Abstract
The BRAINS (Brain Research: Analysis of Images, Networks, and Systems) image analysis software has been in use, and in constant development, for over twenty years. The original neuroimage analysis pipeline using BRAINS was designed as a semi-automated procedure to measure volumes of the cerebral lobes and subcortical structures, requiring manual intervention at several stages in the process. Through use of advanced image processing algorithms the need for manual intervention at stages of image realignment, tissue sampling and mask editing have been eliminated. In addition, inhomogeneity correction, intensity normalization, and mask cleaning routines have been added to improve the accuracy and consistency of the results. The fully automated method, AutoWorkup, is shown in this study to be more reliable (ICC ≥ 0.96, Jaccard index ≥ 0.80 and Dice index ≥ 0.89 for all tissues in all regions) than the average of 18 manual raters. On a set of 1130 good quality scans the failure rate for correct realignment was 1.1%, and manual editing of the brain mask was required on 4% of the scans. In other tests, AutoWorkup is shown to produce measures that are reliable for data acquired across scanners, scanner vendors, and across sequences. Application of AutoWorkup for the analysis of data from the 32-site, multi-vendor PREDICT-HD study yield estimates of reliability to be greater than or equal to 0.90 for all tissues and regions.
Keywords: BRAINS, Automated image analysis, pipeline, volumetric analysis, morphometry, segmentation
Introduction
The BRAINS (Brain Research: Analysis of Images, Networks, and Systems) image analysis software (Andreasen et al., 1996; Magnotta et al., 2002) has a history of development at The University of Iowa for over twenty years.
From early on in the development of the BRAINS software suite, a semi-automated image analysis pipeline was available to analyze anatomical brain scans in a reliable and valid manner. These tools have been reported in detail previously (Andreasen et al., 1996; Harris et al., 1999; Magnotta et al., 1999; Pierson et al., 2002; Powell et al., 2008; Spinks et al., 2002). The analysis is composed of four major steps:
Realign and coregister all scans into the same orientation and resolution,
Perform tissue classification,
Define the borders of the brain and parcellate it into subregions and structures, and
Measure volumes of regions of the brain and various tissues within the defined regions.
Each of the four cortical lobes is evaluated with volume measures for each hemisphere of gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) volumes (Andreasen et al., 1996; Harris et al., 1999). Both total CSF and separate surface and internal CSF measures are available. The methods used in these processes were validated against manually traced cortical lobes and tissue types. In addition, several studies have been undertaken to review the reliability and reproducibility of BRAINS (Agartz et al., 2001; Okugawa et al., 2003b). Finally, through application of artificial neural networks trained on manual exemplars (Magnotta et al., 1999; Powell et al., 2008) measures may also be completed for the brain, caudate, putamen, nucleus accumbens, globus pallidus, thalamus, and hippocampus.
The goals for the development of BRAINS have been to continuously refine the software processes and user experience, and to take advantage of advances in algorithms, software engineering, and computer hardware. Through this effort, the software that was initially developed as a manual image-tracing tool has evolved to include methods for image registration (linear and non-linear), tissue classification, automated labeling, brain extraction, surface generation, and diffusion tensor analysis.
BRAINS has been used to investigate a multitude of populations and disorders, for example, in normal development (Tiemeier et al., 2010), childhood neglect (Bauer et al., 2009), post-traumatic stress disorder (Jatzko et al., 2006), attention deficit hyperactivity disorder (Mackie et al., 2007), autism (Jou et al., 2009), dementia and Alzheimer’s disease (Krueger et al., 2009), temporal lobe epilepsy (Dabbs et al., 2009), anorexia nervosa (McCormick et al., 2008), cleft lip and palette (Weinberg et al., 2009), Huntington disease (Nopoulos et al., 2007; Paulsen et al., 2010), fetal alcohol syndrome (Swayze et al., 1997), drug abuse (Block et al., 2000; O'Leary et al., 2007), alcohol abuse (Hill et al., 2009), phenylketonuria (Pfaendner et al., 2005), and schizophrenia (Crespo-Facorro et al., 2007; Ho et al., 2007; Ho, 2007; Ho and Magnotta, 2010; Okugawa et al., 2003a). The successful use of the BRAINS software suite across many populations and diseases has been a primary motivation for implementing robust tools that can be consistently and easily applied to a wide range of disease states.
In this report we describe the development of a fully automated method for analysis of brain MRI using tools available in BRAINS2. The semi-automated standard workup used until now required the investment of 2 to 6 hours of human time due to the stages that required manual intervention. Historically, manual intervention was necessary to ensure accuracy and validity of all of the data. However, the methods developed in this paper have automated and refined these stages so that the quality of results from the automated method now exceeds those of the methods that depend on manual intervention.
Methods
When developing the automated workup procedure for anatomical brain images (AutoWorkup) a set of guiding principles was identified to govern the development.
Variance in brain tissue measures due to any source other than anatomical differences is to be reduced as much as possible. These sources of variance include scanner characteristics and image artifacts.
Accuracy and validity of the workup are the most important benchmarks. When developing the modules very little compromise will be made in these as a tradeoff with other factors, such as failure rates or speed of completion.
Within the limitations of 1) and 2) above, success rates of each step in the workup should be maximized, ideally 100%. Whenever automated methods require human intervention, time is wasted and unwanted variability is introduced.
When accuracy or success rates of a simple process suffer due to automation, use iterative methods to work gradually towards highly accurate and reproducible results. Using several successively more-accurate iterations of a robust basic process is preferable to creating a complex process with numerous decision points.
Modularize the process so that stages of the workup are easily modified as needed for customization of the analysis. Modifications may extend even to the complete substitution of modules, such as tissue classification, cortical lobe or subcortical structure definition, without major refactoring of the pipeline.
In the rare case of failure at some point in the process, design the flow control to be able to restart at the point of manual intervention to complete the rest of the automated workup methods.
A summary of the stages in AutoWorkup are listed in Table 1.
Table 1.
| Major Steps in Iowa AutoWorkup |
|---|
| Reorient T1 to template |
| Coregister other modalities to T1 |
| Create initial estimate of brain |
| Initial Tissue Classification |
| Create ANN/MUSH brain mask |
| BiasCorrector inhomogeneity correction |
| Intensity Normalization |
| Save out final modal images |
| Final Tissue Classification |
| Create final ANN/MUSH brain mask |
| Measure General Brain/Cortex volumes |
| Subcortical Parcellation |
| Measure Subcortical Volumes. |
Template Images
To provide a basis for coregistration and preliminary segmentation, a template image was created from synthesized average anatomies using inverse-consistent registration (Christensen et al., 2006). The template image was created using scans from patients and healthy controls from the Iowa studies of schizophrenia and were chosen to reflect a wide range of brain and ventricle size and shape. The resulting template image reflects a true average of the subjects used in its creation, with the shape and size being the actual mean for the population. Therefore, the image space is not either MNI or Talairach, but specifically chosen to represent a population average. The final atlas set contains average images of the skull-stripped T1 images, complete T1 images, and brain probability images; masks of the average brain and cerebrum; and parameters defining the Talairach atlas on this set of template images.
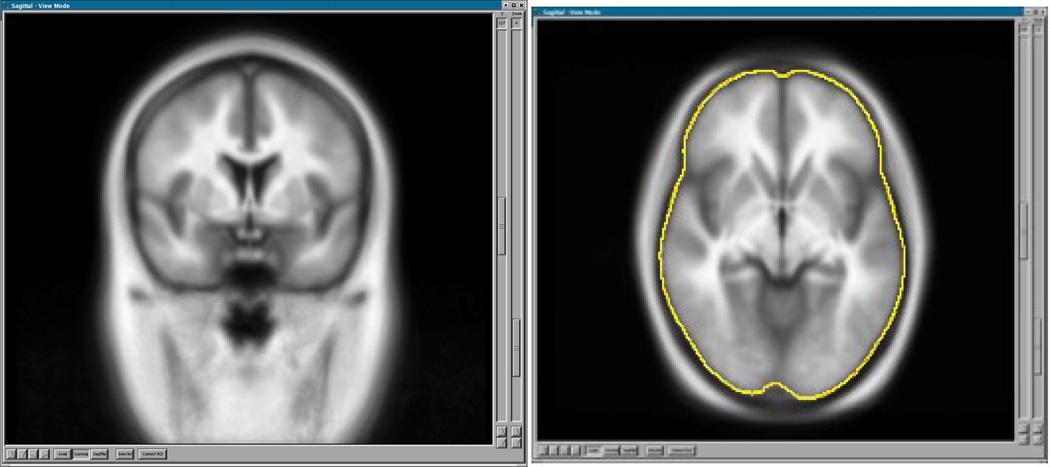
The average T1 and its associated brain mask are shown in Figure 1.
Fig. 1.
The template created from 108 T1 scans. The axial view shows in yellow the outline of the brain mask created by thresholding the brain probability at 0.5.
Spatial alignment
The reorientation of the T1 weighted image is accomplished in a multi-step approach using the BRAINSFit (Johnson et al., 2007) program to coregister the T1 to the 108-subject template image space. First, 3dSkullStrip (Cox, 1996) creates an initial estimate of the brain. Next, the T1 image is intensity normalized to linearly map the intensities of the T1 image within the 3dSkullStrip brain region to extend from 0 to 255. At this point, a new 3dSkullStrip brain mask is created. The brain mask for the subject is co-registered to the template brain mask with BRAINSFit using a six parameter registration to bring the images roughly into alignment, then a nine parameter registration to “fine-tune” the orientation. The use of two stages of registration is employed to reduce the failure rate of the process. In the first coregistration, rigid registration, BRAINSFit initializes using the center of the head to correct the main translational differences. The rigid registration is then completed using a high rotational change in the initial iterations. The second stage of registration, using translation, rotation, and scale, is initialized using the transform from the rigid registration. This allows the maximal fit of the individual brain mask to the template brain mask, reducing minor variability in orientation of the brain. The scale differences between the subject and atlas are used to generate a new atlas image of approximately the same size as the subject. The rigid parameters are extracted from the transform of the nine-parameter registration and utilized to resample the T1-weighted image into its final orientation. Skull stripping is repeated, and the mask is processed to remove small connections to tissues outside the brain. This procedure achieves this by first eroding a mask three-dimensionally by 3 voxels, then finding the largest connected region. This largest connected region is then dilated by 5 voxels three-dimensionally, and the result is intersected with the original mask. This results in the erroneous regions of the brain mask (those that are not connected to the main volume or are connected by a stalk of no more than 6 voxels thickness) being removed from the main volume. Finally, the Talairach parameters (AC and PC points) are resampled from the average template space into the subject’s realigned T1 image space.
Next the T2 image is coregistered to the resampled T1 image using a rigid body registration. The raw PD is then coregistered to the T2 (in case the PD had not been acquired in a dual-echo sequence with the T2). The PD-to-T2 transform is combined with the T2-to-T1 transform to bring it into coregistration with the atlas-aligned T1 weighted image.
The spatial alignment process results in the images being resampled into an orientation with the midsagittal plane in the center of the image space, with AC and PC points on a horizontal line in the midsagittal plane. This reflects a Talairach orientation, but the images are not scaled in any fashion; they retain the actual size of the subject’s brain.
Inhomogeneity correction – including initial tissue sampling, classification, and brain segmentation
The discriminant tissue classification method (Harris et al., 1999) is then used on the T1 and T2 images to create a continuous classified image. Training classes are created from ‘plugs’ created around a number of points randomly chosen within the brain to sample the brain tissue. The mask generated from the T1 weighted image, after alignment with the atlas image, is eroded by 5 mm and serves as the region for picking tissue plugs. After passing tests for minimal variance and coverage, these plugs are grouped by intensity into three classes, or tissue types, using a multidimensional K-means clustering. Multiple samplings of 4000 plugs each are acquired iteratively, with the K-means clustering assigning the plugs to the various classes. This continues until the tissues are represented by a minimum of 2000 white matter plugs, 4000 gray matter plugs, and 200 CSF plugs. The plugs in each class must also represent tissues spanning at least 85% of the brain region in each dimension, and plug picking will continue until this additional condition is met. After the initial plug picking, a region within 5 mm of the brain mask border is isolated for picking exemplars of blood. Choosing voxels within this region that satisfy the criteria of having intensities ranging from 60% to 80% of the grey matter mean on the T1 image and less than 20% of the grey matter mean on the T2 image provides a representation of blood. Using these training classes, a set of discriminant functions is created and the image space is classified, voxel by voxel, into the various tissue types. A continuous-classified image is created, where the intensity of each voxel codes for the likelihood of its being the specific tissue type. An intensity of 10 represents pure cerebrospinal fluid, 130 for pure gray matter, and 250 for pure white matter. Intensities in ranges between these values represent the partial volume effects of that voxel having two different classifications. Blood is given an intensity of 1, and voxels that were not classifiable by the discriminant functions (that is, are not multispectrally similar to CSF, grey matter, white matter or blood) are assigned an intensity of 0.
An automated segmentation algorithm based on an artificial neural network (ANN) is then used to define the brain, including surface CSF, on the continuous tissue classified image (Magnotta et al., 1999). A second brain mask is generated using the maximum uniformity summation heuristic (MUSH) algorithm (Pierson et al., 2009). This algorithm generates a synthetic image by summing the T1 and T2 weighted images in proportions that minimize the variance of the signal intensity within an estimate of the brain compartment. This synthetic image can be thresholded to generate a separate estimate of the brain mask, including CSF. The ANN and MUSH brain masks are then combined using a procedure that uses a reference mask to clean a newly generated mask. Regions that differ by less than 3 voxels thickness, three dimensionally, are left alone on the new mask. Regions that differ by more than that are either filled in or removed to match the reference mask. This allows the majority of a new ANN-generated brain mask to accurately reflect the anatomy of a scan, while those areas that go awry due to “leaking” into the neck or skull can be clipped.
The initial tissue classification process provides tissue intensity profiles and an accurate estimate of the brain to be used for inhomogeneity correction. Using the inverse of the final spatial transforms for each raw image, the brain mask is resampled into the raw image space for each modality. Inhomogeneity correction is then performed on the original raw images (Styner et al., 2000) using a 3rd order Legendre polynomial to model and correct the inhomogeneity. After inhomogeneity correction and intensity normalization, the images are resampled back to their final, atlas based orientation.
Final tissue classification, parcellation, and measurements
Because the images are now corrected for inhomogeneity, a new tissue classification is performed, identical to that described above, using the newly resampled, inhomogeneity corrected, and intensity normalized images. The brain estimate used in this classification for picking tissue exemplars is the brain mask created after the first tissue classification step, since it is a more accurate representation of the intracranial space. After this tissue classification, a new brain mask is again created using a combination of the ANN and MUSH algorithms. The prior estimates of the Talairach landmarks, defining the bounds of the cerebral cortex, are adjusted based on this final brain mask.
Using an atlas-based anatomical identification procedure (Andreasen et al., 1996), tissue volumes for grey matter, white matter, and CSF (surface and internal) are measured for the four cortical lobes (frontal, temporal, parietal, and occipital), ventricles, and subcortical structures. Using an augmented ANN algorithm (Powell et al., 2008), subcortical and cerebellar regions are parcellated. Volumetric measurements of these regions are also obtained.
Application of AutoWorkup
AutoWorkup was applied to several different studies to assess its accuracy, reproducibility, failure rates and reliability, as well as to assess the comparability of results obtained when applying AutoWorkup to data acquired on different scanners and with different scanning sequences.
Reliability between AutoWorkup and Semi-Automated Workup
To evaluate the reliability of AutoWorkup compared to the semi-automated standard workup, a set of 15 scans, of good quality and acquired within the time span of one year on the same scanner, were processed using both the semi-automated method and the fully automated AutoWorkup. Each participant’s data included T1, T2, and PD weighted images collected on a 1.5T GE Signa scanner. The T1 weighted scan was acquired using a 3D spoiled recalled gradient echo sequence with the following scan parameters: TE = 5 ms, TR = 24 ms, flip angle = 40°, NEX = 2, FOV = 26 × 19.2 × 18.6 cm, matrix = 256 × 192 × 124. The PD and T2 weighted scans were acquired using a fast spin-echo sequence with the following parameters: TE = 28/96 ms for the PD and T2 respectively, TR = 3000 ms, slice thickness/gap = 3.0 to 4.0 mm / 0.0 mm, NEX = 1, FOV = 26 cm, matrix = 256 × 192, ETL = 8.
Intra-class correlation, Dice coefficient (Dice, 1945) and Jaccard index (Jaccard, 1912) were calculated for each major region, separately for gray matter, white matter, and CSF (total, internal, and surface). The ICC was calculated through use of restricted maximum likelihood estimates of variance, and is equal to the estimated variance across subjects divided by the sum of all variance estimates (Shrout and Fleiss, 1979). The Dice coefficient is defined as:
where the numerator is twice the number of voxels in the intersection between the tissue masks of the semi-automated and AutoWorkup results for each region and the denominator is the sum of the voxels in each tissue mask. The Jaccard index is calculated as:
where the numerator is the number of voxels in the intersection and the denominator is the number of voxels in the union of the tissue masks in each region for the two methods. Since the resulting images are in different spaces (the semiautomated method centers the image midway between the anterior commissure and posterior commissure, and AutoWorkup centers the images on the anterior commissure), it was necessary to transform the masks from the semiautomated method into the AutoWorkup image space for each scan before these could be calculated.
Evaluation of success rate on a large study
A sample of 1130 scans that had successfully been processed through semi-automated standard workup was processed through AutoWorkup. The sample was composed of participants from the Iowa studies of schizophrenia and included 213 controls and 392 patients. The scans had been acquired over a span of 18 years, with several major scanner software and hardware upgrades occurring during this time. The artifacts present in this sample were quite varied, including compressed dynamic range, wrapping, motion, and a variety of inhomogeneity artifacts. Semi-automated workup on this group of scans had been completed by 13 different raters.
Reliability across scanners
At the time of a scanner replacement in a longitudinal study, a group of 10 participants were scanned on the GE Signa 1.5T scanner using the sequence described above, and also on a Siemens Avanto 1.5T scanner using the following sequence. A coronal T1 FLASH was acquired with TE = 5 ms, TR = 24 ms, flip = 30, FOV = 260 × 260 × 216 mm, Matrix = 256 × 192 × 144, NEX = 2, BW = 160 Hz/pixel. A coronal PD/T2 was acquired with TE = 26/90 ms, TR = 9500 ms, FOV = 260 × 260, Matrix = 256 × 256, NEX = 1, Slice thickness = 3.0 mm, ETL = 5, BW = 100 Hz/pixel. The scans were processed through AutoWorkup successfully. Reliability (ICC, R2), Dice coefficient and Jaccard index were calculated for the cross-scanner comparison, as well as the average differences in volumes between the various measures for the two scanners.
Reliability across scanners and acquisition parameters
The sequence from the GE Signa scanner, above, was acquired on 60 participants, and an additional high-resolution sequence was acquired on a GE/CVi scanner with the following parameters: The T1 weighted scan was acquired using a 3D spoiled recalled gradient echo sequence with the following scan parameters: TE = 6 ms, TR = 20 ms, flip angle = 30°, FOV = 160×160×192 mm, matrix = 256×256×124, NEX = 2. The T2 images were acquired using a 2D fast spin-echo sequence in the coronal plane with the following parameters: TE = 85 ms, TR = 4800 ms, slice thickness/gap = 1.8/0.0 mm, FOV = 160×160 mm, matrix = 256×256, NEX = 3, number of echoes = 8, number of slices = 124. To assess the feasibility of combining data from both the high-resolution and the standard-resolution studies into one, larger study, only the modalities common to both (T1 and T2) were used in the analysis. To further reduce the differences, a preprocessing step was included to down-sample the raw images from the high-resolution sequence to the same resolution as the raw images from the low-resolution sequence. All other stages of AutoWorkup were completed normally.
Application to 3T scanners
A set of 50 scans acquired on an Siemens 3T TrioTim scanner were processed through AutoWorkup. A coronal MPRAGE sequence was acquired with TE = 2.8 ms, TR = 2530 ms, flip = 10, FOV = 256 × 256 × 256 mm, Matrix = 256 × 256 × 256, NEX = 1, BW = 180 Hz/pixel. A 3D T2 was acquired with TE = 404 ms, TR = 4000 ms, FOV = 176 × 227.5 × 260, Matrix = 176 × 256 × 224, NEX = 1, Slice thickness = 1.0 mm, BW = 592 Hz/pixel. To properly handle the higher range of inhomogeneities, the inhomogeneity correction algorithm (BIASCorrector) used a 5th order Legendre polynomial instead of 3rd order, and the blood sampling algorithm was restricted to use only those voxels with intensities on the T2 image that were below 6% of the mean gray matter intensities. No other modifications were attempted.
Application to a large, multi-site study
To evaluate the applicability of the method to multi-site, multi-scanner, longitudinally collected studies, the data from the PREDICT-HD study (Paulsen et al., 2006; Paulsen et al., 2008) were processed using AutoWorkup. Scans of 657 participants from 32 sites, acquired on 32 different scanners (30 GE scanners and 2 Siemens scanners) were included in this part of the study. Participants only received scans at their home site, so data were not available for a rigorous calculation of cross-site reliability.
For this comparison we calculated an “r-square-style” statistic for scanner effects that is generally analogous to the ICC statistics. In this case, the statistic describes the proportion of variance for each structure that is estimated to not be due to intra-scanner variability. It cannot strictly be interpreted as an intraclass or reliability correlation, because different subjects were used at each site. It can, however, be interpreted as an approximation to such statistics, if we assume that the mixture of subjects is approximately similar among sites and scanners. These variances were derived by fitting a random effect model with no fixed effects except an intercept. A random effect for unique scanners was included, and the statistics are derived from variance estimates for the scanner effect and the residual, which can be thought of as the variance due to all other influences. The ratio of variance without scanner effects to total variance is reported as the “Estimated ICC.”
Software
BRAINS is written in C++ and builds upon ITK and VTK. The BRAINS software is currently available for both Linux and Mac OSX platforms from the NITRC website (www.nitrc.org), BRAINS supports a number of standard file formats including NIFTI, allowing the resulting images and masks to be readily utilized by a wide variety of other packages, including SPM, FreeSurfer, AFNI, and FSL. The automated pipeline is instrumented using the TCL scripting language, allowing easy modification and customization.
Results
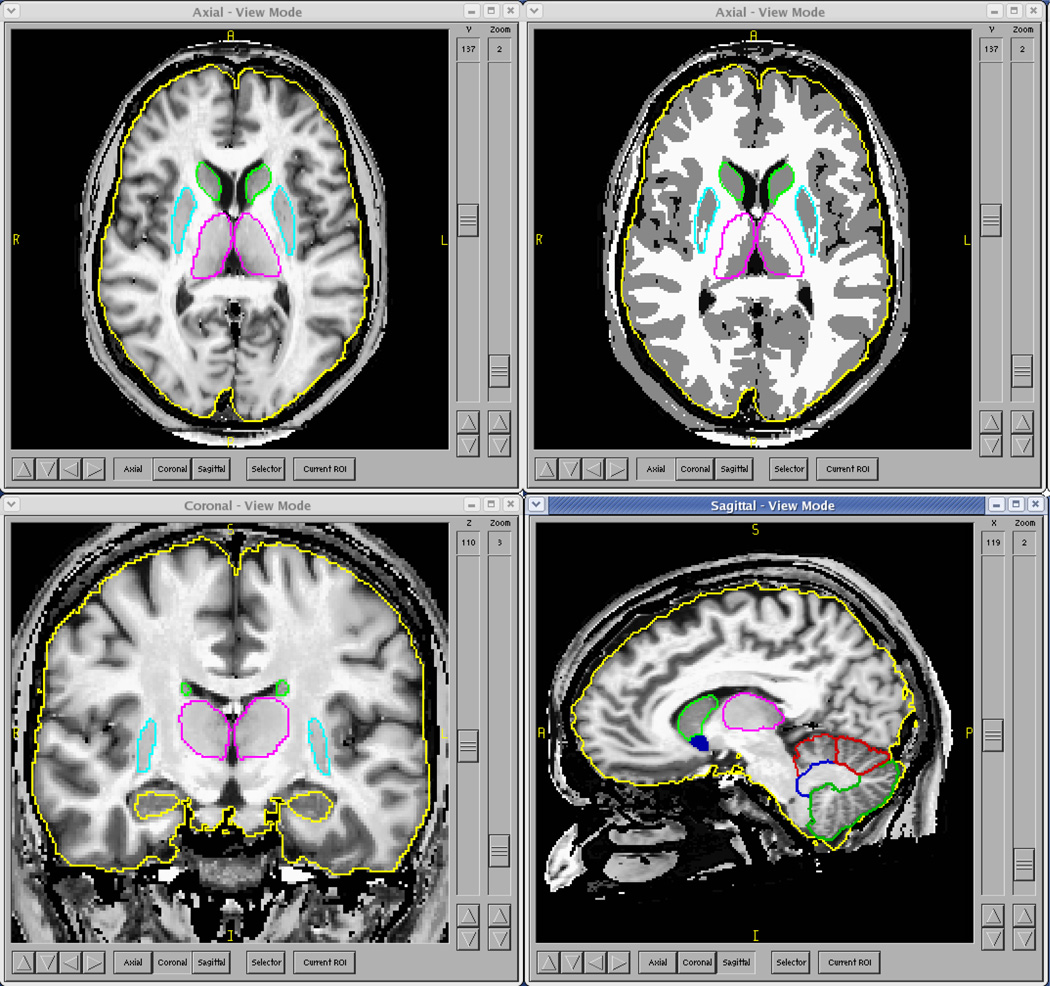
AutoWorkup was applied to several different studies to assess its accuracy, reproducibility, failure rates, and reliability. The simple end user interface for AutoWorkup allowed many scans to be processed simultaneously on a cluster of computers. A single fully automated process completed in an average of 72 minutes per scan. An example of a 3T scan from a Siemens Trio scanner processed through AutoWorkup is shown in Figure 2.
Fig. 2.
Final Review of AutoWorkup results and masks created by use of artificial neural networks. The upper right view shows the discrete-classified image; the other views are displaying the continuous-classified image. The scan was acquired on a 3T Siemens Trio scanner.
Reliability
The results for the reliability study comparing the semi-automated methods of BRAINS workup with AutoWorkup are listed in Tables 2 and 3. The vast majority of the regions have very high intraclass correlation, R2 ≥ 0.99. The lowest was 0.96 for cranial surface CSF (CSF on the entire surface of the brain). The Dice coefficients for gray and white matter ranged from 0.89 to 0.95, and for CSF ranged from 0.72 to 0.89. Jaccard indices for gray matter and white matter ranged from 0.79 to 0.92, and for CSF ranged from 0.72 to 0.85. This set of scans had been used for approximately 10 years for rater validation on the semi-automated workup, so data were available for 18 raters compared to the same gold-standard rater (HK). Average ICC’s for those 18 raters are also shown in Table 2.
Table 2.
Comparison of the reliability of AutoWorkup versus the gold-standard rater, with the average reliabilities of 18 other raters.
| AutoWorkup ICC | Average ICC – 18 | |||
|---|---|---|---|---|
| Raters | ||||
| Region | Left | Right | Left | Right |
| Total Internal CSF | 0.98 | 0.98 | 0.96 | 0.97 |
| Total Surface CSF | 0.96 | 0.96 | 0.91 | 0.93 |
| Total CSF | 0.98 | 0.99 | 0.94 | 0.95 |
| Total GM | 0.99 | 0.99 | 0.97 | 0.97 |
| Total WM | 0.99 | 0.97 | 0.94 | 0.94 |
| Frontal Internal CSF | 0.99 | 0.98 | 0.93 | 0.93 |
| Frontal Surface CSF | 0.99 | 0.98 | 0.93 | 0.94 |
| Frontal CSF | 0.99 | 0.98 | 0.94 | 0.95 |
| Frontal GM | 0.99 | 0.99 | 0.93 | 0.95 |
| Frontal WM | 0.98 | 0.99 | 0.92 | 0.93 |
| Occipital Internal CSF | 0.99 | 0.99 | 0.93 | 0.93 |
| Occipital Surface CSF | 0.97 | 0.97 | 0.74 | 0.59 |
| Occipital CSF | 0.99 | 0.99 | 0.91 | 0.91 |
| Occipital GM | 0.99 | 0.98 | 0.89 | 0.89 |
| Occipital WM | 0.99 | 0.99 | 0.87 | 0.85 |
| Parietal Internal CSF | 0.99 | 0.99 | 0.95 | 0.95 |
| Parietal Surface CSF | 0.99 | 0.99 | 0.88 | 0.91 |
| Parietal CSF | 0.99 | 0.99 | 0.93 | 0.94 |
| Parietal GM | 0.99 | 0.99 | 0.91 | 0.91 |
| Parietal WM | 0.99 | 0.99 | 0.94 | 0.92 |
| Temporal Internal CSF | 0.99 | 0.97 | 0.88 | 0.92 |
| Temporal Surface CSF | 0.99 | 0.98 | 0.83 | 0.78 |
| Temporal CSF | 0.99 | 0.98 | 0.90 | 0.91 |
| Temporal GM | 0.99 | 0.99 | 0.85 | 0.89 |
| Temporal WM | 0.98 | 0.99 | 0.88 | 0.87 |
| Ventricular CSF | 0.99 | 0.99 | 0.99 | 0.99 |
Table 3.
Comparison of the Dice coefficient and the Jaccard index versus the gold-standard rater. Scans from the semiautomated workup were transformed into the AutoWorkup image space to allow voxel-wise comparisons.
| Dice Coefficent | Jaccard Index | |||
|---|---|---|---|---|
| Region | Left | Right | Left | Right |
| Total Internal CSF | 0.87 | 0.87 | 0.81 | 0.82 |
| Total Surface CSF | 0.86 | 0.89 | 0.76 | 0.78 |
| Total CSF | 0.87 | 0.88 | 0.79 | 0.81 |
| Total GM | 0.95 | 0.95 | 0.92 | 0.91 |
| Total WM | 0.92 | 0.93 | 0.89 | 0.90 |
| Frontal Internal CSF | 0.88 | 0.89 | 0.85 | 0.86 |
| Frontal Surface CSF | 0.87 | 0.89 | 0.83 | 0.85 |
| Frontal CSF | 0.88 | 0.89 | 0.84 | 0.85 |
| Frontal GM | 0.93 | 0.93 | 0.86 | 0.86 |
| Frontal WM | 0.94 | 0.95 | 0.87 | 0.88 |
| Occipital Internal CSF | 0.75 | 0.72 | 0.72 | 0.70 |
| Occipital Surface CSF | 0.76 | 0.78 | 0.71 | 0.75 |
| Occipital CSF | 0.76 | 0.75 | 0.71 | 0.72 |
| Occipital GM | 0.92 | 0.91 | 0.83 | 0.82 |
| Occipital WM | 0.89 | 0.90 | 0.79 | 0.82 |
| Parietal Internal CSF | 0.86 | 0.89 | 0.79 | 0.82 |
| Parietal Surface CSF | 0.85 | 0.79 | 0.80 | 0.79 |
| Parietal CSF | 0.86 | 0.85 | 0.80 | 0.81 |
| Parietal GM | 0.93 | 0.94 | 0.87 | 0.86 |
| Parietal WM | 0.94 | 0.94 | 0.88 | 0.89 |
| Temporal Internal CSF | 0.82 | 0.81 | 0.72 | 0.72 |
| Temporal Surface CSF | 0.79 | 0.82 | 0.73 | 0.79 |
| Temporal CSF | 0.81 | 0.81 | 0.72 | 0.75 |
| Temporal GM | 0.95 | 0.94 | 0.89 | 0.88 |
| Temporal WM | 0.91 | 0.90 | 0.82 | 0.85 |
| Ventricular CSF | 0.94 | 0.93 | 0.90 | 0.91 |
Success rate
All of the scans were visually inspected after AutoWorkup was performed. Only 13 of the scans (1.1%) failed to be reoriented or coregistered correctly, and an additional 46 (4%) had brain masks that required editing of more than 25 cc (approximately 2% of total brain volume). Of those that passed visual inspection the root mean square difference between manual and AutoWorkup definition of the anterior commissure was 0.93 mm and of the posterior commissure was 1.2 mm.
Reliability across scanners
In the comparison of the reliability for using two different scanners to assess brain morphometry, as measured using AutoWorkup, the intraclass correlations for the brain measures were all greater than 0.9, except for occipital white matter. The Dice coefficients for gray and white matter ranged from 0.75 to 0.97. Jaccard indices for gray matter and white matter ranged from 0.72 to 0.97.
Results are shown in Table 3. In addition, the percent differences in tissue volumes in each region are reported, with only the occipital gray and white matter exceeding 3% difference.
Reliability across scanners and acquisition parameters
In this reliability comparison the sequences used for acquisition were quite different, and though the scanners were both from the same vendor (General Electric), the scanners were also different. All scans completed processing through AutoWorkup successfully, with minor brain mask trimming needed on four scans. Reliability was calculated for the cross-scanner/cross-sequence comparison, as well as differences in average volumes for each tissue in the main regions of the atlas-based measures. Results are shown in Table 4. As in the previous cross-scanner study, the intraclass correlations were all above 0.9 except for, in this case, occipital gray and white matter. The Dice coefficients for gray and white matter ranged from 0.82 to 0.99. Jaccard indices for gray matter and white matter ranged from 0.75 to 0.99. Percent differences in tissue volumes were generally less than 1%, with occipital white matter and gray matter each near 2%.
Table 4.
A reliability comparison of measures produced by AutoWorkup using standard, trimodal acquisitions from a GE Signa 1.5T scanner, and a Siemens Avanto 1.5T scanner on 10 subjects.
| Region/Tissue | ICC (R2) | Dice Coefficent |
Jaccard Index | % difference (GE – Siemens) |
|---|---|---|---|---|
| Cerebral Tissue | 0.98 | 0.97 | 0.97 | 1.2 |
| Cerebral GM | 0.94 | 0.92 | 0.90 | 1.4 |
| Cerebral WM | 0.99 | 0.97 | 0.94 | 0.02 |
| Frontal GM | 0.96 | 0.92 | 0.86 | −0.82 |
| Frontal WM | 0.99 | 0.93 | 0.88 | −0.92 |
| Temporal GM | 0.90 | 0.88 | 0.84 | 2.7 |
| Temporal WM | 0.94 | 0.90 | 0.86 | −0.21 |
| Parietal GM | 0.96 | 0.92 | 0.86 | −1.2 |
| Parietal WM | 0.94 | 0.90 | 0.88 | 1.7 |
| Occipital GM | 0.91 | 0.87 | 0.82 | −3.24 |
| Occipital WM | 0.79 | 0.75 | 0.72 | 9.0 |
| Surface CSF | 0.97 | 0.93 | 0.83 | 0.4 |
Application to 3T scanners
Of the 50 scans processed through AutoWorkup, only 1 scan was considered a failure. This was due to abnormally large inhomogeneities in the subcortical and occipital regions. These inhomogeneities were not adequately corrected, leading to misclassification of the occipital white matter as gray matter and venous blood.
Application to a large, multi-site study
As described above, a strict reliability estimate through use of an intraclass correlation was unavailable due to the lack of data on different scanners for any one subject. The ICC estimates shown in Table 5 describe the proportion of variance for each structure that is estimated to not be due to intra-scanner variability. For all measures except occipital gray matter the estimated ICC’s are in excess of 0.9. In addition, the standard deviation with and without the estimated effect of scanner are included.
Table 5.
A reliability comparison of measures produced by AutoWorkup using scans acquired on different GE scanners at different resolutions and using different scanning sequences on 60 subjects.
| Region/Tissue |
ICC (R2) |
Dice Coefficent |
Jaccard Index |
% difference (High resolution) – (Low resolution) |
|---|---|---|---|---|
| Cerebral Tissue | 0.99 | 0.99 | 0.99 | 0.25% |
| Cortical GM | 0.97 | 0.96 | 0.95 | −0.21% |
| Cerebral WM | 0.98 | 0.98 | 0.96 | 0.92% |
| Frontal GM | 0.97 | 0.91 | 0.84 | 0.44% |
| Frontal WM | 0.98 | 0.93 | 0.85 | 0.12% |
| Temporal GM | 0.96 | 0.92 | 0.87 | 0.98% |
| Temporal WM | 0.96 | 0.90 | 0.80 | −1.2% |
| Parietal GM | 0.92 | 0.88 | 0.85 | −0.45 |
| Parietal WM | 0.96 | 0.92 | 0.88 | 0.11% |
| Occipital GM | 0.88 | 0.83 | 0.77 | −2.2% |
| Occipital WM | 0.88 | 0.82 | 0.75 | 1.8% |
| Surface CSF | 0.97 | 0.95 | 0.92 | 1.2% |
Discussion
The assessment of AutoWorkup
Through creative use of modern image analysis tools, AutoWorkup has been developed as a valid and highly reliable tool for measurement of brain morphometry.
In the studies to evaluate AutoWorkup it was shown that the process was reliable when compared with semi-automated standard workup as completed by our lab’s expert rater. In fact, in comparison to this expert rater, it is more reliable than the average of 18 trained human raters. AutoWorkup was also shown to have a very high rate of successful completion, approximately 99% for good quality scans with few artifacts.
In these studies AutoWorkup was shown to be efficient, completing the process in an average of 72 minutes per data set. With implementation in a cluster, a large number of scans can be processed in a reasonable time—more than 100 scans per week, per cpu.
In the two studies to evaluate scanner and sequence comparability, AutoWorkup was shown to allow combining of data sets across scanner vendors and acquisition sequences, with relatively high reliability when comparing the measures from the scans acquired on two different scanners and sequences, and relatively minor differences in tissue composition across most regions of the brain. This is also borne out by the analysis of variance for the multi-site PREDICT-HD study, where the estimates of intraclass correlations for each region were very similar to those of the strict cross-scanner, cross sequence reliability studies. It is interesting to note that the one region that consistently was least reliable across all of the studies was the occipital lobe. It may be that this region experiences the most extreme inhomogeneity, which the current algorithm is unable to completely normalize. This is also supported by the results of the 3T scans that were processed; the one scan that was considered a failure had relatively large uncorrected inhomogeneity in the occipital region.
Insight into the developmental goals
In addition to automation of tasks that previously required manual intervention, additional methods were implemented to enhance the face validity and reliability of the results. The “gold standard” used for comparison in this study was the semi-automated, standard workup. To ensure the technicians were able to produce consistent results, formal reliability testing was necessary, such as that described in (Agartz et al., 2001; Okugawa et al., 2003b). These studies show that with trained raters, very little difference would be found in the measures produced by two different raters using the semi-automated method. In the development of AutoWorkup there are additional processes that may have improved the consistency, accuracy, and reliability of AutoWorkup to be at least as good and possibly better than that of the semiautomated methods.
First, reorienting to an atlas orientation may provide a more consistent alignment for the brain, rather than using the anterior and posterior commissures. The alignment with the Talairach brain segmentation atlas may also remain more consistent due to this. This may create better alignment with brain probability maps for the basic ANN, resulting in better brain masks.
In AutoWorkup the use of the maximum uniformity summation heuristic to generate a second estimate of the brain to “clean” the ANN-generated brain mask results in very little need to manually edit the resulting brain masks. For the semi-automated standard workup, every brain mask was cleaned up with manual trimming, whereas only 46 out of 1130 required manual edited for AutoWorkup.
Finally, tissue classification and other procedures may be more stable across scans when the images are inhomogeneity corrected and intensity normalized to optimize the dynamic range within the brain region. In addition, the new ANN for subcortical segmentations requires very little manual trimming (Powell et al., 2008). As a result the entire process for a large group of scans requires a fraction of the manual labor of semi-automated workup, with increased reliability.
Implications for medical imaging research
The use of medical imaging to study the brain for morphometric changes due to neurological and psychiatric disorders has expanded rapidly over the past twenty years. However, imaging results from studies of brain illness obtained from multiple groups often differ due the difficulties in analyzing MR imaging data in a rapid and reliable manner. As summarized in the review article on MRI findings in schizophrenia by Shenton et al. there are a number of brain areas that have positive findings in approximately 60% of the studies and negative findings in the other 40% (Shenton et al., 2001). Regions that have had contradictory findings include the frontal lobe, parietal lobe, temporal lobe, and basal ganglia. Clearly, with this growth in imaging to study the brain there is also an increasing need for reliable tools that automate data analysis.
Several multi-center neuroimaging studies are currently being undertaken, including the Morphology Biomedical Informatics Research Network (MBIRN, structural imaging in Alzheimer's disease), Functional Biomedical Informatics Research Network (fBIRN, functional imaging in schizophrenia), MIND Clinical Imaging Consortium (MCIC, structural and functional imaging in schizophrenia), PREDICT (structural imaging in Huntington disease), and Human Brain Informatics: A Database Project On Schizophrenia (HUBIN, structural imaging in schizophrenia). With the challenges inherent in multisite studies, these will all benefit from rapid, reliable and valid tools for the analysis of the MR data.
There are a number of benefits to using AutoWorkup as compared to the semi-automated standard workup or manual ROI-based analysis of anatomical brain scans.
First, fully automated procedures can produce more consistent results by eliminating rater drift and bias from the analysis. This is especially important for longitudinal studies that take place over a number of years, where personnel turnover is inevitable. As the results from the reliability study show, compared to an average of 18 different human raters, AutoWorkup is more reliable.
Second, there is less startup overhead for an automated process, making it possible for research groups to begin structural imaging research with substantially less investment of start-up time or training.
Third, an automated analysis is fast enough that it allows one to refine or update the methods and repeat the analysis of entire studies in a very short time. For a study that has collected data and measurements for a number of years, AutoWorkup can be run very easily during the data acquisition, even to simply ensure the scan quality is adequate for accurate processing. Then, should there be major updates to AutoWorkup during that time period the data can be reprocessed in a very short period of time. This allows long-term studies to use the most up-to-date methods for eventual publication.
Fourth, for some studies it could be useful to combine several datasets into a single dataset for larger sample sizes, or increased diversity of demographics and disease states. For studies of rare diseases or abnormalities it may be necessary to utilize a multisite design to be able to acquire adequate numbers of participants. For these situations, inhomogeneity correction and intensity normalization, and possibly other stages in AutoWorkup, remove a large amount of the variability in scan features that can affect tissue classification and parcellation. This is clearly supported by the PREDICT-HD imaging data, where the use of 32 different sites and 2 different MRI vendors allowed the measurement of brain volumes with a very low estimated effect due to scanner, and estimates of the standard deviation with and without scanner effects to be very similar.
We did not directly compare the results of the BRAINS automated image analysis pipeline to other software packages (e.g., FreeSurfer, FSL, 3D Slicer). This would be a beneficial area of future work in neuroscience. However, such an effort should be lead by an unbiased investigator providing training and testing data from a variety of scanners. Second, it is likely that the developers of the software packages being compared would obtain the best results.
Future improvements in AutoWorkup will certainly continue as new technologies in image processing become available. BRAINS continues to be developed, with a complete refactoring currently underway that will improve a wide range of new functionality and user options. AutoWorkup is also being implemented in this new version of BRAINS. For the near future improvements will include a new method to more accurately estimate the locations of the AC and PC points, a more robust initial estimate of the brain than 3dSkullStrip, an inhomogeneity correction algorithm that is able to correct more severe inhomogeneities, and continued optimization and validation of the AutoWorkup processing methods for use on high-field and T1-only datasets.
Table 6.
For data acquired in a 32 site study using 32 scanners from 2 vendors, an estimate of the ICC using a ratio of variance not due to scanner effects, compared to the variance due to all effects.
| Region/Tissue |
Estimate of ICC (R2) |
Std. Dev. (cc) | Std. Dev. w/o Scanner effect (cc) |
|---|---|---|---|
| Cerebral Tissue | 0.95 | 132.3 | 128.7 |
| Cortical GM | 0.92 | 61.1 | 58.4 |
| Cerebral WM | 0.97 | 63.0 | 62.1 |
| Frontal GM | 0.92 | 27.9 | 26.8 |
| Frontal WM | 0.96 | 25.9 | 25.3 |
| Temporal GM | 0.90 | 16.9 | 16.0 |
| Temporal WM | 0.97 | 11.1 | 10.9 |
| Parietal GM | 0.92 | 14.1 | 13.5 |
| Parietal WM | 0.97 | 16.4 | 16.1 |
| Occipital GM | 0.85 | 9.5 | 8.8 |
| Occipital WM | 0.92 | 9.0 | 8.7 |
| Surface CSF | 1.00 | 10.7 | 10.7 |
Acknowledgments
This research was supported by funding from grants NS050568 and NS40068 from the National Institute of Neurological Disorders and Stroke and grants MH31593, MH40856, from the National Institute of Mental Health, MHCRC grant MHCRC43271, and CHDI Foundation, Inc. We would like to thank the Iowa MHCRC, the PREDICT-HD sites, all of the study participants, and the National Research Roster for Huntington Disease Patients and Families.
We would especially like to thank the reviewers for their time and suggestions which have helped improve the quality of this manuscript. In addition we would like to recognize and thank Dr. Douglas Langbehn for the statistical analysis of the PREDICT-HD multisite study data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agartz I, Okuguwa G, Nordstrom M, Greitz D, Magnotta V, Sedvall G. Reliability and reproducibility of brain tissue volumetry from segmented MR scans. Eur. Arch. Psychiatry Clin. Neurosci. 2001;251:255–261. doi: 10.1007/pl00007542. [DOI] [PubMed] [Google Scholar]
- Andreasen NC, Rajarethinam R, Cizadlo T, Arndt S, Swayze VW, 2nd, Flashman LA, O'Leary DS, Ehrhardt JC, Yuh WT. Automatic atlas-based volume estimation of human brain regions from MR images. J. Comput. Assist. Tomogr. 1996;20:98–106. doi: 10.1097/00004728-199601000-00018. [DOI] [PubMed] [Google Scholar]
- Bauer PM, Hanson JL, Pierson RK, Davidson RJ, Pollak SD. Cerebellar Volume and Cognitive Functioning in Children Who Experienced Early Deprivation. Biol. Psychiatry. 2009;66:1100–1106. doi: 10.1016/j.biopsych.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block RI, O'Leary DS, Ehrhardt JC, Augustinack JC, Ghoneim MM, Arndt S, Hall JA. Effects of frequent marijuana use on brain tissue volume and composition. Neuroreport. 2000;11:491–496. doi: 10.1097/00001756-200002280-00013. [DOI] [PubMed] [Google Scholar]
- Christensen GE, Johnson HJ, Vannier MW. Synthesizing average 3D anatomical shapes. Neuroimage. 2006;32:146–158. doi: 10.1016/j.neuroimage.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Crespo-Facorro B, Roiz-Santianez R, Pelayo-Teran JM, Gonzalez-Blanch C, Perez-Iglesias R, Gutierrez A, de Lucas EM, Tordesillas D, Vazquez-Barquero JL. Caudate nucleus volume and its clinical and cognitive correlations in first episode schizophrenia. Schizophr. Res. 2007;91:87–96. doi: 10.1016/j.schres.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Dabbs K, Jones J, Seidenberg M, Hermann B. Neuroanatomical correlates of cognitive phenotypes in temporal lobe epilepsy. Epilepsy Behav. 2009;15:445–451. doi: 10.1016/j.yebeh.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice LR. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26:297–302. [Google Scholar]
- Harris G, Andreasen NC, Cizadlo T, Bailey JM, Bockholt HJ, Magnotta VA, Arndt S. Improving tissue classification in MRI: a three-dimensional multispectral discriminant analysis method with automated training class selection. J. Comput. Assist. Tomogr. 1999;23:144–154. doi: 10.1097/00004728-199901000-00030. [DOI] [PubMed] [Google Scholar]
- Hill SY, Wang S, Kostelnik B, Carter H, Holmes B, McDermott M, Zezza N, Stiffler S, Keshavan MS. Disruption of orbitofrontal cortex laterality in offspring from multiplex alcohol dependence families. Biol. Psychiatry. 2009;65:129–136. doi: 10.1016/j.biopsych.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B, Andreasen N, Dawson J, Wassink T. Association between brain-derived neurotrophic factor Val66Met gene polymorphism and progressive brain volume changes in schizophrenia. Am. J. Psychiatry. 2007;164:1890–1899. doi: 10.1176/appi.ajp.2007.05111903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC. MRI brain volume abnormalities in young, nonpsychotic relatives of schizophrenia probands are associated with subsequent prodromal symptoms. Schizophr. Res. 2007;96:1–13. doi: 10.1016/j.schres.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC, Magnotta V. Hippocampal volume deficits and shape deformities in young biological relatives of schizophrenia probands. Neuroimage. 2010;49:3385–3393. doi: 10.1016/j.neuroimage.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard P. The distribution of flora in the alpine zone. New Phytol. 1912;11:37–50. [Google Scholar]
- Jatzko A, Rothenhofer S, Schmitt A, Gaser C, Demirakca T, Weber-Fahr W, Wessa M, Magnotta V, Braus DF. Hippocampal volume in chronic posttraumatic stress disorder (PTSD): MRI study using two different evaluation methods. J. Affect. Disord. 2006;94:121–126. doi: 10.1016/j.jad.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Johnson H, Harris G, Williams K. BRAINSFit: Mutual information registrations of whole-brain 3D images, using the insight toolkit. Insight Journal. 2007 http://hdl.handle.net/1926/1291. [Google Scholar]
- Jou RJ, Minshew NJ, Melhem NM, Keshavan MS, Hardan AY. Brainstem volumetric alterations in children with autism. Psychol. Med. 2009;39:1347–1354. doi: 10.1017/S0033291708004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger CE, Dean DL, Rosen HJ, Halabi C, Weiner M, Miller BL, Kramer JH. Longitudinal Rates of Lobar Atrophy in Frontotemporal Dementia, Semantic Dementia, and Alzheimer's Disease. Alzheimer Dis. Assoc. Disord. 2010;24:43–48. doi: 10.1097/WAD.0b013e3181a6f101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie S, Shaw P, Lenroot R, Pierson R, Greenstein DK, Nugent TF, 3rd, Sharp WS, Giedd JN, Rapoport JL. Cerebellar development and clinical outcome in attention deficit hyperactivity disorder. Am. J. Psychiatry. 2007;164:647–655. doi: 10.1176/ajp.2007.164.4.647. [DOI] [PubMed] [Google Scholar]
- Magnotta VA, Harris G, Andreasen NC, O'Leary DS, Yuh WT, Heckel D. Structural MR image processing using the BRAINS2 toolbox. Comput. Med. Imaging Graph. 2002;26:251–264. doi: 10.1016/s0895-6111(02)00011-3. [DOI] [PubMed] [Google Scholar]
- Magnotta VA, Heckel D, Andreasen NC, Cizadlo T, Corson PW, Ehrhardt JC, Yuh WT. Measurement of brain structures with artificial neural networks: two- and three-dimensional applications. Radiology. 1999;211:781–790. doi: 10.1148/radiology.211.3.r99ma07781. [DOI] [PubMed] [Google Scholar]
- McCormick LM, Keel PK, Brumm MC, Bowers W, Swayze V, Andersen A, Andreasen N. Implications of starvation-induced change in right dorsal anterior cingulate volume in anorexia nervosa. Int. J. Eat. Disord. 2008;41:602–610. doi: 10.1002/eat.20549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nopoulos P, Magnotta VA, Mikos A, Paulson H, Andreasen NC, Paulsen JS. Morphology of the cerebral cortex in preclinical Huntington's disease. Am. J. Psychiatry. 2007;164:1428–1434. doi: 10.1176/appi.ajp.2007.06081266. [DOI] [PubMed] [Google Scholar]
- O'Leary DS, Block RI, Koeppel JA, Schultz SK, Magnotta VA, Ponto LB, Watkins GL, Hichwa RD. Effects of smoking marijuana on focal attention and brain blood flow. Hum. Psychopharmacol. 2007;22:135–148. doi: 10.1002/hup.832. [DOI] [PubMed] [Google Scholar]
- Okugawa G, Sedvall GC, Agartz I. Smaller cerebellar vermis but not hemisphere volumes in patients with chronic schizophrenia. Am. J. Psychiatry. 2003a;160:1614–1617. doi: 10.1176/appi.ajp.160.9.1614. [DOI] [PubMed] [Google Scholar]
- Okugawa G, Takase K, Nobuhara K, Yoshida T, Minami T, Tamagaki C, Magnotta VA, Andreasen NC, Kinoshita T. Inter- and intraoperator reliability of brain tissue measures using magnetic resonance imaging. Eur. Arch. Psychiatry Clin. Neurosci. 2003b;253:301–306. doi: 10.1007/s00406-003-0444-3. [DOI] [PubMed] [Google Scholar]
- Paulsen JS, Nopoulos PC, Aylward E, Ross CA, Johnson H, Magnotta VA, Juhl A, Pierson RK, Mills J, Langbehn D, Nance M the PREDICT-HD Investigators and Coordinators of the Huntington Study Group. Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res. Bull. 2010;82:201–207. doi: 10.1016/j.brainresbull.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Hayden M, Stout JC, Langbehn DR, Aylward E, Ross CA, Guttman M, Nance M, Kieburtz K, Oakes D, Shoulson I, Kayson E, Johnson S, Penziner E the Predict-HD Investigators and Coordinators of the Huntington Study Group. Preparing for preventive clinical trials: the Predict-HD study. Arch. Neurol. 2006;63:883–890. doi: 10.1001/archneur.63.6.883. [DOI] [PubMed] [Google Scholar]
- Paulsen JS, Langbehn DR, Stout JC, Aylward E, Ross CA, Nance M, Guttman M, Johnson S, MacDonald M, Beglinger LJ, Duff K, Kayson E, Biglan K, Shoulson I, Oakes D, Hayden M Predict-HD Investigators and Coordinators of the Huntington Study Group. Detection of Huntington's disease decades before diagnosis: the Predict-HD study. J. Neurol. Neurosurg. Psychiatry. 2008;79:874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaendner NH, Reuner G, Pietz J, Jost G, Rating D, Magnotta VA, Mohr A, Kress B, Sartor K, Hahnel S. MR imaging-based volumetry in patients with early-treated phenylketonuria. AJNR Am. J. Neuroradiol. 2005;26:1681–1685. [PMC free article] [PubMed] [Google Scholar]
- Pierson R, Corson PW, Sears LL, Alicata D, Magnotta V, Oleary D, Andreasen NC. Manual and semiautomated measurement of cerebellar subregions on MR images. Neuroimage. 2002;17:61–76. doi: 10.1006/nimg.2002.1207. [DOI] [PubMed] [Google Scholar]
- Pierson R, Harris G, Johnson H, Dunn S, Magnotta V. Maximize uniformity summation heuristic (MUSH): a highly accurate simple method for intracranial delineation. In: Pluim J, Dawant B, editors. Medical Imaging 2009: Image Processing. Bellingham, WA: SPIE; 2009. [Google Scholar]
- Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. Neuroimage. 2008;39:238–247. doi: 10.1016/j.neuroimage.2007.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton ME, Dickey CC, Frumin M, McCarley RW. A review of MRI findings in schizophrenia. Schizophr. Res. 2001;49:1–52. doi: 10.1016/s0920-9964(01)00163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol. Bull. 1979;86:420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Spinks R, Magnotta VA, Andreasen NC, Albright KC, Ziebell S, Nopoulos P, Cassell M. Manual and automated measurement of the whole thalamus and mediodorsal nucleus using magnetic resonance imaging. Neuroimage. 2002;17:631–642. [PubMed] [Google Scholar]
- Styner M, Brechbuhler C, Szekely G, Gerig G. Parametric estimate of intensity inhomogeneities applied to MRI. IEEE Trans. Med. Imaging. 2000;19:153–165. doi: 10.1109/42.845174. [DOI] [PubMed] [Google Scholar]
- Swayze VW, 2nd, Johnson VP, Hanson JW, Piven J, Sato Y, Giedd JN, Mosnik D, Andreasen NC. Magnetic resonance imaging of brain anomalies in fetal alcohol syndrome. Pediatrics. 1997;99:232–240. doi: 10.1542/peds.99.2.232. [DOI] [PubMed] [Google Scholar]
- Tiemeier H, Lenroot RK, Greenstein DK, Tran L, Pierson R, Giedd JN. Cerebellum development during childhood and adolescence: a longitudinal morphometric MRI study. Neuroimage. 2010;49:63–70. doi: 10.1016/j.neuroimage.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SM, Andreasen NC, Nopoulos P. Three-dimensional morphometric analysis of brain shape in nonsyndromic orofacial clefting. J. Anat. 2009;214:926–936. doi: 10.1111/j.1469-7580.2009.01084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]