Abstract
SVIP (small p97/VCP-interacting protein) was initially identified as one of many cofactors regulating the valosin containing protein (VCP), an AAA+ ATPase involved in endoplasmic-reticulum-associated protein degradation (ERAD). Our previous study showed that SVIP is expressed exclusively in the nervous system. In the present study, SVIP and VCP were seen to be co-localized in neuronal cell bodies. Interestingly, we also observed that SVIP co-localizes with myelin basic protein (MBP) in compact myelin, where VCP was absent. Furthermore, using nuclear magnetic resonance (NMR) and circular dichroism (CD) spectroscopic measurements, we determined that SVIP is an intrinsically disordered protein (IDP). However, upon binding to the surface of membranes containing a net negative charge, the helical content of SVIP increases dramatically. These findings provide structural insight into interactions between SVIP and myelin membranes.
INTRODUCTION
Endoplasmic-reticulum-associated protein degradation (ERAD) targets misfolded proteins for proteasomal degradation. Misfolded substrates are extracted out of the ER membrane by the valosin-containing protein (VCP) complex, and escorted to the 26S proteasome for degradation [2]. VCP, also named p97 or cdc48, executes diverse functions by association of this ATPase with over 40 different adaptor proteins (cofactors). These interactions typically depend on specific protein domains in both VCP and cofactors. One such domain is the VCP-interacting motif (VIM) that has been found in some cofactors, including SVIP (small p97/VCP-interacting protein) and gp78 (ubiquitin ligase) [3;24]. There is a minimal consensus sequence (RX5AAX2R) that defines the VIM motif [24]. VIM-containing cofactors compete with each other for this binding groove to regulate VCP function [2].
We have recently demonstrated that SVIP is almost exclusively localized to the central and peripheral nervous systems [26]. While the SVIP and VCP were co-localized in neuronal cell bodies [26], we here present evidence that SVIP localizes in compact myelin in a manner independent of its interactions with VCP. We have used nuclear magnetic resonance (NMR) spectroscopy and circular dichroism (CD) to characterize the conformation of purified SVIP. We report that SVIP is largely unfolded in solution, but a much more highly helical conformation when it interacts with acidic phospholipid membranes. This discovery reveals conformational adaptability of SVIP that may be critical for SVIP interactions with myelin membranes.
MATERIALS AND METHODS
1. Immunostaining
This technique has been described in a previous publication [1]. In brief, sciatic nerves of adult rats were fixed in 4% paraformaldehyde overnight. Cryostat sections of 10μm thickness were reacted with primary antibodies at 4°C overnight, followed by one-hour incubation with secondary antibodies.
For the teased nerve fiber study, 1 cm segments of sciatic nerve were fixed in 4% paraformaldehyde overnight and separated into individual nerve fibers on a glass slide, followed by immunostaining (see above). The following primary antibodies were used: mouse neurofilament (SMI31R, Covance, 1:500) and rat MBP (MAB386, Millipore, 1:100). SVIP antibodies were a gift from Dr. Shengyun Fang. The SVIP antibody has been described [4]. To verify its specificity, we performed Western blot on lysates of mouse sciatic nerves. It detected SVIP in Svip+/+ sciatic nerves, but showed no immunoreactivity in Svip−/− sciatic nerves (unpublished observation).
2. Western blot
Nerves were homogenized. The protein concentration was determined by BCA protein assay (#23225, Thermo Scientific). The protein samples were loaded into the lanes of an SDS–polyacrylamide gel and transferred to PVDF (polyvinylidene difluoride) membrane after electrophoresis. The membranes were probed with primary antibodies overnight at 4°C, and followed by a secondary antibody conjugated to horseradish peroxidase (Cat# AP307P, Millipore, Billerica, MA). The protein was detected by the chemoilluminescence.
3. Construction of Plasmids
The DNA encoding human SVIP (NCBI Reference Sequence: NM_148893.1) were amplified by PCR from SVIP cDNA plasmid (in pCIneo-vector, a gift from Dr. Shengyun Fang, University of Maryland). The cDNA was sub-cloned into the NdeI/XhoI-cleaved plasmid pET22b(+) (Novagen) to generate a fusion protein with a His6 tag fused at its carboxyl termini. The primers: sense: 5′ catatgatggggctgtgttttc 3′, anti-sense primer: 5′ ctcgagtgaaactgtccacctaag 3′.
To generate the Cys4Ser/Cys7Ser mutant form of SVIP (SVIPM), single-site mutagenesis was carried out to substitute residues Cys4 and Cys7 by a serine using the QuikChange® II XL Kit. Primers: sense 5′ tatgatggggctgagttttcctagtcccggggagtc 3′; anti-sense: 5′gactccccgggactaggaaaactcagccccatcata 3′. DNA sequencing confirmed the mutations.
4. Expression and Purification of SVIP
Human SVIP proteins were expressed in Escherichia coli BL21 (DE3) cells. The expression was induced with 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG). The soluble His6-tagged SVIP proteins were purified on Ni-NTA resin (Qiagen) [20]. Using a Millipore centrifugal filter (<30 kDa) the purified protein was exchanged into buffer-A (25 mM phosphate buffer, 25 mM NaCl, 0.1 mM EDTA, pH 5.0) and concentrated to 0.4–0.6 mM. The purity of the SVIP was confirmed by Tricine-SDS-PAGE (12%, w/v) and the concentration was measured using BCA kits (Pierce). To label the proteins with 15N or with both 15N and 13C for NMR spectroscopic studies, E. coli cells were grown in M9 medium that contained 15NH4Cl (1g/liter) and 13C6-glucose (2.5g/liter) as the sole nitrogen and carbon sources.
Mixtures of SVIPM and lyso-myristoylphatidylcholine (LMPC) or lyso-myristoylphosphatidylglycerol (LMPG) (Avanti, Alabama) micelles were prepared by adding 10% LMPC or LMPG into SVIPM solution to 0.5% w/v. Samples was further incubated overnight before the CD and NMR experiments.
5. NMR Experiments
The purified proteins were prepared in 25mM phosphate buffer (25 mM NaCl and 0.1 mM EDTA at pH 5.0 in a 95% H2O/5% D2O mixture). Unless otherwise specified, all NMR spectra were collected at 298K on a Bruker Avance 600MHz spectrometer equipped with a cryogenic triple resonance probe. 1H-15N HSQC spectra were acquired on 15N-labeled or 15N/13C-labeled samples. The latter sample was also used to collect triple resonance spectra. 15N-separated NOESY and 13C-separated NOESY spectra were collected with a mixing time of 120ms and 100ms respectively. NMR data were processed with NMRPipe [6], or TopSpin (Bruker Biospin Inc., MA) and analyzed using Sparky (Goddard and Kneller, V3.113, 2006, University of California).
6. Circular Dichroism (CD) Measurements
CD experiments were carried out using a Jasco-810 spectropolarimeter. Far-UV (ultraviolet) CD spectra were recorded at wavelengths between 190nm and 260nm using a 0.1cm path-length quartz cuvette. For near-UV, CD spectra were recorded at wavelengths between 260 and 320 nm using a 1 cm path length cell. All CD spectra were the average of three scans after the blank spectrum was subtracted. Protein concentrations were 0.1 and 1 mg/ml for far-UV CD and near-UV CD respectively. The protein solutions contained 10 mM phosphate (K2HPO4-KH2PO4) at pH 5.6. The protein fractional helicity was calculated from the far-UV CD spectrum as described [18].
RESULTS
SVIP is a compact myelin protein
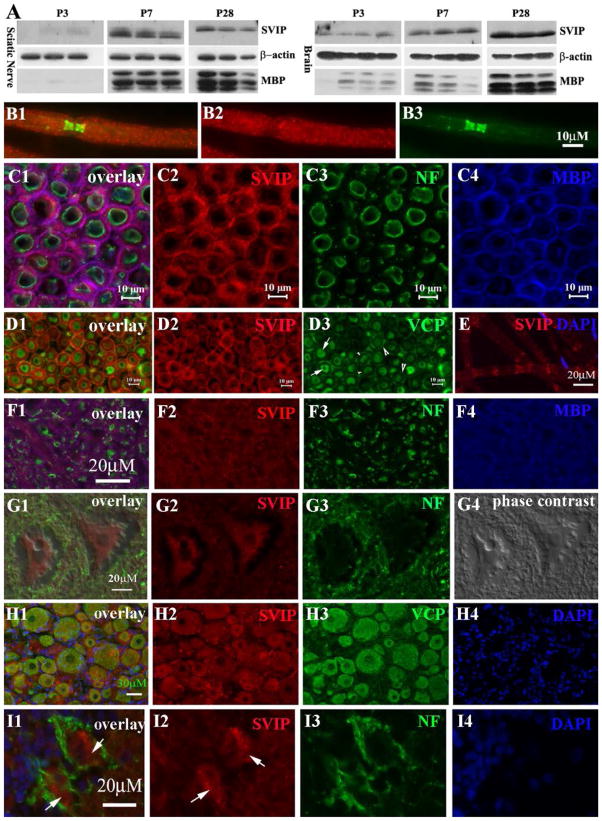
We first examined SVIP localization in detail. Western blot showed that SVIP expression correlated with levels of myelin basic protein (MBP) in the developing nerves. MBP is another compact myelin protein. Levels of both proteins increased during nerve development (Figure 1A).
FIGURE 1. Expression of SVIP in the nervous system.
(A) Mouse tissues were taken at postnatal day 3, 7 and 28 and processed for Western blot analysis. SVIP was barely detectable at P3 and strongly increased at P7 and P28. A similar change was also observed for MBP. (B1–3) Adult rat sciatic nerves were teased into individual fibers and stained with antibodies against SVIP and Caspr, a paranodal marker. SVIP was localized on internodal myelin and weakly in paranodal myelin. (C1–4) SVIP was stained on the cryostat transverse sections of adult rat ventral and dorsal roots. Myelin was labeled by antibodies against MBP (blue color in C4) and axon was labeled by antibodies against phosphorylated neurofilaments (NF; green color in C3). SVIP overlapped well with MBP (pink color in C1), but not neurofilament, suggesting its localization in compact myelin. (D1–3) Cryostat transverse sections of adult rat sciatic nerves were stained with antibodies against SVIP and VCP. VCP was mainly expressed in axons (arrows in D3) and in the abaxonal cytoplasmic space of Schwann cell (large arrowheads in D3), but not in compact myelin (small arrowheads in D3). VCP does not overlap with SVIP in myelin (D1). E. Teased nerve fibers were stained for SVIP. A small fraction of myelinated nerve fibers showed SVIP expression in Schmidt-Lanterman incisures. (F1–4) Cryostat sections of adult rat spinal cord were stained with antibodies against SVIP and MBP. This image was taken from the ventral column (white matter) of the spinal cord. SVIP immunoreactivity (red color in F2) also overlapped with MBP (pink color in F1), but not with axons labeled by neurofilament (green color in F1). (G1–4) SVIP immunoreactivity was also found in the cytoplasm of spinal motor neurons (G2). (H1–4) Immunofluorescence staining was also performed on the cryostat sections of the adult rat dorsal root ganglion. Again, SVIP and VCP were well overlapped in the cytoplasm of neurons. (I1–4) Cryostat sections from the adult rat cerebellum were stained and showed SVIP expression in the cytoplasm of Purkinje cells (arrows in I1–2).
By immunostaining, SVIP was seen in the compact myelin of peripheral and central myelinated nerve fibers, but not in axons. SVIP was well overlapped with MBP (Figure 1B1–C4). Interestingly, in a small fraction of myelinated nerve fibers, SVIP was localized in Schmidt-Lanterman incisures (non-compact myelin) (Figure 1E). This finding was also in line with a study that detected SVIP in myelin extracts using mass spectroscopy [16].
To determine the anatomical relationship between SVIP and VCP, peripheral nerves were double-stained with antibodies against both SVIP and VCP. VCP was distributed in the Schwann cell perinuclear region, abaxonal cytoplasmic space, or axons, but absent in the compact myelin (Figure 1, D1–D3). Thus, SVIP and VCP were not co-localized in the peripheral nerve myelin. However, SVIP was co-localized with VCP in neuronal cell bodies (Figure 1, G1–I4).
These data suggest that SVIP in adult myelin may exert certain biological functions independent of VCP.
A major portion of SVIP is intrinsically disordered in solution
SVIP localization in compact myelin raises questions regarding its potential roles in myelin. Protein structural studies have shown that myelin protein zero (MPZ) interacts homotypically in cis and trans as a mechanism to compact myelin lamina [22].
We first utilized a Jpred (http://www.compbio.dundee.ac.uk/jpred) to predict the secondary structure of SVIP based on amino acid sequence. It predicted two distinct α-helical regions (L18-A37 and V44-S64).
We then investigated the structure of SVIP using solution NMR spectroscopy [15]. The wild-type human SVIP exhibited a high propensity to self-associate. The Cys4Ser/Cys7Ser mutant of SVIP (SVIPM) was resistant to self-association and facilitated purification (Supplementary Figure 1A). A 1H-15N-HSQC NMR spectrum of SVIPM was similar to the spectrum from wild-type SVIP (Supplementary Figure 1B), indicating that the structure of the mutant protein is similar to that of wild type.
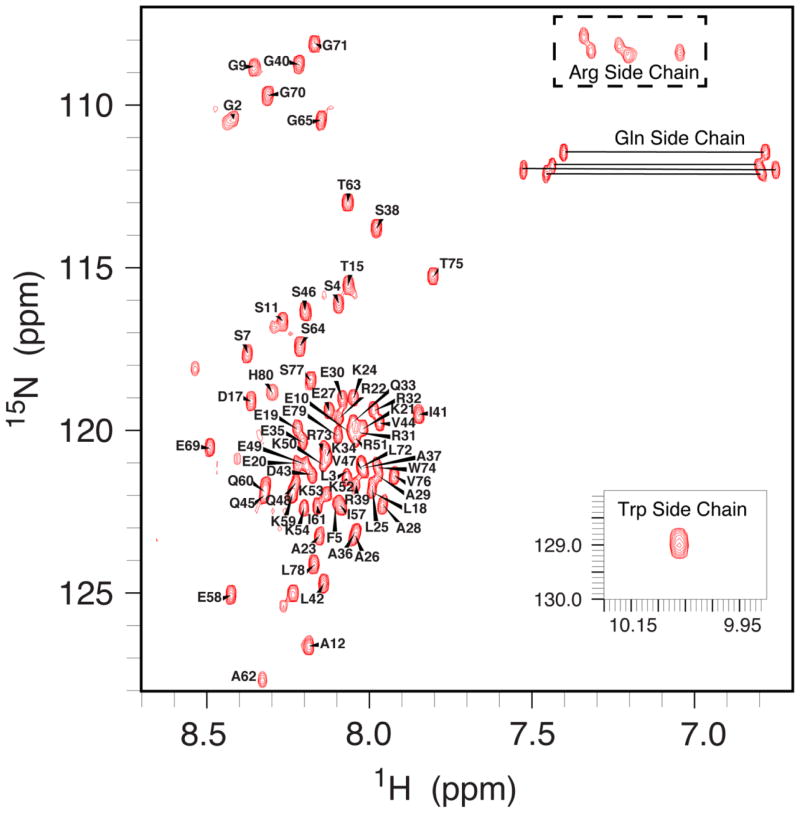
Backbone sequential assignments of SVIPM were completed using data from HSQC-based versions of HNCA, HN(CO)CA, HNCACB and CBCA(CO)NH. Side-chain resonance assignments of SVIP were performed using three-dimensional (3D) HBHA(CBCACO)NH, 15N-NOESY-HSQC, 15N-TOCSY-HSQC, H(C)(CO)NH-TOCSY, (H)C(CO)NH-TOCSY, HCCH-COSY, HCCH-TOCSY, and 13C-NOESY-HSQC data [17;19]. With the exception of prolines, E55, and K56, the backbone resonances were completely assigned (Figure 2). Notice that the N-terminal methionine of SVIPM was retained post-translationally. Based on chemical shifts of G2 (upper left corner in Figure 2), M1 was identified by the following assignments: CA 53.651, CB 30.883, CO 173.761, and HA 4.491.
FIGURE 2.
600 MHz 1H-15N-HSQC NMR spectrum of SVIPM in pH 5.0 buffer-A at 25°C.
Backbone chemical shifts of SVIPM were poorly dispersed (0.7 ppm) in the proton dimension of its 1H-15N HSQC spectrum (Figure 2), indicating that SVIPM was likely an intrinsically disordered protein (IDP) in solution. This conclusion was further supported both by an absence of long-range NOEs between side-chains and by the fact that the near-UV CD spectra was flat on baseline (not shown).
NMR-Determined Secondary Structure of SVIP in Solution
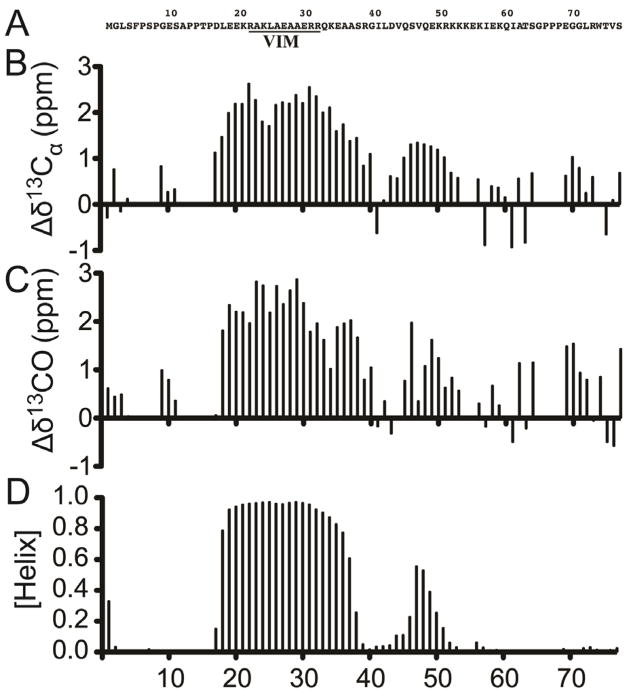
Backbone 13Ca, 13CO and 15NH chemical shifts for SVIPM were analyzed using the chemical shift index (CSI) method [29] and the TALOS+ program [23] to determine secondary structure. The α-helix-forming probability was plotted against residue numbers (Figure 3B-D). A stable α-helix is found around the VIM domain and spans Leu18 through Ala37, consistent with the Jpred prediction. A second helix does appear to be present. However, this helix appears to be much shorter and less stable, spanning only residues 44–51 (Fig 3).
FIGURE 3. Secondary structure of SVIPM.
NMR measurements were carried out on SVIPM at 600 MHz for 1 mM 1H-15N-13C-labeled SVIPM in 50 mM phosphate buffer (pH 5.50; 25°C). (A) Amino acid sequence of SVIPM. The VIM domain is underlined. (B) Chemical shift index (CSI) results: Shown are the deviations of observed 13Cα chemical shift from the random coil values, with these results supporting a stable helix spanning roughly D17-G40 and an unstable helix spanning roughly L44-K51. (C) CSI results for 13CO chemical shifts, which are consistent with the patterns in 3B in terms of indicating helical segments. (D) Results of TALOS+ analysis of the chemical shifted data also confirm that the SVIPM in solution has a stable helix spanning roughly D17-S38 containing the VIM domain and a less stable helix from roughly V44 to K51. In line with findings in 3B-C, TALOS+ analysis indicated that the rest of SVIP C-terminal was largely unstructured. The “missing” resonances were either proline sites or due to extensive line-broadening.
SVIP undergoes a significant increase in structural order in the presence of anionic micelles
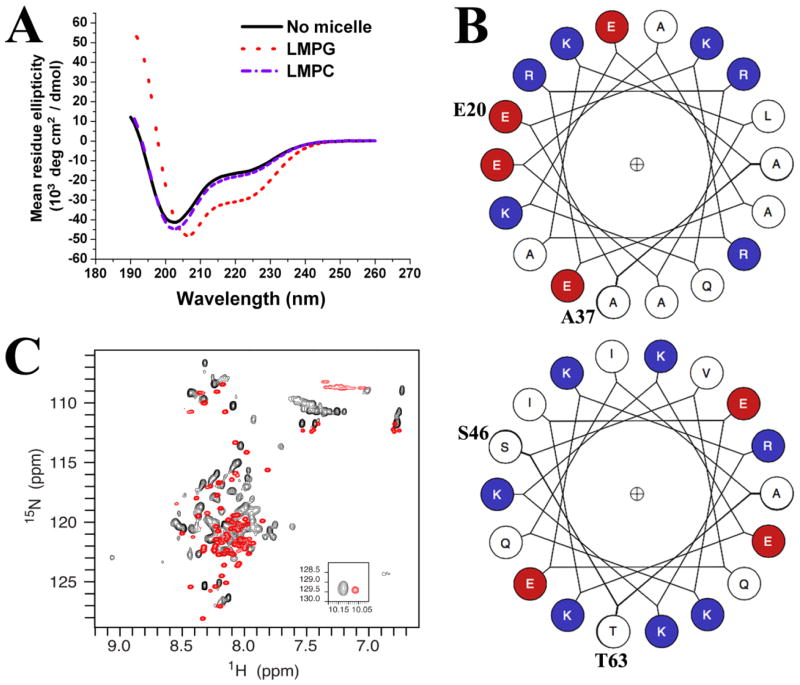
To examine the factors that affected α-helical contents of SVIPM, we performed far-UV CD experiments and analyzed the spectra with K2D3 software [13]. In the absence of detergent, the spectra at pH 5.6 and 298 K is shown in Figure 4A (black curve). This spectrum exhibited a negative minimum near 202 nm and a negative shoulder around 222 nm and was estimated to have only 29% α-helical content (25 residues), fully consistent with the conclusion from the NMR study that SVIPM in solution was a largely unfolded protein with only a modest fraction of α-helix (Figure 4A, black curve). Very similar results were found for detergent-free solutions over a range of pH (pH 5.0, 6.5 and 7.4) and temperatures (298K to 310K). There was also no evidence for coiled-coil helices in the far-UV CD data [8].
FIGURE 4. Negatively-charged model membrane results in a significant increase in the structural order of SVIP.
(A) Far-UV CD spectra of SVIP (0.1 mg/ml) in 10mM phosphate buffer, pH 5.6 in the absence of detergent (black trace), in the presence of zwitterionic LMPC micelles (blue curve), and in the presence of anionic LMPG micelles (red curve). (B) The helix wheels for the putative first (E20-A37) and second (S46-T63) helical domains of SVIPM that are likely present when SVIP is bound to the surface of LMPG micelles. R, K = positively charged residues (Blue); D,E = Negatively charged (Red); Uncharged (White). (C) 1H-15N-HSQC NMR spectrum of 15N-labeled SVIPM in pH 5.0 buffer-A in the absence (red) and presence (black) of LMPG micelles. The LMPG concentration was 0.5% w/v.
To see if the presence of a model membrane results in changes in the SVIPM structure we examined its CD spectrum after the addition of micelles composed of zwitterionic (LMPC) or anionic (LMPG) detergents. Addition of the net neutral charged LMPC micelles had little impact on the secondary structure of SVIPM (blue trace in Fig 4A), suggesting no interaction of the protein with net neutral model membranes. However, when SVIPM was mixed with anionic LMPG micelles, a dramatic change in the far-UV CD spectrum was observed (Fig. 4A, red trace). The α-helicity of SVIPM increased from 29% in detergent-free 10mM phosphate buffer to 72% (55 residues) with LMPG (Supplementary Table 1). This result likely reflects both the retention of the first helix seen for SVIPM in solution (residues 18–37) and a stabilization and extension of the unstable helix seen in solution to encompass the helical span predicted by Jpred (44–64). Whether there are additional helices to account for the additional 14 helical residues or whether these two helices are extended even further than their Jpred spans is not clear.
SVIP does not have a continuous stretch of hydrophobic amino acids and is not expected to be a transmembrane protein, a fact consistent with its apparent lack of interaction with net neutral-charged LMPC micelles. The increase in α-helicity seen in the presence of anionic micelles is suggestive of SVIP interactions with a micelle surface that is driven by favorable electrostatics. We therefore used helical wheels plots [21] to scan the distribution of electrical charges in the two at least partially helical segments (corresponding to residues L18-A37 and V44-G64) (Figure 4B). The first α-helical region possessed predominantly hydrophobic amino acids on one side of the helix, while most hydrophilic amino acids were placed on the other.
The wheel for the second probable helical region appeared to exhibit a face with a net positive-charge of +3. Moreover, the entire helix has a net positive charge of +4. The highly basic nature of this segment has been seen in other IDPs. The clustering of positively charged amino acids presumably generates strong electric repulsion among residues that destabilizes the folding of the protein in solution. However, these positively charged domains may form α-helices during their interaction with acidic phospholipid membrane [25]. Detergent micelles provide a surface that mimics the water-membrane interface in membranes, an environment that is well known to induce ordered secondary and even tertiary structure for some protein segments [28].
Finally, 1H-15N HSQC spectra were acquired in Buffer-A with 0.5% LMPG and without LMPG (Figure 4C). Peaks in the presence of LMPG were much more highly dispersed (spread out) than for the SVIPM in solution. Peaks were broader for the micelle-associated form of the protein, mostly due to the higher molecular weight of the protein-micelle complex relative to the free solution. It also appeared that some peaks were split into a major and a minor (less intense) peak, consistent with there being two conformations of the protein (or two modes of micelle association) that were in slow exchange with each other on the NMR time scale. This finding further supports the adoption by SVIPM of a much higher degree or ordered structure upon association with the anionic micelle surface.
DISCUSSION
Our results demonstrate that SVIP is an IDP in solution. IDPs are defined by exhibiting no tertiary structure and only modest secondary structure in solution. The highly dynamic nature of these molecules makes their structural study challenging. Yet, their biological importance is increasingly recognized (Uversky, 2009).
SVIP undergoes a dramatic conformational change from a largely unstructured structure in water to a much more highly ordered α-helical structure in anionic model membranes. This transition is triggered by anionic micelles but not by other environmental changes, such as temperature, pH value, or zwitterionic detergent micelles. These biochemical features resemble those observed in another compact myelin protein, MBP.
The majority of compact myelin proteins, such as the proteolipid protein (PLP) and myelin protein zero, are transmembrane proteins. One of the key functions of these proteins is to establish myelin compaction, which is the molecular basis for myelin insulation to ensure the successful propagation of action potentials [11]. Myelin compaction has been attributed to homotypic adherence of the transmembrane proteins in cis and trans fashion (bridging juxtaposed membranes) [22].
In contrast, the compact myelin protein MBP does not have transmembrane domains. While there are over 8 isoforms of MBP (5 classic isoforms and 3 Golli isoforms) resulted from alternative splicing of a single mRNA. Classic isoforms of 14- and 18.5kDa are the predominant ones in human compact myelin [7]. Like SVIP, all isoforms of MBP are IDPs in solution. Anionic lipid bilayers induce a dramatic increase in α-helical content in the MBP. This mechanism has been believed to strengthen the tight association between MBP and the cytoplasmic surface of juxtaposed myelin membranes, thereby facilitating the compaction of opposing myelin membrane to form the major dense line [5;10]. Studies in MBP-deficient mice support this notion [14]. MBP deficiency in the central nervous system results in severe white matter disease. Mouse nerves lacking both MPZ and MBP show no major dense line, while mouse nerves deficient in either protein reveal partial or normal myelin compaction [14].
Moreover, the adaptability of MBP conformation permits MBP to interact with a variety of molecules to exert additional functions in myelin, including actin, calmodulin, and tubulin [10].
Our results reveal a new member, SVIP in the family of compact myelin proteins with features of IDP when in solution. Because of these similarities between MBP and SVIP in biochemistry and localization, we speculate that SVIP may also affect myelin via its conformational adaptability upon its association with membranes.
In addition, VCP/SVIP complexes are known to associate with the negatively charged membrane of ER vesicles [2]. Our data reveal that interaction of the unstructured SVIP interaction with lipid membrane promotes additional α-helical content. The N-terminal α-helix containing the VIM motif is bound to VCP with an interaction site that consists predominantly of hydrophobic amino acids [9]. The other face of the first α-helix is net positively charged and would be suitable for interaction with the negatively charged membrane of the ER [27]. The remaining C-terminus of SVIP is also positively charged and likely forms a stable and extended α-helix during its interaction with acidic phospholipids. It is unknown if this specificity toward acidic phospholipids can guide the SVIP to certain membrane microdomains [12]. This mechanism could be important for VCP/SVIP complexes in their association with ER vesicles that are abundantly present in the neuronal bodies.
In summary, we have identified a novel compact myelin protein, SVIP. In addition to its strong expression in compact myelin, a large portion of SVIP reveals no stable structure in solution but dramatically adopts α-helical structure during its interaction with negatively charged lipid membrane. This change of SVIP conformation may be critical to SVIP interactions with myelin membranes.
Supplementary Material
Highlights.
SVIP (small p97/VCP-interacting protein) co-localizes with myelin basic protein (MBP) in compact myelin.
We determined that SVIP is an intrinsically disordered protein (IDP).
The helical content of SVIP increases dramatically during its interaction with negatively charged lipid membrane.
This study provides structural insight into interactions between SVIP and myelin membranes.
Acknowledgments
Authors wish to thank Dr. Shengyun Fang for his generous gifts of antibodies. We also thank Dr. Xuebao Zhang, Ms. Sezgi Arpag and Audra Hamilton for their technical assistance. This research is supported by grants from NINDS (R21NS081364 and R01NS066927 to J.L.; RO1 NS058815 to C.S.), NIH (SIG #1S-10RR025677-01), and VA (B6243R to J.L.). This material is the result of work partially supported with resources and the use of facilities at the VA Tennessee Valley Healthcare System.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Bai Y, Zhang X, Katona I, Saporta MA, Shy ME, O’Malley HA, Isom LL, Suter U, Li J. Conduction block in PMP22 deficiency. J Neurosci. 2010;30:600–608. doi: 10.1523/JNEUROSCI.4264-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballar P, Fang S. Regulation of ER-associated degradation via p97/VCP-interacting motif. Biochem Soc Trans. 2008;36:818–822. doi: 10.1042/BST0360818. [DOI] [PubMed] [Google Scholar]
- 3.Ballar P, Shen Y, Yang H, Fang S. The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. J Biol Chem. 2006;281:35359–35368. doi: 10.1074/jbc.M603355200. [DOI] [PubMed] [Google Scholar]
- 4.Ballar P, Zhong Y, Nagahama M, Tagaya M, Shen Y, Fang S. Identification of SVIP as an endogenous inhibitor of endoplasmic reticulum-associated degradation. J Biol Chem. 2007;282:33908–33914. doi: 10.1074/jbc.M704446200. [DOI] [PubMed] [Google Scholar]
- 5.Baran C, Smith GS, Bamm VV, Harauz G, Lee JS. Divalent cations induce a compaction of intrinsically disordered myelin basic protein. Biochem Biophys Res Commun. 2010;391:224–229. doi: 10.1016/j.bbrc.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 6.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 7.Fulton D, Paez PM, Campagnoni AT. The multiple roles of myelin protein genes during the development of the oligodendrocyte. ASN Neuro. 2010;2:e00027. doi: 10.1042/AN20090051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenfield NJ, Hitchcock-DeGregori SE. Conformational intermediates in the folding of a coiled-coil model peptide of the N-terminus of tropomyosin and alpha alpha-tropomyosin. Protein Sci. 1993;2:1263–1273. doi: 10.1002/pro.5560020809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanzelmann P, Schindelin H. The structural and functional basis of the p97/valosin-containing protein (VCP)-interacting motif (VIM): mutually exclusive binding of cofactors to the N-terminal domain of p97. J Biol Chem. 2011;286:38679–38690. doi: 10.1074/jbc.M111.274506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harauz G, Libich DS, Polverini E, Vassall A. The classic basic protein of myelin-conserved structural motifs and the dynamic molecular barcode involved in membrane adhersion, protein-protein interactions, and pathogenesis in multiple sclerosis. In: Dunn Ben M., editor. Advances in Protein and Peptide Science. Bentham Science Publishers; Oak Park, IL, USA: 2013. [Google Scholar]
- 11.Hartline DK, Colman DR. Rapid conduction and the evolution of giant axons and myelinated fibers. Curr Biol. 2007;17:R29–R35. doi: 10.1016/j.cub.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 12.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 13.Louis-Jeune C, Andrade-Navarro MA, Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. 2011 doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 14.Martini R, Mohajeri MH, Kasper S, Giese KP, Schachner M. Mice doubly deficient in the genes for P0 and myelin basic protein show that both proteins contribute to the formation of the major dense line in peripheral nerve myelin. J Neurosci. 1995;15:4488–4495. doi: 10.1523/JNEUROSCI.15-06-04488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagahama M, Suzuki M, Hamada Y, Hatsuzawa K, Tani K, Yamamoto A, Tagaya M. SVIP is a novel VCP/p97-interacting protein whose expression causes cell vacuolation. Mol Biol Cell. 2003;14:262–273. doi: 10.1091/mbc.02-07-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patzig J, Jahn O, Tenzer S, Wichert SP, de Monasterio-Schrader P, Rosfa S, Kuharev J, Yan K, Bormuth I, Bremer J, Aguzzi A, Orfaniotou F, Hesse D, Schwab MH, Mobius W, Nave KA, Werner HB. Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. J Neurosci. 2011;31:16369–16386. doi: 10.1523/JNEUROSCI.4016-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pervushin K. Impact of transverse relaxation optimized spectroscopy (TROSY) on NMR as a technique in structural biology. Q Rev Biophys. 2000;33:161–197. doi: 10.1017/s0033583500003619. [DOI] [PubMed] [Google Scholar]
- 18.Rohl CA, Baldwin RL. Comparison of NH exchange and circular dichroism as techniques for measuring the parameters of the helix-coil transition in peptides. Biochemistry. 1997;36:8435–8442. doi: 10.1021/bi9706677. [DOI] [PubMed] [Google Scholar]
- 19.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders CR, Ismail-Beigi F, McEnery MW. Mutations of peripheral myelin protein 22 result in defective trafficking through mechanisms which may be common to diseases involving tetraspan membrane proteins. Biochemistry. 2001;40:9453–9459. doi: 10.1021/bi010894f. [DOI] [PubMed] [Google Scholar]
- 21.Schiffer M, Edmundson AB. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967;7:121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro L, Doyle JP, Hensley P, Colman DR, Hendrickson WA. Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin. Neuron. 1996;17:435–449. doi: 10.1016/s0896-6273(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 23.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stapf C, Cartwright E, Bycroft M, Hofmann K, Buchberger A. The general definition of the p97/valosin-containing protein (VCP)-interacting motif (VIM) delineates a new family of p97 cofactors. J Biol Chem. 2011;286:38670–38678. doi: 10.1074/jbc.M111.274472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uversky VN. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28:305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Ballar P, Zhong Y, Zhang X, Liu C, Zhang YJ, Monteiro MJ, Li J, Fang S. SVIP induces localization of p97/VCP to the plasma and lysosomal membranes and regulates autophagy. PLoS One. 2011;6:e24478. doi: 10.1371/journal.pone.0024478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinstein JN, Blumenthal R, van RJ, Kempf C, Klausner RD. Charge clusters and the orientation of membrane proteins. J Membr Biol. 1982;66:203–212. doi: 10.1007/BF01868495. [DOI] [PubMed] [Google Scholar]
- 28.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 29.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.