Abstract
Background
Sterol esterases and lipases are enzymes able to efficiently catalyze synthesis and hydrolysis reactions of both sterol esters and triglycerides and due to their versatility could be widely used in different industrial applications. Lipases with this ability have been reported in the yeast Candida rugosa that secretes several extracellular enzymes with a high level of sequence identity, although different substrate specificity. This versatility has also been found in the sterol esterases from the ascomycetes Ophiostoma piceae and Melanocarpus albomyces.
Results
In this work we present an in silico search of new sterol esterase and lipase sequences from the genomes of environmental fungi. The strategy followed included identification and search of conserved domains from these versatile enzymes, phylogenetic studies, sequence analysis and 3D modeling of the selected candidates.
Conclusions
Six potential putative enzymes were selected and their kinetic properties and substrate selectivity are discussed on the basis of their similarity with previously characterized sterol esterases/lipases with known structures.
Keywords: Hydrolases, Models, In silico, Fungal genomics, Candida rugosa, Ophiostoma piceae
Background
Lipases, also known as triacylglycerol lipases (EC. 3.1.1.3), act on ester bonds of several compounds, with acylglycerols as their natural substrates. These enzymes are able not only to catalyze under aqueous conditions the hydrolysis of triglycerides to free fatty acids, diglycerides and monoglycerides but also to carry out synthesis and trans- and interesterification reactions in the presence of organic solvents. Lipases are produced by animals, plants and microorganisms, but the latter have gained special interest in the industry due to their stability, selectivity or wide substrate specificity. Some of the most commercially important lipases belong to yeasts, such as Candida rugosa (synonym C. cylindracea) and Candida antarctica, or filamentous fungi, such as Aspergillus niger, Humicola lanuginosa, Mucor miehei, and Rhizopus species [1,2]. Sterol esterases (EC. 3.1.1.13), also known as cholesterol esterases, act on esters of sterols and long-chain fatty acids. The mammalian cholesterol esterases are the best known for their role in lipid metabolism and cholesterol absorption [3,4] but these proteins have also been reported in filamentous fungi, yeast and bacteria [5,6].
Both kinds of enzymes, lipases and sterol esterases, belong to the α/β-hydrolase superfamily and some of them show broad substrate specificity, including triglycerides and water insoluble sterol esters. For example, C. rugosa secretes a variety of closely related enzymes, commercialized as lipases or sterol esterases. Although at least three of them (Lip1, Lip2 and Lip3) display activity on both triglycerides and cholesteryl oleate, they differ in their specificity [7]. In the case of sterol esterases, this promiscuity has been reported for the enzymes characterized in Ophiostoma piceae[8] and Melanocarpus albomyces[9].
Lipases are generally strongly activated by water–lipid interfaces [10]. The active site of these enzymes presents a hydrophobic cavity covered by an amphipathic loop named “lid” or “flap” and most of them show interfacial activation, a phenomenon that occurs by the movement of the flap caused by a conformational change on this helical element, making the enzyme’s active site more accessible to the substrate [11]. This property has been considered as a distinguishing feature between lipases and sterol esterases although lipases lacking a flap [12] and sterol esterases showing interfacial activation have been reported [13,14]. These structural elements have been exhaustively studied in C. rugosa lipases and sterol esterases to understand the substrate specificity of the different isoenzymes. The five extracellular enzymes (Lip1–Lip5) characterized in this yeast have 534 amino acids, present a high level of sequence identity (77–88%) but show differences on pI and putative N-glycosilation sites [15]. The structural comparisons of three of them (Lip1, Lip2, and Lip3) revealed several amino acid changes affecting the flap, the substrate-binding pocket and the hydrophobic tunnel, that could be responsible for the differences in their catalytic properties [7]. This family of lipases forms the so called C. rugosa lipase-like family (abH03.01), which comprises proteins of relatively large molecular masses (>60 kDa) that contain a GESAG sequence located in the middle of the polypeptide chains, corresponding to the position 222 in C. rugosa Lip3, one of the enzymes described in more detail at the biochemical and structural levels [7]. The Ser in this sequence functions as a catalytic residue and constitutes a ‘catalytic triad’ together with the conserved Glu and His residues that are presumed to facilitate the hydrolysis [16]. These enzymes also contain the sequence GGGF involved in the oxyanionic hole (position 137 in C. rugosa Lip3), which allows the substrate entry into the catalytic pocket. Most of these characteristics are present in the O. piceae sterol esterase, which shows more than 40% sequence identity with C. rugosa lipases and similar substrate-binding sites, as suggested by its structural model based on the crystal structures of C. rugosa Lip3 [17].
Nowadays, with the development of new molecular techniques such as massive DNA sequencing, the genomes of an enormous number of organisms can be studied in a short time. In this sense, the Joint Genome Institute (JGI) from the US department of energy (DOE) was pioneer in this kind of projects, and more than 128 genomes from different fungi with potential biotechnological interest are currently accessible in its website (http://www.jgi.doe.gov/). Bioinformatics approaches to analyze these genomes allow the finding of new enzymes taking into account the analysis of conserved motifs in the available DNA sequences [18]. Furthermore, molecular modeling plays a key role in structural biology. Current methods to study protein structure are very interesting to discover enzymes with improved catalytic properties and activities [19].
In the present work we carried out a bioinformatics screening of public fungal genomes deposited at JGI to explore the presence of genes encoding sterol esterases/lipases from the genomes of environmental microorganisms, as a strategy to find novel enzymes. The candidates were selected taking into account the conserved motifs detected in versatile lipases and sterol esterases described in yeast or filamentous fungi. The kinetic properties of the new putative enzymes are discussed on the basis of their three-dimensional model structure, built from the crystal structures of C. rugosa lipases.
Methods
Genomes screening
Seven sterol esterases/lipases with wide substrate versatility and potential industrial application, as C. rugosa lipases (Lip1, 2, 3, 4 and 5) and sterol esterases from O. piceae (OPE) [8] and M. albomyces[9], were selected. The search for conserved motifs in the amino acidic sequence was carried out using MEME (http://meme.sdsc.edu/meme/intro.html). In all 7 sequences the motifs GESAG (forming the catalytic shoulder of the catalytic Ser) and GGGF (forming the oxyanionic hole) were found. These motifs are already described in the “The Lipase Engineering Database” for the lipases of the C. rugosa lipase-like family [20,21].
To search for genes codifying this kind of proteins in the public genomes from http://www.jgi.doe.gov/[22], the automatically predicted proteins from all 128 fungal genomes containing the terms “esterase” (4786 sequences) or “lipase” (6180 sequences) were downloaded using the Advanced Search option at the JGI web-site. From this pool of sequences (10976 redundant sequences) the ones containing the conserved motifs GESAG and GGGF were selected using the Bioedit 7.1.3 software.
Phylogenetic analysis
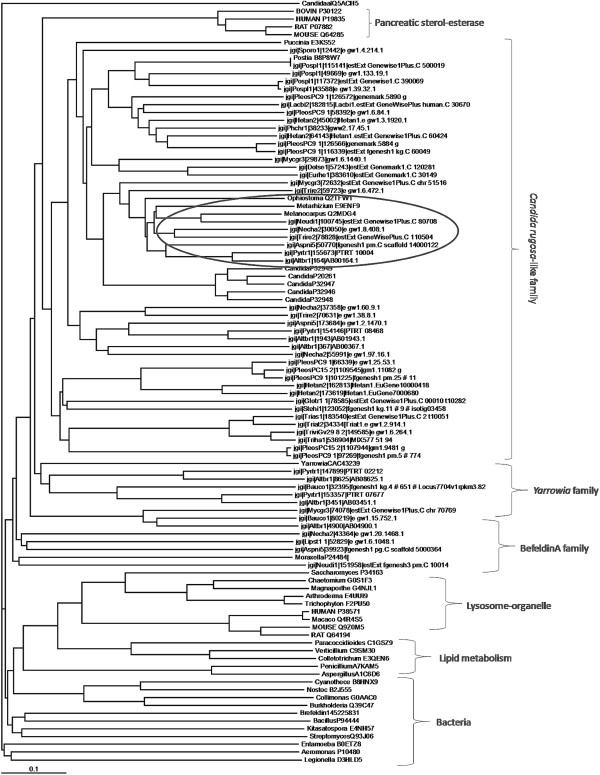
A phylogenetic tree was built using the 56 protein sequences selected in the previous section and representative sequences of different lipase and sterol esterase families: C. rugosa-like (ascomycetes and basidiomycetes), Yarrowia family, brefeldin family, cellular organelles, pancreas and bacteria (accession numbers in Figure 1). An un-rooted tree was created using MUSCLE multiple sequence alignment and the Maximum-Likelihood method. Treeview 1.6.6 was used to draw the trees. Sequences grouping with the versatile lipases and sterol esterases from the C. rugosa-like family were selected for further analyses.
Figure 1.
Phylogenetic tree of putative protein sequences of fungal sterol esterase/lipases. Phylogenetic tree of 56 putative protein sequences of sterol esterase/lipases from fungal genomes, and representative sequences of the C. rugosa-like family, Yarrowia family, Brefeldin family, cellular organelles, pancreatic sterol esterases, and bacterial esterases. The 6 selected candidates are marked with an oval.
Sequence analysis
Sequences from the 6 candidates selected in the previous section were compared by means of BLAST against the NR (NCBI) and “The Lipase Engineering Data Project” (LED) databases. Sequence alignments were also carried out versus C. rugosa Lip3 sequence using Clustal X 1.8, and putative signal peptides in each sequence were predicted using SignalP 4.0 [23].
Structural models
A three-dimensional model of the selected putative proteins was generated using the programs implemented by the automated protein homology-modeling server SWISS-MODEL (Swiss Institute of Bioinformatics [24]). In all cases the selected template, with higher homology to the putative sequence, was C. rugosa Lip3 (Parent PDB 1llf, Chain B). The 6 models matched all the sequence length excepting the putative signal peptide.
The models were exhaustively analyzed using PyMol 1.1 (http://pymol.org/). Putative intramolecular tunnels were modeled from the catalytic serine using Caver 2.0 v0.003 [25].
Results and discussion
This study shows an interesting bioinformatics approach to find new sterol esterases/lipases from the genomes of environmental microorganisms thanks to the natural engineering process driven by evolution.
Identification of conserved motifs in lipase/sterol esterase enzymes and search in public microbial genomes
After the analysis of the protein sequences described as lipase/sterol-esterase with versatile activity against triglycerides and sterol esters, two conserved motifs described for α/β hydrolases of the C. rugosa-like family were identified: GGGF and GESAG.
The search of putative proteins automatically annotated as sterol esterase or lipase in the 128 public genomes deposited in the JGI returned only 56 sequences from 26 different genomes.
Phylogenetic analysis
Figure 1 shows a phylogenetic tree with the 56 putative enzymes from the different genomes analyzed, as well as representative sterol-esterases/lipases from different fungal families. Twelve putative proteins from the JGI genomes grouped with the lipases from the lipolitic yeast Yarrowia[26] or the Bacillus subtilis Brefeldin A family [27], typical enzymes with high affinity for triglycerides whose ability to hydrolyze sterol esters has not been reported. On the other hand, this figure shows 44 of the candidate proteins grouped with the C. rugosa-like family, and 6 out of these putative proteins were selected to continue the study because they were the most closely related to the proteins from C. rugosa (P20261, P32946, P32947, P32948 and P32949) [7,15], O. piceae (Q2TFW1) [8], and M. albomyces[9] with activity on both triglycerides and sterol esters. The 6 candidate proteins belong to different organisms but all of them are plant-associated fungi (Table 1). Species from the genus Neurospora are known for their ability to colonize the wood from the trees after forest fires, [28]. The plant pathogenic Nectria haematococca has also been related to the degradation of aromatic recalcitrant compounds, [29,30]. Trichoderma species are frequently isolated from forest or agricultural soils, and are known for their ability to produce enzymes of industrial relevance [31]. Aspergillus niger secretes a wide array of hydrolytic and oxidative enzymes [32]. Pyrenophora tritici-repenti is a plant pathogen causing a disease called “tan spot” that mainly affects wheat [33]. Finally, the genus Alternaria comprises many common saprophytic and plant pathogenic species [34].
Table 1.
Candidate sequence analysis
| Selected candidates (putative protein JGI accession number) | Fungal species | CDS length | Homology against NCBI | Homology against LED | % of similitude with Lip3 |
|---|---|---|---|---|---|
| jgi|Neudi1|100745| |
Neurospora discreta |
1737 nt |
75% with Q2MDG4 |
75%with Q2MDG4 |
42% |
| jgi|Necha2|30050| |
Nectria haematococca |
1764 nt |
64% with Q2MDG4 |
62% with Q2MDG4 |
39% |
| jgi|Trire2|78828| |
Trichoderma reesei |
1713 nt |
59% with Q2MDG4 |
59%with Q2MDG4 |
40% |
| jgi|Aspni5|50770| |
Aspergillus niger |
1689 nt |
64% with Q5XTQ4 |
57% with Q5XTQ4 |
40% |
| jgi|Pyrtr1|155673| |
Pyrenophora tritici-repentis |
1683 nt |
62% with Q5XTQ4 |
62% with Q5XTQ4 |
46% |
| jgi|Altbr1|164|AB00164 | Alternaria brassicicola | 1686 nt | 64% with Q5XTQ4 | 61% with Q5XTQ4 | 44% |
The ecological role of the extracellular lipases/esterases in these fungi can be related to the initial degradation of the epicuticular waxes and cuticle, which consist of a mixture of long-chain fatty acids, aldehydes, alkanes, primary and secondary alcohols, ketones and wax esters [35]. The deterioration of these external layers makes the plant cell-wall polysaccharides more accessible as carbon source for the plant-associated fungi, or facilitates the colonization by pathogenic species [36,37].
Candidate sequences analysis
The six candidate sequences as putative versatile enzymes with lipase and sterol esterase activity were compared against NCBI and LED databases using Blastp. In all cases, the highest score matched with both a sterol esterase precursor from M. albomyces[9] and the lipase from Botryotinia fuckeliana[38], with sequence identities between 57 and 75% (Table 1). The sequence identity of each candidate with the C. rugosa Lip3, used as a model protein, is shown in Table 1. All these data correlate with the results from the phylogenetic analysis (Figure 1).
Additional file: 1 Figure S1 shows the sequence alignment of Lip3 against the versatile lipase/esterase candidates, highlighting the predicted signal peptides at the N-terminal of the putative proteins consisting in 15 to 22 amino acids, the 2 conserved Cys residues forming disulfide bonds and delimiting the lid region, the oxyanionic hole (GGGF), and the catalytic triad (Ser-Gln-His). An N-glycosylation site is present in all the sequences analyzed, except for jgi|Trire2|78828|.
Moreover, it is known that the number of hydrophobic amino acids at the lid region is related to enzyme-substrate specificity [39]. Lipase isoenzymes from C. rugosa share over 80% sequence identity but diverge in the sequence of the lid. In the active enzyme conformation, the open lid participates in the substrate-binding site and contributes to substrate recognition. The substitution of the lid from isoenzyme Lip1 for that of Lip3 was sufficient to confer to Lip1 cholesterol esterase activity [39]. Additional file: 1 Figure S1 shows the differences in the residues located at the lid region of the putative proteins. The number of hydrophobic residues is 14 in Necha2 and Trire2, 13 in Altbr1, 12 in Pyrtr1 and Neudi1, and 11 in Aspni5, while Lip3 only contains 10. The number of hydrophobic residues in the lid of the different lipases produced by C. rugosa is higher in the isoforms that are more active on sterol esters than on triglycerides: Lip2 > Lip3 > Lip1 (12, 10, and 8 hydrophobic residues, respectively) [7,15]. According to this data, the O. piceae sterol esterase, with 13 hydrophobic residues in lid region, has been reported to possess higher activity on sterol esters than C. rugosa Lip3 [17]. Based on these observations it can be inferred that Necha2, Trire2, Pyrtr1, and Altbr1 may have high affinity for sterol esters, while Neudi1 and Aspni5 could have intermediate characteristics.
Molecular modeling analysis of selected candidates
Molecular models of the selected putative candidate sequences were generated using SWISS-MODEL server and in all cases the template automatically selected was Lip3 from C. rugosa (pdb 1llfB and 1cle), one of the versatile lipases better characterized [5-7]. Figure 2 displays the comparison of the 3D structures obtained, showing the typical α/β-hydrolase conformation [40] with a lid region formed by an α-helix which, in most lipases, allows a conformational change in the presence of the substrate improving its accessibility to the active site [11]. A superimposition of the residues from the catalytic triad in the 6 candidate models and the Lip3 structure, showed an identical geometry in all cases, with the catalytic Ser close to the mouth of the substrate pocket (data not shown).
Figure 2.
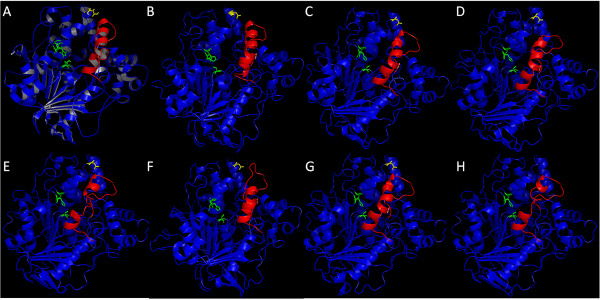
Tridimensional structures/models of the proteins in its open conformation. (A) Lip3 from C. rugosa. Models, based in C. rugosa Lip3 structure, from (B) OPE and the six selected putative proteins (C) Altbr1, (D) Aspni5, (E) Necha2, (F) Neudi1, (G) Pyrtr1 and (H) Trire2. Lid region is indicated in red, the three catalytic residues are highlighted in green and the residue for a putative glycosylation site is marked in yellow.
The percentage of hydrophobic residues at a distance of 4 Å or shorter from the predicted tunnels was calculated for each model. Trire2 had 11 hydrophobic residues out of 22 in the tunnel area (50%), similar to Aspni5 that had 9 out of 18 (50%) and Pyrtr1, 11 out of 21 (52%). However, Neudi1 presented a higher number of these residues 11 out of 20 (55%) and Necha2 showed even higher number, 13 out of 22 (59%). Altbr1 had less hydrophobic residues 8 out of 22 (36%). In the case of C. rugosa lipases, Lip1 had 9 out of 21 (42%), Lip2 presented 12 out of 25 (48%) and Lip3 showed 11 out of 24 (46%), being Lip2 and 3 the ones with the highest affinity for sterol esters. OPE, with higher affinity than any Lip on sterol esters, has 10 hydrophobic residues out of 21 (48%) in the tunnels area. Since all the selected putative enzymes, except Altbr1, presented more hydrophobic residues than OPE and Lip2 and 3, this could be indicative of its increased affinity for sterol esters.
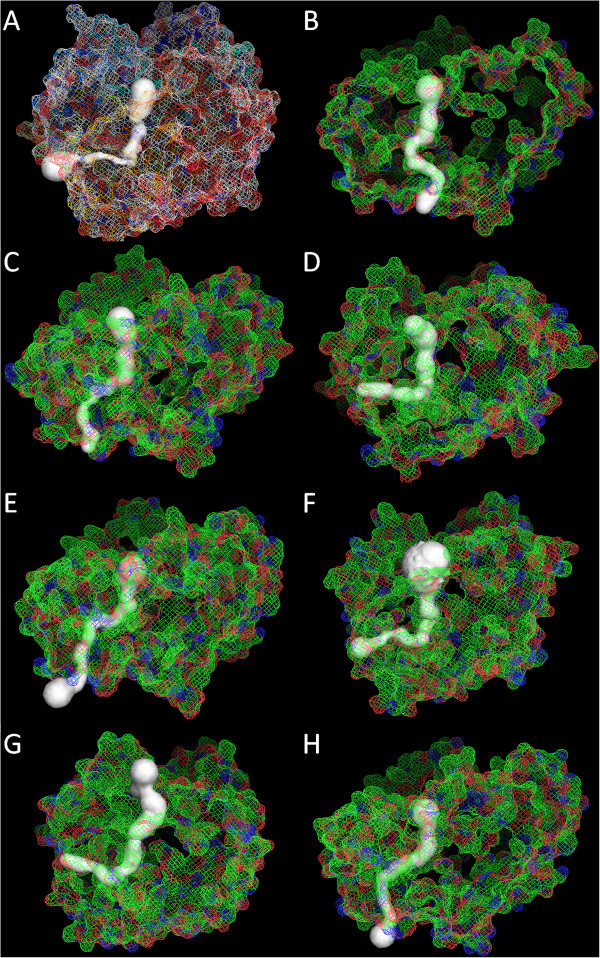
The model of the internal tunnels in each structure was made using Caver 2.0 (Figure 3). In all cases, a big tunnel coinciding with the substrate binding pocket and a second putative tunnel that may be an exit for the release of the reaction products can be observed. This hypothesis was already proposed for C. rugosa Lip1 as a possible mechanism responsible for the high catalytic efficiency of this enzyme [41]. The length and shape of these tunnels differ for each model and the residues forming the tunnels can also vary. Lip3 has a prominent entry tunnel and a bended exit tunnel, similar to that reported for Lip1 [41], while the O. piceae sterol esterase model presented a broad straight tunnel opposed to the substrate pocket, which could contribute to its reported higher catalytic efficiency as compared with Lip3 [17].
Figure 3.
Models of the intramolecular tunnels. Internal tunnels in each structure were modeled using Caver 2.0. (A) Lip3 (model structure: PDB 1llf, Chain B), (B) OPE, (C) Altbr1, (D) Aspni5, (E) Necha2, (F) Neudi1, (G) Pyrtr1 and (H) Trire2.
Neudi1, Pyrt1 and Aspni5 showed a prominent entry tunnel and a bended exit tunnel, similar to Lip3 structure (Figure 3A, D, F and G). Altbr1, Necha2 and Trire2 presented a broad straight tunnel opposed to the substrate pocket, in this case more similar to that found in O. piceae sterol esterase (Figure 3B, C, E and H). As mentioned before, Altbr1 has the smallest hydrophobic residues content in the tunnel, indicating that this characteristic is not strictly related to its shape, and that it could be a very efficient enzyme oxidizing triglycerides. All these data suggest that those putative enzymes with OPE-like tunnels could be more efficient than those with C. rugosa-like tunnels, allowing a faster entry of the substrate and exit of the products, and the hydrolysis of big substrates.
Conclusions
Here we present a useful strategy to identify enzymes with potential biotechnological interest by means of genomes mining. The strategy combines search of conserved motifs, phylogenetic analysis, sequence homology studies and protein model analysis. This strategy allowed identifying 6 candidate sequences (Table 1). The six putative proteins selected were automatically annotated at JGI server except for Aspni5 that has been manually annotated as a putative sterol esterase with homology to the enzyme from O. piceae. In this study we inferred the potential characteristics of the putative enzymes based on their model structures. Our analysis, derived from the percentage of hydrophobic residues in the lid region, suggests that the affinity towards sterol esters of all the candidate proteins selected could be superior to that of the C. rugosa model lipases. In addition, the putative proteins Altbr1, Necha2, and Trire2 may also have higher efficiency against this substrate than C. rugosa lipases due to their exit tunnel conformation. However, further studies on the expression and characterization of these new proteins will be necessary to corroborate their potential biotechnological or industrial interest.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JB participated in the overall conceptual and experimental design of this study, analysis and interpretation of results, as well as in drafting the manuscript. AP participated in its design and coordination and helped to draft the manuscript. MJM participated in the analysis and interpretation of results, and in drafting and revising the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Sequence alignment of the putative proteins against Lip3. Sequence alignment of model protein Lip3 from C. rugosa against the six putative sterol esterase/lipases selected from fungal genomes. The lid region is written in red. The predicted signal peptides are highlighted in green, cysteines in blue, the oxyanionic hole in yellow, the catalytic triad residues (Ser-Gln-His) are underlined and written in orange and the conserved surrounding residues are underlined. A putative N-glycosylation site is highlighted in magenta.
Contributor Information
Jorge Barriuso, Email: jbarriuso@cib.csic.es.
Alicia Prieto, Email: aliprieto@cib.csic.es.
Maria Jesus Martínez, Email: mjmartinez@cib.csic.es.
Acknowledgments
This work was supported by the Spanish projects BIO2012-3637, PRI-PIBAR-2011-1402 and S-2009AMB-1480. J. Barriuso thanks the financial support from the JAE-DOC CSIC program. The sequence data were produced by the US Department of Energy Joint Genome Institute (JGI) in collaboration with the user community (http://www.jgi.doe.gov/). The authors thank C. Kubicek, T. Jeffries, A. Pisabarro and D. Cullen, coordinators of non free JGI genomes, their permission for analyzing the sequences used in this study.
References
- Saxena RK, Ghosh PK, Gupta R, Davidson WS, Bradoo S, Gulati R. Microbial lipases: Potential biocatalysts for the future industry. Current Sci. 1999;77:101–115. [Google Scholar]
- Jaeger KE, Reetz MT. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 1998;16:396–403. doi: 10.1016/S0167-7799(98)01195-0. [DOI] [PubMed] [Google Scholar]
- Moreau RA, Hicks KB. The in vitro hydrolysis of phytosterol conjugates in food matrices by mammalian digestive enzymes. Lipids. 2004;39:769–776. doi: 10.1007/s11745-004-1294-3. [DOI] [PubMed] [Google Scholar]
- Brown AW, Hang J, Dussault PH, Carr TP. Plant sterol and stanol substrate specificity of pancreatic cholesterol esterase. J Nutr Biochem. 2010;21:736–740. doi: 10.1016/j.jnutbio.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Wawrzak Z, Pletnev VZ, Li N, Kaiser R, Pangborn W, Jörnvall H, Erman M, Duax WL. Structure of uncomplexed and linoleate-bound Candida cylindracea cholesterol esterase. Structure. 1995;3:279–288. doi: 10.1016/S0969-2126(01)00158-7. [DOI] [PubMed] [Google Scholar]
- Rahim MA, Sih CJ. Microbial steroid esterases. Methods Enzymol. 1969;15:675–684. [Google Scholar]
- Mancheño JM, Pernas MA, Martínez MJ, Ochoa B, Rua ML, Hermoso JA. Structural insights into the lipase/esterase behavior in the Candida rugosa lipases family: crystal structure of the lipase 2 isoenzyme at 1.97A resolution. J Mol Biol. 2003;332:1059–1069. doi: 10.1016/j.jmb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Calero-Rueda O, Plou FJ, Ballesteros A, Martínez AT, Martínez MJ. Production, isolation and characterization of a sterol esterase from Ophiostoma piceae. BBA Proteins Proteomics. 2002;1599:28–35. doi: 10.1016/S1570-9639(02)00378-3. [DOI] [PubMed] [Google Scholar]
- Kontkanen H, Tenkanen M, Reinikainen T. Purification and characterisation of a novel steryl esterase from Melanocarpus albomyces. Enzyme Microb Technol. 2006;39:265–273. doi: 10.1016/j.enzmictec.2005.10.013. [DOI] [Google Scholar]
- Pernas MA, Lopez C, Rua ML, Hermoso J. Influence of the conformational flexibility on the kinetics and dimerisation process of two Candida rugosa lipase isoenzymes. FEBS Lett. 2001;501:87–91. doi: 10.1016/S0014-5793(01)02630-8. [DOI] [PubMed] [Google Scholar]
- Cygler M, Schrag JD, Ergan F. Advances in structural understanding of lipases. Biotechnol Gen Eng Rev. 1992;10:143–184. doi: 10.1080/02648725.1992.10647887. [DOI] [PubMed] [Google Scholar]
- Verger R. ‘Interfacial activation’ of lipases: Facts and artifacts. Trends Biotechnol. 1997;15:32–38. doi: 10.1016/S0167-7799(96)10064-0. [DOI] [Google Scholar]
- Svendsen A, Borch K, Barfoed M, Nielsen TB, Gormsen E, Patkar SA. Biochemical properties of cloned lipases from the Pseudomonas family. Biochim Biophys Acta. 1995;1258:9–17. doi: 10.1016/0005-2760(95)00117-u. [DOI] [PubMed] [Google Scholar]
- Kontkanen H, Tenkanen M, Fagerström R, Reinikainen T. Characterisation of steryl esterase activities in commercial lipase preparations. J Biotechnol. 2004;108:51–59. doi: 10.1016/j.jbiotec.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Lotti M, Tramontano A, Longhi S, Fusetti F, Brocca S, Pizzi E, Alberghina L. Variability within the Candida rugosa lipases family. Protein Eng. 1994;7:531–535. doi: 10.1093/protein/7.4.531. [DOI] [PubMed] [Google Scholar]
- Cygler M, Schrag JD, Sussman JL, Harel M, Silman I, Gentry MK, Doctor BP. Relationship Between Sequence Conservation and 3-Dimensional Structure in A Large Family of Esterases, Lipases, and Related Proteins. Protein Sci. 1993;2:366–382. doi: 10.1002/pro.5560020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Rueda O, Barba V, Rodriguez E, Plou F, Martínez AT, Martínez MJ. Study of a sterol esterase secreted by Ophiostoma piceae: Sequence, model and biochemical properties. Biochim Biophys Acta. 2009;1794:1099–1106. doi: 10.1016/j.bbapap.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Ferrer M, Martinez-Abarca F, Golyshin PN. Mining genomes and ‘metagenomes’ for novel catalysts. Curr Opin Biotechnol. 2005;16:588–593. doi: 10.1016/j.copbio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Hui DY, Howles PN. Carboxyl ester lipase: structure-function relationship and physiological role in lipoprotein metabolism and atherosclerosis. J Lipid Res. 2002;43:2017–2030. doi: 10.1194/jlr.R200013-JLR200. [DOI] [PubMed] [Google Scholar]
- Fischer M, Pleiss J. The Lipase Engineering Database: a navigation and analysis tool for protein families. Nucleic Acid Res. 2003;31:319–321. doi: 10.1093/nar/gkg015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss J, Fischer M, Peiker M, Thiele C, Schmid RD. Lipase engineering database -Understanding and exploiting sequence-structure-function relationships. J Mol Catal B-Enz. 2000;10:491–508. doi: 10.1016/S1381-1177(00)00092-8. [DOI] [Google Scholar]
- Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, Otillar R, Poliakov A, Ratnere I, Riley R, Smirnova T, Rokhsar D, Dubchak I. The Genome Portal of the Department of Energy Joint Genome Institute. Nucleic Acid Res. 2012;40:D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Meth. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Kuenzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acid Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medek P, Benes P, Sochor J. Computation of tunnels in protein molecules using Delaunay triangulation. J WSCG. 2007;15:107–114. [Google Scholar]
- Pignede G, Wang HJ, Fudalej F, Gaillardin C, Seman M, Nicaud JM. Characterization of an extracellular lipase encoded by LIP2 in Yarrowia lipolytica. J Bacteriol. 2000;182:2802–2810. doi: 10.1128/JB.182.10.2802-2810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Jacobson DJ, Powell AJ, Dettman JR, Saenz GS, Barton MM, Hiltz MD, Dvorachek WH, Glass NL, Taylor JW, Natvig DO. Neurospora in temperate forests of western North America. Mycologia. 2004;96:66–74. doi: 10.2307/3761989. [DOI] [PubMed] [Google Scholar]
- Rodríguez A, Perestelo F, Carnicero A, Regalado V, Pérez R, de la Fuente G, Falcón MA. Degradation of natural lignins and lignocellulosic substrates by soil-inhabiting fungi imperfecti. FEMS Microbiol Ecol. 1996;21:213–219. doi: 10.1111/j.1574-6941.1996.tb00348.x. [DOI] [Google Scholar]
- Verdin A, Sahraoui ALH, Durand R. Degradation of benzo[a]pyrene by mitosporic fungi and extracellular oxidative enzymes. Int Biodeterior Biodegrad. 2004;53:65–70. doi: 10.1016/j.ibiod.2003.12.001. [DOI] [Google Scholar]
- Xu Q, Singh A, Himmel ME. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr Opin Biotechnol. 2009;20:364–371. doi: 10.1016/j.copbio.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Maeda RN, Serpa VI, Rocha VAL, Mesquita RAA, Santa Anna LMM, de Castro AM, Driemeier CE, Pereira N, Polikarpov I. Enzymatic hydrolysis of pretreated sugar cane bagasse using Penicillium funiculosum and Trichoderma harzianum cellulases. Process Biochem. 2011;46:1196–1201. doi: 10.1016/j.procbio.2011.01.022. [DOI] [Google Scholar]
- Chu C, Chao S, Friesen T, Faris J, Zhong S, Xu S. Identification of novel tan spot resistance QTLs using an SSR-based linkage map of tetraploid wheat. Mol Breeding. 2010;25:327–338. doi: 10.1007/s11032-009-9335-2. [DOI] [Google Scholar]
- Sharma P, Deep S, Gothandapani S, Sharma M, Kalia P, Chowdappa P. Pathogenic and genetic diversity in Alternaria brassicae and Alternaria brassicicola causing black leaf spot of cauliflower in India. Phytopathol. 2011;101:S164. [Google Scholar]
- Juniper BE, Jeffree CE. Plant Surfaces. London: Edward Arnold; 1983. [Google Scholar]
- de Vries RP, Poulsen CH, Kroon PA, vanden Heuvel RHH, Faulds CB, Williamson G, vanden Hombergh JPTW, Visser J. The faeA genes from Aspergillus niger and Aspergillus tubingensis encode ferulic acid esterases involved in degradation of complex cell wall polysaccharides. App Environ Microbiol. 1997;63:4638–4644. doi: 10.1128/aem.63.12.4638-4644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss RP. Composition and enzymatic activity of the extracellular matrix secreted by germlings of Botrytis cinerea. App Environ Microbiol. 1999;65:404–408. doi: 10.1128/aem.65.2.404-408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis H, Pfiffi S, Hahn M. Molecular and functional characterization of a secreted lipase from Botrytis cinerea. Mol Plant Pathol. 2005;6:257–267. doi: 10.1111/j.1364-3703.2005.00280.x. [DOI] [PubMed] [Google Scholar]
- Brocca S, Secundo F, Ossola M, Alberghina L, Carrea G, Lotti M. Sequence of the lid affects activity and specificity of Candida rugosa lipase isoenzymes. Protein Sci. 2003;12:2312–2319. doi: 10.1110/ps.0304003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M, Dijkstra BW. α/β Hydrolase fold enzymes: the family keeps growing. Curr Opin Struc Biol. 1999;9:732–737. doi: 10.1016/S0959-440X(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Foresti ML, Ferreira ML. Computational approach to solvent-free synthesis of ethyl oleate using Candida rugosa and Candida antarctica B lipases. I. Interfacial activation and substrate (ethanol, oleic acid) adsorption. Biomacromolecules. 2004;5:2366–2375. doi: 10.1021/bm049688o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of the putative proteins against Lip3. Sequence alignment of model protein Lip3 from C. rugosa against the six putative sterol esterase/lipases selected from fungal genomes. The lid region is written in red. The predicted signal peptides are highlighted in green, cysteines in blue, the oxyanionic hole in yellow, the catalytic triad residues (Ser-Gln-His) are underlined and written in orange and the conserved surrounding residues are underlined. A putative N-glycosylation site is highlighted in magenta.