Abstract
How does BRCA1’s evolutionarily conserved E3 ligase activity contribute to DNA damage responses? Genetically engineered cells containing a BRCA1 RING domain mutation have been used to identify Claspin as a new target of BRCA1 E3 ligase activity in response to specific forms of DNA damage.
The primary cause of hereditary breast and ovarian cancer syndrome is heterozygous germline mutation of the breast cancer early onset genes BRCA1 and BRCA2. Both BRCA gene products are essential for efficient DNA double strand break (DSB) repair mediated by homologous recombination (HR). In addition, BRCA1 acts to integrate the activities of several protein partners during the response to DSBs and contributes to DNA damage-induced checkpoint activation in part through promoting ATR-dependent phosphorylation of checkpoint kinase 1 (CHK1). Within the context of heterozygous BRCA patients, tumors lose the wild-type allele, motivating synthetic lethal therapeutic approaches that exploit the tumor-specific HR deficiency [1–4]. A fundamental understanding of BRCA-directed DNA repair mechanisms therefore has clear implications for the effective design and implementation of DNA-damaging chemotherapeutics strategies.
The BRCA1 protein is organized into two main functional domains. The amino-terminal region contains a RING domain that imparts E3 ubiquitin ligase activity, and the carboxy-terminal part of the protein contains two BRCT (BRCA1 C-terminal) repeats that bind to a phosphorylated serine present within a consensus SPXF motif in binding partners [5,6]. Mutations resulting in highly penetrant breast and ovarian cancers affect either of these two domains. Because many pathogenic mutations in the amino-terminal BRCA1 RING domain affect its interaction with the stoichiometric binding partner BARD1, the contributions of BRCA1 E3 ligase activity to DNA damage responses and tumor suppression have until recently remained enigmatic. The advent of genetically engineered cells and mouse models has begun to shed light on this important topic. Ludwig, Baer and colleagues have generated a mouse model in which a single amino acid substitution (I26A) within the RING domain renders BRCA1 E3 ligase inactive by disrupting interaction with E2 enzymes, while leaving intact its ability to heterodimerize with BARD1 [7,8]. Surprisingly, BRCA1 I26A cells, both in culture and in mice, are not deficient in homology-directed repair of DSBs and do not display sensitivity to DNA inter-strand crosslinking (ICL) agents. Furthermore, BRCA1 I26A mice are not tumor prone. However, knock-in of a cancer-causing BRCA1 RING domain allele, BRCA1 C61G, that disrupts E3 ligase activity and BARD1 interaction does lead to DNA repair deficiency and cancer susceptibility [9]. Collectively, these findings suggest that BRCA1 E3 ligase activity is dispensable for its tumor suppressor and genome integrity functions, while interaction with BARD1 is the more relevant target of pathogenic RING domain mutations. It should be noted, however, that embryonic stem cells carrying the I26A mutation accumulate cytogenetic rearrangements at a higher rate than control cells when subjected to the ICL agent mitomycin C (MMC) [7]. Interestingly, pathogenic BRCA1 RING domain mutations have been described that, like I26A, selectively disrupt interaction with E2 enzymes while leaving BARD1 interaction intact [10,11]. Moreover, BRCA1 I26A mice are smaller and male mice are infertile [8], implying that the evolutionarily conserved E3 activity contributes to at least a subset of BRCA1 functions.
In this issue of Current Biology, Sato et al. [12] report experiments in genetically engineered chicken DT-40 cells that identify Claspin as a new target of BRCA1 E3 ligase activity. The authors provide evidence that BRCA1-mediated ubiquitination of Claspin is required for responses to topoisomerase poisons, but not to MMC. Claspin is a reported BRCA1-interacting partner and is required along with several other DNA damage response mediator proteins, including BRCA1, to promote ATR-dependent phosphorylation and activation of CHK1 [13]. The current study brings forth evidence that BRCA1 selectively ubiquitinates Claspin in response to topoisomerase inhibitors, increasing the stability of the protein and its association with chromatin. By using a ‘hit and run’ strategy, the authors engineered the DT40 chicken B cell line with a BRCA1 RING domain valine 26 to alanine mutation (V26A), recapitulating the I26A change that had been previously knocked into the murine BRCA1 locus. Phosphorylation and thereby activation of CHK1 was selectively compromised in V26A cells treated with Camptothecin (CPT) or other topoisomerase inhibitors, as was Claspin ubiquitination, Rad51 foci formation, sister chromatid exchange and cellular resistance to CPT. Surprisingly, these responses were specific to topoisomerase inhibitors since treatment of V26A cells with MMC did not result in detectable DNA damage response impairment.
Topoisomerase inhibitors and ICL agents both result in replication fork stalling, an event that requires subsequent intervention by the claspin–CHK1 pathway for its resolution (Figure 1). It is thus somewhat unexpected that BRCA1 E3 ligase activity would be selectively required for responses to only one of these agents. MMC acts as a highly potent alkylating agent and reacts with nucleophiles present within DNA bases, resulting in DNA inter-strand and intra-strand crosslinks as well as monoalkylation products. ICL damage is initially recognized by Fanconi Anemia proteins and subsequently processed to DSBs that necessitate homology-directed recombination repair by BRCA1 and BRCA2. While BRCA1 E3 ligase activity is seemingly dispensable for ICL repair, both BRCA1 and Claspin are required for FANCD2 assembly into subnuclear foci [14,15]. On the other hand, Camptothecin (CPT) is a selective inhibitor of the topoisomerase type 1B enzymes that relax DNA during replication and transcription. CPT stabilizes the Top1–DNA intermediates and prevents DNA religation, thereby creating DSBs that require HR repair in S phase. Camptothecin treatment activates the ATM–CHK2 axis and is also responsible for CHK1 phosphorylation by ATR to arrest the cell cycle and promote DNA repair [16].
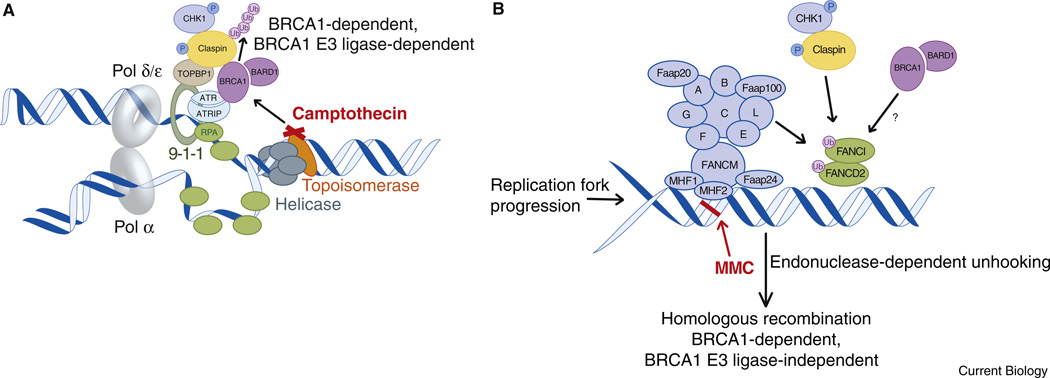
Figure 1. Differential DNA damage responses to Camptothecin and Mitomycin C treatment during replication.
(A) Topoisomerase poisons such as camptothecin induce replication-associated DNA double strand breaks and trigger a DNA damage response, which includes BRCA1-dependent ubiquitination of Claspin and activation of CHK1. (B) Forms of DNA damage involving inter-strand crosslinks are recognized by several Fanconi Anemia proteins, culminating in the monoubiquitination of the FANCD2/FANCI complex. Endo-nuclease processing of crosslinks is a prerequisite for subsequent repair of the lesion by homologous recombination. Claspin and BRCA1 also participate in promoting FANCD2 activation and BRCA1 is required for the later steps of homologous recombination after DNA double strand break formation.
The specific requirement for BRCA1 E3 ligase activity in response to CPT but not MMC begs several questions. How is it that BRCA1 ubiquitinates Claspin in response to topoisomerase inhibitors but not DNA crosslinks? Moreover, why does Claspin require ubiquitination for its stable association with chromatin only after DNA damage arising from topoisomerase inhibition? Perhaps a closer examination of the responses to each agent will reveal differences that could account for the selective requirement for BRCA1 E3 activity (Figure 1). One possibility is that BRCA1 is placed within proximity to Claspin at early stages of topoisomerase-induced DNA damage, but not during ICL repair, which requires endonucleolytic cleavage of crosslinks to initiate DSB responses. This difference and the requirement for different protein complexes during the initial recognition of each lesion could potentially account for the specific requirement for BRCA1 E3 ligase activity in the context of topoisomerase inhibitor-induced DSBs. It is interesting to note that differential requirements for BRCA1 are observed in response to poly(ADP) ribose polymerase inhibitors and ICL agents in mouse cells [17], thus invoking different BRCA1-dependent mechanisms to each response.
While the findings from this study await further investigation in additional cell lines and in vivo systems, they have several potential clinical implications. For example, BRCA1 mutant tumors may respond differently to topisomerase inhibitors in comparison to ICL agents in a manner that depends on where the BRCA1 mutation is located. Additionally, resistance mechanisms to each agent in tumors may not be equivalent. Finally, the studies by Sato et al. [12] emphasize the power of genetic systems to uncover additional complexity within cellular DNA damage responses and our ever-evolving understanding of how BRCA1 contributes to this process.
References
- 1.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 2.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 3.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 4.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 5.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 6.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 7.Reid LJ, Shakya R, Modi AP, Lokshin M, Cheng JT, Jasin M, Baer R, Ludwig T. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl. Acad. Sci. USA. 2008;105:20876–20881. doi: 10.1073/pnas.0811203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shakya R, Reid LJ, Reczek CR, Cole F, Egli D, Lin CS, deRooij DG, Hirsch S, Ravi K, Hicks JB, et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science. 2011;334:525–528. doi: 10.1126/science.1209909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drost R, Bouwman P, Rottenberg S, Boon U, Schut E, Klarenbeek S, Klijn C, van der Heijden I, van der Gulden H, Wientjens E, et al. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell. 2011;20:797–809. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 10.Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D, 3rd, Fukuda M, Ohta T, Klevit R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris JR, Pangon L, Boutell C, Katagiri T, Keep NH, Solomon E. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum. Mol. Genet. 2006;15:599–606. doi: 10.1093/hmg/ddi476. [DOI] [PubMed] [Google Scholar]
- 12.Sato K, Sundaramoorthy E, Rajendra E, Hattori H, Jeyasekharan AD, Ayoub N, Schiess R, Aebersold R, Nishikawa H, Sedukhina A, et al. The BRCA1 E3 ligase triggers Claspin ubiquitylation, CHK1 activation and homology-diretced repair during a subset of DNA-damage responses. Curr. Biol. 2012;22:1659–1666. doi: 10.1016/j.cub.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 13.Lin SY, Li K, Stewart GS, Elledge SJ. Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc. Natl. Acad. Sci. USA. 2004;101:6484–6489. doi: 10.1073/pnas.0401847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guervilly JH, Mace-Aime G, Rosselli F. Loss of CHK1 function impedes DNA damage-induced FANCD2 monoubiquitination but normalizes the abnormal G2 arrest in Fanconi anemia. Hum. Mol. Genet. 2008;17:679–689. doi: 10.1093/hmg/ddm340. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 16.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 17.Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]