Summary
Drosophila neuroblasts (NBs) have emerged as a model for stem cell biology that is ideal for genetic analysis but is limited by the lack of cell-type-specific gene expression data. Here, we describe a method for isolating large numbers of pure NBs and differentiating neurons that retain both cell-cycle and lineage characteristics. We determine transcriptional profiles by mRNA sequencing and identify 28 predicted NB-specific transcription factors that can be arranged in a network containing hubs for Notch signaling, growth control, and chromatin regulation. Overexpression and RNA interference for these factors identify Klumpfuss as a regulator of self-renewal. We show that loss of Klumpfuss function causes premature differentiation and that overexpression results in the formation of transplantable brain tumors. Our data represent a valuable resource for investigating Drosophila developmental neurobiology, and the described method can be applied to other invertebrate stem cell lineages as well.
Graphical Abstract
Highlights
► Purification of fly neuroblasts and their daughter cells by FACS ► Identification of a hypothetical transcriptional network for self-renewal ► Klumpfuss is a regulator of neuroblast identity and self-renewal
Knoblich and colleagues used fluorescence-activated cell sorting to purify Drosophila larval neuroblasts and their direct differentiating daughter cells. These essentially pure populations of stem cells and differentiated cells were used to unravel the transcriptome of neuroblasts by mRNA sequencing and to uncover a hypothetical transcriptional network for stem cell self-renewal. Gain- and loss-of-function analysis of these transcription factors identified Klumpfuss (Drosophila homolog of the EGR family of transcriptional regulator) as a tumor inducer in Drosophila neural stem cells.
Introduction
Stem cells can generate a huge variety of different cell types during development and replace damaged or dying cells during tissue homeostasis. For this purpose, they have to remain in an undifferentiated state and maintain their stem cell identity over a series of cell divisions. At the same time, stem cells need to generate more differentiated cells that ultimately undergo terminal differentiation. It is important to understand how the balance between self-renewal and differentiation is regulated in a stem cell lineage, especially given that a disturbance of this balance can result in tissue degeneration or tumorigenesis. Identifying the regulatory transcriptional networks that maintain self-renewal capacity in stem cells and the mechanisms that alter those networks in a subset of daughter cells will be a critical step in addressing this issue.
Drosophila larval neuroblasts (NBs) have been used extensively as a model system for stem cell biology (Chia et al., 2008; Doe, 2008; Knoblich, 2008). These neural stem cells undergo repeated rounds of asymmetric cell division, and in the larval central brain, two types of NBs can be distinguished based on their division mode (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008). Type I NBs can be identified by the expression of the transcription factors (TFs) Deadpan (Dpn) and Asense (Ase) (Bowman et al., 2008). Type I NBs divide into a larger cell that maintains NB properties and a smaller ganglion mother cell (GMC) that generates two postmitotic neurons/glia. Type II NBs do not express Ase, but also divide asymmetrically into a self-renewing NB and a smaller intermediate neural progenitor (INP) cell. The INP undergoes a maturation phase and first turns on Ase, followed by the reexpression of Dpn. INPs have the capacity to divide asymmetrically multiple times, generating GMCs that then give rise to neurons/glia cells through a terminal division (Izergina et al., 2009; Boone and Doe, 2008; Bowman et al., 2008). This modified lineage allows type II NBs to produce up to 450 neurons, whereas type I NBs typically generate only 110 neurons (Bello et al., 2008).
In both NB lineages, different cell fates are established via the unequal distribution of the cell fate determinants Numb, Prospero (Pros), and Brat (Chia et al., 2008; Doe, 2008; Knoblich, 2008). During mitosis, these factors locate in a cortical crescent on the basal side of the NB/INP, and upon cytokinesis they segregate into the smaller daughter cell. In this cell, Numb inhibits the Notch signaling pathway, whereas the TF Pros represses cell-cycle genes and activates neuronal differentiation genes (Choksi et al., 2006). Brat can act as a translational repressor (Sonoda and Wharton, 2001), but to date, its function in NBs is unclear. Mutation of any of these three factors disturbs the balance between self-renewal and differentiation. In brat mutant larvae, the smaller daughter cell fails to mature into a functional INP and instead continues to express NB markers (Bello et al., 2006; Betschinger et al., 2006; Bowman et al., 2008; Lee et al., 2006b). This leads to an excess of NB-like cells that divide in an uncontrolled manner (Knoblich, 2010; Reichert, 2011) and results in the formation of transplantable tumors, which can be propagated indefinitely by serial injection into the abdomen of host flies. Transplanted tumors become aneuploid and start to invade other tissues, ultimately resulting in the death of the host fly (Caussinus and Gonzalez, 2005; Gonzalez, 2007). Similar tumors can be observed in pros or numb mutants (Bello et al., 2006; Bowman et al., 2008; Choksi et al., 2006). In contrast to brat, however, mutations in these genes can also lead to tumor formation in type I NB lineages (Lin et al., 2010; Bello et al., 2006).
Although the machinery that segregates cell-fate determinants during mitosis is fairly well understood, it is currently unclear how the combined action of these determinants leads to differentiation (Doe, 2008; Knoblich, 2008; Reichert, 2011). Multiple genetic screens for NB lineage defects have identified a huge number of potential regulators (Neumüller et al., 2011; Slack et al., 2006; Sousa-Nunes et al., 2009), but we still do not know the regulatory network that controls self-renewal in NBs or how this network might be modified by the segregating determinants. A recently developed genome-wide library of transgenic Drosophila RNA interference (RNAi) lines (Dietzl et al. (2007) allows large numbers of genes to be tested in a cell-type-specific manner, and identifying all genes that are expressed in the various cell types would be an important step in the identification of this network. In contrast to other stem cell systems, however, it is currently not possible to isolate Drosophila NBs in large numbers, and thus their transcriptome is not known. Several techniques have been developed to circumvent this technical limitation. One such technique is TU-tagging, which uses an enzyme-based assay to specifically modify and label newly synthesized mRNA in only certain cell types, allowing their subsequent purification and analysis (Miller et al., 2009). RNA-modifying enzymes can be expressed in NBs with the use of the UAS/Gal4 system. However, any technique that employs this system will be limited by the fact that both Gal4 and the expressed target genes will be inherited by both NB daughters, resulting in significant spill-over into the differentiating population. Alternatively, RNA can be isolated from tumor brain tissue, which is enriched for NB-like cells, and compared with wild-type brains, which are mostly made up of neurons (Carney et al., 2012). Although this approach has identified a significant number of NB-specific genes, its specificity is limited and it cannot be used to characterize certain cell subpopulations or to compare wild-type with tumor mutant NBs.
Here, we used fluorescence-activated cell sorting (FACS) to purify large numbers of NBs and neurons from Drosophila larval brains. We found that FACS purification did not affect viability or lineage characteristics, and we used these very pure populations to characterize the NBs and neuronal transcriptomes by mRNA sequencing. This led us to propose a hypothetical transcriptional network for self-renewal in NBs, and we tested the functional relevance of the identified TFs with overexpression and knockdown studies. In addition to known factors, we identified the TF Klumpfuss (Klu) as a regulator of self-renewal whose overexpression results in the formation of transplantable brain tumors. Our data form the basis for future functional studies that will take the functional redundancy of the identified factors into account, and may shed light on the mechanisms by which stem cells can lose their growth control and become tumorigenic.
Results and Discussion
FACS Isolation of Larval NBs
For transcriptional profiling of larval NBs, we wanted to isolate the cells in large numbers by FACS sorting. FACS sorting has been used successfully in Drosophila to purify embryonic cell populations (Cumberledge and Krasnow, 1994; Shigenobu et al., 2006), adult ovarian stem (Kai et al., 2005) and follicle cells (Calvi and Lilly, 2004; Bryant et al., 1999), hemocytes (Tirouvanziam et al., 2004), and posterior wing imaginal disc cells (Neufeld et al., 1998). Typically, green fluorescent protein (GFP) is expressed in a tissue-specific manner using the UAS/Gal4 system, and cells are sorted based on their fluorescence signal. In NBs, however, this method is not applicable because the equal inheritance of Gal4 results in GFP expression in both daughter cells. Furthermore, no Gal4 line exists that is specific and sufficiently strong for type I NBs and does not express in the optic lobes.
Because cell size and GFP expression levels differ greatly between NBs and GMCs/neurons, we FACS sorted for cell size using forward scattering and for fluorescence intensity. We marked NB lineages with the type I lineage-specific ase-Gal4 line, which drives expression of a strong nuclear GFP, UAS-stingerGFP (Barolo et al., 2000). In addition, we used a low-pressure FACS protocol to ensure cell survival. Finally, we recorded a very large number of sorting events on a logarithmic scale to account for the low frequency and large size of the NBs. Using this protocol (summarized in Figure 1A; see Experimental Procedures for details), we were able to identify a population of large, strongly GFP-positive cells (NBs) and less-defined populations of small cells with weaker GFP signal (differentiated cells; Figure 1A).
Figure 1.
Pure Populations of Larval NBs and Differentiated Cells Can Be Obtained by FACS
(A) After dissection of larval CNS, tissue was disrupted and the cell suspension was subjected to quality control experiments (cell culture) or FACS sorting. Plotting GFP intensity (vertical axis) to cell size (horizontal axis) shows a small population of large cells with high GFP signal (NBs) and a population of smaller cells with a lower GFP signal (differentiated cells). FACS sorted cells were either subjected to quality control (cell culture or qRT-PCR analysis) or to paired-end Illumina Solexa mRNA sequencing.
(B) Immunofluorescence staining for the NB marker aPKC, the differentiation marker ELAV and DAPI of an unsorted cell suspension (left column), FACS-sorted NBs (middle column), and neurons (right column). NBs are large aPKC-positive cells with a strong nuclear GFP signal (yellow arrowheads in left column), whereas their differentiated sibling cells are small, with a weaker GFP signal and stain positive for ELAV. The FACS-sorted NBs and neurons stain only for aPKC and ELAV, respectively (see single channels).
Genotypes are ase-Gal4, UAS-stingerGFP. Scale bars: 20 μm.
We stained unsorted cultures and FACS-sorted cells for specific NB and neuronal markers. In unsorted cell suspensions, large aPKC-positive cells with a strong GFP signal (Figure 1B, yellow arrowheads, NBs), as well as smaller ELAV-positive cells (differentiated cells), can be detected. FACS sorting of these cultures resulted in an essentially pure population of aPKC-, Dpn- (data not shown), and Miranda (Mira)-positive NBs (data not shown, but see Figure 2), and ELAV-negative NBs (98.9% NBs, 1.1% neurons; n = 3, p value 6 × 10−7, Student’s t test). The size of these cells corresponds well to the described size of NBs in vivo, and is clearly larger than that of INPs. Neurons can be distinguished from GMCs and INPs by the absence of Mira expression and their smaller size. Sorting and investigation of all observed cell populations revealed that only the population indicated in the FACS plot in Figure 1A (differentiated cells) was devoid of larger, Mira-positive cells, and contained small ELAV- and Pros-positive cells (data not shown) but never any NBs. Because no specific GMC markers are available, we cannot exclude the possibility that our neuronal population also contained very few GMCs or INPs. To exclude the presence of glia cells in the sorted populations, we stained unsorted cell cultures and FACS-sorted cells with the glial marker Repo. We could detect Repo-positive glia cells in dissociated brains but never in FACS-sorted populations (data not shown). On the basis of these experiments, we conclude that we were able to sort very pure populations of larval NBs and their more-differentiated daughter cells.
Figure 2.
FACS-Sorted NBs Express Correct Markers and Divide Asymmetrically
(A) NB in a dissociated cell culture in telophase shows localization of aPKC to the apical and Miranda (Mira) to the basal cortex.
(B) FACS-sorted NBs in metaphase (yellow arrowheads, left panel) and interphase (white arrowheads, left panel), or telophase (middle panel) asymmetrically localize aPKC (left panel) or Mira (middle panel) to the apical and basal cortex, respectively. The right panel shows FACS-sorted NBs arrested in mitosis with colchicine, which display correct localization of aPKC and Mira (n = 2). Metaphase DNA in (A) and (B) is marked with DAPI.
(C) Stills from a movie (see also Movie S1) of an NB (yellow arrow) in an unsorted dissociated culture showing multiple rounds of asymmetric divisions. GMCs (yellow arrowhead) also divide to give rise to two neurons (white arrowhead).
(D) Stills from a movie (see also Movie S2) of a FACS-sorted NB showing multiple rounds of asymmetric divisions. GMCs divide terminally to give rise to two neurons.
(E) Quantification of cell cycle lengths from ten NBs show that the first and second divisions of sorted NBs, as well as the division of the first GMC, are only slightly affected, whereas later divisions are delayed compared with unsorted NBs.
Three subsequent divisions, and the time point of the first GMC division, of ten NBs from three independent experiments each were measured; p = 0.39 (first NB), 0.02 (second NB), 0.0001 (third NB), and 0.92 (GMC), n.s. = not significant (Student’s t test).
(F) FACS-sorted Dpn-positive NBs cultured for 5.5 hr show cortical localization of Pros (arrows) and multiple smaller Pros-positive, Dpn-negative GMCs.
Genotypes are ase-Gal4, UAS-stingerGFP. Scale bars in (A-D): 12 μm; in (B) right panel and in (F): 20 μm.
FACS-Sorted Larval NBs Are Alive
To test cell viability and lineage, we analyzed the ability of dissociated NBs to localize cell-fate determinants asymmetrically before and after FACS sorting. Consistent with previous publications (Ceron et al., 2006), dissociated mitotic NBs in metaphase (data not shown) or telophase showed correct localization of Mira (Figure 2A), Pros (data not shown), Numb (data not shown; all basal), or aPKC (Figure 2A) and Pins (data not shown; both apical) before FACS sorting. After FACS sorting, the ability of sorted NBs to localize proteins asymmetrically was unchanged (Figure 2B). Of importance, upon arrest of FACS-sorted NBs in mitosis by colchicine, 79% of those cells showed the typical localization of aPKC and Mira to opposite sides (Figure 2B). Thus, FACS sorting does not affect the viability or mitotic activity of larval NBs.
To verify the lineage of FACS-sorted NBs, we performed live imaging on cultured NBs before and after FACS sorting (Figures 2C and 2D; see also Movies S1 and S2). Both cell populations underwent multiple rounds of asymmetric cell divisions, and always gave rise to smaller GMCs that divided terminally into two differentiating neurons. Quantification of cell-cycle lengths (see Extended Experimental Procedures) showed that the first two NB divisions, as well as the division of the GMC, were only very slightly affected by FACS sorting, although subsequent divisions were somewhat delayed (Figure 2E). Antibody staining of these cultures revealed that NBs and GMCs continued to express the markers Dpn and Pros, respectively, and that the ability of NBs to localize Pros asymmetrically was unimpaired even after 5.5 hr of cell culture (Figure 2F). Taken together, our results indicate that the FACS sorting procedure does not introduce any detectable modification of NB properties.
Extended Experimental Procedures.
Cell Dissociation and FACS
Third instar larva were collected and washed once each in phosphate-buffered saline (PBS) and 70% ethanol, and dissected in supplemented Schneider’s medium (10% fetal bovine serum, 2% Pen/Strep, 0.02 mg/mL insulin, 20 mM glutamine, 0.04 mg/mL glutathione, Schneider’s medium; GIBCO). Larval brains were collected and washed in Rinaldini solution (Ceron et al., 2006) on ice, and incubated in this solution with the addition of 1 mg/mL collagenase I and papain (Sigma Aldrich) for 1 hr at 30°C. Brains were washed twice each in Rinaldini solution and supplemented Schneider’s medium, and disrupted manually with a pipette tip in 200 μl supplemented Schneider’s medium. The tissue pieces were forced through a cell-strainer FACS tube (BD Falcon) and then either plated on poly-D-lysin-hydrobromide-coated glass-bottom dishes (MatTek; Ceron et al., 2006) or subjected to FACS sorting.
Larval NBs and neurons were sorted with a FACSAriaIII machine (BD) with a 100 μm nozzle and low pressure (20 psi) according to cell size and GFP intensity. We were able to reproducibly sort 100–150 NBs and 18,000–25,000 neurons per brain, depending on the age of the larva. The purities of the sorted cell populations were reproducibly high. For cell stainings, cells were sorted on coated glass-bottom dishes and immunostaining was carried out as described previously for larval brains (Betschinger et al., 2006).
For colchicine experiments, cells were subjected to a 16 hr treatment of 25 μM colchicine at room temperature.
For RNA isolation, NBs were sorted directly in TRIzol LS reagent (Invitrogen). Neurons were sorted in 1.5 ml Eppendorf tubes, spun at 3000 rpm for 15 min at 4°C, and after removal of the supernatant, TRIzol LS was added.
Microscopy and Live imaging
Images were acquired on either LSM510, 780 microscopes (Carl Zeiss GmbH) or a TCS SP5 microscope (Leica GmbH).
Live imaging of cells in culture was performed on a Perkin Elmer Ultra View Vox confocal spinning disc on an Axio Observer microscope (Carl Zeiss GmbH). Cells were kept in Schneider’s medium in a glass-bottom petri dish and left to settle for 2 hr after dissociation. NBs were directly FACS-sorted onto the petri dish, and also left to settle for 2 hr before live imaging. The interval for picture recording was set to 3 min and multiple positions were monitored. Cell-cycle lengths were measured from the nuclear breakdown of NBs/GMCs until the next nuclear breakdown.
qPCR Analysis of FACS-Sorted NBs and Neurons
Comparisons of qPCR data and RNA-Seq were done using SYBRGreen (BioRad) on a BioRad CFX96 cycler using the following primer pairs:
mira CCCAATTGGAGCTGGACAACA, GGTGTTCCCAGCAGAGAGG
dpn CGCTATGTAAGCCAAATGGATGG, CTATTGGCACACTGGTTAAGATGG
wor CAGTAATGGTGAAGAGGAGGAG, GATTAATAAATGGCCGGTGGTTG
numb CGAGACCAAGGGCCTGATAG, ATCCCGGCATATGTAGCTGAAG
ase CAGTGATCTCCTGCCTAGTTTG, GTGTTGGTTCCTGGTATTCTGATG
elav CGCACCATTCGGAGCAATAAT, AGGCAATGATAGCCCTTGTGG
brat GTGGTTAGTGGCGCTGGAG, GGATAGATAGTGGCCGAAAGC
pros GCTGTCACCGAAGGCATCAAG, GAAGAACTCCCGCAGAGTCG
Sample Preparation and RNA-Seq
Per experiment, total RNA from 200,000–250,000 FACS-sorted NBs and 28–35 million neurons was isolated by TRIzol purification (Invitrogen) and quality was assessed on a bioanalyzer (Agilent). The obtained RNA was enriched for poly(A) plus mRNA (Dynabeads mRNA purification kit; Invitrogen), fragmented for 2.5 min at 94°C (200 mM Tris pH 8.2, 500 mM KOAc, 150 mM MgOAc), and subjected to first-strand cDNA synthesis (based on a protocol developed by Mortazavi et al. (2008). MiniQuick spin DNA columns (Roche) were used to eliminate dNTPs and enzymes. To generate second-strand cDNA, 1x second-strand buffer (Invitrogen), 200 nM final of ATP, CTP, GTP, and UTP (5′), 20 U of Escherichia coli DNA ligase (Invitrogen), 40 U of polymerase I (Invitrogen), and 4 U of E. coli RNase H (Invitrogen) were added to the first-strand cDNA and incubated for 2 hr at 16°C. Double-stranded cDNA was purified using the MinElute Reaction Cleanup Kit (QIAGEN), and quantified using the Quant-iT Picogreen dsDNA assay in a NanoDrop ND-3300 fluorospectrometer.
Library preparation was done using a modified protocol from Illumina with NEBNext DNA sample Prep Reagent kits (NEB). Double-stranded cDNA was end-repaired, poly(A) was added, and adapters were ligated to DNA fragments. After size selection (200–600 bp) and UDGase-treatment for strand specificity, adaptor-modified DNA fragments were enriched by PCR, and the amount and quality were assessed by qPCR and bioanalyzer (Agilent), respectively. After each step, reactions were cleaned using the QIAquick PCR Purification Kit (QIAGEN), and 76-bp paired-end sequencing was performed on GAIIX or Hiseq2000 machines.
The strand-specific paired-end reads were screened for ribosomal RNA by alignment with BWA (v0.6.1; Li et al., 2009) against known rRNA sequences (RefSeq). The insert statistics were estimated by aligning the remaining reads uniquely to the transcriptome and calculating the mean insert length and SD. The rRNA-subtracted paired-end reads were aligned with TopHat (v1.4.1; Trapnell et al., 2009) against the Drosophila melanogaster genome (FlyBase r5.44) and a maximum of six mismatches. Introns between 20 and 150,000 bp were allowed, based on FlyBase statistics. The maximum of multi hits was set to one, and both InDel and microexon searches were enabled. Additionally, a gene model was provided as GTF (FlyBase r5.44). snRNA, rRNA, tRNA, snoRNA, and pseudogenes were masked for downstream analysis. Aligned reads in valid pairs were subjected to FPKM estimation with Cufflinks (v1.3.0; Trapnell et al., 2010; Roberts et al., 2011). In this step, bias detection and correction were performed, and only those fragments that were compatible with FlyBase annotation (r5.44) were allowed and counted toward the number of mapped hits used in the FPKM denominator. Furthermore, the aligned reads were counted with HTSeq, and the polyA-containing transcripts were subjected to differential expression analysis with DESeq (v1.8.3; Anders and Huber, 2010). Parameters were chosen to match the DESeq publication. GO annotation is from flybase (r5.44), and analysis was done using BINGO, a plugin (Maere et al., 2005) for cytoscape (http://www.cytoscape.org).
Alternative Splicing
Aligned reads of the technical replicates were merged into one biological replicate (Li et al., 2009). The different biological replicates were analyzed with cuffdiff (v1.3.0; Trapnell et al., 2010). The reads were bias corrected, and only reads that were compatible with the annotation (r5.44) were considered. Significant splice variant switches (FDR < 0.01) were retrieved with a custom R script.
Microarray Analysis and Network Inference
Microarray data were obtained from the FlyAtlas (Chintapalli et al., 2007) project and supplemented with some of our own data (Neumüller et al., 2011, and unpublished data). All network inference and microarray analyses were performed in R using Bioconductor packages. Raw probe intensities were normalized using RMA (Irizarry et al., 2003). The CLR algorithm (Faith et al., 2007) was used to infer putative regulatory interactions at a standardized difference score (z-score) cutoff of six.
Generation of Overexpression Constructs
Total RNA was extracted from third instar larval brains and transcribed into first-strand cDNA using oligo(dT) primers and SuperScriptIII (Invitrogen). Coding sequences, including the stop codon, were amplified using the following specific primer pairs:
Bigmax CACCATGAGCGATAATAACAACGCGTTG, TCAGCTGAAACCCTCGCTCG
Coop CACCATGGAGATGGCCTACAATGCGT, TCAGCCAAACTCCAGTTCCTC
CG4553 CACCATGACTGCCCGCATGATCCAA, CTATTTATCACCACTGGCTCTG
CG4570 CACCATGGTTCTCGCTCCCGAAGTT, CTAGCAGAACTCGGTCTGATC
CG6701 CACCATGACAAAAAATAAAAAGAACATAATCAACAAT, TTATGAAATGATACAGTTGAATTTTTTGTTC
CG6769 CACCATGTCGCACTTCACCTGCCT, TTAGATCAGAACCTGTGCACGATA
CG10565 CACCATGACGAGCGGTACGGTAGC, TCATTTGACCGCCGCCTGTG
CG13897 CACCATGGACATGCACAGGCTGATCT, TCAGTTCCAATTCGTGGAGCTTG
CG15715 CACCATGGCACGTGGACACCAGAA, TCAGACCTCCTTCAGCTCCT
CG31875 CACCATGTCGGCACGCAAGGAGAA, TCATCTATGGATGGCCTGTTGG
crc CACCATGAGCACCTATATATTTATGCAAGC, CTAGCGCTTGCGTTCATGGTA
dpn CACCATGGATTACAAAAACGATATTAATTCCG, CTACCACGGCCTCCAAGC
HLHmγ CACCATGTCGTCGCTACAAATGTCCG, CTACCAGGGACGCCAGAC
ken CACCATGAAAGAGTTTCAAAGAATGTTGATGTT, CTATTCGCGCAGATTCTTTGTCA
king-tubby CACCATGGAGGCCTACATCCGGC, TCACTCGCAGGCTATTTTGCCA
klu CACCATGACGATGGCAGAAGGCAC, TTAGGCGCTCTCCGTCTTGA
l(2)k10201 CACCATGGATTCGGAAGCTGCGGG, CTAATCCAGGATGTCGTTAATGG
mod CACCATGGCCCAAAAGAAAGCCGTCA, TAAAATCTTGCCCTTTTAACAAACGAT
mTTF CACCATGATTAGAAGCCTTCTGCG, TCATCCTTCTGATACACTTTG
Rel CACCATGAACATGAATCAGTACTACGACC, TCAAGTTGGGTTAACCAGTAGGG
Ssb-c31a CACCATGCCCAAAACAAAGAAGAAGGATT, TTAATTCTCGATCGCGCGGGT
su(Hw) CACCATGAGTGCCTCCAAGGAGGG, TCAAGCTTTCTCTTGTTCGCCTA
TFAM CACCATGATCTACACCACAACACTGATG, CTATATATCTTTGGAGGCCAGCG
wek CACCATGGGAGTTCCCACAAGCGA, CTAATCCTGTTTGGCCTTGGCC
wor CACCATGGATAAACTCAAGTACAGCCG, TTAATAAATGGCCGGTGGTTGCA
After gel purification, entry clones were generated using the pENTR TOPO cloning kit (Invitrogen). Destination clones were generated with the Gateway System (Invitrogen) using a Gateway pUASt attB vector (kindly provided by Konrad Basler), and targeted insertion into fly embryos was performed as described previously (Groth et al., 2004; Bischof et al., 2007).
Generation of klu shmiR
We developed our own software to predict efficient shRNAs with minimal off-targets (available on request). The synthesized oligos were annealed and cloned into the WALIUM20 vector according to protocols of The Transgenic RNAi Project (flyrnai.org). The following primers were used for klu:
AATTCGCTGATGCTGGCAAGTACATCAATATGCTTGAATATAACTATTGATGTACTTGCCAGCATCAACTG
CTAGCAGTTGATGCTGGCAAGTACATCAATAGTTATATTCAAGCATATTGATGTACTTGCCAGCATCAGCG
Larval NB Transcriptome
To determine the transcriptome of purified NBs and neurons, we isolated polyA-mRNA and generated libraries that were sequenced by 76-bp paired-end Illumina mRNA sequencing (mRNA-Seq). At least two independent biological samples were analyzed for each cell type, and technical replicates were analyzed to address reproducibility. All rRNA reads were removed by alignment against known rRNA sequences (RefSeq) and the remaining paired-end reads were aligned against the D. melanogaster genome (FlyBase r5.44), allowing a maximum of six mismatches and an intron size of 20 bp to 150 kb. Pseudogenes, snRNA, rRNA, tRNA, and snoRNA were masked for downstream analysis. Gene expression was estimated as the number of fragments per kilobase of combined exon length (according to gene models in FlyBase r5.44) per one million of total mapped reads (FPKM value). For a detailed description of the bioinformatics analysis, see Extended Experimental Procedures.
Our RNA-Seq data revealed a total of 3,532 genes that were differentially expressed between NBs and neurons (assuming a false discovery rate [FDR] of 0.01, p < 0.01; see also Table S1). The data showed that 1830 (52%) of these genes were upregulated in neurons, and 1702 (48%) were upregulated in NBs. Of interest, a previous comparison of human dopaminergic neurons with progenitor cells (Marei et al., 2011) revealed a similar ratio of up- and downregulated genes (47.5% of differentially regulated genes upregulated in progenitors, and 52.5% upregulated in neurons). A gene ontology (GO) term analysis of our results (Table 1; see also Tables S2 and S3) revealed that processes such as metabolism, cell cycle, DNA replication, and ribosome biogenesis are primarily upregulated in NBs. Not surprisingly, these GO terms were also found to be overrepresented in brat mutant tumors (Carney et al., 2012). Cell communication, signal transduction, neuron differentiation, and axonogenesis are overrepresented in neurons, consistent with what is known about functional regulation in differentiated neurons.
Table 1.
Overrepresented GO Terms in NBs and Neurons
| Larval NBs |
Larval Neurons |
||
|---|---|---|---|
| GO Term (GO ID) | Corr. p Value | GO Term (GO ID) | Corr. p Value |
| Metabolic process (8152) | 9.63 × 10−41 | Cell communication (7154) | 1.62 × 10−76 |
| Mitotic cell cycle (278) | 1.18 × 10−40 | Signaling (23052) | 4.32 × 10−76 |
| Mitotic spindle organization (7052) | 6.06 × 10−34 | Response to stimulus (50896) | 5.08 × 10−54 |
| Microtubule cytoskeleton organization (226) | 9.70 × 10−33 | Signal transduction (7165) | 1.46 × 10−53 |
| Cell-cycle process (22402) | 5.86 × 10−29 | Neuron projection morphogenesis (48812) | 7.75 × 10−46 |
| DNA replication (6260) | 1.25 × 10−28 | Neuron projection development (31175) | 1.00 × 10−45 |
| Cellular biosynthetic process (44249) | 3.48 × 10−28 | Axonogenesis (7409) | 5.58 × 10−45 |
| M phase (279) | 5.56 × 10−27 | Generation of neurons (48699) | 5.58 × 10−45 |
| Microtubule-based process (7017) | 1.26 × 10−25 | Neuron differentiation (30182) | 1.12 × 10−44 |
| Ribonucleoprotein complex biogenesis (22613) | 1.70 × 10−25 | Axon guidance (7411) | 7.36 × 10−40 |
| Ribosome biogenesis (42254) | 1.96 × 10−25 | Chemotaxis (6935) | 2.09 × 10−39 |
| Macromolecule metabolic process (43170) | 5.91 × 10−25 | Response to chemical stimulus (42221) | 8.21 × 10−32 |
| Neurogenesis (22008) | 8.01 × 10−25 | Regulation of signaling (23051) | 1.97 × 10−30 |
| Gene expression (10467) | 3.52 × 10−24 | Locomotion (40011) | 2.21 × 10−30 |
| DNA metabolic process (6259) | 5.53 × 10−23 | Nervous system development (7399) | 2.22 × 10−28 |
GO term analysis revealed processes expected for growing and dividing cells (e.g., metabolism, cell cycle, DNA replication, and ribosome biogenesis) to be enriched in NBs. Cell communication, signal transduction, neuron differentiation, and axonogenesis are overrepresented in neurons. Corr, Corrected.
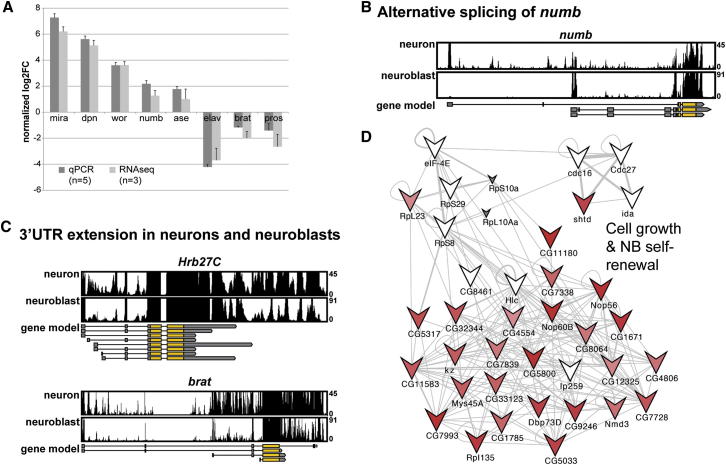
To assess the quality of our data, we verified some of the known NB and neuronal markers by quantitative reverse transcriptase (qRT)-PCR from FACS-sorted larval NBs and neurons (Figure 3A). We were able to show the upregulation of the genes mira, dpn, wor, numb, and ase, as well as the downregulation of ELAV, brat, or pros in NBs. In all cases, the two methods showed the same trend of transcriptional regulation, although qRT-PCR detected higher fold changes in transcription levels. Thus, we have obtained high-quality data showing the transcriptional differences between a Drosophila stem cell population and its sister cells.
Figure 3.
Bioinformatic Analysis of Transcriptome Data
(A) Expression levels of FACS-sorted NBs obtained by qRT-PCR and RNA-Seq data for known NB- and neuron-specific genes correlate (n denotes the number of experiments, error bars represent SD).
(B) Numb transcript is alternatively spliced. The gene model from flybase (r5.44) is indicated, numb-RB is specifically expressed in neurons (upper track), numb-RA in NBs (lower track).
(C) Hrb27c and brat are shown as examples of genes with 3′ UTR extensions. RNA-Seq tracks for neurons (upper track) and NBs (lower track) are shown. Note that for both genes the read coverage extends the 3′ UTR annotated in flybase (r5.44).
(D) A network of genes involved in cell growth and NB self-renewal, resulting in loss or underproliferation of NBs knockdown phenotypes (Neumüller et al., 2011). Correlation with gene expression data shows that 71% of genes are significantly upregulated in NBs. “V” node shape denotes underproliferation. Small gray nodes are genes that are not expressed in NBs or neurons. Red nodes are genes expressed significantly higher in NBs (the strength of the color indicates fold change levels). White nodes are genes expressed at the same level in NBs and neurons. See also Figure S1 for an expanded network of asymmetric cell division.
Alternative Splicing and 3′ UTR Extension
In addition to total transcript levels, RNA-Seq allows for the detection of splicing isoforms. This may be relevant for NB biology, because a previous genome-wide RNAi screen (Neumüller et al., 2011) identified splicing as one of the important processes in NBs, and RNA metabolism, transcription, and splicing are among the processes that are transcriptionally upregulated in NBs (Table S2). In total, we found 69 genes to be alternatively spliced between NBs and neurons (Table S4). These included alternatively spliced genes such as longitudinals lacking (lola) (Neumüller et al., 2011), but also many genes for which tissue-specific alternative splicing has not been described. The known cell-fate determinant numb is among the alternatively spliced genes. Its different isoforms are due to alternative initiation, which results in differences in the coding sequence, and not due to alternative splicing of the pre-mRNA. We found that the isoform numb-RA is primarily expressed in NBs, whereas differentiated neurons express an alternative isoform (numb-RB; Figure 3B). Of interest, a previous deletion analysis of numb showed that numb-RA, but not numb-RB, can segregate asymmetrically in NBs (Knoblich et al., 1997). Because Numb binds to α-Adaptin, which was shown to be required for presynaptic vesicle recycling (González-Gaitán and Jäckle, 1997), we speculate that Numb-PB could participate in this process in mature neurons.
We also used our RNA-Seq data to address the cell-type specificity of 3′ UTR elongation, a phenomenon that was recently described in the Drosophila nervous system by Hilgers et al. (2011) and Smibert et al. (2012). In both Drosophila embryos and the larval central nervous system (CNS), a large set of transcripts are extended beyond the predicted end of the 3′ UTR. These authors proposed that this confers complex regulation by miRNAs or RNA-binding proteins in a tissue-specific manner. Our data indicate that 40 of the 400 genes described by Smibert et al. (2012) are more highly expressed in NBs, and 357 of these genes are upregulated in neurons (Table S5). 3′ UTR extension was detected in both NBs and neurons in all cases in which transcripts could be detected in both samples despite the differential expression. This was particularly evident for Hrb27c and brat, two genes that were previously reported to display 3′ UTR extension (Smibert et al., 2012; Figure 3C). Thus, alternative splicing (but not 3′ UTR extension) seems to be one of the processes that are differentially regulated between NBs and neurons.
Integrating Transcriptional and Phenotypic Data
The transgenic RNAi technology developed by Dietzl et al. (2007) has allowed for genome-wide RNAi screening in a tissue-specific manner. We used our transcriptome data to correlate functional data from a genome-wide RNAi screen in larval NBs (Neumüller et al., 2011) with gene expression data. In addition to a huge number of other genes, this screen identified a set of 38 genes that cause NB loss or size reduction when knocked down in NBs, and have been arranged in a potential network for growth and self-renewal. As expected, we observed a tight correlation between these phenotypic data and our gene expression results. All but two genes in the network are expressed in NBs; 71% are significantly upregulated in NBs, and therefore may be responsible for the enhanced growth rate in NBs compared with GMCs and neurons (Figure 3D). In addition to restricting functional data, our transcriptome data can also be used to expand functional regulatory networks from RNAi screens. For example, starting from a set of known regulators, genome-wide RNAi data have been used to generate a functional network for asymmetric cell division. This network can now be expanded by differentially expressed genes with previously reported protein interactions to members of the existing network (see also Figure S1) and could form the basis for further studies to increase our understanding of neural stem cell biology.
Figure S1.
Transcriptome Information Extending the Phenotype-Based Network of Asymmetric Cell Division, Cell Growth, and NB Self-Renewal, Related to Figure 3
The joined phenotypic network of asymmetric cell division, cell growth, and NB self-renewal as described by Neumüller et al. (2011) was expanded by adding strongly linked genes with available transcriptome data and five or more interaction partners in the phenotypic network. Interaction data were extracted from DroID (v5; http://www.droidb.org), STRING (v7.0 and v8.2), and BioGRID (v2.0.40), including experimental, genetic text-mining, and interolog interaction data.
Genes are shown as nodes and the color of the nodes reflects the differential expression of genes, with red denoting higher expression in NBs, blue indicating higher expression in neurons, and white indicating no differential expression. For differentially expressed genes, the color intensity reflects the strength of the log-fold change. Approximately 12 genes from the phenotypic network were not expressed; they are shown in gray and small size. The shape of the nodes denotes phenotypes from the RNAi screen (Neumüller et al., 2011). The V shape is underproliferation, triangle is overproliferation, diamond is over- and underproliferation, hexagon is “other” (e.g. GFP aggregates), and ellipse is no phenotype. The white border and transparent colored nodes indicate genes from the original network, and black borders and solid node color denote genes with which the network was expanded. Thus, this network could form the basis for further studies and might help increase our understanding of neural stem cell biology.
Hypothetical Transcriptional Network for NB Self-Renewal
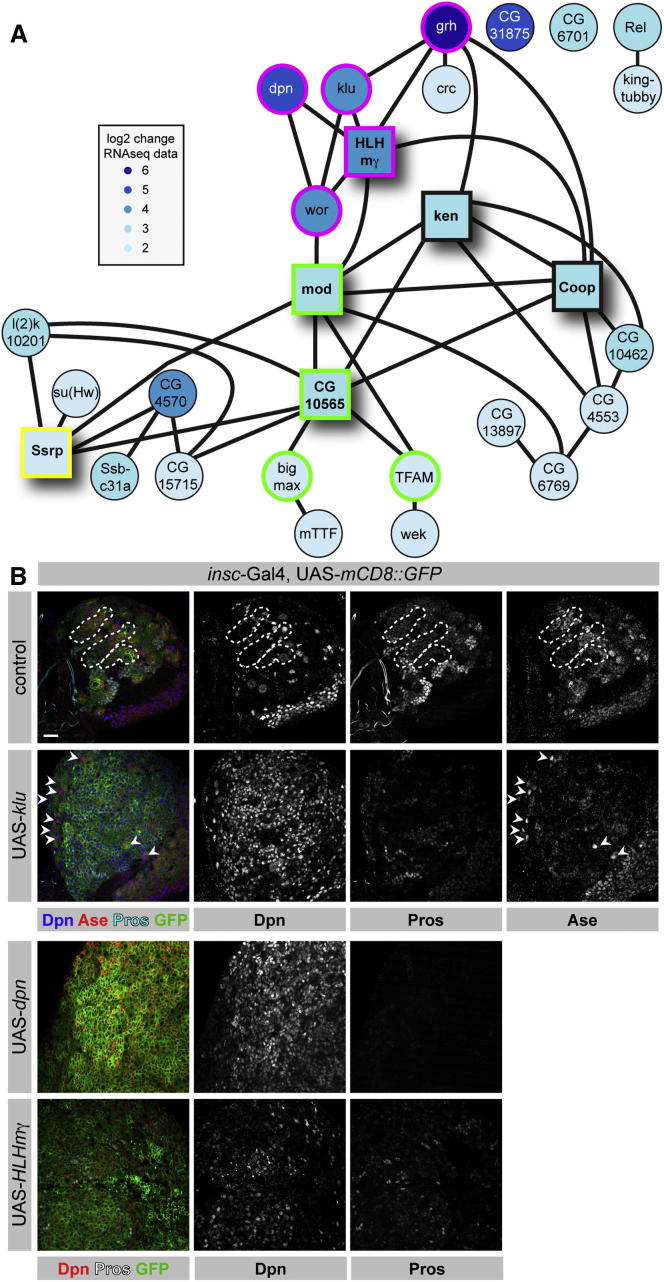
Unlike their differentiating sibling cells, NBs regrow to their original size after each division and maintain their identity over many cell divisions. Therefore, NBs must express a regulatory transcriptional network that is highly robust over time but can be rapidly and irreversibly modified by Numb, Pros, and Brat. Previous loss-of-function experiments revealed a surprising level of redundancy among the known TFs acting in NBs (San-Juán and Baonza, 2011; Zacharioudaki et al., 2012). Therefore, we used our transcriptome data to identify a complete set of predicted TFs that are highly expressed in NBs and strongly downregulated during differentiation. In total, 28 TFs match our criteria (FDR 0.01, logFC > 3, FPKM value > 15). Assuming that functionally related TFs are more likely to be coexpressed, we used stage- and tissue-specific microarray data (Chintapalli et al., 2007) and the context likelihood of relatedness algorithm (Faith et al., 2007) to infer putative regulatory interactions (see Extended Experimental Procedures for more information).
The resulting hypothetical network contains six hubs that are connected to more than five genes in the network (Figure 4A). The first hub is HLHmγ, a direct nuclear target of the Notch signaling pathway (Almeida and Bray, 2005). HLHmγ connects to Dpn, and both were shown to act redundantly in controlling NB self-renewal (Zacharioudaki et al., 2012). It also connects to Wor and Klu, which have been linked to Notch signaling by both coexpression and functional studies. Finally, it connects to Grh, which can regulate proliferation in NBs (Cenci and Gould, 2005; Almeida and Bray, 2005). Surprisingly, Grh is actually a negative regulator of HLHmγ (Almeida and Bray, 2005), but it has been proposed that such paradoxical elements are frequent components of transcriptional circuits and can maintain homeostatic concentrations or contribute to robust regulation (Hart et al., 2012). Thus, HLHmγ is a major hub for transcriptional activation immediately downstream of Notch.
Figure 4.
Klumpfuss is an NB Identity Factor and Part of a Hypothetical Transcriptional Network for NB Self-Renewal
(A) Hypothetical transcriptional network of 28 strong and differentially expressed TFs in NBs (FDR 0.01, log2FC > 3, FPKM value > 15) based on correlative Drosophila microarray expression data. Genes involved in growth control (mod, CG10565, bigmax, and TFAM), genes downstream of or regulated by Notch signaling (HLHmγ, dpn, wor, klu, and grh), and the chromatin remodeler Ssrp are marked in green, magenta, and yellow, respectively. Hubs are indicated with squares.
(B) Larval brains overexpressing klu, dpn, HLHmγ, and control stained for Dpn, Pros, and Ase (Ase only shown for control and UAS-klu). Ectopic expression of these genes leads to overproliferation of Dpn-positive cells at the expense of differentiating cells (loss of Ase/Pros staining). White arrowheads indicate Ase-positive type I NBs in the klu overexpression brain. See also Figure S2 for Klu overexpression with the type II lineage-specific PntP1-Gal4, which also leads to tumor formation.
Scale bars: 20 μm.
HLHmγ and Wor are connected to two other hubs that consist of genes involved in ribosome biogenesis and growth control. Mod is the Drosophila homolog of Nucleolin, the major nucleolar protein of growing eukaryotic cells that is thought to play a role in rRNA transcription and ribosome assembly (Srivastava and Pollard, 1999). CG10565 is the fly ortholog of MPP11, a chaperone of the DNA-J family that is involved in ribosome assembly and has been implicated in the regulation of cell growth (Otto et al., 2005; Jaiswal et al., 2011). In NBs, CG10565 connects to Bigmax and TFAM, two factors that may be involved in growth control because they are direct downstream targets of the major growth regulators Myc and Max (Orian et al., 2003). The fourth interesting hub is Ssrp, a chromatin regulator that has been identified based on its RNAi overproliferation phenotype (Neumüller et al., 2011). Surprisingly, Ssrp RNAi causes gain rather than loss of NBs, and may therefore maintain a chromatin state that allows differentiation. Finally, Ken and Coop have been implicated in Jak/STAT (Arbouzova et al., 2006) and wingless signaling (Song et al., 2010), respectively. Both of these pathways are linked to Notch signaling, although no such functional link has been described in NBs.
Thus, our hypothetical transcriptional network provides potential explanations for several aspects of NB biology. For example, it could explain the direct effect of Notch signaling on cell growth that has been described in Drosophila NBs (Song and Lu, 2011).
Klumpfuss Overexpression Causes Transplantable Tumors
To test the functional relevance of the identified TFs, we performed overexpression studies. Assuming that key factors need to be downregulated during differentiation, we generated overexpression constructs for all 28 NB-specific TFs and expressed them in type I and type II NBs, as well as in GMCs and INPs, using insc-Gal4. Although most factors did not cause an overproliferation phenotype, overexpression of HLHmγ, Dpn, and Klu resulted in a strong expansion of the NB pool (Figure 4B). Because the Dpn and HLHmγ overexpression phenotypes have been described for larval NBs (Zacharioudaki et al., 2012; San-Juán and Baonza, 2011), we focused on Klu.
Klu is a conserved Zn finger TF of the early growth response (EGR) family. Antibody staining reveals that Klu is highly expressed in type I NBs but is rapidly downregulated in the Ase+ GMCs (Figure 5A). In type II lineages, Klu is expressed in the NB but is lost from immature INPs. It is not expressed in Ase+, Dpn− INPs, but reappears with Dpn in mature INPs and disappears again when the GMCs are formed (Figure 5A).
Figure 5.
Immature INPs Reacquire NB Identity upon Ectopic Expression of Klumpfuss
(A) In type II lineages (left column), Klu is expressed in primary NBs, reexpressed together with Dpn in Ase-positive mature INPs (see also single magenta/GFP channels, and schematic drawing) and turned off in GMCs/neurons. In type I NBs, Klu is expressed in primary NBs but, similarly to Dpn, not in GMCs or neurons (magenta/GFP in right column).
(B) GFP-marked fragments from Klu overexpression brain tumors transplanted into the abdomens of host flies resulted in tumors in the adult hosts (filled green abdomen) 9 days after transplantation. Extracted tissue (right panels) stained with Mira and ELAV shows that the tumor tissue consists mostly of NBs.
(C) As in wild-type type II lineages (left panels), mitotic (pH3 positive) NBs overexpressing Klu (right panels) show asymmetric localization of aPKC (apical marker, upper panels) or Mira (basal marker, lower panels).
(D) Time-controlled induction of klu expression reveals blocked maturation of INPs and their dedifferentiation into Dpn-positive NB-like cells (white arrowheads, lower panels). In control type II lineages (upper panels), two to three Ase and Dpn negative immature INPs (white arrows) and three to four Ase-positive, Dpn-negative mature INPs (yellow arrowheads) are located next to the Dpn-positive, Ase-negative primary NB (asterisk). Lower panels show type II lineage after 19h of klu overexpression. Ase- and Dpn-negative immature INPs (white arrows) can be seen close to the primary NB (asterisk). Ectopic NB-like cells expressing Dpn, but not Ase, are found close to the primary NB (white arrowheads). See also schematic drawing for phenotype description. Smaller panels show blowups of the marked area in the left panels. Two subsequent focal planes are shown for control and klu overexpression (ten brains from two independent experiments were investigated).
See also Figure S3 for Klu overexpression with the mature INP specific R9D11-Gal4, which does not lead to tumor formation.
Scale bars: 20 μm.
Klu overexpression causes an excess of Dpn-positive cells at the expense of Pros-positive GMCs and neurons (Figure 4B). To test whether the overexpression of Klu results in tumor formation, we transplanted fragments from control or Klu overexpressing larval brains into the abdomen of female host flies. Although none of the control transplants resulted in tumors, 52 of 88 (59%) of the adult hosts transplanted with tissue from brains overexpressing Klu developed tumors 9 days after transplantation (Figure 5B). Antibody staining of extracted tumor tissue revealed that the tumors consisted mostly of Mira- and Dpn-positive NBs (data not shown), with fewer differentiating cells that were positive for Pros (data not shown) or ELAV (Figure 5B). Thus, our data show that upregulation of Klu prevents differentiation and causes tumor formation in the Drosophila brain.
To test whether the tumors originated from type I or type II lineages, we used the type II lineage-specific PointedP1-Gal4 (PntP1-Gal4) driver (Zhu et al., 2011). Expression of Klu from PntP1-Gal4 resulted in tumors (see Figure S2) that were indistinguishable from those induced by insc-Gal4. Together with the fact that these tumors are negative for the type I NB marker Ase, these results demonstrate that Klu overexpression causes overproliferation in type II lineages.
Figure S2.
Overexpression of klu with PntP1-Gal4 Results in Tumor Formation, Related to Figure 4
Expression of klu from PntP1-Gal4 resulted in tumors that showed an excess of ectopic NB-like cells at the expense of differentiating GMCs or neurons.
Klu Overexpression Causes Dedifferentiation of Immature INPs
Klu overexpression could induce tumor formation by either disrupting asymmetric cell division or causing dedifferentiation of INPs or GMCs. To test for asymmetric cell division, we used the apical marker aPKC and the basal marker Mira. In similarity to wild-type NBs, aPKC and Mira are localized to opposite sides of the cell cortex in dividing pH3-positive Klu overexpressing NBs (Figure 5C), indicating that asymmetric cell division is not affected. To test for dedifferentiation, we used a temperature shift assay to overexpress Klu in a time-controlled manner (Figure 5D). In control type II lineages, two to three Ase−, Dpn− immature INPs (white arrows), and three to four Ase+, Dpn− mature INPs (yellow arrowheads) are formed close to the Dpn+, Ase− primary NB (marked with an asterisk) before Dpn turns on and completes the maturation of INPs (Bowman et al., 2008). After 19 hr of inducing Klu overexpression at 29°C, one to three Ase−, Dpn− immature INPs (white arrows) can be seen close to the primary NB (asterisk), which is still recognizable at this stage. However, Ase expression is never initiated, indicating that maturation does not proceed. Instead, ectopic Dpn+, Ase− NBs can be seen close to the primary NB (white arrowheads). Therefore, immature INPs never undergo maturation when Klu expression continues. Instead, these cells revert back into an NB-like cell (Figure 5D; see schematic drawings), leading to tumor formation.
To test whether Klu overexpression can still induce dedifferentiation in mature INPs, we used the Gal4 line R9D11-Gal4, which drives expression in mature Ase+, Dpn− INPs (Weng et al., 2010). In contrast to PntP1-Gal4 or insc-Gal4, Klu overexpression using R9D11-Gal4 does not result in an overproliferation phenotype (see also Figure S3). We conclude that the transition from immature to mature INPs requires the downregulation of Klu expression. Continued expression of Klu leads to dedifferentiation of INPs into an NB-like fate and the formation of a brain tumor.
Figure S3.
Overexpression of klu in Mature INPs Does Not Cause a Tumor, Related to Figure 5
Overexpression of klu using R9D11-Gal4, which starts expressing in mature Ase-positive, Dpn-negative INPs (Weng et al., 2010), does not result in an over-proliferation phenotype.
Klu Is Required for NB Growth and Self-Renewal
Surprisingly, previous RNAi screens have not identified any of the TFs in our network as causing loss of NBs. Some caused lethality (crc, TFAM, ken, and CG15715; Neumüller et al., 2011), but considering that the NB number and size were not reported to be affected, this may be due to off-target effects or secondary phenotypes caused by expression of RNAi in other tissues. For Klu, the predicted quality of existing RNAi lines is low (81 off-targets predicted; http://www.stockcenter.vdrc.at), and therefore we generated a shimR line (Haley et al., 2008). Expression of this shmiR line resulted in a complete absence or strong reduction of Klu antibody staining in NBs (data not shown), and we used it to test for the klu loss-of-function phenotype in NBs.
Normally, eight type II lineages are detected per brain lobe, but this number is highly reduced upon klu RNAi expression, and on average, only one of these lineages remains (Figures 6A and 6B). Similarly, the number of type I NBs is lower, although in this case the average number is only reduced by 12% (Figure 6C). In addition, klu RNAi results in a reduction of NB size (Figure 6D). This phenotype is not due to apoptosis, as p35 expression cannot rescue the loss of NBs (data not shown), or to symmetric NBs divisions, since we never found more than one Dpn+ cell. Thus, our data indicate that klu is essential for maintaining type II NB fate and, to a lesser extent, for growth and self-renewal in type I NBs.
Figure 6.
Klumpfuss Is Required for NB Self-Renewal and Growth
(A) Klumpfuss knockdown by RNAi causes loss of type II lineages. Larval brains are stained for Dpn and Pros, and type II lineages (left) and areas with loss of type II lineages (right panel) are marked.
(B) Quantification of number of type II NBs upon klu knockdown by RNAi. On average, only one type II lineage can still be identified (n denotes the number of brains counted, error bars represents SD; p value 7.7 × 10−31, Student’s t test).
(C) Quantification of the number of type I NBs upon klu knockdown by RNAi shows a reduction in total type I NBs per brain hemisphere (n denotes the number of brains counted, error bars represents SD; p value 2.7 × 10−5, Student’s t test).
(D) Quantification of type I NBs cell size in the larval central brain shows that loss of klu function leads to a reduction in NB size (n denotes the number of NBs measured).
(E) MARCM loss of function clones of kluR51 induced 30-48 hr AEL (right panel) show loss of primary NBs (right panel, type II lineage is shown) compared with wild-type MARCM clones (left panel). Larval brains were stained for Dpn and Pros, and the locations of NBs are indicated with asterisks.
See also Figure S4 for the transheterozygous combination of mutant alleles kluG410 and kluR51, which recapitulates the klu clonal and shmiR loss of NB phenotypes.
Scale bars: 15 μm.
To exclude off-target effects, we attempted to recapitulate those phenotypes in kluR51 mutant MARCM clones (Klein and Campos-Ortega, 1997). When the clones were induced 50–68 hr after egg laying (AEL), we could not find any previously observed NB number or size phenotype. When the clones were induced 30–48 hr AEL, however, NB size was reduced (Figure 6D), and 10.7% of the mutant clones (n = 374) displayed an NB loss phenotype (Figure 6E). We further confirmed the loss of NB phenotypes by crossing two mutant alleles for Klu transheterozygously (kluG410/kluR51; Figure S4).
Figure S4.
Transheterozygous Combination of Two Klu Mutant Alleles Causes Loss of Type I and Type II NBs, Related to Figure 6
The transheterozygous combination of the two mutant alleles kluG410 and kluR51 causes a loss of type II lineages (A) and a reduction of type I NBs by 15% (B).
(n denotes number of brains counted, error bars represents SD, p = 0.01(Student’s t test)).
We used our klu shmiR line in a time-shift assay and found the same sensitive phase for Klu loss (data not shown). Either Klu functions in the NB only at a specific time-point or the observed phenotype develops only some time after knockdown or MARCM-induced loss of Klu. This could be due to protein perdurance, although we could not detect any residual protein in any of the two experiments by immunofluorescence (data not shown). Taken together, our results identify a regulator of self-renewal that had not been previously described in Drosophila larval NBs.
Klu is a member of the EGR family of transcriptional regulators, which is conserved in vertebrates. EGR genes contain three conserved Zn finger motifs, can act as both positive and negative regulators of transcription, and are implicated in regulation of proliferation and cell growth. The biological function of EGR-1 has also been connected to human cancers (Thiel and Cibelli, 2002). Our identification of Klu as a tumor inducer in Drosophila NBs may help to further clarify the role of these genes in tumor formation.
Conclusions
Previous studies of Drosophila NBs have provided important insights into the mechanism of asymmetric cell division. Despite this progress, however, the transcriptional circuits that control self-renewal and act downstream of segregating determinants have remained elusive. Functional redundancy has already been demonstrated for some TFs acting in NBs (Zacharioudaki et al., 2012) and may have hindered the genetic identification of others. Our transcriptional data lay the foundation for a more targeted search for functional interconnections between redundantly acting factors. The hypothetical transcriptional network we have identified provides testable hypotheses for how Notch signaling might connect to the machinery for growth regulation. It suggests that Notch target genes control other TFs that regulate ribosome biogenesis and assembly in a surprisingly direct manner. With the use of our established method to purify larval NBs, and the development of new techniques for chromatin immunoprecipitation even from limited numbers of cells, it will become technically feasible to test the proposed network. The ease with which individual TFs can be removed by RNAi may ultimately allow us to understand the architecture of this network in unprecedented detail.
Experimental Procedures
Fly Strains, RNAi, and Clonal Analysis
The fly strain kluG410 (Klein and Campos-Ortega, 1997) was used. RNAi lines were obtained from the Vienna Drosophila RNAi Center (Dietzl et al., 2007), and TriP lines were obtained from the Bloomington Stock Center. ShmiR lines for klu and overexpression lines for all 28 TFs were generated in this study (see Extended Experimental Procedures).
The following Gal4-driver lines were used: ase-Gal4, UAS-stingerGFP (Zhu et al., 2006, Barolo et al., 2000), UAS-dicer2; MZ1407(insc)-Gal4, UAS-mCD8::GFP (Neumüller et al., 2011), UAS-mCD8::GFP;; PointedP1-Gal4 (Zhu et al., 2011), and R9D11-Gal4 (Weng et al., 2010).
Embryonically derived phenotypes (to prevent embryonic lethality) and UAS constructs (for time-course experiments) were expressed with tub-Gal80ts, reared at 18°C, and then shifted to 29°C for the time indicated in the respective figures/experiments. All other transgenes were expressed at 25°C for 24 hr and then shifted to 29°C for 5 days.
MARCM clones derived from FRT2A, klu212lR51C (amorphic; Klein and Campos-Ortega, 1997) were induced as described by Lee and Luo (2001).
Antibodies and Immunohistochemistry
The following antibodies were used: guinea pig anti-Dpn (1:1000, courtesy of J. Skeath; Lee et al., 2006a), rat anti-Ase (1:50), mouse anti-Pros (1:100, MR1A, Developmental Studies Hybridoma Bank [DSHB]), mouse anti-Repo (1:100, 8D12, DSHB), rabbit anti-Klu (1:200; Klein and Campos-Ortega, 1997), rabbit anti-Mira (1:200; Betschinger et al., 2006), rabbit anti-aPKC (1:500; Santa Cruz Biotechnology), mouse anti-PH3 (1:1000; Cell Signaling Technology), rat anti-ELAV (1:100 cell culture, 1:30 tissue, 7E8A10; DSHB) and rabbit anti-Mira 1:100 (kind gift from Y.N. Jan). Immunohistochemistry experiments were performed as previously described (Betschinger et al., 2006). Cultured cells were fixed in 8% paraformaldehyde (PFA) in 0.1% PBT-X for 15 min.
For transplanted tumors, abdomens of host flies were fixed in 2% PFA and nuclei were labeled with Toto-3 iodide (Molecular Probes).
Cell Dissociation, FACS, Sample Preparation, and RNA Sequencing
Third instar larvae were dissected and dissociated, and cell cultures were subjected to FACS sorting (FACSAriaIII, BD), live imaging (see Extended Experimental Procedures), immunostaining, or cell-cycle arrest experiments. Total RNA from sorted cells was isolated using TRIzol reagent (Invitrogen), and poly(A)mRNA was fragmented and transcribed into first- and second-strand complementary DNA (cDNA; using dUTP; Invitrogen). The library was prepared using a modified Illumina protocol: second-strand cDNA was digested with UDGase, thereby conferring strand specificity, and paired-end sequencing was performed on either GAIIX or Hiseq2000. For details regarding sample preparation, live imaging, colchicine treatment, data processing and analysis, qRT-PCR, and bioinformatics tools used for GO term, alternative splicing, network, and cluster analyses, see Extended Experimental Procedures.
Transplantation of Larval Brains
Crosses of UAS-Dicer2; MZ1407-Gal4, UAS-mCD8::GFP with the control or UAS-klu were set up at 29°C, and after 5–6 days, transplantations of GFP-positive larval brain pieces were performed as previously described (Caussinus and Gonzalez, 2005) with minor modifications.
Acknowledgments
We thank Elif Eroglu for generating the rabbit anti-Dpn and rat anti-Ase antibodies; all members of the Knoblich laboratory for discussions; Thomas Klein, Y.N. Jan, the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, and the Developmental Studies Hybridoma Bank for fly stocks and reagents; TriP, Harvard Medical School, for providing plasmid vectors; Gerald Schmauss, Gabriele Stengl, Thomas Lendl, and Nina Corsini for help with the FACS; and Pawel Pasierbek for imaging support. C.B. was supported by an EMBO Long-Term Fellowship. Work in J.A.K.’s laboratory is supported by the Austrian Academy of Sciences, the Austrian Science Fund, the EU FP7 network EuroSystems, and an advanced grant from the European Research Council. Work in H.R.’s laboratory is supported by Swiss National Fonds grant NFP63.
Published online: August 9, 2012
Footnotes
Supplemental Information includes Extended Experimental Procedures, four figures, five tables, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.07.008.
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
Accession Numbers
The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE38764.
Supplemental Information
Table containing a complete list of genes differentially expressed in NBs and neurons with FDR = 0.01; 1,830 (52%) of these genes were upregulated in neurons, and 1,702 (48%) were expressed higher in NBs.
Table containing a complete list of enriched GO terms in NBs only from genes with FPKM value > 1, fold change > 1, and p > 1.
Table containing a complete list of enriched GO terms in neurons only from genes with FPKM value > 1, fold change > 1, and p > 1.
Table containing alternatively spliced genes with at least two differentially expressed isoforms in NBs and neurons. See Experimental Procedures for details.
Table containing 40 genes with an extended 3′ UTR that are expressed higher in NBs and 357 that are upregulated in neurons. 3′ UTR extension is not NB or neuron specific.
NBs in an unsorted cell culture divide asymmetrically multiple times. The first- and second-born GMCs divide terminally to give rise to neurons. Genotype: ase-Gal4, UAS-stinger::GFP
FACS-sorted NBs divide asymmetrically multiple times. The first- and second-born GMCs divide terminally to give rise to differentiated neurons. Genotype: ase-Gal4, UAS-stingerGFP
References
- Almeida M.S., Bray S.J. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 2005;122:1282–1293. doi: 10.1016/j.mod.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Arbouzova N.I., Bach E.A., Zeidler M.P. Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr. Biol. 2006;16:80–88. doi: 10.1016/j.cub.2005.11.033. [DOI] [PubMed] [Google Scholar]
- Barolo S., Carver L.A., Posakony J.W. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29 doi: 10.2144/00294bm10. 726, 728, 730, 732. [DOI] [PubMed] [Google Scholar]
- Bello B., Reichert H., Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- Bello B.C., Izergina N., Caussinus E., Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J.A. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Boone J.Q., Doe C.Q. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 2008;68:1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S.K., Rolland V., Betschinger J., Kinsey K.A., Emery G., Knoblich J.A. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant Z., Subrahmanyan L., Tworoger M., LaTray L., Liu C.R., Li M.J., van den Engh G., Ruohola-Baker H. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proc. Natl. Acad. Sci. USA. 1999;96:5559–5564. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi B.R., Lilly M.A. Fluorescent BrdU labeling and nuclear flow sorting of the Drosophila ovary. Methods Mol. Biol. 2004;247:203–213. doi: 10.1385/1-59259-665-7:203. [DOI] [PubMed] [Google Scholar]
- Carney T.D., Miller M.R., Robinson K.J., Bayraktar O.A., Osterhout J.A., Doe C.Q. Functional genomics identifies neural stem cell sub-type expression profiles and genes regulating neuroblast homeostasis. Dev. Biol. 2012;361:137–146. doi: 10.1016/j.ydbio.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E., Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- Cenci C., Gould A.P. Drosophila Grainyhead specifies late programmes of neural proliferation by regulating the mitotic activity and Hox-dependent apoptosis of neuroblasts. Development. 2005;132:3835–3845. doi: 10.1242/dev.01932. [DOI] [PubMed] [Google Scholar]
- Ceron J., Tejedor F.J., Moya F. A primary cell culture of Drosophila postembryonic larval neuroblasts to study cell cycle and asymmetric division. Eur. J. Cell Biol. 2006;85:567–575. doi: 10.1016/j.ejcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Chia W., Somers W.G., Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J. Cell Biol. 2008;180:267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V.R., Wang J., Dow J.A. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Choksi S.P., Southall T.D., Bossing T., Edoff K., de Wit E., Fischer B.E., van Steensel B., Micklem G., Brand A.H. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Cumberledge S., Krasnow M.A. Preparation and analysis of pure cell populations from Drosophila. Methods Cell Biol. 1994;44:143–159. doi: 10.1016/s0091-679x(08)60911-5. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Doe C.Q. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Faith J.J., Hayete B., Thaden J.T., Mogno I., Wierzbowski J., Cottarel G., Kasif S., Collins J.J., Gardner T.S. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 2007;8:462–472. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- González-Gaitán M., Jäckle H. Role of Drosophila α-adaptin in presynaptic vesicle recycling. Cell. 1997;88:767–776. doi: 10.1016/s0092-8674(00)81923-6. [DOI] [PubMed] [Google Scholar]
- Haley B., Hendrix D., Trang V., Levine M. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev. Biol. 2008;321:482–490. doi: 10.1016/j.ydbio.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart Y., Antebi Y.E., Mayo A.E., Friedman N., Alon U. Design principles of cell circuits with paradoxical components. Proc. Natl. Acad. Sci. USA. 2012;109:8346–8351. doi: 10.1073/pnas.1117475109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Perry M.W., Hendrix D., Stark A., Levine M., Haley B. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc. Natl. Acad. Sci. USA. 2011;108:15864–15869. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izergina N., Balmer J., Bello B., Reichert H. Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 2009;4:44. doi: 10.1186/1749-8104-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal H., Conz C., Otto H., Wölfle T., Fitzke E., Mayer M.P., Rospert S. The chaperone network connected to human ribosome-associated complex. Mol. Cell. Biol. 2011;31:1160–1173. doi: 10.1128/MCB.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai T., Williams D., Spradling A.C. The expression profile of purified Drosophila germline stem cells. Dev. Biol. 2005;283:486–502. doi: 10.1016/j.ydbio.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Klein T., Campos-Ortega J.A. klumpfuss, a Drosophila gene encoding a member of the EGR family of transcription factors, is involved in bristle and leg development. Development. 1997;124:3123–3134. doi: 10.1242/dev.124.16.3123. [DOI] [PubMed] [Google Scholar]
- Knoblich J.A. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Knoblich J.A. Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A., Jan L.Y., Jan Y.N. The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc. Natl. Acad. Sci. USA. 1997;94:13005–13010. doi: 10.1073/pnas.94.24.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Lee C.Y., Robinson K.J., Doe C.Q. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Lee C.Y., Wilkinson B.D., Siegrist S.E., Wharton R.P., Doe C.Q. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Lin S., Lai S.L., Yu H.H., Chihara T., Luo L., Lee T. Lineage-specific effects of Notch/Numb signaling in post-embryonic development of the Drosophila brain. Development. 2010;137:43–51. doi: 10.1242/dev.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marei H.E., Althani A., Afifi N., Michetti F., Pescatori M., Pallini R., Casalbore P., Cenciarelli C., Schwartz P., Ahmed A.E. Gene expression profiling of embryonic human neural stem cells and dopaminergic neurons from adult human substantia nigra. PLoS ONE. 2011;6:e28420. doi: 10.1371/journal.pone.0028420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.R., Robinson K.J., Cleary M.D., Doe C.Q. TU-tagging: cell type-specific RNA isolation from intact complex tissues. Nat. Methods. 2009;6:439–441. doi: 10.1038/nmeth.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T.P., de la Cruz A.F., Johnston L.A., Edgar B.A. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Neumüller R.A., Richter C., Fischer A., Novatchkova M., Neumüller K.G., Knoblich J.A. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell. 2011;8:580–593. doi: 10.1016/j.stem.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A., van Steensel B., Delrow J., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W., Cowley S.M., Yost C. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H., Conz C., Maier P., Wölfle T., Suzuki C.K., Jenö P., Rücknagel P., Stahl J., Rospert S. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc. Natl. Acad. Sci. USA. 2005;102:10064–10069. doi: 10.1073/pnas.0504400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert H. Drosophila neural stem cells: cell cycle control of self-renewal, differentiation, and termination in brain development. Results Probl. Cell Differ. 2011;53:529–546. doi: 10.1007/978-3-642-19065-0_21. [DOI] [PubMed] [Google Scholar]
- Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Juán B.P., Baonza A. The bHLH factor deadpan is a direct target of Notch signaling and regulates neuroblast self-renewal in Drosophila. Dev. Biol. 2011;352:70–82. doi: 10.1016/j.ydbio.2011.01.019. [DOI] [PubMed] [Google Scholar]
- Shigenobu S., Arita K., Kitadate Y., Noda C., Kobayashi S. Isolation of germline cells from Drosophila embryos by flow cytometry. Dev. Growth Differ. 2006;48:49–57. doi: 10.1111/j.1440-169X.2006.00845.x. [DOI] [PubMed] [Google Scholar]
- Slack C., Somers W.G., Sousa-Nunes R., Chia W., Overton P.M. A mosaic genetic screen for novel mutations affecting Drosophila neuroblast divisions. BMC Genet. 2006;7:33. doi: 10.1186/1471-2156-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Miura P., Westholm J.O., Shenker S., May G., Duff M.O., Zhang D., Eads B.D., Carlson J., Brown J.B. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 2012;1:277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Goetze S., Bischof J., Spichiger-Haeusermann C., Kuster M., Brunner E., Basler K. Coop functions as a corepressor of Pangolin and antagonizes Wingless signaling. Genes Dev. 2010;24:881–886. doi: 10.1101/gad.561310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Lu B. Regulation of cell growth by Notch signaling and its differential requirement in normal vs. tumor-forming stem cells in Drosophila. Genes Dev. 2011;25:2644–2658. doi: 10.1101/gad.171959.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J., Wharton R.P. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Nunes R., Chia W., Somers W.G. Protein phosphatase 4 mediates localization of the Miranda complex during Drosophila neuroblast asymmetric divisions. Genes Dev. 2009;23:359–372. doi: 10.1101/gad.1723609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Pollard H.B. Molecular dissection of nucleolin’s role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- Thiel G., Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J. Cell. Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- Tirouvanziam R., Davidson C.J., Lipsick J.S., Herzenberg L.A. Fluorescence-activated cell sorting (FACS) of Drosophila hemocytes reveals important functional similarities to mammalian leukocytes. Proc. Natl. Acad. Sci. USA. 2004;101:2912–2917. doi: 10.1073/pnas.0308734101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M., Golden K.L., Lee C.Y. dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila. Dev. Cell. 2010;18:126–135. doi: 10.1016/j.devcel.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharioudaki E., Magadi S.S., Delidakis C. bHLH-O proteins are crucial for Drosophila neuroblast self-renewal and mediate Notch-induced overproliferation. Development. 2012;139:1258–1269. doi: 10.1242/dev.071779. [DOI] [PubMed] [Google Scholar]
- Zhu S., Lin S., Kao C.F., Awasaki T., Chiang A.S., Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
- Zhu S., Barshow S., Wildonger J., Jan L.Y., Jan Y.N. Ets transcription factor Pointed promotes the generation of intermediate neural progenitors in Drosophila larval brains. Proc. Natl. Acad. Sci. USA. 2011;108:20615–20620. doi: 10.1073/pnas.1118595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed]
- Bader, G. D., and Hogue, C. W. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2. [DOI] [PMC free article] [PubMed]
- Bischof, J., Maeda, R.K., Hediger, M., Karch, F., and Basler, K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312–3317. [DOI] [PMC free article] [PubMed]
- Ceron, J., Tejedor, F.J., and Moya, F. (2006). A primary cell culture of Drosophila postembryonic larval neuroblasts to study cell cycle and asymmetric division. Eur. J. Cell Biol. 85, 567–575. [DOI] [PubMed]
- Enright, A. J., Van Dongen, S., and Ouzounis, C. A. (2002). An efficient algorithm for largescale detection of protein families. Nucleic Acids Res. 30, 1575–1584. [DOI] [PMC free article] [PubMed]
- Groth, A.C., Fish, M., Nusse, R., and Calos, M.P. (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782. [DOI] [PMC free article] [PubMed]
- Irizarry, R.A., Bolstad, B.M., Collin, F., Cope, L.M., Hobbs, B., and Speed, T.P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed]
- Li, R., Yu, C., Li, Y., Lam, T.W., Yiu, S.M., Kristiansen, K., and Wang, J. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. [DOI] [PubMed]
- Maere, S., Heymans, K., and Kuiper, M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. [DOI] [PubMed]
- Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. [DOI] [PubMed]
- Neumüller, R.A., Richter, C., Fischer, A., Novatchkova, M., Neumüller, K.G., and Knoblich, J.A. (2011). Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8, 580–593. [DOI] [PMC free article] [PubMed]
- Trapnell, C., Pachter, L., and Salzberg, S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed]
- Trapnell, C., Williams, B.A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M.J., Salzberg, S.L., Wold, B.J., and Pachter, L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed]
- Weng, M., Golden, K.L., and Lee, C.Y. (2010). dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila. Dev. Cell 18, 126–135. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table containing a complete list of genes differentially expressed in NBs and neurons with FDR = 0.01; 1,830 (52%) of these genes were upregulated in neurons, and 1,702 (48%) were expressed higher in NBs.
Table containing a complete list of enriched GO terms in NBs only from genes with FPKM value > 1, fold change > 1, and p > 1.
Table containing a complete list of enriched GO terms in neurons only from genes with FPKM value > 1, fold change > 1, and p > 1.
Table containing alternatively spliced genes with at least two differentially expressed isoforms in NBs and neurons. See Experimental Procedures for details.
Table containing 40 genes with an extended 3′ UTR that are expressed higher in NBs and 357 that are upregulated in neurons. 3′ UTR extension is not NB or neuron specific.
NBs in an unsorted cell culture divide asymmetrically multiple times. The first- and second-born GMCs divide terminally to give rise to neurons. Genotype: ase-Gal4, UAS-stinger::GFP
FACS-sorted NBs divide asymmetrically multiple times. The first- and second-born GMCs divide terminally to give rise to differentiated neurons. Genotype: ase-Gal4, UAS-stingerGFP