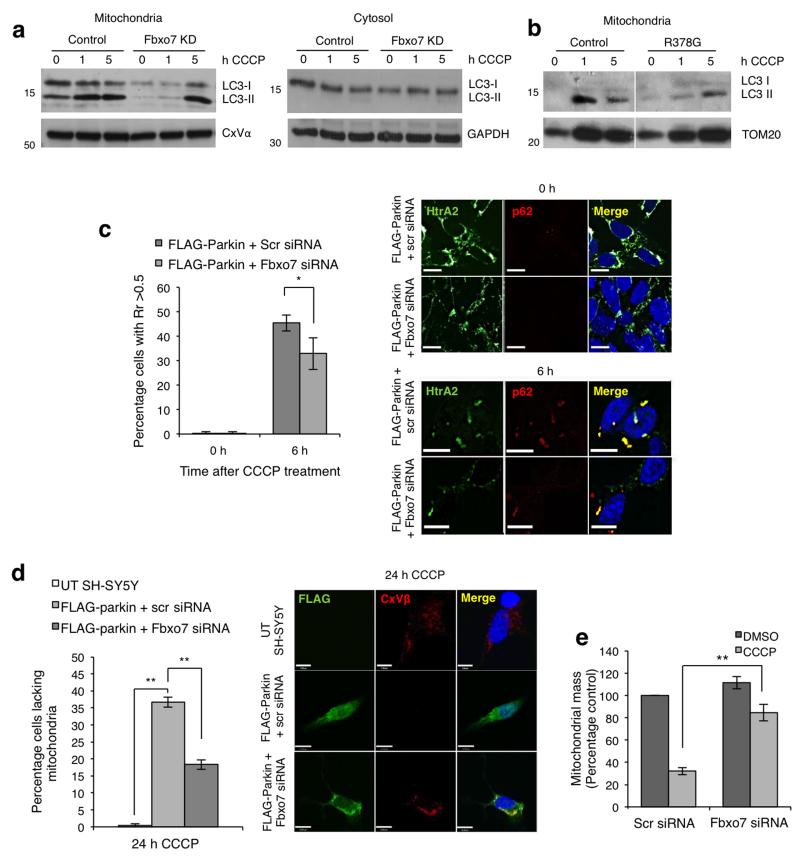
Parkin recruitment to the mitochondria has been previously shown to result in autophagic clearance of depolarised mitochondria following prolonged treatment with CCCP1. Since Fbxo7 appears to participate in Parkin recruitment to the mitochondria, we further investigated its effect on subsequent mitophagy. During induction of autophagy LC3-I is lipidated to its mature form LC3-II, which localises to the autophagosome as it engulfs the substrate for degradation33. Immunoblotting revealed that in control cells CCCP treatment results in a progressive increase in LC3-II at 1 and 5 h in mitochondrial but not cytosolic fractions, whereas in stable Fbxo7 knockdown SH-SY5Y cells formation of LC3-II at the mitochondria is substantially delayed (Fig. 7a). Similarly, the ratio of LC3-II to LC3-I was substantially lower in the mitochondrial fraction of patient fibroblasts compared to controls (Fig. 7b). LC3-II may be recruited to the mitochondria by the adaptor protein p62, which is also recruited to depolarised mitochondria in a Parkin-dependent manner4,34,35. Analysis of p62 localisation by both immunofluorescence and immunoblotting revealed that this accumulation was substantially reduced in cells transfected with Fbxo7 siRNA compared to controls (Fig. 7c and Supplementary Fig. S8a), suggesting that Fbxo7 participates in p62-dependent mitophagy.
Figure 7. Fbxo7 is important for mitophagy.
a, Treatment with CCCP (10 μM) results in an increase in LC3-II in the mitochondrial but not the cytosolic fraction of cells stably expressing the empty shRNA vector (control), and this is delayed in stable Fbxo7 knockdown SH-SY5Y cells. b, As in (a), an accumulation of LC3-II was observed in the mitochondrial fraction of healthy control fibroblasts following 1 and 5 h CCCP treatment, but this was reduced in fibroblasts from a patient carrying the R378G mutation. Western blots were performed a minimum of three times. c, Mitochondrial accumulation of p62 following CCCP treatment is inhibited by Fbxo7 siRNA. FLAG-Parkin overexpressing SH-SY5Y cells were transfected with scrambled (scr) or Fbxo7 siRNA as indicated and treated with either DMSO or CCCP (10 μM) for 6 h. Colocalisation of p62 with HtrA2, a mitochondrial marker, was assessed by Pearson’s correlation co-efficient (Rr) on a cell by cell basis. Histogram shows the percentage of cells in which Rr was greater than 0.5. Data are represented as mean ± S.E.M., * p < 0.05. Scale bar, 10 μm. d, Mitophagy was analysed in untransfected (UT) SH-SY5Y cells or in stable FLAG-Parkin overexpressing SH-SY5Y cells transfected with either scrambled (scr) or Fbxo7 siRNA. Histogram indicates the percentage of cells with no remaining mitochondria following 24 h treatment with CCCP (10 μM) for each condition. Complex V β subunit (CxVβ) was used as a mitochondrial marker. Data are presented as mean ± S.E.M., ** p < 0.01. Scale bar, 10 μm. e, Mitochondrial mass was measured in FLAG-Parkin overexpressing SH-SY5Y cells transfected with scrambled (scr) or Fbxo7 siRNA and treated for 24 h with either DMSO or CCCP (10 μM). For representative images, see Supplementary Fig. S8d. Full-length blots are presented in Supplementary Figure S9.
A decrease in levels of LC3-II may be explained by a decrease in autophagy or an increase in autophagic turnover36. We attempted to monitor autophagic flux by analysing p62 recruitment to mitochondria after treatment with CCCP and/or bafilomycin. However, we were unable to detect any significant increase in p62 recruitment when FLAG-Parkin SH-SY5Y cells were treated with CCCP and bafilomycin compared to those treated with CCCP alone (data not shown), indicating that this approach may not be appropriate to monitor mitophagy in this model. To specifically investigate mitochondrial clearance, we therefore employed an immunofluorescent assay as has been previously reported1,4. Consistent with these reports, a substantial proportion of SH-SY5Y cells overexpressing FLAG-Parkin were found to have no remaining mitochondria following 24 h CCCP treatment, while untransfected cells retained their mitochondria (Fig. 7d). This mitochondrial disappearance could be inhibited by bafilomycin (Supplementary Fig. S8b), which inhibits the lysosomal degradation of proteins, resulting in an accumulation of undegraded LC3-II in the mitochondrial fraction of SH-SY5Y cells following CCCP treatment (Supplementary Fig. S8c). This inhibition thus confirms that the mitochondrial disappearance observed is due to mitophagy in these cells. The percentage of cells with no remaining mitochondria following CCCP treatment was significantly lower in FLAG-Parkin overexpressing cells transfected with Fbxo7 siRNA compared to those transfected with scrambled siRNA (Fig. 7d). These data were further confirmed in live cells by measuring mitochondrial mass after 24 h CCCP treatment, using DsRed-Mito as an independent mitochondrial marker and calcein to measure cytosolic volume (Fig. 7e; for representative images, see Supplementary Fig. S8d). These data indicating Fbxo7 can promote mitophagy, coupled with the ability of Fbxo7 to substitute for Parkin in Drosophila, prompted us to test whether Fbxo7 could promote mitophagy in the absence of Parkin in mammalian cells. However, overexpression of Fbxo7 in HeLa cells, which do not express Parkin, failed to induced mitophagy following 24 h CCCP (data not shown). Taken together these results indicate that in mammalian cells Fbxo7 acts to promote mitophagy in a Parkin-dependent manner but is insufficient to induce mitophagy in the absence of Parkin.
Discussion
In the last few years, several studies have identified a molecular pathway for the clearance of damaged mitochondria involving the Parkinson’s disease-associated proteins PINK1 and Parkin5. Our data now show that a third protein associated with Parkinson’s disease, Fbxo7, also plays a role in this pathway. We find that upon CCCP treatment, cytosolic Fbxo7 relocalises to the mitochondria. Furthermore, Fbxo7 interacts physically and genetically with Parkin and the loss of Fbxo7 expression results in a significant inhibition of Parkin recruitment to depolarised mitochondria and subsequent mitophagy.
Importantly, our data show that three Parkinson’s disease-causing mutations in Fbxo7 (T22M, R378G and R498X) interfere with this function in both flies and mammalian cells. The T22M mutation was found to prevent the interaction of Fbxo7 with Parkin, and was consequently unable to rescue Parkin translocation to the mitochondria. The R378G mutation located close to the F-box domain (Fig. 1a) impairs ubiquitination of Mfn1 but not the ability of Fbxo7 to recruit Parkin to damaged mitochondria, indicating that Fbxo7’s role in the PINK1-Parkin mitophagy pathway is not limited to the recruitment of Parkin but that it also participates in substrate ubiquitination. The R378G mutation was previously shown to impair Fbxo7’s interaction with Skp1 and through it, the SCF complex37, raising the possibility that Fbxo7 may facilitate Parkin-dependent ubiquitination in an SCF-dependent manner. Finally, the R498X truncation is unable to recruit Parkin to the mitochondria, indicating an involvement of the C-terminal proline rich region of the protein in this step. This domain is involved in forming protein-protein interactions but our data show that it does not participate in the interaction with Parkin, suggesting that another protein(s) interacting with this domain of Fbxo7 is important for recruitment of Parkin to the mitochondria.
The precise mechanism by which Parkin, a cytosolic protein, translocates to depolarised mitochondria remains poorly defined despite a clear dependence on the mitochondrial localisation of PINK12-4. We have identified an MTS in the N-terminus of Fbxo7 and find that this is essential for its recruitment of Parkin to the mitochondria, raising the possibility that Fbxo7 may participate in the physical translocation of Parkin to mitochondria. In support of this hypothesis, when both proteins were overexpressed in cells the interaction was found to occur in both cytosolic and mitochondrial fractions.
It is worth noting Drosophila parkin mutants completely lack Parkin protein, ruling out the possibility that the observed rescue by human Fbxo7 could arise from an increased recruitment of residual parkin to the mitochondria and suggesting that Fbxo7 may cooperate with another E3 ubiquitin ligase which remains to be identified. The finding that none of the three pathogenic mutants were able to rescue loss of parkin mutants in vivo further supports the idea that this activity to maintain mitochondria homeostasis is defective in PARK15 patients.
We find that Fbxo7 functions downstream of PINK1 in mammalian cells since Fbxo7 relocation was PINK1 dependent, but that Fbxo7 overexpression fails to rescue the effects of PINK1 silencing in mammalian cells or in Drosophila. This failure to rescue loss of PINK1 may be because Fbxo7’s role in the pathway absolutely requires PINK1 activity, for example if phosphorylation of either Fbxo7 itself or its substrate(s) by PINK1 is necessary for Fbxo7 function at the mitochondria. PINK1 has been reported to phosphorylate Parkin but not Fbxo738, suggesting that the latter hypothesis may be more likely.
In conclusion, this study demonstrates for the first time that Fbxo7 participates in Parkin recruitment to damaged mitochondria, Mfn1 ubiquitination and mitophagy. This finding places further emphasis on the importance of mitochondrial turnover in neuroprotection and as a contributing factor in Parkinson’s disease.
Online methods
In silico sequence analysis
Sequences were examined for putative mitochondrial targeting sequences using PSORT39 (http://psort.hgc.jp/form2.html), Mitoprot40 (http://ihg.gsf.de/ihg/mitoprot.html), Target P41 (http://www.cbs.dtu.dk/services/TargetP/) and Predotar36 (http://urgi.versailles.inra.fr/predotar/predotar.html).
Cell culture and construct generation
SH-SY5Y neuroblastoma and U2OS osteosarcoma cells were obtained from ECACC and were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 4.5 g/L glucose and supplemented with 10% heat inactivated foetal calf serum (FCS) and 5 mM glutamine in a humidified chamber at 37°C with 5% CO2. Transient Fbxo7 knockdown was achieved by transfecting the cells with a pool of four siRNA constructs (siGenome, Dharmacon), while clonal Fbxo7 knockdown SH-SY5Y and U2OS cell lines were produced by retroviral infection with two independent miR30-based short hairpin constructs against either bp 387-404 or bp 444-460 of Fbxo7 isoform 1 (constructs generated in-house42). Silencing was maintained in stable cell lines by the addition of 1 μg/mL puromycin to the growth medium. Where indicated, cells were treated with 10 μM CCCP for the indicated times before harvesting lysates or fixing for immunostaining.
Drosophila S2R+ cells were cultured in Schneider’s medium (Invitrogen) supplemented with 5% fetal calf serum (Sigma) and 1% penicillin-streptomycin (Invitrogen-Gibco). Cells were transfected using Effectene reagent (Qiagen) following manufacturer’s instructions and collected after 24-48hrs.
Constructs for the expression of untagged Fbxo7 and T7-tagged full-length and truncated Fbxo7 have been described previously20. Deletion of the Ubl domain was achieved by PCR amplification of the region corresponding to 89-522 and cloned into pcDNA3 containing T7 or FLAG epitopes at the N-terminus. T22M, R378G and R498X mutations were introduced by site directed mutagenesis. Mutations in the MTS (R2D and R6W) were introduced by PCR and the product subcloned into pcDNA3. WT or mutant Fbxo7-HA was subcloned from the appropriate untagged construct and inserted into pcDNA3. The FLAG-Parkin construct and overexpressing SH-SY5Y cell line were kind gifts from Dr Helen Ardley (Leeds Institute of Molecular Medicine) and have been described previously43. GST-Parkin and ΔN-PINK1 expression vectors were generated by subcloning full-length Parkin or PINK1(104-581) into pGEX-KG. The GST fusion to the Ubl domain of Fbxo7 was done by PCR amplification and subcloning in pGEX-KG. The PINK1-HA construct was obtained through collaboration with Dr Emma Deas (UCL Institute of Neurology) and has been described previously5. The PINK1-myc construct has been described previously44.
Patient fibroblasts
With ethical approval (project number 07/H0720/161) and informed consent, punch biopsies were taken from the upper arm of a control and an affected individual with the Fbxo7 R378G mutation using a standard technique. This tissue was dissected and the fibroblasts cultured in DMEM containing L-glutamine supplemented with 10% FCS.
Immunoprecipitation and immunoblotting
Immunoprecipitation and immunoblotting methods were described previously44. Antibodies used were as follows: anti-FLAG M2 beads (Sigma A2220); EZview Red anti-HA affinity gel (Sigma E6779); anti-Fbxo7 (described previously20, 1:1000 for WB; or Aviva ARP43128, 1:2000 for WB); anti-HA (Roche 11867423001, 1:5000 for WB); anti-Parkin (Cell Signalling 4211, 1:1000 for WB); anti-Mfn1 (Abcam ab57602, 1:1000 for WB); anti-T7 (Novagen 69522-3, 1:5000 for WB); anti-Myc (Cell Signalling 2272, 1:1000); anti-Complex Vα (1:5000, Abcam ab14730); anti-GAPDH (Abcam ab8245, 1:2000); anti-LC3 (Novus NB100-2220, 1:1000); anti-p62 (Abcam ab56416, 1:5000); anti-Mfn (Drosophila) (described previously8). Where indicated, whole mitochondria were isolated by centrifugation45, except where Fbxo7 relocalisation was assayed by immunoblotting, in which case mitochondria were isolated as described10. Mitochondrial and cytosolic fractions were tested for cross-contamination by immunoblotting using Apotrack antibody cocktail (Abcam ab110415, 1:1000) to confirm that GAPDH was exclusively cytosolic and complex Vα and pyruvate dehydrogenase were exclusively mitochondrial. In vitro protein interactions were performed as described previously37. In brief, GST-tagged proteins were expressed in FB810 E. coli and immobilised on glutathione-Sepharose beads. These were incubated for 4 h with recombinant proteins that were produced by in vitro translation (IVT) in reticulocyte lysates (TNT T7 Quick Coupled Transcription/Translation System; Promega). Bound proteins were analysed by immunoblotting for the indicated proteins. Data collection and analysis were not performed blind to the conditions of the experiments.
Immunofluorescence
Cells were fixed with 4% PFA/PBS solution for 10 min at room temperature, then permeabilised using 0.5% Triton X-100/PBS solution for 5 min. Samples were blocked for 30 min in 10% FCS/PBS solution before addition of the primary antibody for 2 h in blocking solution. Antibodies used were: mouse anti-FLAG (Sigma F3165, 1:2000); rabbit anti-FLAG (Sigma F7425, 1:5000); rat anti-HA (Roche 11867423001, 1:500); rabbit anti-HtrA2 (Cell Signaling 2176, 1:1000); mouse anti-complex Vβ subunit (Abcam ab14730, 1:500) and mouse anti-p62 (BD Transduction Labs 610833, 1:500). The appropriate secondary antibodies (AlexaFluor 488, 568 and 633 secondary antibodies from Invitrogen, 1:2000) were added in 10% FCS/PBS for 30 min, then cell nuclei were stained with 1 μM DAPI for 5 min.
To quantify accumulation of Parkin/PINK1 at the mitochondria or mitochondrial clearance by mitophagy, cells were scored visually by an observer who was blinded to the identity of each coverslip. In order to score positively for mitochondrial accumulation, a cell must exhibit a clear mitochondria distribution in the FLAG/HA stain with negligible fluorescence in the cytosol (see representative images, Fig. 2c and 5a). In order to score positively for mitochondrial clearance a cell must contain no staining for the integral mitochondrial protein CxVβ (see representative images, Fig. 7d). In all cases a minimum of 100-150 cells were scored per coverslip and each experiment was performed at least three times. Where indicated in the text, these results were confirmed by repeating the experiment in cells stably overexpressing FLAG-Parkin or PINK1-HA, then quantifying the colocalisation of FLAG or HA with mitochondrial markers by Pearson’s correlation co-efficient (Rr) on a cell-by-cell basis using the software Colocalizer Pro (CoLocalization Research Software,www.colocalizer.com). The percentage of cells in which Rr exceeded 0.5 was then calculated for a minimum of 100-150 cells per coverslip.
Drosophila brains were dissected from 30-day old flies and immunostained with anti-tyrosine hydroxylase (Immunostar Inc. 22491) as described previously25. Brains were imaged with an Olympus FV1000 confocal with SIM-scanner on a BX61 upright microscope. Tyrosine hydroxylase-positive neurons were counted under blinded conditions.
Measurement of mitochondrial mass
To measure mitochondrial mass (mitochondrial volume as a percentage of cellular volume) in live cells, FLAG-Parkin overexpressing SH-SY5Y cells were transfected with DsRed-Mito and either scrambled or Fbxo7 siRNA and treated with CCCP for 24 h. The media was then replaced with HBSS containing 5 μM calcein-AM (Molecular Probes, Invitrogen) plus 0.005% pluronic for 30 min at room temperature. Cells were washed twice with fresh HBSS before Z-stack confocal images were taken using a Zeiss 710 vis CLSM equipped with a META detection system and a 63× oil immersion objective, with an excitation/emission of 495/515 nm for calcein and 558/583 nm for DsRed-Mito. Images were analysed using Volocity image analysis software (PerkinElmer) to measure mitochondrial and cytosolic volume.
Drosophila stocks and procedures
Drosophila were raised under standard conditions at 25°C on agar, cornmeal and yeast food. park25 mutants have been described previously24. PINK1B9 mutants26 were provided by J. Chung (KAIST). w1118 and da-GAL4 strains were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). UAS-Fbxo7 lines were constructed by cloning the entire human Fbxo7 open reading frame (isoform 1, WT, mt-MTS and pathogenic variants or isoform 2) into pUAST-attB vector (BestGene Inc.). Site-directed integration was targeted to attP site ZH-51C (Bloomington stock, 24482). WT Fbxo7 was also cloned into pUAST vector for germline transformation by random insertion to allow variable expression levels (numbered Tg lines). For all integration events, multiple independent lines were isolated and assessed. Flight and climbing assays were performed as previously described24. For all the experiments, unless otherwise indicated, a mixture of genders were used. Data collection and analysis were not performed blind to the conditions of the experiments unless otherwise indicated.
Fertility assay
At least 30 single 3-4 days old male flies were mated with 2-3 virgin w1118 female flies reared on normal food. After 5-7 days of mating, the fraction of fertile flies was determined by the presence of larvae46.
Histology
Tissue sectioning and TEM: Thoraces were prepared from 5-day old adult flies and treated as previously described24. Semi-thin sections were then taken and stained with Toluidine blue, while ultra-thin sections were examined using a TEM (FEI tecnai G2 Spirit 120KV).
Light microscopy imaging and scanning electron microscopy
Light microscopy imaging was assessed using a Nikon motorized SMZ stereo zoom microscope fitted with 1x Apo lens. Extended focus images were then generated using Nikon Elements software, using the same settings for all the genotypes. Flies were anaesthetised with CO2 during the process. Scanning electron microscopy (SEM) was performed according to a standard protocol47. All animals of a given genotype displayed essentially identical phenotypes and randomly selected representative images are shown. Images were taken using a SEM microscope (Philips XL-20 SEM).
Mitochondrial morphology assay
Double-stranded RNAs (dsRNAs) were prepared using the MEGA script kit (Ambion). Primers used to generate dsRNAs are described as follows: Control - C. elegans (ZK686.3) – TGATTGTCGCCAACTCTCAC and GTGGGTTCAGCTTCTTTCCA; parkin – CATGTCACCTTGCGACAATC and ACCTGTATCGACTGCTTCCG; PINK1 – CAATTAACCCGCATCCAATC and TCCAAGTCATCGATGGTGAA. 1.5 μg dsRNA probes were spiked into μ-Slide 8-well imaging chambers (Ibidi, 80826). A total of 80,000 Drosophila SR2+ cells were seeded in serum-free Schneider’s Drosophila medium (Invitrogen Gibco, 21720024) and incubated at 25°C for 1 h. Equal volumes of 2× FBS-containing Schneider’s Medium (Sigma-Aldrich F4135) volumes of 2× FBS-containing Schneider’s Medium (Sigma-Aldrich F4135-500ML) was added to each well and cells were incubated for a further 24 h. At that time, cells were transfected using Effectene (Qiagen, 301425) according to manufacturers’ methods with pAct-PPA::GFP plasmid plus either pAc5.1 empty vector or pAc5.1::Fbxo7, at a ratio of 1:3. After 3 days, 50 nM MitoTracker© Red CMXRos (Invitrogen, M7512) and 2 μ g/mL Hoechst 33342 (Invitrogen) was added for 15 minutes, and then replaced with fresh media for imaging. At least 10 live images per condition were taken on a Deltavision RT system (Applied Precision, Inc. Washington) using an Olympus 60× PlanApoN (1.42 NA) objective. All images were coded and scored with the experimenter blind to the treatment. Cells were scored on a cell-by-cell basis using a 4-point scale where 1=fragmented, 2=wild-type, 3=tubular and 4=hyper-fused (clumped).
Statistical analysis
To analyse the effect of Fbxo7 siRNA compared to control on Parkin translocation, PINK1 accumulation, Mfn1 ubiquitination, p62 recruitment and mitochondrial mass, p-values were calculated using an unpaired homoscedastic t-test, where n is taken to be the number of independent experiments (at least three in all cases). In all cases homoscedasticity was first confirmed using an F-test. Where multiple groups were compared, statistical significance was calculated by one-way ANOVA with a post-hoc Bonferroni correction. This test was applied to: the rescue of Parkin translocation by WT and mutant Fbxo7; Drosophila flight and climbing assays; survival of dopaminergic neurons; mitochondrial morphology in Drosophila; and immunofluorescent analysis of mitophagy. All tests performed were two-sided, and normality was assumed although not formally tested. All statistical significance was calculated at p = 0.05, using GraphPad Prism 5. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications1,4. For all the analysis, samples were collected and processed simultaneously and therefore no randomization was appropriate.
Supplementary Material
Acknowledgements
We would like to thank Mohadeseh Mehrabian and Elahe Elahi for obtaining patient biopsies, Helen Ardley for donating the FLAG-Parkin construct and cell line, Kira Holmström and Adrian Isaacs for helpful discussions and Matthew Gegg for technical advice. We thank Chris Hill for technical assistance within the University of Sheffield Electron Microscopy Facility. We thank the Herman Steller lab for kind provision of the ntc mutant lines.
The work was funded by a Wellcome/MRC Parkinson’s Disease Consortium grant to UCL/IoN, the University of Sheffield and the MRC Protein Phosphorylation Unit at the University of Dundee (grant number WT089698), an MRC Career Development Award (G0700183; H.P-F.), the MEFOPA project funded through the European Union FP7 research program (A.J.W.), an ERC Starting Grant (no. 309742; A.J.W). This research was supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. V.S.B. is funded by the MRC Doctoral Training Account. D.E.N. is funded by the BBSRC (BB/F012764/1; BB/J007846/1). The Wellcome Trust is acknowledged for support of the Sheffield Light Microscopy Facility (GR077544AIA). The MRC Centre for Developmental and Biomedical Genetics is supported by Grant G070091.
Footnotes
The authors declare no competing financial interests.
References
- 1.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–83. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 5.Deas E, Wood NW, Plun-Favreau H. Mitophagy and Parkinson’s disease: the PINK1-parkin link. Biochim Biophys Acta. 2011;1813:623–33. doi: 10.1016/j.bbamcr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin SM, et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–42. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitworth AJ, et al. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson’s disease factors Pink1 and Parkin. Dis Model Mech. 2008;1:168–74. doi: 10.1242/dmm.000109. discussion 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–23. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gegg ME, et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–70. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka A. Parkin-mediated selective mitochondrial autophagy, mitophagy: Parkin purges damaged organelles from the vital mitochondrial network. FEBS Lett. 2010;584:1386–92. doi: 10.1016/j.febslet.2010.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rakovic A, et al. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS One. 2011;6:e16746. doi: 10.1371/journal.pone.0016746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarraf SA, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013 doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–80. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shojaee S, et al. Genome-wide linkage analysis of a Parkinsonian-pyramidal syndrome pedigree by 500 K SNP arrays. Am J Hum Genet. 2008;82:1375–84. doi: 10.1016/j.ajhg.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Fonzo A, et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–5. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 17.Paisan-Ruiz C, et al. Early-onset L-dopa-responsive parkinsonism with pyramidal signs due to ATP13A2, PLA2G6, FBXO7 and spatacsin mutations. Mov Disord. 2010;25:1791–800. doi: 10.1002/mds.23221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–19. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 19.Hsu JM, Lee YC, Yu CT, Huang CY. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis by a proline-rich region. J Biol Chem. 2004;279:32592–602. doi: 10.1074/jbc.M404950200. [DOI] [PubMed] [Google Scholar]
- 20.Laman H, et al. Transforming activity of Fbxo7 is mediated specifically through regulation of cyclin D/cdk6. EMBO J. 2005;24:3104–16. doi: 10.1038/sj.emboj.7600775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuiken HJ, et al. Identification of F-box only protein 7 as a negative regulator of NF-kappaB signalling. Journal of cellular and molecular medicine. 2012;16:2140–9. doi: 10.1111/j.1582-4934.2012.01524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirk R, et al. Structure of a conserved dimerization domain within the F-box protein Fbxo7 and the PI31 proteasome inhibitor. J Biol Chem. 2008;283:22325–35. doi: 10.1074/jbc.M709900200. [DOI] [PubMed] [Google Scholar]
- 23.Chang YF, Cheng CM, Chang LK, Jong YJ, Yuo CY. The F-box protein Fbxo7 interacts with human inhibitor of apoptosis protein cIAP1 and promotes cIAP1 ubiquitination. Biochem Biophys Res Commun. 2006;342:1022–6. doi: 10.1016/j.bbrc.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 24.Greene JC, et al. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–83. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitworth AJ, et al. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc Natl Acad Sci U S A. 2005;102:8024–9. doi: 10.1073/pnas.0501078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 27.Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 28.Bader M, et al. A conserved f box regulatory complex controls proteasome activity in Drosophila. Cell. 2011;145:371–82. doi: 10.1016/j.cell.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bader M, Arama E, Steller H. A novel F-box protein is required for caspase activation during cellular remodeling in Drosophila. Development. 2010;137:1679–88. doi: 10.1242/dev.050088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–8. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Exner N, et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci. 2007;27:12413–8. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song S, et al. Characterization of PINK1 (PTEN-induced Putative Kinase 1) Mutations Associated with Parkinson Disease in Mammalian Cells and Drosophila. The Journal of biological chemistry. 2013;288:5660–72. doi: 10.1074/jbc.M112.430801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding WX, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. The Journal of biological chemistry. 2010;285:27879–90. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SJ, et al. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO reports. 2010;11:226–32. doi: 10.1038/embor.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson DE, Laman H. A competitive binding mechanism between SKP1 and exportin 1 (CRM1) controls the localization of a subset of F-box proteins. J Biol Chem. 2011 doi: 10.1074/jbc.M111.220079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondapalli C, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open biology. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–86. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 41.Small I, Peeters N, Legeai F, Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–90. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 42.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature genetics. 2005;37:1289–95. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 43.Ardley HC, et al. Inhibition of proteasomal activity causes inclusion formation in neuronal and non-neuronal cells overexpressing Parkin. Mol Biol Cell. 2003;14:4541–56. doi: 10.1091/mbc.E03-02-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plun-Favreau H, et al. The mitochondrial protease HtrA2 is regulated by Parkinson’s disease-associated kinase PINK1. Nat Cell Biol. 2007;9:1243–52. doi: 10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- 45.Samali A, Cai J, Zhivotovsky B, Jones DP, Orrenius S. Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of jurkat cells. EMBO J. 1999;18:2040–8. doi: 10.1093/emboj/18.8.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tain LS, et al. Drosophila HtrA2 is dispensable for apoptosis but acts downstream of PINK1 independently from Parkin. Cell Death Differ. 2009;16:1118–25. doi: 10.1038/cdd.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan W, Ashburner M, Hawley RS. Drosophila Protocols. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.