Abstract
We have investigated the role of two selected amino acids, glycine and arginine, on damage induced to a short chain of single stranded DNA, the tetramer GCAT, during 1 eV electron exposure. At this energy, DNA has a high cross section for DNA damage via exclusively dissociative electron attachment. Surprisingly, at low ratios of glycine : GCAT, an increase in the total fragmentation yield is observed, whilst at higher ratios, glycine and arginine appear to protect DNA from the direct action of electrons. In addition, binding energies were calculated by molecular modelling of the interactions between these amino acids and either nucleobases or nucleotides. These binding energies appear to be related to the ability of amino acids to protect DNA against low energy electron damage.
Introduction
The damaging effect of ionizing radiation, i.e. photons (X-rays and gamma rays) and particulate radiation (electrons, protons, neutrons, beta and alpha particles) has been recognized for many years. These primary quanta can generate a large number (~5 × 104) of secondary electrons per 1 MeV of energy deposited.1 The energy distribution of these electrons, when ejected from a water medium is peaked below 10 eV.2,3 Low energy electrons (LEE) also dominate the experimental secondary electron emission distribution from biomolecular targets exposed to different energies of primary radiation.4 Due to the complexity of the radiation induced processes in cellular environment, the contribution of secondary electrons to the damage in living matter still remains largely unquantified. However, their ability to damage isolated DNA molecules under high vacuum conditions is very well established5 showing resonant structures in the yield of DNA strand breaks measured after LEE exposure (with maxima at around 1 and 10 eV). At 1 eV the collisional process responsible for DNA damage is dissociative electron attachment (DEA), which leads to single strand breaks in the sugar–phosphate backbone.6–8 It has been both experimentally observed9 and theoretically10 demonstrated that the incoming electron is initially attached to base π* orbitals to form a shape resonance. The electron is afterwards transferred and localised on the phosphate group. The formation of this local transient anion then results in cleavage of sugar–phosphate σ* (C–O) bond. At higher incident electron energies (ca. 10 eV), the formation of both single or double strand breaks in DNA has been reported.11 In this case, the incoming electron is bound by the positive electron affinity of an electronically excited state of the nucleobase and forms a core-excited resonance. Such transient anions can then decay through different channels, one of which being the DEA channel which leads to the fragmentation of the molecules.12
To date most of these conclusions concerning damage induced by LEE have been derived from several experimental studies on different types of DNA molecules such as plasmid DNA or single and double stranded oligonucleotides with different chain lengths. It is, however, reasonable to ask how these processes change if DNA molecules are bound or in close proximity to other constituent cellular molecules, e.g. proteins or ionic salt. The studies on direct damage to complexes of positively charged polypeptides with DNA induced by X-rays showed that the degree of DNA radioprotection increases with a molar ratio of polypeptides.13 These polypeptide–DNA complexes provide a better mimic of the cellular situation, where DNA is bonded to histones via ionic interactions. Moreover, recently it has been also shown that organic ions can prevent alterations in DNA induced by 10 eV electrons.14
The first attempt to study the interaction of a protein–DNA complex with LEE (3 eV) has been reported very recently.15 In this study, LEE exposure caused much lower damage to the DNA molecule in the complexes in comparison to that without a bound protein. It was suggested that besides the physical shielding of DNA by proteins, they can also reduce DNA damage due to the molecular interaction.
It is known that the majority of amino acid–base interactions can be applied across all protein–DNA complexes.16 Thus, in our approach to model damage to DNA–protein complexes induced by LEE and also to get more insight into the role of specific amino acids upon exposure through their molecular interactions to DNA, we have studied tetramer oligonucleotides containing all four nucleobases: adenine (A), cytosine (C), guanine (G) and thymine (T) with two amino acids: glycine (Gly) and arginine (Arg). The physical and chemical properties of these two amino acids differ considerably, e.g. glycine contains in its structure the non-polar aliphatic group, while arginine has a positively charged group. Their molecular interactions with DNA bases have also a different nature: glycine interacts mainly via van der Waals interactions and arginine via hydrogen bonds.16
Irradiation with electrons of 1 eV was chosen to simplify the effects of multiple electron scattering in our samples as well as our understanding of the mechanism involved in electron collision processes with the complexes; i.e. 1 eV electrons can only induce single strand breaks in DNA via DEA.6–8 Additionally, our computational modelling has revealed binding energies and sites for particular amino acid–nucleobase/nucleotide pairs, which have served to explain the observed differences in the experimental results with the two amino acids.
Experimental
The tetramer oligos (GCAT) were purchased from Alpha DNA, Montreal, QC, purified by HPLC with standard deviation of 10% and then desalted. The oligos were dissolved in deionised water without adding any salt, so that the negative charge on one of the oxygen atoms of the phosphate group is mainly counterbalanced by a proton (H+ from H2O). The amino acid solution was added to the DNA solution with a concentration of 0.01 mg ml−1 in order to achieve the required molar ratio of a mixture. The amino–GCAT films were deposited onto a chemically clean tantalum sheet (1 cm × 0.6 cm). The samples were then frozen at 90 K and lyophilized with a hydrocarbon-free sorption pump operating at a pressure of about 1 Pa for 2 h. The lyophilized GCAT samples formed a film with a thickness of about 3 nm, three monolayers (ML) of single stranded DNA. With an increase of amino acids the thickness of films increased, the average thickness of films was estimated from the volume of deposited DNA or/and amino acids solutions at a known density of glycine (1.16 g cm−3), arginine (1.1 g cm−3) and DNA (1.7 g cm−3).17 The average thickness for particular ratios and amino acid–GCAT mixtures are presented in Table 1.
Table 1.
The averaged film thickness for GCAT and amino acid–GCAT samples in nanometres (nm) and monolayers (ML) with a constant amount of oligonucleotides in the samples (100 ng)
| Amino acid : nucleotide molar ratio | Film thickness (in nm or ML)
|
|
|---|---|---|
| Glycine | Arginine | |
| 0 : 1 | 3 | 3 |
| 0.5 : 1 | 3.6 | 4.4 |
| 1 : 1 | 4.2 | 5.8 |
| 2 : 1 | 5.4 | 8.6 |
| 3 : 1 | 6.6 | 11.4 |
| 4 : 1 | 7.8 | 14.2 |
The lyophilized samples were directly transferred into an ultra-high vacuum chamber and evacuated for at least 6 h. The DNA–amino acid film was exposed to an electron beam at a background pressure of 10−5 Pa at room temperature and at a fixed incident electron current of 5 nA (3.12 ×1010 electrons s−1). A constant incident electron energy of 1 eV was maintained during the irradiation time of 10 min. This exposure time was in the linear regime of a dose response curve (not shown here) for a fixed electron beam flux in order to exclude any multiple events producing more than one damaged oligonucleotide. The energy resolution of the beam was 0.5 eV full width at half-maximum. The area of the electron beam was adjusted to be the same as that of the sample (5 mm).
After irradiation the samples were resuspended in 10 μL of high purity water and then analyzed by high performance liquid chromatography with UV detection (HPLC-UV). The HPLC consisted of a Waters Alliance system equipped with a refrigerated autosampler, a solvent delivery module and a dual wavelength (210 and 260 nm) absorbance detector. The products formed were separated using an ODS-AQ column (150 × 6 mm), kept at 303 K, using a linear gradient from 1 to 10% acetonitrile in buffer containing 25 mM ammonium acetate pH 5.7 over an interval of 60 min and a flow rate of 1 mL min−1.
This technique was successfully used in previous studies on LEE induced damage to short oligonucleotides.18–20 In the case of GCAT oligonucleotides, each tetramer has 16 possible sites of cleavage, i.e. 12 sites of phosphodiester bond breaks and 4 sites of N-glycosidic bond breaks, as it was illustrated elsewhere.19 The rupture of C–O and P–O backbone bonds of GCAT leads to the formation of smaller fragments such as nucleosides, mononucleotides, dinucleotides and trinucleotides with or without a terminal phosphate group, whilst the rupture of glycosidic bonds results in unaltered base release. This gives a total of 32 possible products formed from the irradiated tetramer. The quantification of yields for particular fragments carries an error due to a rich chromatogram spectrum consisted of many overlapping peaks (the examples of GCAT spectra are shown in ref. 18 and 19). Therefore in the present studies the total fragmentation yield was estimated as a ratio of the sum of the peak areas which corresponds to the amount of all products that arise from LEE irradiation to the total peak area for the GCAT peak. At least 10 samples for each ratio of amino acid : GCAT complexes were analyzed and the mean value of the total fragmentation yields was calculated. The estimated error does not exceed 20% and is mainly due to nonuniformity in lyophilized films.
Computational modelling
The binding energies for amino acid–nucleobase/nucleotide pairs were calculated by applying universal forcefield (UFF) and Dreiding forcefield theories combined with the consistent charge equilibration method. The UFF method is able to predict the structure and energies of a variety of organic molecules21 and the Dreiding forcefield is based on hybridization rules.22 Both forcefield theories can be applied to very large biomolecules, while ab initio molecular orbital calculations are limited to smaller systems due to limitations in our computational resources.
The geometry optimization were calculated by using Forcite and DMol3 (GGA and BLYP functional and TNP basis set) and binding energies were calculated by Blends, all modules are incorporated in Materials Studio from Accelrys Software Inc.23 Before performing binding energy calculations, the input structures of the two amino acids, the four bases and four nucleotides were optimized with a high degree of accuracy. The Blends module was used to obtain the binding energy distribution and the lowest energy configurations for amino acid–base/nucleotide pairs. The Blends module combines a modified Flory–Huggins model and molecular simulation techniques to calculate compatibility of binary mixtures.23 The energy distribution was calculated by generating a large number of pair configurations in which van der Waals surface of the two molecules are in contact. The van der Waals interactions and hydrogen bonding are described by the Lennard-Jones potential, whilst electrostatic interactions were described by atomic monopoles and a screened Coulombic term.
Results and discussion
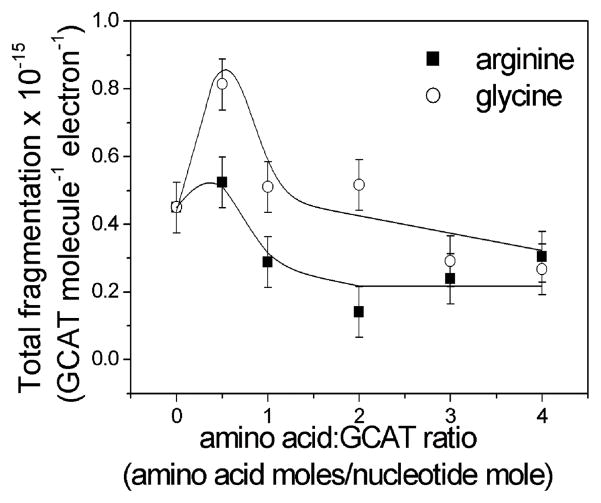
In this work, the yields of fragmentation are presented in terms of the total number of damaged molecules per GCAT molecule in the sample, divided by the number of electrons delivered to the sample. The total fragmentation yields were measured for different molar ratios of Arg : GCAT and Gly : GCAT, i.e. 0.5 : 1, 1 : 1, 2 : 1, 3 : 1 and 4 : 1, where the amount of GCAT remained constant (100 ng). In the case of the Arg–GCAT pair the total fragmentation yield decreases with higher ratios of amino acids in the mixture reaching the plateau above the ratio of 2 : 1. Both amino acids show protection levels against 1 eV electrons estimated around 1.5 for molar ratios above 2 : 1. A similar dependence was observed for the total base release, which can be correlated with DNA strand breaks, from polypeptide–DNA films irradiated by X-rays.13 Studies on the radioprotection of DNA from direct damage by polypeptides have shown that poly-L-lysine protected DNA by a factor of approximately 2.5, while poly-L-arginine protected by a factor of 1.8.
The dependence of GCAT damage in irradiated Gly–GCAT films prepared with a ratio below 2 : 1 as well as for a thin layer of films, below 4 ML, shows a very different behaviour than the tetramer in irradiated Arg–GCAT films (Fig. 1 and 2). After irradiation, the total fragmentation of oligonucleotides for Gly–GCAT films with a ratio of 0.5 : 1 is doubled, whereas in Arg–GCAT films with a ratio of 0.5 : 1 the increase is not significant and within the experimental error. It should be realized, however, that these sensitizations are relative to pure GCAT films, because in a GCAT film, a given molecule is surrounded with other similar molecules, which may or may not have an effect on induced damage. In other words, our measurements are with reference to a GCAT molecular environment. Since the yields in Fig. 1 and 2 are expressed per GCAT molecule, the present results do not depend on the number of GCAT molecules in the film, but depend on changes of the molecular environment. That is, the molecule–molecule interactions in the GCAT–GCAT film are different than those between GCAT and arginine or glycine. However, we must take into account the increasing film thickness with molar concentration of amino acids in our experiments.
Fig. 1.
Total fragmentation yields induced by 1 eV electrons as a function of amino acid : GCAT ratios for arginine (■) and glycine (○). The data points represent the average of at least 10 independent experiments. The error bars represent the standard derivation of 1 eV electron exposures for a given molar ratio.
Fig. 2.
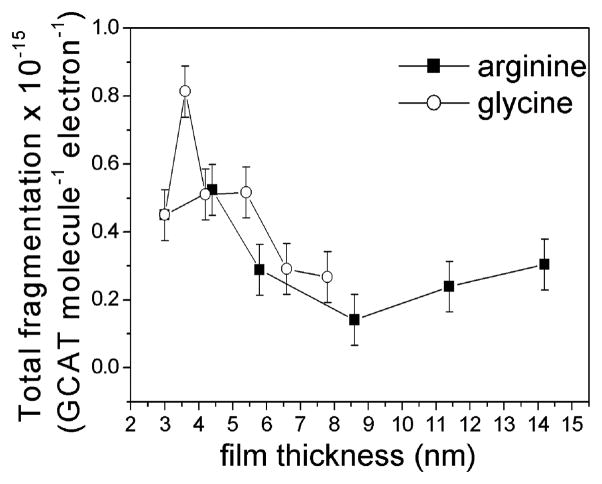
Total fragmentation yields induced by 1 eV electrons as a function of film thickness for arginine–GCAT (■) and glycine–GCAT (○) films. The data points represent the average of at least 10 independent experiments. The error bars represent the standard derivation of 1 eV electron exposures for a given molar ratio.
Fig. 2 shows the total fragmentation yields of oligonucleotides as a function of film thickness for Gly–GCAT and Arg–GCAT films. In both cases a decrease in the damage for higher coverage is observed above thickness of 4 nm (4 ML). For the mixed film of GCAT and glycine the total fragmentation yield is, on average, slightly above that for arginine at corresponding thicknesses, which suggests that the damage induced to the oligonucleotides is not only dependent on thickness, but also on a molecule interacting with GCAT. As the film thickness increases multiple electron scattering within the film also increases as well as electron energy losses. This multiple scattering effect is severe at energies above the electronic excitation threshold, where a single energy loss event can reduce considerably the electron’s energy and hence its scattering and attachment cross sections.
For 1 eV electrons only vibrational excitation is possible that reduces the energy of electrons by values of the order of 0.1 eV. Although this value is small, it may have an effect on the yields, if multiple scattering is intense.
Previous studies have shown that DNA has a strong shape resonance at 0.8 eV.6,7 In our experiment, this resonance has been observed to cause a DEA peak at 0.5 eV in GCAT film. Hence as multiple scattering increases and electrons decrease their energy from 1 to 0.5 eV, their probability to form the 0.5 eV resonance increases as well as the cross section for DEA. In other words, considering that the thermalization distance of LEE in DNA24 and in solid organic films25 is about 12 nm, we expect the damage to increase below a thickness of at least 6 nm if the film were made of only DNA molecules. From another point of view, gas-phase studies on electron attachment to several amino acids (i.e. glycine,26–31 alanine,26,29,30–32 proline,26,29,33,34 tryptophan,26,29,35 phenylalanine,26,26,29,30 cysteine,36 valine37 and leucine38) have all revealed strong resonance structure for the formation of anions at around 1.2 eV. Considering the induced polarization potential in organic matter to be of the order of 1 eV,39 the gas-phase 1.2-eV shape resonance is expected to lie around 0.2 eV in condensed films. Therefore, we can conjecture that, as the content of amino acids increases in the mixed films, the damage to amino acids via DEA increases producing more radicals that induce damage of GCAT. Hence, the total fragmentation yield of GCAT should increase as a function of film thickness up to larger thicknesses of the order of the thermalization distance of electrons in arginine films (i.e., ~10 nm). This hypothesis is contrary to the results of Fig. 2, which shows for both amino acids a decrease in total fragmentation yield up to 9 nm. We can therefore conclude that at these very low energies (≤1 eV), multiple scattering does not contribute appreciably to the thickness dependence of the fragmentation yields, which is thus controlled by the amino acid–GCAT interactions. Thus, the results of Fig. 1 beyond the ratios 2 : 1 for Gly : GCAT and 0.5 : 1 for Arg : GCAT mixtures are interpreted to be due to protection of oligonucleotides by the amino acids.
At the glycine : GCAT ratio of 0.5, however, the increased damage to GCAT (or sensitization) could be due to the strong production of H• radicals by electron impact on glycine. At 1 eV, the only operating channel is the dehydrogenation of the molecule with production of a mobile H• radical. In most of the previous studies on DEA to amino acids, the formation of the hydrogen radical was assigned to the electron attachment into the π* orbital which is coupled to the dissociative σ*(O–H) orbital of the carboxyl group, whilst recent R-matrix calculations have shown that the DEA to glycine involves the capture of the excess electron directly into the σ*(O–H) orbital.40 Therefore, the increase of GCAT damage for glycine–oligonucleotide pairs, at low molar ratios can be ascribed to the 1 eV DEA to glycine followed by the interaction of H• radicals generated in this process.
It is known that H• radicals are one of the most abundant species, besides OH• radicals and hydrated electrons, produced as radiation products of the aqueous solution.41 However, recent Monte Carlo simulations have demonstrated that the hydrogen radical interactions with DNA components, i.e. nucleobases and deoxyribose are not significant.42 Most of the H• radicals are positioned outside the first hydration shell in these simulations, and diffuse away from the target molecule, and the H• attack efficiency is calculated to be less then 1%.42 Whilst experimental studies have shown that the H• radical, due to its electrophilic property, interacts preferentially with C–C double bonds and with electron-rich sites of molecules;43 however, little is known about the direct effect of H• on the phosphate group or the DNA molecule. Very early studies on dry samples of DNA bombarded by atomic hydrogen have showed that the most of modifications are on base groups, and only about 20% are on deoxyribose groups.44
The H• radical formed can also react with amino acids and cause their fragmentation.45,46 Thus the decrease of the total yield of degradation of GCAT samples for higher concentrations of amino acid can be due to the higher possibility of hydrogen radical interactions with amino acids than oligonucleotides. Amino acids are also known to be effective radical scavengers, but mainly for oxygen species.47,48
Clearly according to Fig. 1, the level of protection of GCAT from direct damage caused by LEE is higher in the case of arginine. This could be related to a stronger molecular interaction between arginine and GCAT than in the case of glycine and GCAT.
In order to explore this hypothesis further, the binding energies for nucleobase–amino acid and nucleotide–amino acid pairs were calculated by using UFF and Dreiding force-field. The binding energy was calculated for each pair and the average total energies for molecular interactions between the two compounds are presented in Table 2. In addition, the components of total energy for each pair were also calculated by using the Dreiding forcefield (Table 3).
Table 2.
The total binding energy (in kcal mol−1) for amino acid–nucleobase/nucleotide pairs calculated by applying universal force field and Dreiding theories combined with the consistent charge equilibration method. These values are averages of 107 energy samples calculated for a temperature of 298 K
| Glycine
|
Arginine
|
|||
|---|---|---|---|---|
| UFF | Dreiding | UFF | Dreiding | |
| A | −1.052 | −0.712 | −1.476 | −0.928 |
| C | −0.958 | −0.651 | −1.349 | −0.863 |
| G | −1.059 | −0.732 | −1.478 | −0.966 |
| T | −0.998 | −0.694 | −1.387 | −0.928 |
| pdA | −1.229 | −0.842 | −1.635 | −1.097 |
| pdC | −1.233 | −0.854 | −1.636 | −1.121 |
| pdG | −1.243 | −0.872 | −1.641 | −1.141 |
| pdT | −1.207 | −0.839 | −1.621 | −1.106 |
Table 3.
The energy (in kcal mol−1) of three components of the total energy, i.e. van der Waals, electrostatic and H-bonding for amino acid–nucleobase/nucleotide pairs calculated by applying Dreiding forcefield combined with the consistent charge equilibration method. These values are averages of 107 energy samples calculated for a temperature of 298 K
| Glycine
|
Arginine
|
|||||
|---|---|---|---|---|---|---|
| vdW | Electr. | H-bond | vdW | Electr. | H-bond | |
| A | −0.706 | 0.004 | −0.010 | −0.928 | 0.009 | −0.009 |
| C | −0.641 | 0.004 | −0.014 | −0.847 | −0.004 | −0.012 |
| G | −0.723 | 0.005 | −0.015 | −0.947 | −0.006 | −0.013 |
| T | −0.683 | 0.002 | −0.013 | −0.907 | −0.008 | −0.012 |
| pdA | −0.825 | −0.006 | −0.011 | −1.099 | 0.014 | −0.012 |
| pdC | −0.835 | −0.007 | −0.012 | −1.118 | 0.010 | −0.013 |
| pdG | −0.853 | −0.006 | −0.013 | −0.137 | 0.009 | −0.014 |
| pdT | −0.828 | 0.001 | −0.012 | −1.094 | 0.000 | −0.013 |
Previous calculations for 20 amino acids and the four nucleo-bases revealed that two-thirds of all interactions between these molecules involve van der Waals contacts.16 This is in good agreement with our modelling as is shown in Table 3. Other interactions within the complex formation are due to electrostatic interactions and hydrogen bonds. For arginine the number of van der Waals contacts was reported to be twice as great for guanine than for other nucleobases, whereas the number of hydrogen bonds differs for each nucleobase; guanine interacts the most, then thymine, adenine and cytosine. In our modelling the highest total binding energy of Arg–nucleobase pairs is also for guanine with estimated values of −1.478 and −0.966 kcal mol−1 from UFF and Dreiding, respectively. In the case of glycine the largest number of van der Waals contacts was observed for thymine.16 Previous simulations have shown that arginine forms more interactions with the DNA nucleobases than glycine.16 Thus arginine interacts more strongly, which is in good agreement with our prediction based on calculated binding energies.
Table 2 also shows the binding energies for both amino acids with the four nucleotides: adenosine monophosphate (pdA), cytidine monophosphate (pdC), guanosine monophosphate (pdG) and thymidine monophosphate (pdT). The average values for the binding energy for Arg–nucleotide and Gly–nucleotide pairs are −1.633 and −1.228 kcal mol−1 in the case of UFF and −1.116 and −0.852 kcal mol−1 in the case of Dreiding, respectively. It is also worth noting that the binding energies for amino acid–nucleotide pairs is higher than those for nucleobases. This can be correlated with the larger number of amino acid interactions which are made with the sugar-phosphate backbone of the DNA.16
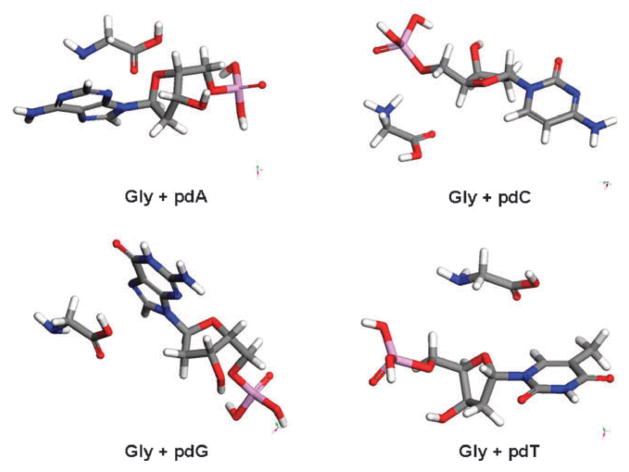
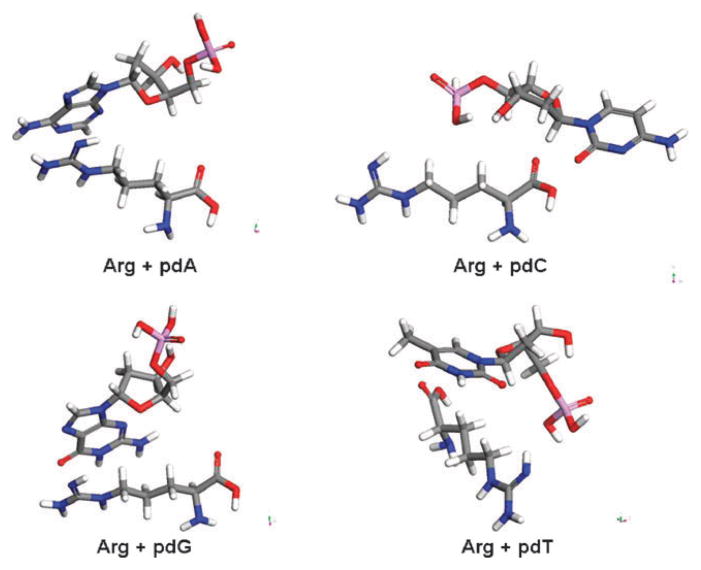
In addition, Fig. 3 and 4 display the molecular orientations of amino acid–nucleotide pairs. As can be seen in these figures arginine forms more interaction sites than glycine with the nucleotides. Thus, according to calculations, the protection level of GCAT in the case of arginine should be higher than that of glycine for the same dose of electron irradiation at all lower molar ratios of amino acid : GCAT, that is in agreement with experiments (Fig. 1).
Fig. 3.
The lowest energy configurations for all nucleotides and glycine (Gly) pairs obtained by using Dreiding forcefield.
Fig. 4.
The lowest energy configurations for all nucleotides and arginine (Arg) pairs obtained by using Dreiding.
Conclusions
The damage to DNA induced by ionizing radiation can occur through several different reaction pathways such as electron transfer and radical attack. The type of damage, as well the amount of damage induced depends upon the presence of other molecular components in close proximity to DNA, particularly the proteins. Therefore from the point of view of radiation damage to DNA, it is important to study the role of proteins bound to DNA. Most interactions between proteins and DNA occur via amino acids. To study these specific interactions the present studies have been focused on irradiation of two amino acids interacting with a short chain of DNA. Low energy electrons were used as the incident particles, since they are the most abundant secondary species produced by ionizing radiation and they allow to study the damage induced by a specific mechanism (i.e., DEA).
For low molar ratios of glycine : GCAT, the degree of total fragmentation of oligonucleotides is higher than that for pure GCAT. This is most likely due to the H• radicals which are produced via DEA to amino acids. A further increase in the number of amino acids in the samples shows a protective role of both glycine and arginine against LEE irradiation. The shielding of GCAT by amino acids can be explained by the relatively strong interactions between molecules, mainly with van der Waals character.
The binding energies for amino acid–nucleobase/nucleotide pairs were also calculated and reveal a stronger interaction for arginine:GCAT and thence arginine with DNA than for glycine, which is in good agreement with other molecular modelling results.
In the future, it would be interesting to explore the yield and reaction channels occurring over a wider range of energies of electron irradiation. Moreover, the influence of other amino acids found in natural proteins would provide a better understanding of protein–DNA radiolysis at the molecular level.
Acknowledgments
This research was supported by the Canadian Institutes of Health Research. S. P. gratefully acknowledges financial support from Engineering and Physical Sciences Research Council EPSRC in the form of a Postdoctoral Fellowship (EP/D067138/1). L.S. would like to thank the European Commission for financial support via a Marie Curie Incoming Fellowship
Notes and references
- 1.International Commission on Radiation Units and Measurements. ICRU Report 31 ICRU. Washington, DC: 1979. [Google Scholar]
- 2.Cobut V, Frongillo Y, Patau JP, Goulet T, Fraser MJ, Jay-Gerin JP. Radiat Phys Chem. 1998;51:229. [Google Scholar]
- 3.Pimblott SM, La Verne JA. Radiat Phys Chem. 2007;76:1244. [Google Scholar]
- 4.Moretto-Capelle P, Le Padellec A. Phys Rev A: At, Mol, Opt Phys. 2006;74:062705. [Google Scholar]
- 5.Sanche L. Chem Phys Lett. 2009;474:1. [Google Scholar]
- 6.Martin F, Burrow PD, Cai ZL, Cloutier P, Hunting D, Sanche L. Phys Rev Lett. 2004;93:068101. doi: 10.1103/PhysRevLett.93.068101. [DOI] [PubMed] [Google Scholar]
- 7.Panajotovic R, Martin F, Cloutier P, Hunting D, Sanche L. Radiat Res. 2006;165:452. doi: 10.1667/rr3521.1. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Y, Hunting DJ, Ayotte P, Sanche L. Phys Rev Lett. 2008;100:198101. doi: 10.1103/PhysRevLett.100.198101. [DOI] [PubMed] [Google Scholar]
- 9.For a review, see: Sanche L. In: Radiation Induced Molecular Phenomena in Nucleic Acid. Shukla MK, Leszczynski J, editors. ch 19 Springer; Dordrecht: 2008.
- 10.Berdys J, Skurski P, Simons J. J Phys Chem B. 2004;108:5800. [Google Scholar]
- 11.Boudaiffa B, Cloutier P, Hunting D, Huels MA, Sanche L. Science. 2000;287:1658. doi: 10.1126/science.287.5458.1658. [DOI] [PubMed] [Google Scholar]
- 12.Ptasińska S, Sanche L. Phys Chem Chem Phys. 2007;9:1730. doi: 10.1039/b616619a. [DOI] [PubMed] [Google Scholar]
- 13.Roginskaya M, Bernhard WA, Razskazovskiy Y. Radiat Res. 2006;166:9. doi: 10.1667/RR3571.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dumont A, Zheng Y, Hunting D, Sanche L. J Chem Phys. 2010;132:045102. doi: 10.1063/1.3298895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomun T, Skalický T. Chem Phys Lett. 2008;453:101. [Google Scholar]
- 16.Luscombe NM, Laskowski RA, Thornton JM. Nucleic Acids Res. 2001;29:2860. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fasman GD. Handbook of Biochemistry and Molecular Biology. 3. CRC; Boca Raton, FL: 1995. [Google Scholar]
- 18.Zheng Y, Cloutier P, Hunting DJ, Sanche L, Wagner JR. J Am Chem Soc. 2005;127:16592. doi: 10.1021/ja054129q. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Y, Cloutier P, Hunting DJ, Wagner JR, Sanche L. J Chem Phys. 2006;124:064710. doi: 10.1063/1.2166364. [DOI] [PubMed] [Google Scholar]
- 20.Li ZJ, Zheng Y, Cloutier P, Sanche L, Wagner JR. J Am Chem Soc. 2008;130:5612. doi: 10.1021/ja077601b. [DOI] [PubMed] [Google Scholar]
- 21.Rappé AK, Casewit CJ, Colwell KS, Goddar WA, III, Skiff WM. J Am Chem Soc. 1992;114:10024. [Google Scholar]; Casewit CJ, Colwell KS, Rappé AK. J Am Chem Soc. 1992;114:10035. [Google Scholar]; Casewit CJ, Colwell KS, Rappé AK. J Am Chem Soc. 1992;114:10046. [Google Scholar]
- 22.Mayo SL, Olafson BD, Goddard WA., III J Phys Chem. 1990;94:8897. [Google Scholar]
- 23.Accelrys Software Inc. Materials Studio, v4.4. 2008. [Google Scholar]
- 24.Cai ZL, Dextraze ME, Cloutier P, Hunting D, Sanche L. J Chem Phys. 2006;124:024705. doi: 10.1063/1.2141505. [DOI] [PubMed] [Google Scholar]
- 25.Sanche L. J Chem Phys. 1979;71:4860. [Google Scholar]
- 26.Aflatooni K, Hitt B, Gallup GA, Burrow PD. J Chem Phys. 2001;115:6489. doi: 10.1063/1.3319751. [DOI] [PubMed] [Google Scholar]
- 27.Gohlke S, Rosa A, Illenberger E, Brüning F, Heuls MA. J Chem Phys. 2002;116:10164. [Google Scholar]
- 28.Ptasinska S, Denifl S, Abedi A, Scheier P, Märk TD. Anal Bioanal Chem. 2003;377:1115. doi: 10.1007/s00216-003-2254-x. [DOI] [PubMed] [Google Scholar]
- 29.Scheer AM, Mozejko P, Gallup GA, Burrow PD. J Chem Phys. 2007;126:174301. doi: 10.1063/1.2727460. [DOI] [PubMed] [Google Scholar]
- 30.Vasil’ev YV, Figard BJ, Voinow VG, Barofsky DF, Deinzer ML. J Am Chem Soc. 2006;128:5506. doi: 10.1021/ja058464q. [DOI] [PubMed] [Google Scholar]; Vasil’ev YV, Figard BJ, Barofsky DF, Deinzer ML. Int J Mass Spectrom. 2007;268:106. [Google Scholar]
- 31.Abouaf R. Chem Phys Lett. 2008;451:25. [Google Scholar]
- 32.Ptasinska S, Denifl S, Candori P, Matejcik S, Scheier P, Märk TD. Chem Phys Lett. 2005;403:107. [Google Scholar]
- 33.Abdoul-Carime H, Illeneberg E. Chem Phys Lett. 2004;397:309. [Google Scholar]
- 34.Sulzer P, Alizadeh E, Mauracher A, Märk TD, Scheier P. Int J Mass Spectrom. 2008;277:274. [Google Scholar]
- 35.Abdoul-Carime H, Gohlke S, Illenberger E. Chem Phys Lett. 2005;402:497. [Google Scholar]
- 36.Abdoul-Carime H, Gohlke S, Illenberger E. Phys Chem Chem Phys. 2004;6:161. [Google Scholar]
- 37.Papp P, Urban J, Matejcik S, Stano M, Ingólfsson O. J Chem Phys. 2006;125:204301. doi: 10.1063/1.2400236. [DOI] [PubMed] [Google Scholar]
- 38.Abdoul-Carime H, König-Lehmann C, Kopyra J, Farizon B, Farizon M, Illenberg E. Chem Phys Lett. 2009;477:245. [Google Scholar]
- 39.Chu SC, Burrow PD. Chem Phys Lett. 1990;172:17. [Google Scholar]
- 40.Gallup GA, Burrow PD, Fabrikant II. Phys Rev A. 2009;79:042701. [Google Scholar]
- 41.Health Risks From Exposure to Low Levels of Ionizing Radiation: BEIR VII, Phase 2. National Academy Press; 2006. [PubMed] [Google Scholar]
- 42.Aydogan B, Bolch WE, Swarts SG, Turner JE, Marshall DT. Radiat Res. 2008;169:223. doi: 10.1667/RR0293.1. [DOI] [PubMed] [Google Scholar]
- 43.von Sontag C. Free-Radical-Induced DNA Damage. Springer-Verlag; Berlin: 2006. [Google Scholar]
- 44.Holroyed RA, Glass JW. Radiat Res. 1969;39:758. [PubMed] [Google Scholar]
- 45.Snipes W, Schmidt J. Radiat Res. 1966;29:194. [PubMed] [Google Scholar]
- 46.Holroyd R, Glass J, Riesz P. Radiat Res. 1970;44:59. [PubMed] [Google Scholar]
- 47.Clausen MR, Skibsted LH, Stagsted J, Agric J. Food Chem. 2009;57:2912. doi: 10.1021/jf803449t. [DOI] [PubMed] [Google Scholar]
- 48.Khan MA, van Dyk J, Yeung IWT, Hill RP. Radiother Oncol. 2003;66:95. doi: 10.1016/s0167-8140(02)00325-0. [DOI] [PubMed] [Google Scholar]