In Trypanosoma brucei, homologous recombination (HR) is important for antigenic variation and is widely exploited for genetic manipulation. Thus, parameters that influence HR are of considerable interest. We have evaluated the effect of target site transcription on HR in bloodstream-form T. brucei. Strains were created with a conditional RNA polymerase (RNAP) I promoter driving transcription of an HR-target integrated within chromosomal DNA. Gene targeting efficiency was measured in the absence and presence of tetracycline-induced target-site transcription. The results indicate that transcription stimulates HR more than three-fold.
HR predominates over non-homologous end joining (NHEJ) in T. brucei and participates in the main mode of antigenic variation [1,2]. HR is also routinely and widely exploited to manipulate genes and other sequences and to integrate (conditional) expression cassettes into the genome, particularly for experiments involving RNA interference or expression of tagged proteins. A few different targets have been used for the latter approaches but non-transcribed ribosomal RNA (RRNA) spacer loci are probably the most popular. To facilitate reproducible targeting and circumvent problems with position effects, we tagged one of 15-20 spacer loci with a unique target sequence [3]. An unexpected but welcome outcome was that the tagged locus was reproducibly targeted more efficiently than un-tagged loci. One possible explanation is transcriptional stimulation of HR at the tagged target. This arises from the fact that, along with the tag, an RNAP-I promoter was integrated to drive selectable marker gene expression; RNAP-I naturally transcribes certain protein-coding genes in T. brucei including the single active Variant Surface Glycoprotein (VSG) gene [4].
Because HR predominates in T. brucei, the efficiency of stable integrative transformation has proven to be a useful measure of HR. The approach involves electroporation with a linear DNA construct containing any one of several resistance markers. Powerful antibiotic selection is then used to eliminate non-transformed cells. Transformation efficiency is low in bloodstream form cells, <10% of electroporated cells survive and, for many targets, <10−6 survivors are transformed [5]. Efficiency can be increased ~8-fold using particle bombardment rather than electroporation [6] and long-term cultured insect-stage cells are transformed at ten to 100-fold higher efficiency [7]. The reason for this latter difference is not known. Apart from the HR machinery itself [8,9], a number of other parameters can influence HR. Homology adjacent to double-strand breaks is clearly important. Circular and linear DNA are taken up by cells equally well but circular DNA or linear DNA with non-homologous sequence at either end reduce HR [10,11]. Target-sequence divergence also has a negative impact [8] and although there is some evidence that the length of homology correlates with HR efficiency [11], a study using targets between 80-400 bp, directed towards transcriptionally silent loci, indicated no significant difference [7]. HR is not increased by using >5 μg of plasmid DNA [5] and is independent of target site copy number when a single transcriptionally active and several transcriptionally silent loci are compared [7].
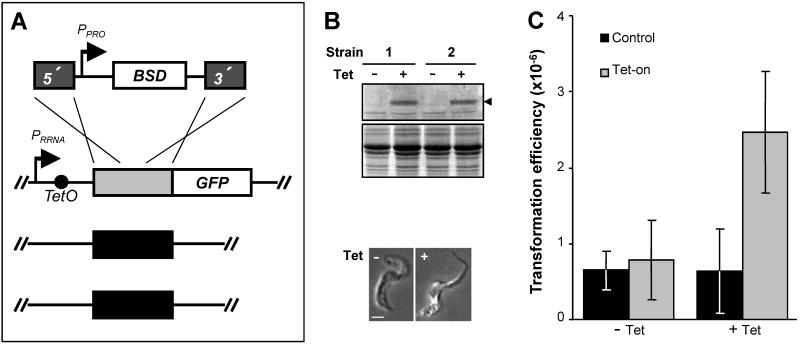
Although target site transcription is not required for HR in T. brucei, transcription does stimulate HR in yeast [12] and human cells [13]. To examine the relationship between transcription and HR in T. brucei, we established an assay where target transcription is the only altered parameter. The assay was based on HR between a chromosomal target and a donor cassette introduced into bloodstream form cells by electroporation. Plasmid-based molecules capable of stable, episomal propagation are yet to be reported in bloodstream form cells so recombination in vivo is a pre-requisite for stable antibiotic resistance in this life-cycle stage. We established strains with HR targets downstream of a tetracycline (Tet)-inducible RRNA promoter (Fig. 1A). The cassette was integrated at a tagged RRNA spacer locus to ensure reliable, inducible transcription [3]. Selectable marker transcription was constitutive and independent of HR-target transcription at this locus. For the HR target, we chose a T. brucei sequence encoding a dispensable mitochondrial protein [14]. Selection of a T. brucei sequence meant that the native sequences (T. brucei is diploid) would serve as critical internal control targets where RNAP-II transcription should remain constant. We chose a gene encoding a dispensable protein so there would be no selection against native locus targeting and a mitochondrial protein so there was little chance of interference with nuclear recombination processes. We also fused the ectopic HR-target to a Green Fluorescent Protein (GFP) gene such that GFP was expressed in the single mitochondrion following Tet-induction. This allowed us to confirm conditional expression at the target locus (Fig. 1B) and also facilitated scoring of HR at the native versus ectopic targets; clones in which the ectopic target participates in HR can no longer express GFP (Fig. 1). The donor cassette consists of terminal recombination targets flanking a BSD selectable marker. A promoter (PPRO, another RNAP-I promoter) is an essential component of this cassette because it ensures selectable marker expression regardless of the transcriptional status of the target. Thus, although transcription apparatus may be recruited and donor sequence transcribed prior to recombination, any impact on HR should be Tet-independent.
Figure 1.
Transcription stimulates homologous recombination.
A. The assay strains were established by integrating a cassette containing a Tet-on RRNA promoter (PRRNA) driving transcription of the HR-target (GeneDB ID: Tb927.8.3140) fused to a GFP reporter. TetO, Tet operator. We used 2T1 cells that express TetR:BLE and have a tagged RRNA spacer [3]. The schematic illustrates the HR-assay construct (top) and all three HR-targets in the assay strain, the ectopic Tet-on target (light-grey, transcribed by RNAP-I) and both native targets (black, transcribed by RNAP-II, bottom). The donor construct contains a procyclin promoter (PPRO) to drive expression of the blasticidin S deaminase (BSD) selectable marker which is flanked by target-homologous segments 189 (5′) and 216 bp (3′) in length. The expected double-cross-over is indicated and deletes an 83 bp (NcoI / NdeI) segment from the target. T. brucei strains were grown and manipulated in vitro as previously described [3]. Electroporation was with a BioRad Gene Pulser set at 1.4 kV and 25μF without a pulse controller.
B. The assay strains (1 and 2) express GFP in the presence of Tet (1 μg ml−1 for 24 h) as shown by western blotting (top panel, arrowhead) and immunofluorescence analysis (bottom panel, strain 1 is shown) carried out using rabbit anti-GFP as previously described [14]. A lower molecular weight, cross-reactive band on the western blot indicates equal loading and an equivalent coomassie-stained control is shown below the blot. The immunofluorescence images are merged with the phase images. Scale bar: 5μm.
C. Summary of key data from Table 1. Two independent strains were analysed in duplicate experiments and transformation efficiency is expressed as the proportion of cells that survive electroporation (see Table 1). The black and light-grey bars represent HR at the native and ectopic targets respectively. Tet was added 24 h prior to electroporation and maintained during manipulation. Survivorship was determined1 (see Table 1), blasticidin selection (10 μg ml−1) was applied and cultures were distributed in 24-well plates 6 h following electroporation. Wells containing transformed cultures were counted on day six (see Table 1) at which point it is easy to discriminate between positive and negative wells. None of the twenty plates analysed had more than 25% positive wells indicating that the vast majority represented clones. GFP expression was assessed by immunofluorescence in 83 of 120 cultures (see B and Table 1) and were scored as ectopic or native target recombination when GFP negative or positive respectively (see A and Table 1). Error bars, one standard deviation.
The assay itself consisted of a series of electroporation experiments in strains with HR-target transcription either repressed or induced (Fig. 1). We measured transformation efficiency and used loss of GFP expression to measure integration at the ectopic target. Of 120 transformed clones, 83 were screened for the ability to express GFP (Table 1). The results (Fig. 1C) indicate no change in HR at the control loci (GFP-positive clones) and more than three-fold increase in HR at the transcribed locus (GFP-negative clones). Fifteen-fold and three to twenty-fold transcription-stimulated HR have been reported in yeast [12] and human cells [13] respectively.
Table 1. Gene targeting data.
| Strain | Experiment a | Tet | Survivorship b (%) |
Transformed clones |
GFP negative |
Transformation efficiency (×10−6) c | ||
|---|---|---|---|---|---|---|---|---|
| EctopicGFP− | NativeGFP+ | Total | ||||||
| 1 | 1 | No | 5.1 | 4 | 2/4 | 0.39 | 0.39 | 0.79 |
| 1 | 2 | No | 9.3 | 10 | 3/10 | 0.32 | 0.75 | 1.07 |
| 2 | 1 | No | 4.2 | 10 | 6/10 | 1.43 | 0.95 | 2.38 |
| 2 | 2 | No | 8.1 | 12 | 8/12 | 0.99 | 0.49 | 1.48 |
|
| ||||||||
| Mean +/−SD [sum] | 6.7 +/−2.4 | 9 +/−3.5 [36] | 52% +/−16 [19] | 0.78 +/−0.52 | 0.65 +/−0.25 | 1.43 +/−0.7 | ||
|
| ||||||||
| 1 | 1 | Yes | 6.9 | 25 | 12/12 | 3.63 | 0.00 | 3.63 |
| 1 | 2 | Yes | 8.1 | 23 | 10/12 | 2.37 | 0.47 | 2.84 |
| 2 | 1 | Yes | 6.6 | 21 | 7/12 | 1.85 | 1.32 | 3.18 |
| 2 | 2 | Yes | 5.4 | 15 | 8/11 | 2.02 | 0.76 | 2.78 |
|
| ||||||||
| Mean +/−SD [sum] | 6.8 +/−1.1 | 21 +/−4.3 [84] | 79% +/−18 [66 d] | 2.47 +/−0.80 | 0.64 +/−0.55 | 3.11 +/−0.39 | ||
Each experiment consisted of five rounds of electroporation, each with 10 μg of donor construct (cut with BamHI/XhoI) and 2×107 cells.
Proportion of cells that survive electroporation.
Transformed clones / cell that survives electroporation.
47 out of 84 clones were screened. This is the adjusted figure.
There are a number of possible explanations for increased HR within transcribed DNA. Strand-invasion and/or DNA lesions can initiate HR [15]. Transcription may increase accessibility to the HR machinery or promote strand invasion through topological changes that create a more accessible structure or lead to the generation of regions of single-stranded DNA. Transcribed DNA may also be more prone to lesions introduced by nucleases involved in transcription or DNA replication.
We report a modest increase in targeting efficiency that has potential technical application. Low efficiency transformation limits the application of forward-genetic approaches but also renders reverse-genetics approaches more cumbersome. This effect has already been exploited to facilitate increased throughput in 2T1 cells. In these cells, a downstream target is constitutively transcribed by RNAP-I at a tagged RRNA locus [3]. We routinely obtain several transformed clones from a single electroporation experiment with these cells and now have an explanation for this improved output.
It is also worth considering how our findings might impact on T. brucei biology. RNAP-I and II transcribed domains may be more prone to recombination at stages of the cell and life-cycle when they are actively transcribed. This may be particularly important at the single active VSG locus transcribed by RNAP-I only in bloodstream-form cells [4]. High rates of HR have been reported at VSG loci and this is known to be important for accessing the large reservoir of sub-telomeric VSGs and VSG fragments available for antigenic variation [16]. It will be necessary to directly assess the effect of transcription on VSG recombination but our results indicate that recombination involving the active VSG could be favoured.
Acknowledgements
We are grateful to Martin Taylor for comments on the manuscript. This work was funded by the Wellcome Trust (069909/079457).
Footnotes
The volume added to each cuvette has a major impact on survivorship. We recommend 420 μl in a 2mm gap cuvette to cover the electrodes and prevent short-circuit. Larger volumes increase survivorship but not the number of transformed clones, probably because diminished charge is delivered above the electrodes.
References
- [1].Barry JD. The relative significance of mechanisms of antigenic variation in African trypanosomes. Parasitol Today. 1997;13:212–8. doi: 10.1016/s0169-4758(97)01039-9. [DOI] [PubMed] [Google Scholar]
- [2].Taylor JE, Rudenko G. Switching trypanosome coats: what’s in the wardrobe? Trends Genet. 2006;22:614–20. doi: 10.1016/j.tig.2006.08.003. [DOI] [PubMed] [Google Scholar]
- [3].Alsford S, Kawahara T, Glover L, Horn D. Tagging a T. brucei RRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Mol Biochem Parasitol. 2005;144:142–8. doi: 10.1016/j.molbiopara.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gunzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, Chung HM, Lee PT, Lee MG. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell. 2003;2:542–51. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Carruthers VB, van der Ploeg LH, Cross GA. DNA-mediated transformation of bloodstream-form Trypanosoma brucei. Nucleic Acids Res. 1993;21:2537–8. doi: 10.1093/nar/21.10.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hara T, Yasuda K, Fukuma T. Effective gene transfer into Trypanosoma brucei bloodstream forms by particle bombardment. Mol Biochem Parasitol. 2002;119:117–9. doi: 10.1016/s0166-6851(01)00384-x. [DOI] [PubMed] [Google Scholar]
- [7].Wickstead B, Ersfeld K, Gull K. The frequency of gene targeting in Trypanosoma brucei is independent of target site copy number. Nucleic Acids Res. 2003;31:3993–4000. doi: 10.1093/nar/gkg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bell JS, McCulloch R. Mismatch repair regulates homologous recombination, but has little influence on antigenic variation, in Trypanosoma brucei. J Biol Chem. 2003;278:45182–8. doi: 10.1074/jbc.M308123200. [DOI] [PubMed] [Google Scholar]
- [9].McCulloch R, Barry JD. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999;13:2875–88. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].ten Asbroek AL, Mol CA, Kieft R, Borst P. Stable transformation of Trypanosoma brucei. Mol Biochem Parasitol. 1993;59:133–42. doi: 10.1016/0166-6851(93)90014-o. [DOI] [PubMed] [Google Scholar]
- [11].ten Asbroek AL, Ouellette M, Borst P. Targeted insertion of the neomycin phosphotransferase gene into the tubulin gene cluster of Trypanosoma brucei. Nature. 1990;348:174–5. doi: 10.1038/348174a0. [DOI] [PubMed] [Google Scholar]
- [12].Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–30. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- [13].Thyagarajan B, Johnson BL, Campbell C. The effect of target site transcription on gene targeting in human cells in vitro. Nucleic Acids Res. 1995;23:2784–90. doi: 10.1093/nar/23.14.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alsford S, Kawahara T, Isamah C, Horn D. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation Mol. Microbiol. 2007 doi: 10.1111/j.1365-2958.2006.05553.x. In press. [DOI] [PubMed] [Google Scholar]
- [15].Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Robinson NP, Burman N, Melville SE, Barry JD. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol Cell Biol. 1999;19:5839–46. doi: 10.1128/mcb.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]