Abstract
Besides helping to maintain a reducing intracellular environment, the thioredoxin (Trx) system impacts bioenergetics and drug-metabolism. We show that hepatocyte-specific disruption of Txnrd1, encoding Trx reductase-1 (TrxR1), causes a metabolic switch in which lipogenic genes are repressed and periportal hepatocytes become engorged with glycogen. These livers also overexpress machinery for biosynthesis of glutathione and conversion of glycogen into UDP-glucuronate; they stockpile glutathione-S-transferases and UDP-glucuronyl-transferases; and they overexpress xenobiotic exporters. This realigned metabolic profile suggested that the mutant hepatocytes might be preconditioned to more effectively detoxify certain xenobiotic challenges. Hepatocytes convert the pro-toxin acetaminophen (APAP, paracetamol) into cytotoxic N-acetyl-p-benzoquinone imine (NAPQI). APAP defenses include glucuronidation of APAP or glutathionylation of NAPQI, allowing removal by xenobiotic exporters. We found that NAPQI directly inactivates TrxR1, yet Txnrd1-null livers were resistant to APAP-induced hepatotoxicity. Txnrd1-null livers did not have more effective gene expression responses to APAP challenge; however their constitutive metabolic state supported more robust GSH biosynthesis-, glutathionylation-, and glucuronidation-systems. Following APAP challenge, this effectively sustained the GSH system and attenuated damage.
Keywords: Bioenergetics, Drug metabolism, Redox biology, Thioredoxin reductase
Introduction
Two major antioxidant systems maintain the reduced intracellular environment: the thioredoxin (Trx) system and the glutathione (GSH) system. These each fuel a wide and highly overlapping spectrum of enzymatic reduction reactions, including protein cysteine-disulfide reduction, ribonucleotide reduction, peroxidase reactions, and others. To drive these reactions, electrons from reduced nicotinamide adenine dinucleotide-phosphate (NADPH) are used by either a Trx-reductase (TrxR) or glutathione reductase (Gsr) to reduce the oxidized forms of Trx or glutathione (GSSG), respectively [1, 2].
The Trx system also participates in aspects of metabolism that may not directly involve transfer of electrons from NADPH and are not redundant with GSH system activities. For example, a predominant phenotype of mammalian cells or organs lacking cytoplasmic TrxR1 is induction of drug-metabolism pathways [3–6]. Moreover, an antagonist of the Trx system, Trx-interacting protein 1 (Txnip, also called TBP-2 or VDUP1), has been shown to affect bioenergetics. Upon binding to and stabilizing Trx1, Txnip protein inhibits adipogenesis [7]. Txnip-null mice exhibit increased muscle glycogen content under fasting [8] and increased adipogenesis [9]. Inhibition of glycolysis is correlated to Txnip upregulation in tumors [10]; Txnip has been proposed to function as a sensor of glycolytic flux [11]; and human TXNIP is upregulated in diabetic conditions [12].
Previously we reported the resting liver transcriptome of mice having complete hepatocyte-specific disruption of a conditional-null allele of the Txnrd1 gene, encoding cytoplasmic TrxR1 [3]. Down-regulated transcripts in the Txnrd1-null livers include mRNAs for stearoyl-coenzyme-A desaturase, fatty acid binding protein 5, glucokinase, fatty acid desaturase 2, fatty acid synthase, ATP citrate lyase, and acyl-CoA thioesterase 1. Repression of these lipogenic mRNAs suggests the metabolic profile of TrxR1-deficient livers might be realigned away from lipid biogenesis. By contrast, most up-regulated mRNAs in Txnrd1-null livers are not associated with bioenergetics, but rather, encode drug-metabolism enzymes, in particular those on the Nrf2/Keap1 pathway. The Nrf2/Keap1 pathway typically provides a rapid cytoprotective response to acute environmental or oxidative stress [13, 14]. In unstressed conditions, Nrf2 is constitutively synthesized and bound by Keap1, which is a cysteine-rich protein that targets Nrf2 for proteasomal degradation. Stress-induced oxidation of Keap1 cysteines allows Nrf2 to escape proteasomal targeting, dimerize with Maf1, and induce transcription of target genes bearing antioxidant-response elements (ARE) [15, 16]. Nrf2-response genes include drug metabolism phase I oxidases, e.g., cytochrome P450s (Cyps); phase II conjugases, e.g., glutathione-S-transferases (Gsts); and phase III exporters, e.g., ATP-binding cassette-C (Abcc) proteins [17, 18]. In addition, some genes for GSH biosynthesis and some components of the Trx pathway are induced by Nrf2 [3, 6]. Nrf2/Keap1 pathway activation in Txnrd1-null livers is associated with in vivo occupancy of Nrf2 protein on AREs of Nrf2 target genes [3]. Chronic hepatic activation of Nrf2 target genes also occurs in mice with liver-specific disruption of Keap1 or of the autophagy-related gene Atg5 [19, 20], and this is associated with increased resistance to acetaminophen (paracetamol, N-acetyl-p-aminophenol, APAP) [19, 20].
APAP is a widely used analgesic and a classic pro-hepatotoxin. Whereas APAP, itself, is not cytotoxic, hepatocyte-specific Cyps oxidize APAP into the highly toxic N-acetyl-p-benzoquinone imine (NAPQI) [21]. At low doses, hepatic APAP is conjugated to either sulfonate by sulfotransferases or to glucuronate by UDP-glucuronosyl transferases (UGT), which constitute the hepatocyte’s first lines of defense against APAP toxicity. Conjugation allows excretion by Abcc 2–4; however, the sulfonation and glucuronidation pathways are generally overwhelmed by even moderate APAP doses [22, 23]. In particular, the capacity of the UGT system is limited by hepatic carbohydrate reserves [24], and this has been correlated with increased APAP susceptibility following nutritional deprivation [25]. Excess APAP is converted into NAPQI [21, 26]. At pharmacological doses of APAP, NAPQI is rapidly conjugated to GSH both spontaneously (k = 3.2×104 M−1 s−1) and by glutathione-S-transferases (Gsts; kcat/Km = 8×105 to 3×107 M−1 s−1 for different Gsts) [21, 27] and effectively exported from hepatocytes by Abccs [22, 23]. The Gst pathway, although only acting after formation of NAPQI, normally has a much higher capacity to respond to APAP overdose. The clinical standard of care for APAP overdose is administration of N-acetylcysteine [21], which is a rapidly processed precursor that promotes increased biosynthesis of GSH in the face of NAPQI-driven depletion [28]. At high doses of APAP, however, GSH becomes depleted and unconjugated NAPQI accumulates to cytotoxic levels, leading to hepatocyte necrosis. APAP overdose accounts for most acute liver failure cases in the U.S. and Western Europe, and has nearly a 30% mortality rate [21].
In this paper we show that hepatocyte-specific Txnrd1-null mice exhibit constitutive shifts in several metabolic pathways that, in combination, make the liver refractory to APAP challenge. At a bioenergetic level, TrxR1-deficient livers accumulate increased glycogen, whose levels normally limit the capacity of the UGT-detoxification pathway. At a drug-metabolism level, these livers overexpress enzymes for (1) conversion of glycogen into UDP-glucuronate, (2) conjugation of glucuronate to APAP, (3) reversion of NAPQI back into APAP, (4) biosynthesis of GSH, (5) conjugation of GSH to NAPQI, and (6) export of conjugated xenobiotics from the hepatocyte. These findings identify TrxR1 as a regulator that integrates the hepatocyte’s reductive metabolism/redox homeostasis system with its bioenergetics and cytoprotective drug metabolism systems.
Materials and methods
Reagents
Except as indicated, all general reagents were of molecular biology or higher grade and were purchased from standard laboratory suppliers including Fisher, VWR, MP Biomedicals, and Sigma. Acetaminophen was purchased from Spectrum Chemical Corp. (U.S.P. grade, #AC100). Fixatives, stains, reagents, and embedding materials for electron microscopy were purchased from Electron Microscopy Sciences, Inc. or Ted Pella, Inc.
Mouse lines and care conditions
All animal care and use protocols were reviewed and approved by the Montana State University Institutional Animal Care and Use Committee. The Txnrd1null allele is a true null that cannot generate TrxR1 protein; the Txnrd1cond allele is a functionally wild-type (Txnrd1+) allele that coverts to Txnrd1null upon exposure to Cre [29]. Basal characterization of mice bearing these allelic combinations has been reported [3, 30, 31]. All experiments used young adult (8- to 20-week old) male mice of the indicated genotypes. Mice were maintained on a 14:10 h light:dark cycle with unrestricted access to feed (Picolabs #5058) and water. APAP challenges were administered between 7:00 and 8:30 am. All renewable resources in this study are available for un-restricted non-profit research unless specifically restricted by another party.
Enzyme assays and western blots
Assays for TrxR, Trx, and GSH+GSSG from livers were performed as described previously [30]. Serum ALT levels were determined using the Catachem, Inc. system (#C164-0B) as per the manufacturer’s protocols. Livers were cleared of blood by cardiac saline perfusion and portal drainage, snap-frozen in liquid nitrogen, and stored at −80°C. Western blots were performed as described previously [3, 30] using rabbit antiserum raised against full-length mouse Sec-to-Cys TrxR1 or full-length mouse Trx1, both of which were kindly provided by G. Merrill at Oregon State University. For enzyme assays, rat TrxR1 and its Sec-to-Cys variant were expressed in E. coli and purified using a 2’, 5’-ADP Sepharose column (GE Healthcare) as described previously [32]. Enzymatic activities of wild-type and the Sec-to-Cys mutant TrxR1 were determined using 5,5’-dithiobis-2-nitrobenzoic acid (DTNB) as a model substrate, with formation of product monitored as the increase in absorbance at 412 nm (ξ412 nm = 13.6 mM−1 cm−1). In all other assays, the oxidation of NADPH was monitored as the decrease in absorbance at 340 nm (ξ340 nm = 6.22 mM−1 cm−1). Enzymatic reactions were calculated and plotted with the GraphPad Prism 5 software.
Histology and electron microscopy
For H&E staining, samples were harvested into 10% buffered formalin, fixed at room temperature for 12–72 h, embedded in paraffin, and 5-µmeter sections were prepared. For periodic acid Schiff’s (PAS) staining, samples were harvested into 6:1 of ethanol to glacial acetic acid overnight and paraffin embedded. Sections were incubated in fresh 1% periodic acid (Alpha Aesar, #30472-18) at room temperature, washed, and reacted with fresh Schiff’s reagent (EMD #6073-71) at 60–65°C for 5 min. Samples were counter-stained with hematoxylin and mounted. For TEM, livers were perfused with 3 ml sterile saline followed by 5 ml 10% neutral-buffered formalin. Liver was diced into ~ 2 mm cubes, and fixed for 12–18 h in Karnovsky’s fixative (3% glutaraldehyde, 3% formaldehyde, 0.2 M cacodylate, pH 7.4). Samples were then rinsed 2×10 min in 0.1 M Na/KPO4, pH 7.2, post-fixed in 2% OsO4, pH 7.4, and rinsed as before. Blocks were dehydrated, embedded in Spurr’s low-viscosity medium (Ted Pella, Inc.) by the manufacturer’s protocols, and 60- to 90-n meter sections were prepared with a diamond knife, mounted on grids, and stained with uranyl acetate and lead chloride as per the supplier’s protocols (Electron Microscopy Science, Inc.). Images were taken on a Zeiss LEO 912 transmission electron microscope using a digital image capture system with an electronically embedded scale-bar.
Light microscopy images were taken under bright-field conditions at room temperature in Permount on a Nikon Eclipse 80i microscope, a Nikon DS Ri1 digital camera, and Nikon NIS Elements BR acquisition software.
Glycogen analyses
Glycogen purification by alkaline digestion and ethanol precipitation followed classical protocols [33]. Liver pieces (0.1 g) were hydrolyzed in 10-volumes of 30% KOH in a boiling water bath for 1h, cooled to room temperature, and glycogen was precipitated with 2-volumes of ethanol on ice for ≥ 1 h. Glycogen was collected by centrifugation, washed with 70% ethanol, resuspended in 1 ml of 10 mM Tris-Cl, pH 7.5, 0.5 mM EDTA, and stored frozen. For turbidity assays, samples were thawed, mixed to homogeneity, and two-100 µl portions of each sample were removed. One portion from each liver was adjusted to 2 mM CaCl2 and digested with 1 unit of α-amylase (Tokyo Chemical Industry, Inc., #A0447) at 37°C for 1h to convert glycogen to glucose. α-amylase-digested and undigested samples were diluted to 1 ml with water and precipitated with 2 ml ethanol in glass test tubes on ice for 30 min. Precipitated glycogen was resuspended by vortexing and tubes were photographed against a dark background. To quantify glycogen, samples or a standard curve of bovine glycogen (Sigma #G0885) were digested with 0.01 unit/µl α-amylase, incubated for 1 h at 37°C to convert glycogen to glucose, and reactions were stopped with 2 µl 0.5 M EDTA. Glucose content was determined using Sigma’s glucose assay system (#GAGO20).
RNA isolation, transcriptome analyses, and semi-quantitative RT-PCR
Total RNA was prepared from freshly harvested liver using the Ambion Ribopure system (#AM1924). RNA was resuspended in water, evaluated for quality by denaturing gel electrophoresis, and equivalent portions (10 µg) of each were used to generate a fragmented oligo-(dT)-primed biotinylated cRNA using the Ambion MessageAmp-II Biotin Enhanced (#AM1791) system. Biotinylated cRNAs were hybridized to Affymetrix mouse 430 2.0A expression arrays. Data collection and analysis was as described previously [3]. Three biological replicates (separate animals) were used for each condition and data is shown for each animal. As described previously [3], signals were considered reliable if the average raw hybridization signal for one of the conditions within a comparison was ≥ 100 units. mRNA levels were considered different between conditions if average signals differed by ≥ 2-fold and P values were ≤ 0.05. For semi-quantitative RT-PCR, RNA was prepared and quality-validated as above, and 5.0 µg of each RNA sample was used to generate an oligo-(dT)-primed first-strand cDNA. An equal portion of each cDNA (1.0 µl) was used in each PCR reaction, as described previously [30]. Signal intensities were plotted and the slope of linear amplification represented the relative mRNA level. Primer pairs used for RT-PCRs were described previously [3, 29, 30].
Results
Metabolic switch in livers of mice lacking hepatocytic TrxR1
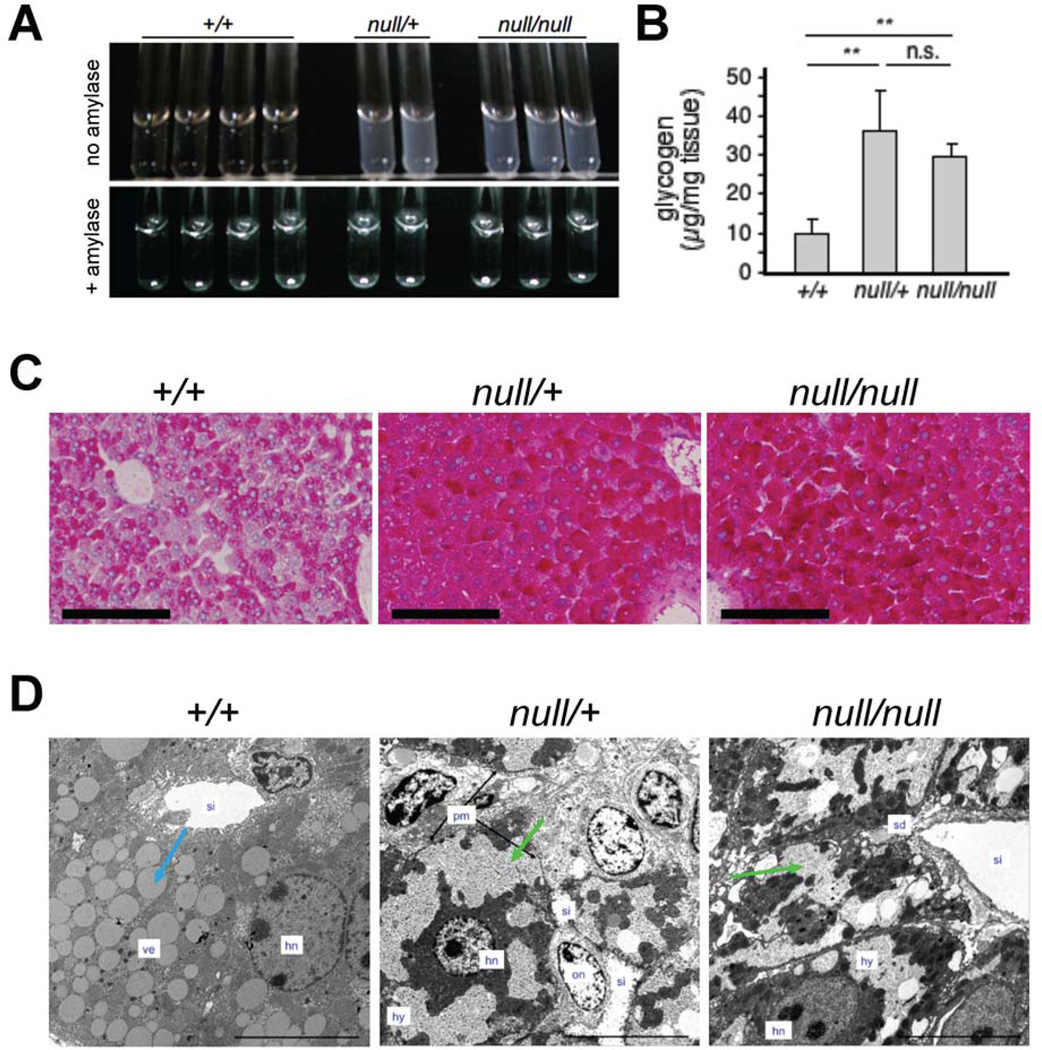
The transcriptome of resting TrxR1-deficient livers [3] suggested that their bioenergetic profile might be substantially different than those in normal livers. Although glycogen levels vary diurnally [34, 35], circadian gene expression is indistinguishable between wild-type and Txnrd1null/null livers [3]. To assess glycogen levels, alkaline digests were here prepared from equal amounts of +/+, null/+, and null/null livers harvested at 10:00 am. Based on the turbidity of ethanol-precipitates of these lysates and the loss of this turbidity in α-amylase-treated control samples, null/null and null/+ livers were found to have substantially more glycogen than did +/+ livers (Figure 1A). Analyses of the glucose content of α-amylase-digested samples showed that null/null and null/+ livers contained 3.0- and 3.5-fold more glycogen per mass of tissue, respectively, than did the +/+ livers (Figure 1B), and this was reflected in the intensity of staining of liver sections by periodic acid-Schiff’s reagent (PAS; Figure 1C). This difference in glycogen content was not overtly apparent in sections stained by hematoxylin and eosin (H&E) [3]; however transmission electron microscopy (TEM) revealed that, whereas periportal hepatocytes in control livers contained abundant cytoplasmic lipid vesicles (“ve”), periportal hepatocytes in null/+ and null/null livers contained fewer vesicles, but instead exhibited vast tracts of granular hyaloplasm (“hy”) (Figure 1D). These cells resembled the glycogen-engorged hepatocytes that have been reported in some forms of hereditary glycogenesis [36] or have been induced in some rodent models [37]. Hyaloplasm was not membrane-delimited. The down-regulation of mRNAs for lipogenic functions [3], the decrease in lipid-filled vesicles, and the increased glycogen stores observed in TrxR1-deficient livers indicates that loss of hepatocyte TrxR1 causes a metabolic switch that favors glycogen accumulation and, by corollary, that TrxR1 in hepatocytes drives a more lipogenic metabolic profile.
Fig. 1.
High accumulation of glycogen in Txnrd1null/null and Txnrd1null/+ hepatocytes. (A) Glycogen-dependent turbidity in extracts of livers harvested at 10:00 am (upper panel) or in parallel samples treated with α-amylase (lower panel). (B) Quantification of glycogen in resting livers as in (A). Bars on scatter plot show mean ± SEM. (C) PAS-staining of glycogen on liver sections. Scale bars = 100 µmeters. (D) Transmission electron microscopy of hepatocytes within representative periportal regions of livers harvested at 10:00 am. Blue arrow denotes a typical electron-semi-opaque vesicle; green arrows denote granular hyaloplasm. Labels: hn, hepatocyte nucleus; hy, granulated hyaloplasm; pm, plasma membrane; on, oval cell nucleus; sd, space of Disse; si, sinusoid; ve, vesicles. Original magnification 800X; scale bars represent 10 µmeters.
Acute APAP challenge diminishes hepatic Trx and TrxR activities in normal mice
APAP challenge causes rapid depletion of hepatic glycogen and GSH [38, 39]; however, to our knowledge, the impact of APAP on the Trx pathway has not been reported. Mice were treated with 100 or 1000 mg/kg APAP (LD50 ~ 338 mg/kg) and, after 4 h, levels of GSH+GSSG and of Trx or TrxR activities, were measured in liver homogenates. Besides depleting hepatic GSH+GSSG levels to near-zero, high-dose APAP challenge caused 10-fold and 4-fold decreases in total TrxR or Trx activities, respectively (I).
Direct inhibition of TrxR1 activity by NAPQI
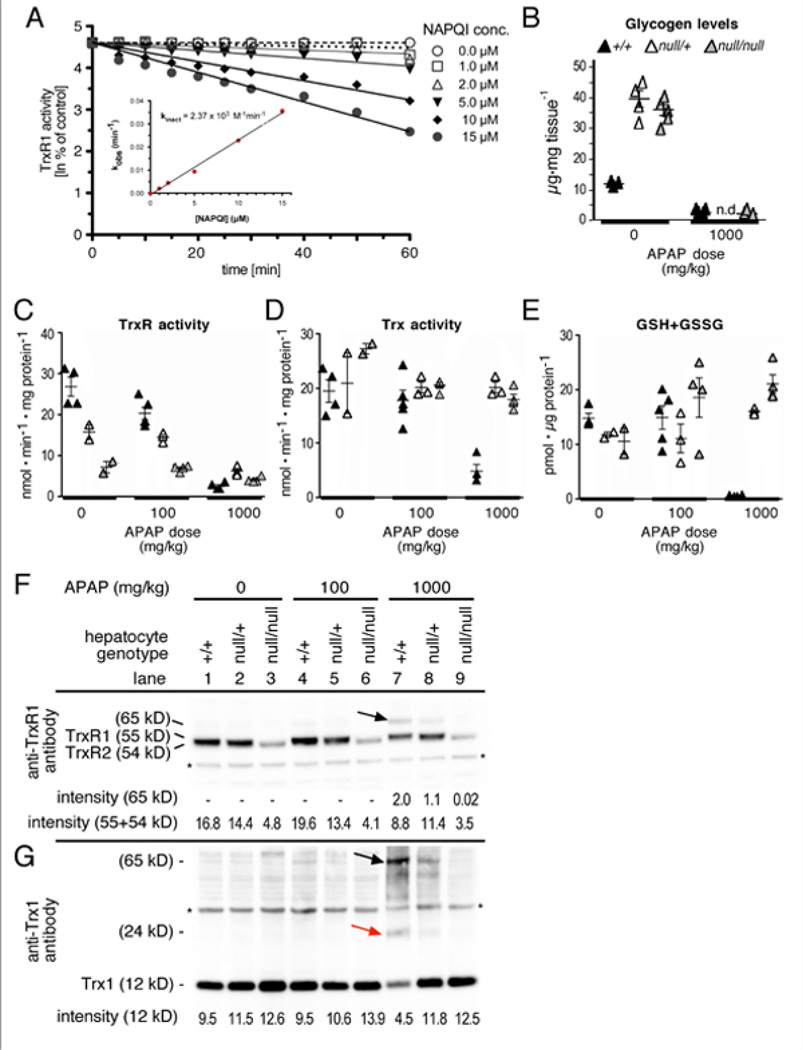
To investigate the mechanisms underlying loss of hepatic TrxR activity in APAP-challenged mice, we tested whether APAP, its precursor acetanilide, its UGT-generated glucuronate conjugate p-acetamidophenyl-β-D-glucuronide (p-AAPG), or its Cyp-generated oxidation product NAPQI could directly target TrxR1. TrxR1 activity was unaffected by APAP, acetanilide, or p-AAPG (Figures S1A, B), indicating these compounds were neither substrates nor inhibitors of the enzyme. By contrast, NAPQI caused irreversible inhibition of Trx reduction by TrxR1 with a second order rate constant of 2.37×103 M−1 min−1 (Figure 2A), yet only partial inhibition of juglone reduction by TrxR1 (Figures S1C, D). Importantly, TrxR1 variants lacking the Sec residue still support NADPH-dependent juglone reduction [40]. Our results indicate NAPQI is a potent inhibitor of TrxR1 and that this inhibition is likely mediated through direct targeting of the Sec residue in the enzyme.
Fig. 2.
NAPQI inhibits TrxR activity in vitro and APAP disrupts the Trx and GSH pathways in vivo. (A) Direct inactivation of TrxR1 by NAPQI. Reduced TrxR1 (0.4 µM) was incubated with NAPQI and aliquots were assayed for TrxR1 activity by the DTNB reduction assay yielding apparent first-order inactivation constants (kobs). A theoretical second-order rate inactivation constant (kinact) was determined by plotting kobs against NAPQI concentration (inset). Effects of other APAP metabolites on TrxR1 is presented in Figure S1A&B and effects of NAPQI on Sec-Cys TrxR1 is presented in Figure S1C. (B-D) Effects of APAP challenge on hepatic Trx- and GSH-pathways in +/+, null/+, and null/null livers. Analyses were as in Table I using mice with livers of the genotypes indicated. Columns at right indicate mean values and SEM (n = 4 for each condition). Numerical data and statistical analyses are presented in Figure S1H-J. (E) TrxR1 western blots. Immunoreactive bands at 54, 55 and 65 kDa were detected. The TrxR1 antibody cross-reacts with the mitochondrial TrxR2 giving rise to the 54 kDa band. The 65 kDa band also reacted with Trx1 antibody and is likely a TrxR1-Trx1 heterodimer (black arrow). Protein levels are integrated with enzyme activity levels in Figure S1D. (F) Trx western blots. Immunoreactive bands at 12, 24 and 65 kDa, corresponding to the Trx1 monomer (12 kDa), a likely covalently linked dimer (24 kDa, red arrow), and a likely TrxR1-Trx1 heterodimer (65 kDa, black arrow) were detected. Asterisks indicate non-specific bands.
Mice having genetic disruption of hepatocyte TrxR1 expression are refractory to further APAP-induced loss of TrxR or Trx activities, and to GSH-depletion
In an unchallenged state, mice lacking hepatocyte TrxR1 are overtly normal, maintain a normal GSH redox state, and show no evidence of hepatic oxidative stress [3, 30, 31] (Figures S2A, B). The total TrxR activity in Txnrd1null/null livers is not zero, which can be explained by a combination of activity derived from mitochondrial TrxR2 as well as TrxR1 expression in non-hepatocyte cells of the liver, which are not affected by the conditional knockout [3, 30]. Although treatment of wild-type mice with 100 or 1000 mg/kg APAP resulted in loss of 24% or 91% of the total liver TrxR activity after 4 h as compared to untreated livers, respectively (Figures 2B, S2D), the same doses in null/+ livers resulted in loss of only 8% and 61% of TrxR activity compared to untreated heterozygous levels, and in null/null livers, these doses resulted in TrxR activity loss of only 3% and 42% compared to untreated mutant mice, respectively (Figure 2B; S2D). This suggested that mice in which hepatocyte TrxR1 was chronically diminished were resistant to further acute inhibition of hepatic TrxR activity by APAP.
Both the Trx activity and the GSH levels in either null/+ or null/null livers were also highly refractory to APAP-induced depletion at 4 h (Figures 2C&D; S2E&F). Most strikingly, although GSH levels in wild-type animals were depleted to near-zero at 4 h following 1000 mg/kg challenge, both the heterozygous and the homozygous mutant livers had significantly increased GSH levels at this time (Figure S2F).
Western blots revealed that, for both Trx1 and TrxR1, the diminution of hepatic enzyme activities following APAP challenge was greater than was the decrease in protein levels (Figure 2E&F). Moreover in +/+ livers, APAP treatment caused shifts in the molecular weights of anti-Trx- and anti-TrxR-reactive bands that were consistent with it inducing DTT-resistant fusion of Trx1 to TrxR1, but this did not occur in either null/+ or null/null livers (Figures 2E&F, black and red arrows in lane 7; Figure S3). Thus, for both Trx and TrxR, it appeared that APAP induced inhibition of the activities of the enzymes uniquely in wild-type livers, and this may have, in part, been due to irreversible binding of Trx to TrxR. In combination, these results indicate that APAP challenge disrupts both the GSH and the Trx systems in normal liver, but that null/+ or null/null livers are refractory to these effects.
The increase in GSH levels seen at 4 h post APAP-challenge in the TrxR1-deficient livers, as well as the maintenance of TrxR and Trx activities in these livers, was strikingly different from the precipitous GSH depletion and the TrxR- and Trx-activity diminution seen in normal mice (Figures 2C-E). To determine whether this resulted from sustained levels or from more rapid recovery of depleted pools of these critical redox-active species, we measured their levels at time points between 0 and 4 h following high-dose challenge of mice having +/+ , null/+, or null/null livers. Results showed that, in +/+ livers, GSH pools fell to near-zero within 1 h of APAP challenge and remained near-zero throughout the time-course (Figure 3A, green line). TrxR activity decreased 2.5-fold by 1 h, and then continued to decline out to 4 h, whereas Trx activity showed slower inhibition following APAP challenge (Figure 3A, blue and red lines, respectively). Both null/+ and null/null livers underwent a similar precipitous decline in GSH levels by 1–2 h, but levels rapidly recovered to resting or higher levels by 4 h (Figure 3B&C, green lines). For TrxR and Trx, the null/null livers and, to an intermediate level the null/+ livers, exhibited continuously sustained activities compared to unchallenged levels (Figures 3C&B, blue and red lines, respectively). These results indicate that TrxR-deficient mice more robustly recover GSH levels and they are resistant to loss of TrxR or Trx activities following APAP challenge.
Fig. 3.
null/null and null/+ livers more rapidly recover from GSH depletion, are resistant to TrxR and Trx activity inhibition, and consume more glycogen following high-dose APAP challenge compared to wild-type livers. (A) Mice with hepatocytes of the indicated genotype were treated with 1000 mg/kg APAP, sacrificed, perfused, and livers were harvested at the indicated time-points thereafter. Levels of TrxR activity (blue diamonds), Trx activity (red squares), or GSH+GSSG (green triangles) were measured as in Fig. 2, above, and, to ease comparisons within each genotype, were presented as the percentage of the 0 h (untreated) value. Values represent mean + SEM. (B) Glycogen consumption following APAP challenge. Mice with hepatocytes of the indicated genotpes were treated with 1000 mg/kg APAP, harvested at the indicated times thereafter, and liver sections were stained with PAS as in Figure 1C. Abbreviations: p, portal triad; cv, central vein. Scale bars represent 100 µmeters. Ultrastructural changes on hepatocytes following APAP challenge are shown in Figure S2.
In the TrxR1-deficient livers, like in wild-type controls, high-dose APAP challenge caused rapid loss of hepatic glycogen stores, which initiated centrolobularly and spread periportally (Figure S2C and data not shown). At 4 h, controls and TrxR1-deficient livers both showed zonal clearance of 75–100% of the glycogen by PAS staining (Figure 3B). Because TrxR1-deficient livers had ~ 3-fold more glycogen than control livers prior to challenge, their APAP-induced glycogen consumption was ~ 3-fold greater (Figure S2C).
TrxR1-deficient livers are refractory to APAP-induced pathology
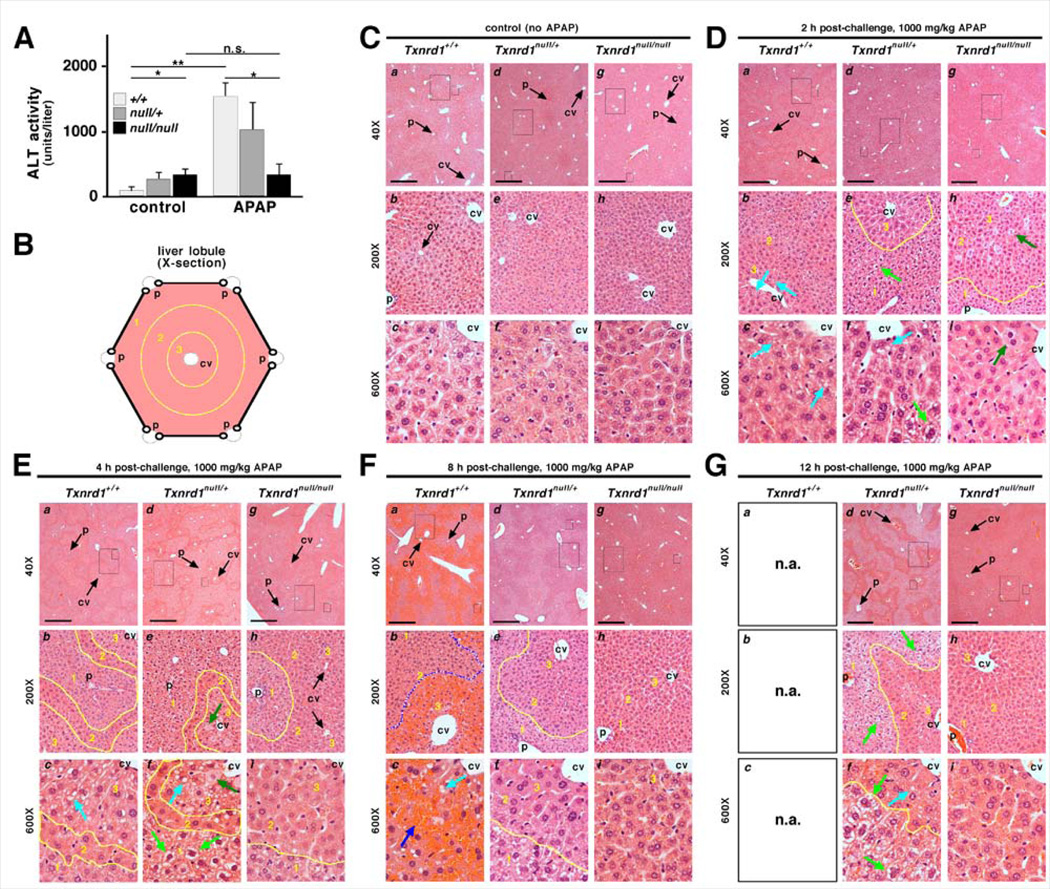
Following APAP challenge in the mutant as compared to control livers, the attenuated impact on levels of Trx and TrxR activities, and the more robust recovery of levels of GSH and glycogen, suggested that the mutant livers might sustain less pathological response. Consistent with this prediction, treatment with 1000 mg/kg APAP caused a 20-fold increase in serum alanine transaminase (ALT) levels in wild-type mice by 4 h, whereas it had no significant effect on ALT levels in the homozygous mutants (Figure 4A). Genotype-specific differences in behavioral responses to APAP-challenge were also evident during the experiment. At 2 and 4 h post challenge with 1000 mg/kg APAP, all animals were lethargic. By 8 h, wild-type animals had deteriorated to a state of complete inactivity, unresponsiveness, and self-isolation, and were not maintained beyond this time. In stark contrast, by 8 h mice with null/null livers had resumed activity and showed no overt signs of being adversely affected. Mice with null/+ livers remained lethargic at 8 h, but this was less severe than the state exhibited by wild-type animals, suggesting the heterozygous animals had an intermediate pathological response to APAP challenge.
Fig. 4.
Mice with null/null and null/+ hepatocytes resist APAP-induced liver damage. (A) Serum ALT levels 4 h after challenge with 0 or 1000 mg/kg APAP (mean + SEM). (B) Architecture of a liver lobule showing central vein (cv) and portal triads (p). Lobular zones 1 – 3 are delineated by yellow lines. (C-G) Representative sections from animals of indicated genotypes harvested at indicated times after challenge with 1000 mg/kg APAP. For each set, sub-panels are labeled as follows: a-c, wild-type; d-f, null/+; g-i, null/null. Rectangles in low-magnification images (4X objective; a, d, and g; scale bars = 500 µm) demarcate areas shown in medium (20X; b, e, and h) and high (60X; c, f, and i) magnification. Some central veins (cv) and portal tracts (p) are indicated. (C) Saline-treated controls. (D) Livers 2 h after APAP challenge. null/+ and null/null hepatocytes in zone 1 had an “open” morphology (e.g., pale green arrows in panels e, f). In +/+ (b, c) and null/+ (f) livers, but not in null/null livers (i), some zone 3 hepatocytes showed unstained vacuoles (aqua arrows). In null/null livers, a few weakly stained necrotic binucleate hepatocytes were seen in zones 2 and 3 (dark green arrows in h, i). (E) Zone-specific staining 4 h after APAP challenge. +/+ and null/+ zone 2 cells stained darkly, like untreated hepatocytes; zone 1 cells had an “open” morphology (pale green arrows); and zone 3 cells were highly vacuolated (aqua arrows). By contrast, null/null zone 2 and 3 hepatocytes stained darkly and were difficult to distinguish, like in untreated livers; zone 1 hepatocytes had lighter staining, delineating this zone (yellow line, h, i). (F) Livers 8 h after exposure to APAP. Wild-type livers (a, b, c) showed severe centrolobular hemorrhagic congestion (delineated with blue line in b). Portal hepatocytes no longer showed the open morphology. null/+ livers resembled null/null at 4 h, with dark staining of zone 2 and 3 hepatocytes appearing normal, yet with pale open morphology in zone 1 allowing its distinction (yellow lines in e, f). null/null (g, h, i) was nearly indistinguishable from untreated controls (compare to panel C). (G) Livers 12 h after exposure to APAP. Wild-type mice were moribund by 8 h and were sacrificed prior to 12 h (n.a., not analyzed). null/null livers (g, h, i) showed little to no overt histopathology. null/+ livers typically retained an open morphology in zone 1 (light green arrows) and vacuolization in some zone 3 hepatocytes (aqua arrow).
We next examined the histopathology of livers from animals at 0, 2, 4, 8, and 12 h following APAP challenge. Figure 4B depicts the cross-sectional architecture of a liver lobule, with zone 3 being centrolobular and associated with venous circulation, zone 1 being periportal and associated with arterial circulation, and zone 2 being intermediate. APAP-treated wild-type livers showed classical progression from centrolobular vacuolization at 2 h (Fig, 4D, frames a-c, zone 3, aqua arrows) to severe sinusoidal congestion by 8 h (Figure 4F, frames a-c, zone 3, blue arrow). In contrast, whereas both the heterozygous (frames d-f) and homozygous-mutant livers (frames g-i) showed some histological response to APAP challenge, including scattered necrosis of large often bi-nucleate hepatocytes at early time-points in zones 2 and 3 (Figures 4D&E, dark green arrows), centrolobular necrosis was minimal in these livers (e.g., green arrows in panels d-i) and did not result in congestion as seen in wild-type livers (Figure 4F). Interestingly, at 4 h in heterozygous and homozygous-mutant livers, zone 1 hepatocytes had a pale or “open” cytoplasmic morphology (Figure 4E, pale green arrows) that was distinct from the often similarly pale morphology arising from the vacuoles in zone 3 hepatocytes (compare pale-green arrows in zone 1 to aqua arrows in zone 3; see below). The open zone 1 morphology in heterozygous livers was often stable throughout the time-course and did not presage necrosis in this region (e.g., pale-green arrows or regions circumscribed by yellow lines in Figures 4F&G, panels d-f). In the case of homozygous mutant livers, this zone 1 morphology was only transient and reverted to the untreated morphology by 8–12 h (Figures 4E-G, panels g-i; see below). Morphologically, zone 2 hepatocytes in all genotypes deviated least from their untreated state following APAP challenge (e.g., zone 2 regions circumscribed by yellow lines in Figures 4D-G) such that they often appeared as an isthmus of normal morphology within a sea of pale hepatocytes (e.g., Figure 4E, panels a-f; Figure 4G, panels d-f).
Ultrastructural examination 4 h after challenge with 1000 mg/kg APAP revealed that, whereas wild-type hepatocytes developed classical APAP cytopathology including formation of large vacuoles and loss of outer membrane integrity in zone 3 hepatocytes [41], this effect was less pronounced in null/+ hepatocytes and rarely seen in null/null hepatocytes (Figure S4). These large vacuoles correlated with the “open” morphology of zone 3 hepatocytes in wild-type livers following APAP challenge (Figure 4E aqua arrows). Interestingly, the extensive hyaloplasm seen in unchallenged periportal null/+ and null/null hepatocytes was retained; however the electron-dense granularity was greatly diminished (Figure S4). This loss of hyaloplasmic granularity correlated with the “open” morphology of zone 1 hepatocytes noted by H&E staining (Figure 4E green arrows) and likely resulted from APAP-induced consumption of the glycogen stores (Figures 3BS2C).
APAP resistance in TrxR1-deficient hepatocytes reflects the preconditioned state of hepatocytes and not a more effective gene expression response to the challenge
Previous studies reported that APAP challenge of normal mice induces a hepatic Nrf2 response [42–44]. Unchallenged TrxR1-deficient mice have a constitutively active Nrf2 pathway [3]; however, it was unclear how this compared to the APAP-induced response in wild-type mice, or whether the TrxR1-deficient livers might exhibit further induction of a cytoprotective response in response to APAP challenge. Therefore, we measured the resting and APAP-induced transcriptomes in wild-type and null/null livers. To avoid artifacts arising from the severe pathological responses caused by high-dose challenge, sub-histopathological challenge conditions (100 mg/kg APAP for 8 h) were used.
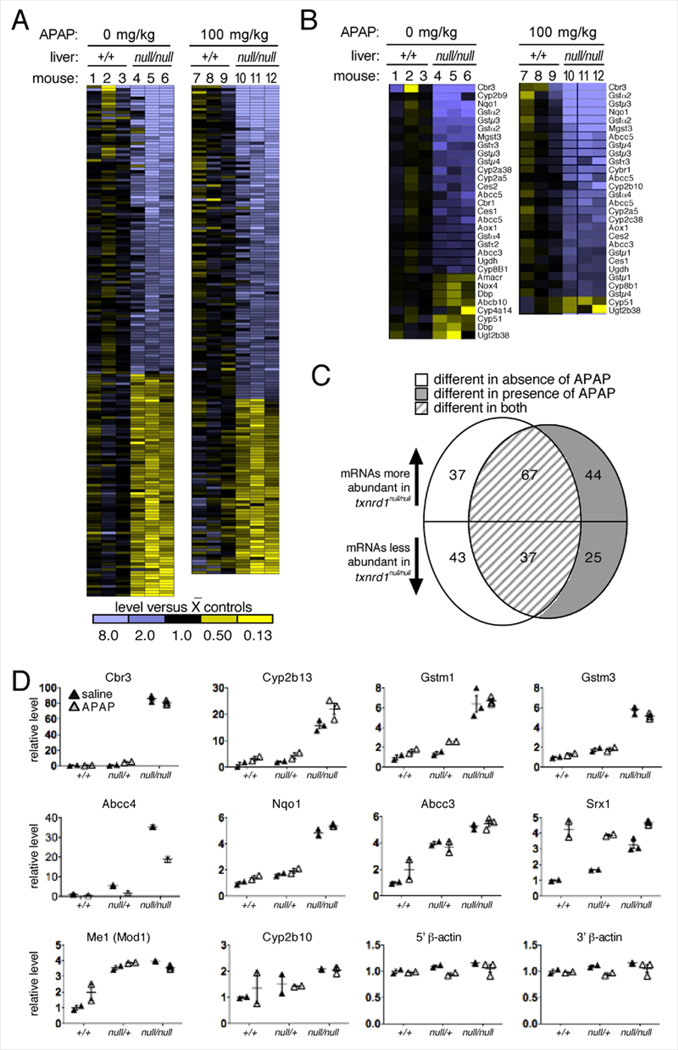
Both in the absence or presence of sub-lethal APAP challenge, the wild-type and mutant livers exhibited strikingly different transcriptome profiles (Figure 5A, compare lanes 1–3 with 4–6 for non-challenged livers, or lanes 7–9 with 10–12 for APAP-challenged livers: Table S1). As reported previously [3], the untreated null/null livers were characterized by an Nrf2 response and a shift from a lipogenic to a glycogenic bioenergetic profile (Figure 5B; Table S2). Reflecting their robust recovery of GSH following APAP challenge, mutant livers showed 1.76-fold higher levels of mRNA encoding the catalytic subunit of glutamate-cysteine ligase (Gclc). Importantly, in both genotypes, the transcriptome response to APAP challenge was very subtle (Figure 5C; Tables S1-S3). These results indicated that both the bioenergetic and the drug-metabolism responses to APAP-challenge were minor compared to the constitutive transcriptome changes induced by genetic disruption of TrxR1 (Figure 5C). Most notably, in response to APAP challenge, wild-type livers either did not alter, or subtly down-regulated, a set of potentially beneficial drug-metabolism mRNAs (GstA1, GstA2, GstM3, and Cbr3) that were among the most strongly up-regulated mRNAs in untreated mutant livers (Table S3). This suggested that the chronic drug-metabolism capacity induced in hepatocytes by disruption of Txnrd1 was more potent and more expansive than was the acute response induced by APAP challenge in livers of either genotype.
Fig. 5.
Genotype-specific impacts of 8 h low-dose APAP challenge on hepatic transcriptome. (A) Heat diagrams of all differentially abundant mRNAs between +/+ and null/null livers 8 h after challenge with 0 (mice 1–6) or 100 mg/kg (7–12) APAP. The average signal value for the three wild-type animals under each treatment condition was defined as “black” on the heat scale, and variance from this value is indicated by yellow (lower values) or blue (higher values). Quantitative data is presented in Table S1. (B) mRNAs encoding drug metabolism enzymes, primarily on the Nrf2/Keap1 pathway, identified based on KEGG designations. Quantitative data is presented in Table S2. (C) Comparison of the effects of Txnrd1 disruption and APAP treatment on the transcriptome of mouse livers. Quantitative data is presented in Table S3. (D) Sentinel mRNA levels in +/+, null/+, or null/null livers. All data are given as relative to the mean value for the saline-challenged (control) wild-type livers, and are presented as a linear multiplier (fold-difference) on the Y-axis. Bars show mean and SEM.
Although heterozygous livers showed similar GSH recovery and glycogen engorgement as did null/null livers, they exhibited greater pathology (see above). To further investigate this, we measured levels of sentinel mRNAs in null/+ livers that were differentially expressed between +/+ and null/null livers. Results showed that, whereas some of these mRNAs were expressed at similar levels in +/+ and null/+ livers (mRNAs encoding Cbr3, Cyp2b13, and GstM1), for others the heterozygotes were either intermediate (Abcc4, GstM3, Nqo1, Srx1, and Cyp2b10) or behaved more similarly to the null/null livers (Abcc3 and Me1) (Figure 5D). Only one of the mRNAs tested, that encoding sulfiredoxin-1 (Srx1), showed a strong response to APAP challenge in the wild-type livers (Figure 5D). This further indicated that, regardless of the TrxR1-status of the hepatocytes, the transcriptome response to APAP was subtle compared to the transcriptome response to genetic disruption of either one or two copies of Txnrd1.
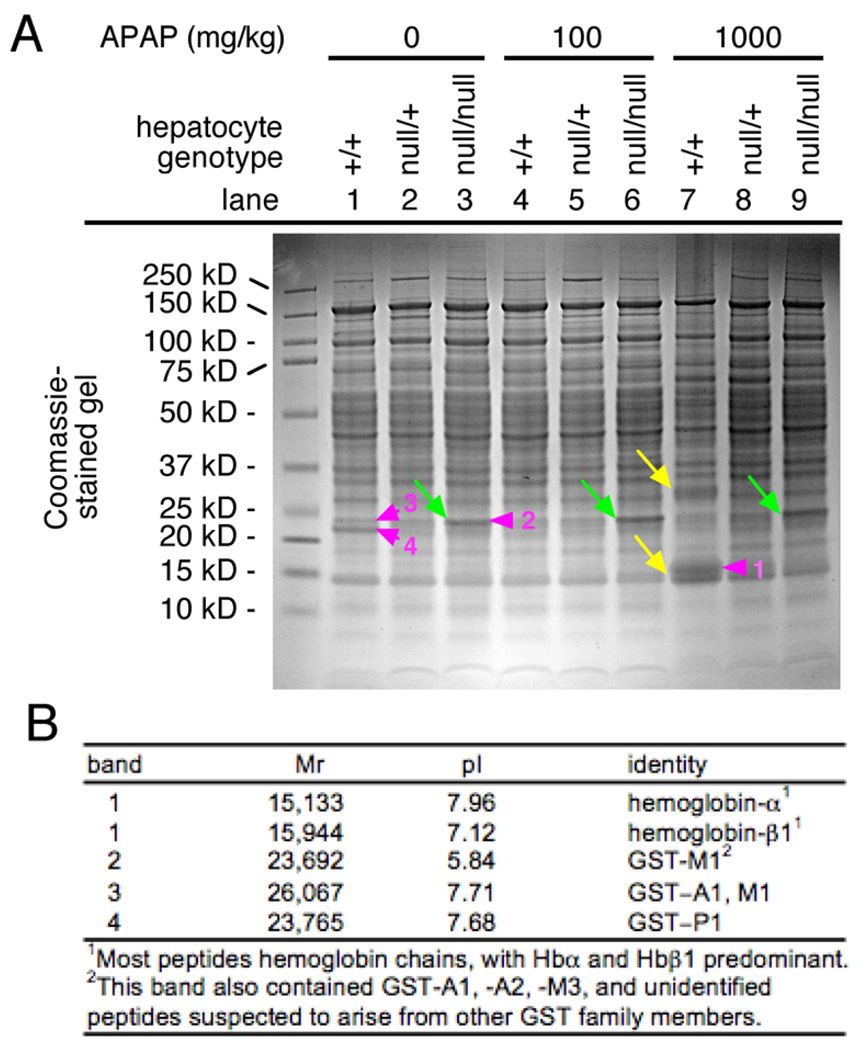
Total liver lysates of +/+, null/+, or null/null livers from mice that were challenged 4 h earlier with 0, 100, or 1000 mg/kg APAP, were separated by SDS-gel electrophoresis and stained with Coomassie. The null/null lysates, uniquely, showed a strong protein band at 23–25 kDa under all APAP-treatment conditions (Figure 6A, green arrows). Most other characteristics of the profiles were similar in all conditions with the exception of the 1000 mg/kg APAP-treated wild-type samples, which showed numerous quantitative and qualitative differences from the other samples, including a predominant cluster of bands around ~ 15 kDa (Figure 6A, yellow arrows). The bands indicated with numbered violet arrowheads were isolated and their major components were identified by mass spectrometry (MS)(Figure 6B). The band at 23–25 kDa that was enriched in homozygous mutant livers contained Gsts, with GstP1 and Gst 1, – 2, and/or ~ 3 predominating (band 2). In wild-type liver, GstP1, at a slightly lower molecular weight, was most abundant (band 4). This was consistent with our previous identification of the bands by pull-down on GSH-agarose beads and by western blotting as containing Gsts, most likely of the A- or M-classes in mutant liver, and P-class in wild-type liver. It was also consistent with our observation that homozygous mutants overexpress mRNAs encoding A- and M-class, but not P-class, GSTs [3]. The cluster of bands at ~ 15 kDa in the APAP-treated wild-type liver contained hemoglobin-α and -β chains (band 1). The presence of hemoglobin chains in lane 7 was consistent with high-dose challenge of +/+ livers, uniquely, causing hemorrhagic congestion (Figure 4F). No identified proteins in any of the APAP-treated samples suggested that APAP, itself, had induced a substantial protective response. The profile of the heterozygous samples differed from those of the wild-type samples in that they did not contain the hemoglobin chains, and they differed from those of the homozygous mutants in that they did not over-accumulate A-and M-class GSTs.
Fig. 6.
Genotype-specific impacts of 8 h low- or high-dose APAP challenge on hepatic protein profiles. (A) Coomassie-stained gel. Genotypes, treatments, and molecular sizes of markers are indicated. Green arrows indicate a predominant band that was more abundant in all null/null livers than in null/+ or +/+ livers, yellow arrows denote two bands that predominant only in APAP-treated +/+ livers. Violet arrowheads indicate species that were isolated and analyzed by MS (see below). (B) MS Identification of predominant species from protein gel in (A).
Discussion
The results of this study carry two major implications. First, TrxR1 was shown to be a determinant of the global metabolic state of the liver. Genetic deletion of Txnrd1 triggered a metabolic switch that favored glycogen- over lipid-accumulation and high expression of drug-metabolism enzymes. Second, TrxR1 was identified as a high-affinity target for direct inactivation by NAPQI, which might contribute to APAP-induced hepatotoxicity. Interestingly, even though APAP treatment was shown to inhibit hepatic TrxR activity in vivo, mice in which hepatocytes were genetically TrxR1-deficient were refractory to APAP-induced hepatotoxicity. Our results indicate that a chronic shift in hepatocyte metabolic processes resulting from genetic disruption of Txnrd1 preconditions an APAP-resistant state in the liver, whereas the acute loss of TrxR1 activity caused by APAP challenge, itself, is insufficient to induce resistance.
TrxR1 regulates the metabolic state of liver
To our knowledge, this is the first study to show a direct link between Txnrd1 status and a metabolic switch between a glycogen-storage versus a lipid-storage phenotype. Disruption of TrxR1 switched liver bioenergetics toward glycogenesis, indicating that TrxR1 activity biases the liver toward lipogenesis. As possible mechanisms for this switch, a potential role of Txnip should be considered (see Introduction). Txnip has been shown to participate in determining the metabolic state of several cell types and systems [7–10], and has been correlated with metabolic switches in others [11, 12]. It is thus possible that the metabolic switch caused by hepatocytic Txnrd1 deletion is a secondary effect related to TrxR1’s influence, via reduction of Trx1, on Txnip. Alternatively, it is possible that TrxR1 and Txnip each independently mediate metabolism through Trx1. Resolving these mechanisms is outside the scope of the present study but clearly warrants future investigation. Here, we can conclude that genetic modulation of the Txnrd1 status in mouse liver has similarly pronounced effects on the metabolic state as have been identified in mouse models with Txnip deletion.
APAP inhibits both the GSH- and the Trx-systems in normal liver
High dose APAP exposure rapidly depletes liver GSH reserves, which will disrupt all hepatic GSH-dependent redox reactions. However, the redox functions of the GSH system are highly overlapping with those of the Trx system, and, indeed, unchallenged hepatocytes in vivo remain overtly normal in the absence of either GSH, Gsr, or TrxR1 [3, 30, 31, 45, 46]. By contrast, short-term depletion of hepatic GSH with buthionine sulfoximine in mice genetically lacking hepatocytic TrxR1 inhibits hepatocyte DNA replication [30], indicating that at least some essential redox reactions require either the GSH- or the Trx-system for activity. In this study, we show that high dose APAP challenge inhibits hepatic TrxR and Trx activities in wild-type livers. This indicates that, under these conditions, both the GSH- and the Trx-system are inhibited and, as a corollary, that all redox reactions requiring either one of these systems will be disrupted. Potential impacts of this on hepatotoxicity are discussed below.
Interplay between the metabolic state of TrxR1-deficient liver and susceptibility to APAP toxicity
Fasting exacerbates APAP hepatotoxicity, and this has been correlated to depleted glycogen stores [39]. However, to our knowledge, it has not been previously shown that glycogen engorgement correlates with APAP resistance. Indeed, the capacity of the UGT pathway is normally insufficient to provide substantial protection against even low-dose APAP challenge [22]. Importantly, the substrate for UGT is APAP, which is not cytotoxic, so the UGT pathway preempts formation of NAPQI, the cytotoxic metabolite. TrxR1-deficient livers contained glycogen levels of ~ 40 µg/mg tissue (Figures 1B, S2C). This correlates to ~ 60 mg, or ~ 0.33 mole-equivalents of glucose or glucuronate, in a 1.5 g liver, and this was consumed very rapidly upon challenge with 1000 mg/kg APAP (Figure 3B, S2C). This APAP dose is equivalent to 30 mg, or 0.2 moles, in a 30 g mouse. Although we do not know how much glycogen is going toward energetic demands versus conjugation, the molar-ratio of consumed glycogen to administered APAP in these animals was high enough that glucuronidation could have contributed substantially to detoxification, thereby effectively curtailing cellular formation of NAPQI.
Interactions of APAP with drug metabolism enzymes in null/null or null/+ livers
Although null/null and null/+ livers accumulated similar levels of glycogen, the null/+ livers exhibited greater pathology following APAP challenge. This can be explained by the different degrees to which drug metabolism pathways were activated in each genotype. For example, null/null hepatocytes but not null/+ hepatocytes overexpress the mRNA for Nqo1 (Figure 5D), an enzyme that catalyzes the conversion of NAPQI into APAP [47]. APAP and NAPQI are in equilibrium in APAP-challenged hepatocytes, with Cyps catalyzing the formation of APAP into NAPQI and Nqo1 playing a protective role by reversing this reaction. Increasing Nqo1 activity will shift this equilibrium toward APAP, thereby lowering cytoplasmic NAPQI concentrations [22, 47]. Although mRNAs encoding several Cyps were up-regulated in null/+ and null/null livers (Figure 5, Table S1), these did not include Cyp family members that convert APAP into NAPQI [48–50]. Compared to +/+ livers, both null/null and null/+ livers overexpress Abcc3, yet only null/null livers also overexpress Abcc4 (Figure 5D). As a result, livers of both genotypes likely exhibit augmented elimination of APAP metabolites, with null/null livers being more effective at this than null/+ livers. In addition, we found that the null/null livers have an augmented GSH biosynthetic pathway, increased steady-state GSH levels, and they exhibit very high accumulation A- and M-class GST mRNAs and proteins, all of which should bolster glutathionylation-mediated export of NAPQI. In summary, the constitutive drug metabolism profile of the TrxR1-deficient livers, as compared to wild-type livers, will be particularly proficient at preventing cytosolic accumulation of NAPQI by (1) more effectively eliminating APAP before it becomes NAPQI and (2) shunting any NAPQI that does form either back into APAP and out of the cell by glucuronidation-medated export, or directly out of the cell via glutathionylation-mediated export. Although it is outside the scope of the current study to distinguish the relative contributions of each pathway to APAP detoxification in these mice, the observation that GSH levels rebound to resting or higher levels within 4 h of high-dose APAP challenge in both null/null and null/+ livers attests to the efficacy with which the overall metabolic shift in TrxR1-deficient livers allows them to resist APAP-induced GSH depletion and hepatotoxicity.
Factors contributing to NAPQI-induced cytotoxicity
The mechanisms of NAPQI toxicity remain incompletely understood. Because other means of GSH depletion are not cytotoxic [45], GSH depletion, alone, cannot account for toxicity. Since NAPQI is highly electrophilic, cysteine residues will react with it at rates that will depend on the accessibility and relative reactivity of individual residues. As such, active site cysteines on molecules like GSH or Trx will be preferred targets. The even more reactive Sec residue on TrxR1 [6] should be particularly susceptible to NAPQI. Here we show that NAPQI is indeed highly effective at inhibiting TrxR1 through its Sec residue, and that APAP-treated wild-type mice undergo marked losses of Trx and TrxR activities. Recent studies have shown that NAPQI also inhibits the mitochondrial superoxide dismutase (MnSOD), presaging mitochondrial oxidative damage [51]. MnSOD inhibition likely also results from the high reactivity of the enzyme’s active site, which is expected to make it a favorable NAPQI target. Thus, NAPQI disrupts both the GSH- and the Trx-systems, which are the cell’s two major NADPH-driven antioxidant systems, and it impedes mitochondrial detoxification of reactive oxygen species (ROS), all of which likely contribute to hepatotoxicity.
Recent biochemical studies have shown that, upon reacting with certain electrophilic drugs, TrxR1 can adopt novel activities. For example, like we here show for NAPQI, cisplatin reacts with the TrxR1 Sec residue; it inhibits its Trx-reductase activity but not its juglone-reductase activity; and it induces irreversible binding of TrxR1 to Trx1 [52]. Some other electrophilic compounds that react with the TrxR1 Sec residue can, in the presence of certain small molecule substrates, induce the enzyme to become a cytotoxic NADPH-oxidase, entitled a “SecTRAP” (Sec-compromised thioredoxin reductase-derived pro-apoptotic protein) that can generate large amounts of ROS [40]. Whereas it is unknown whether NAPQI-bound TrxR1 in hepatocytes might similarly generate ROS, it is intriguing to consider that TrxR1-deficient hepatocytes might be refractory to APAP not only as a consequence of their glycogen stores and augmented drug-metabolism capacity, but also as a consequence of not having TrxR1 converted into a SecTRAP. Further studies will be required to test this possibility.
Finally, our study reveals a surprisingly integrated network of metabolic systems in hepatocytes, with TrxR1 playing a key role in feed-forward cross-talk between systems. The importance of this is emphasized by the synergistic roles that various metabolic pathways play in protecting hepatocytes during APAP exposure. Clinical APAP remediation currently focuses on augmenting GSH production [53]. However it is known that nutritional deficiency is a compromising factor in APAP sensitivity [39], and the increase in APAP-induced glycogen consumption that we see in TrxR1-deficient livers suggests that bioenergetic modulation might provide an additional means to abrogate the hepatotoxic effects of APAP. In the future, combinatorial therapies that sustain GSH production, augment glucuronidation-based detoxification, and possibly prevent SecTRAP formation might lead to improved survival upon APAP overdose.
Supplementary Material
Table I.
Four hour APAP challenge on wild-type mice
| GSH+GSSG activity |
TrxR activity |
Trx activity |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| APAP dose | 0 mg/kg | 100 mg/kg | 1000 mg/kg | 0 mg/kg | 100 mg/kg | 1000 mg/kg | 0 mg/kg | 100 mg/kg | 1000 mg/kg |
| Value (SEM)1 | 14.81 (0.89) | 14.91 (2.14) | 0.68 (0.07) | 26.32 (2.34) | 20.34 (1.50) | 2.43 (0.48) | 19.48 (2.08) | 17.77 (1.90) | 4.83 (1.23) |
| % of control | - | 101% | 4.5%** | - | 75.9%* | 9.1%** | - | 91.2% | 24.8%** |
Units for GSH+GSSG levels are nmol · g−1 protein; units for TrxR and Trx activity are both nmol · min−1 · mg−1 protein.
P ≤ 0.05;
P ≤ 0.01
Acknowledgements
We thank J. Kundert, S. Brumfield, K. McInnerney, U. Hellman, M. Cebula, E. Bostrom, and M. McLoughlin for technical assistance; G. Merrill and E. Suvorova for antibodies or other contributions to this study; and the MSU 2010 VTMB422 students for participating in analyses as a part of their curriculum. Work was supported by grants from the U.S. NCI and NIA to EES, Karolinska Institutet, Swedish Research Council (Medicine) and Swedish Cancer Society to ESJA, an appointment from MAES to EES, an NSF-REU scholarship to TAM, and an appointment from HHMI to MRC. Additional support was provided by the MSU IID Department and an NIH-CoBRE grant to MSU.
Abbreviations
- Abcc
ATP-binding cassette subfamily C multidrug-resistance transporters
- ALT
alanine transaminase
- Aox1
aldehyde oxidase-1
- APAP
acetaminophen (paracetamol)
- BSO
buthionine sulfoximine
- Cre
bacteriophage P1 cyclization recombinase
- CYP
cytochrome P450
- DTNB
5,5’-dithiobis(2-nitrobenzoic acid)
- DTT
dithiothreitol
- EDTA
ethylenediamine tetraacetic acid
- loxP
recognition sites for Cre
- GSH
reduced glutathione
- Gsr
glutathione reductase
- GSSG
oxidized glutathione disulfide
- GST
glutathione-S-transferase
- H&E
hematoxylin and eosin
- kDa
kilo-Dalton
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MS
mass spectrometry
- NAC
N-acetyl cysteine
- NAPQI
N-acetyl-p-benzoquinone imine
- Nqo1
NADPH-quinine oxidase
- p-APDG
p-acetamidophenyl-β-D-glucronide
- p-APDGS
p-acetamidophenyl-β-D-glucronide sodium salt
- ROS
reactive oxygen species
- SDS
sodium dodecylsulfate
- SDS-PAGE
SDS-polyacrylamide gel electrophoresis
- Sec
selenocysteine
- SEM
standard error of the mean
- SULT
sulfotransferase
- Trx
thioredoxin mRNA or protein
- TrxR
thioredoxin reductase mRNA or protein
- Txnip
thioredoxin-interacting protein
- txnrd1
thioredoxin reducatse-1 gene
- UGDH
uridine diphosphate-glucose dehydrogenase
- UGT
uridine diphosphate-glucuronate transferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
Array data, NCBI-GEO, accession #GSE37874.
Conflict of interest
The authors declare they have no conflict of interest.
References
- 1.Arnér ESJ, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 2.Berndt C, Lillig CH, Holmgren A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am J Physiol Heart Circ Physiol. 2007;292:H1227–H1236. doi: 10.1152/ajpheart.01162.2006. [DOI] [PubMed] [Google Scholar]
- 3.Suvorova ES, Lucas O, Weisend CM, Rollins MF, Merrill GF, Capecchi MR, Schmidt EE. Cytoprotective Nrf2 pathway is induced in chronically txnrd 1-deficient hepatocytes. PLoS One. 2009;4:e6158. doi: 10.1371/journal.pone.0006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locy ML, Rogers LK, Prigge JR, Schmidt EE, Arnér ES, Tipple TE. Thioredoxin reductase inhibition elicits Nrf2-mediated responses in Clara cells: implications for oxidant-induced lung injury. Antioxid Redox Signal. 2012;17:1407–1416. doi: 10.1089/ars.2011.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandal PK, Schneider M, Kolle P, Kuhlencordt P, Forster H, Beck H, Bornkamm GW, Conrad M. Loss of thioredoxin reductase 1 renders tumors highly susceptible to pharmacologic glutathione deprivation. Cancer Res. 2010;70:9505–9514. doi: 10.1158/0008-5472.CAN-10-1509. [DOI] [PubMed] [Google Scholar]
- 6.Arnér ESJ. Focus on mammalian thioredoxin reductases--important selenoproteins with versatile functions. Biochim Biophys Acta. 2009;1790:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Chutkow WA, Lee RT. Thioredoxin regulates adipogenesis through thioredoxininteracting protein (Txnip) protein stability. J Biol Chem. 2011;286:29139–29145. doi: 10.1074/jbc.M111.267666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andres AM, Ratliff EP, Sachithanantham S, Hui ST. Diminished AMPK signaling response to fasting in thioredoxin-interacting protein knockout mice. FEBS Lett. 2011;585:1223–1230. doi: 10.1016/j.febslet.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chutkow WA, Birkenfeld AL, Brown JD, Lee HY, Frederick DW, Yoshioka J, Patwari P, Kursawe R, Cushman SW, Plutzky J, Shulman GI, Samuel VT, Lee RT. Deletion of the alpha-arrestin protein Txnip in mice promotes adiposity and adipogenesis while preserving insulin sensitivity. Diabetes. 2010;59:1424–1434. doi: 10.2337/db09-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JL, Merl D, Peterson CW, Wu J, Liu PY, Yin H, Muoio DM, Ayer DE, West M, Chi JT. Lactic acidosis triggers starvation response with paradoxical induction of TXNIP through MondoA. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu FX, Chai TF, He H, Hagen T, Luo Y. Thioredoxin-interacting protein (Txnip) gene expression: sensing oxidative phosphorylation status and glycolytic rate. J Biol Chem. 2010;285:25822–25830. doi: 10.1074/jbc.M110.108290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P, Saxena R, Ladd C, Schulze PC, Mazzini MJ, Jensen CB, Krook A, Bjornholm M, Tornqvist H, Zierath JR, Ridderstrale M, Altshuler D, Lee RT, Vaag A, Groop LC, Mootha VK. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi A, Ohta T, Yamamoto M. Unique function of the Nrf2-Keap1 pathway in the inducible expression of antioxidant and detoxifying enzymes. Methods Enzymol. 2004;378:273–286. doi: 10.1016/S0076-6879(04)78021-0. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen T, Nioi P, Pickett CB. The nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aleksunes LM, Manautou JE. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol Pathol. 2007;35:459–473. doi: 10.1080/01926230701311344. [DOI] [PubMed] [Google Scholar]
- 18.Klaassen CD, Slitt AL. Regulation of hepatic transporters by xenobiotic receptors. Curr Drug Metab. 2005;6:309–328. doi: 10.2174/1389200054633826. [DOI] [PubMed] [Google Scholar]
- 19.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 20.Ni HM, Boggess N, McGill MR, Lebofsky M, Borude P, Apte U, Jaeschke H, Ding WX. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol Sci. 2012;127:438–450. doi: 10.1093/toxsci/kfs133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinson JA, Roberts DW, James LP. Mechanisms of acetaminophen-induced liver necrosis. Handb Exp Pharmacol. 2010:369–405. doi: 10.1007/978-3-642-00663-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zamek-Gliszczynski MJ, Hoffmaster KA, Nezasa K, Tallman MN, Brouwer KL. Integration of hepatic drug transporters and phase II metabolizing enzymes: mechanisms of hepatic excretion of sulfate, glucuronide, and glutathione metabolites. Eur J Pharm Sci. 2006;27:447–486. doi: 10.1016/j.ejps.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Homolya L, Varadi A, Sarkadi B. Multidrug resistance-associated proteins: Export pumps for conjugates with glutathione, glucuronate or sulfate. Biofactors. 2003;17:103–114. doi: 10.1002/biof.5520170111. [DOI] [PubMed] [Google Scholar]
- 24.Thurman RG, Kauffman FC. Sublobular compartmentation of pharmacologic events (SCOPE): metabolic fluxes in periportal and pericentral regions of the liver lobule. Hepatology. 1985;5:144–151. doi: 10.1002/hep.1840050128. [DOI] [PubMed] [Google Scholar]
- 25.Price VF, Schulte JM, Spaethe SM, Jollow DJ. Mechanism of fasting-induced suppression of acetaminophen glucuronidation in the rat. Adv Exp Med Biol. 1986;197:697–706. doi: 10.1007/978-1-4684-5134-4_64. [DOI] [PubMed] [Google Scholar]
- 26.Gillette JR, Nelson SD, Mulder GJ, Jollow DJ, Mitchell JR, Pohl LR, Hinson JA. Formation of chemically reactive metabolites of phenacetin and acetaminophen. Adv Exp Med Biol. 1981;136(Pt B):931–950. [PubMed] [Google Scholar]
- 27.Miners JO, Drew R, Birkett DJ. Mechanism of action of paracetamol protective agents in mice in vivo. Biochem Pharmacol. 1984;33:2995–3000. doi: 10.1016/0006-2952(84)90599-9. [DOI] [PubMed] [Google Scholar]
- 28.Corcoran GB, Todd EL, Racz WJ, Hughes H, Smith CV, Mitchell JR. Effects of N-acetylcysteine on the disposition and metabolism of acetaminophen in mice. J Pharmacol Exp Ther. 1985;232:857–863. [PubMed] [Google Scholar]
- 29.Bondareva AA, Capecchi MR, Iverson SV, Li Y, Lopez NI, Lucas O, Merrill GF, Prigge JR, Siders AM, Wakamiya M, Wallin SL, Schmidt EE. Effects of thioredoxin reductase-1 deletion on embryogenesis and transcriptome. Free Radic Biol Med. 2007;43:911–923. doi: 10.1016/j.freeradbiomed.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prigge JR, Eriksson S, Iverson SV, Meade TA, Capecchi MR, Arnér ESJ, Schmidt EE. Hepatocyte DNA replication in growing liver requires either glutathione or a single allele of txnrd1. Free Radic Biol Med. 2012;52:803–810. doi: 10.1016/j.freeradbiomed.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rollins MF, van der Heide DM, Weisend CM, Kundert JA, Comstock KM, Suvorova ES, Capecchi MR, Merrill GF, Schmidt EE. Hepatocytes lacking thioredoxin reductase 1 have normal replicative potential during development and regeneration. J Cell Sci. 2010;123:2402–2412. doi: 10.1242/jcs.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Q, Antholine WE, Myers JM, Kalyanaraman B, Arnér ES, Myers CR. The selenium-independent inherent pro-oxidant NADPH oxidase activity of mammalian thioredoxin reductase and its selenium-dependent direct peroxidase activities. J Biol Chem. 2010;285:21708–21723. doi: 10.1074/jbc.M110.117259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Der Vies J. Two methods for the determination of glycogen in liver. Biochem J. 1954;57:410–416. doi: 10.1042/bj0570410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roesler WJ, Khandelwal RL. Diurnal variations in the activities of the glycogen metabolizing enzymes in mouse liver. The International journal of biochemistry. 1985;17:81–85. doi: 10.1016/0020-711x(85)90089-8. [DOI] [PubMed] [Google Scholar]
- 35.McVerry P, Kim KH. Diurnal rhythm of rat liver glycogen synthetase. Biochem Biophys Res Commun. 1972;46:1242–1246. doi: 10.1016/s0006-291x(72)80108-6. [DOI] [PubMed] [Google Scholar]
- 36.Suriawinata AA, Thung SN. Liver pathology : an atlas and concise guide. New York: Demos Medical; 2011. [Google Scholar]
- 37.Foster CL. The Demonstration of Glycogen in Liver Cells Fixed in Osmium Tetroxide. Q J Microsc Sci. 1960;101 273-&. [Google Scholar]
- 38.Mitchell JR, Jollow DJ, Potter WZ, Gillette JR, Brodie BB. Acetaminopheninduced hepatic necrosis. IV. Protective role of glutathione. J Pharmacol Exp Ther. 1973;187:211–217. [PubMed] [Google Scholar]
- 39.Price VF, Miller MG, Jollow DJ. Mechanisms of fasting-induced potentiation of acetaminophen hepatotoxicity in the rat. Biochem Pharmacol. 1987;36:427–433. doi: 10.1016/0006-2952(87)90346-7. [DOI] [PubMed] [Google Scholar]
- 40.Anestal K, Prast-Nielsen S, Cenas N, Arnér ESJ. Cell death by SecTRAPs: thioredoxin reductase as a prooxidant killer of cells. PLoS ONE. 2008;3:e1846. doi: 10.1371/journal.pone.0001846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farber JL, Gerson RJ. Mechanisms of cell injury with hepatotoxic chemicals. Pharmacol Rev. 1984;36:71S–75S. [PubMed] [Google Scholar]
- 42.Priyadarsiny P, Khattar SK, Malik R, Udupa V, Seshaiah A, Rahman S, Shingatgeri VM, Bora RS, Saini KS. Differential gene expression analysis of a known hepatotoxin, N-acetyl-p-amino-phenol (APAP) as compared to its non-toxic analog, N-acetyl-mamino- phenol (AMAP) in mouse liver. The Journal of toxicological sciences. 2008;33:163–173. doi: 10.2131/jts.33.163. [DOI] [PubMed] [Google Scholar]
- 43.Jeong SY, Lim JS, Park HJ, Cho JW, Rana SV, Yoon S. Effects of acetaminophen on hepatic gene expression in mice. Physiological chemistry and physics and medical NMR. 2006;38:77–83. [PubMed] [Google Scholar]
- 44.Reilly TP, Bourdi M, Brady JN, Pise-Masison CA, Radonovich MF, George JW, Pohl LR. Expression profiling of acetaminophen liver toxicity in mice using microarray technology. Biochem Biophys Res Commun. 2001;282:321–328. doi: 10.1006/bbrc.2001.4576. [DOI] [PubMed] [Google Scholar]
- 45.Griffith OW, Meister A. Origin and turnover of mitochondrial glutathione. Proc Natl Acad Sci U S A. 1985;82:4668–4672. doi: 10.1073/pnas.82.14.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogers LK, Tamura T, Rogers BJ, Welty SE, Hansen TN, Smith CV. Analyses of glutathione reductase hypomorphic mice indicate a genetic knockout. Toxicol Sci. 2004;82:367–373. doi: 10.1093/toxsci/kfh268. [DOI] [PubMed] [Google Scholar]
- 47.Moffit JS, Aleksunes LM, Kardas MJ, Slitt AL, Klaassen CD, Manautou JE. Role of NAD(P)H:quinone oxidoreductase 1 in clofibrate-mediated hepatoprotection from acetaminophen. Toxicology. 2007;230:197–206. doi: 10.1016/j.tox.2006.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong H, Haining RL, Thummel KE, Rettie AE, Nelson SD. Involvement of human cytochrome P450 2D6 in the bioactivation of acetaminophen. Drug Metab Dispos. 2000;28:1397–1400. [PubMed] [Google Scholar]
- 49.Guo GL, Moffit JS, Nicol CJ, Ward JM, Aleksunes LA, Slitt AL, Kliewer SA, Manautou JE, Gonzalez FJ. Enhanced acetaminophen toxicity by activation of the pregnane X receptor. Toxicol Sci. 2004;82:374–380. doi: 10.1093/toxsci/kfh286. [DOI] [PubMed] [Google Scholar]
- 50.Thummel KE, Lee CA, Kunze KL, Nelson SD, Slattery JT. Oxidation of acetaminophen to N-acetyl-p-aminobenzoquinone imine by human CYP3A4. Biochem Pharmacol. 1993;45:1563–1569. doi: 10.1016/0006-2952(93)90295-8. [DOI] [PubMed] [Google Scholar]
- 51.Agarwal R, MacMillan-Crow LA, Rafferty TM, Saba H, Roberts DW, Fifer EK, James LP, Hinson JA. Acetaminophen-induced hepatotoxicity in mice occurs with inhibition of activity and nitration of mitochondrial manganese superoxide dismutase. J Pharmacol Exp Ther. 2011;337:110–116. doi: 10.1124/jpet.110.176321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prast-Nielsen S, Cebula M, Pader I, Arnér ESJ. Noble metal targeting of thioredoxin reductase - covalent complexes with thioredoxin and thioredoxin-related protein of 14 kDa triggered by cisplatin. Free Radic Biol Med. 2010 doi: 10.1016/j.freeradbiomed.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 53.James LP, McCullough SS, Lamps LW, Hinson JA. Effect of N-acetylcysteine on acetaminophen toxicity in mice: relationship to reactive nitrogen and cytokine formation. Toxicol Sci. 2003;75:458–467. doi: 10.1093/toxsci/kfg181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.