Abstract
The transposons of the Bari family are mobile genetic elements widespread in the Drosophila genus. However, despite a broad diffusion, virtually no information is available on the mechanisms underlying their mobility. In this paper we report the functional characterization of the Bari elements transposition system. Using the Bari1 element as a model, we investigated the subcellular localization of the transposase, its physical interaction with the transposon, and its catalytic activity. The Bari1 transposase localized in the nucleus and interacted with the terminal sequences of the transposon both in vitro and in vivo, however, no transposition activity was detected in transposition assays. Profiling of mRNAs expressed by the transposase gene revealed the expression of abnormal, internally processed transposase transcripts encoding truncated, catalytically inactive transposase polypeptides. We hypothesize that a post-transcriptional control mechanism produces transposase-derived polypeptides that effectively repress transposition. Our findings suggest further clues towards understanding the mechanisms that control transposition of an important class of mobile elements, which are both an endogenous source of genomic variability and widely used as transformation vectors/biotechnological tools.
Introduction
DNA-based transposable elements, also referred as terminal-inverted-repeat elements, Class II transposable elements or transposons sensu strictu, use a DNA-mediated mode of transposition, that sets them apart from retrotransposons and retroposons that move via RNA intermediates. Cut-and-paste DNA transposons are grouped into at least 17 superfamilies [1], and rely on a self-encoded transposase to catalyze the transposition reaction.
The transposable elements related to the Tc1 and mariner families (MLE) (IS630-Tc1-mariner or ItmDx[D/E superfamily] [2] constitute the largest group of cut- and-paste Class II transposons. They are mobile elements of up to 2 Kbp in size, able to move within eukaryotic genomes, and in some cases can constitute high proportions of the genomes they inhabit. These elements usually contain a single, intron-less transposase-encoding gene, typically flanked by two short terminal inverted repeats (TIRs) 19 to 40 bp in length. This relatively simple design combined to a self-encoded transposase able to catalyze all the transposition steps in vitro [3] make the transposition of Tc1/mariner like elements [4] independent of host factors and could explain why these elements have a such widespread occurrence. In fact Tc1 elements require just two TIRs separated by a DNA fragment, a transposase source, and magnesium cations as cofactors for transposition [5] [6].
The Bari elements belong to the IR-DR group of the Tc1 lineage, comprising elements with TIRs ranging from 200 to 250 bp in length. This group also includes other Drosophila elements such as S [7], Minos [8], and Paris [9], and the reconstructed fish transposon Sleeping Beauty (SB) [10], which encode transposases containing a functional bipartite nuclear localization signal (NLS_BP), two HTH motifs in the N-terminal region and an acidic DD34E triad in the C-terminal region [11] [12] [13]. However, the protein motifs of the transposase encoded by Bari-like elements have not been functionally characterized. The TIRs of these elements possess three direct repeats (DRs) called the outer DR (ODR), the middle DR (MDR) and the inner DR (IDR), that are the putative binding sites for the transposase and are necessary for the transposition of autonomous elements [14] [15] [13]. The presence of three DRs is not uncommon; in fact, sequence comparison of TIRs belonging to four Tc1-like elements (Sleeping Beauty, Paris, S, and Minos) has revealed a third conserved DR between ODR and IDR [16] [13].
Although three related Bari subfamilies (Bari1, Bari2 and Bari3) differing in structural organization and potential transposition autonomy are known to exist in different Drosophila species [16], [17], most of the information about the transposition activity of these elements is limited to Bari1 elements in D. melanogaster. Bari1 elements were found arranged in an 80-copies array in the heterochromatic h39 region of the second chromosome of D. melanogaster [18] [19] [20], and highly polymorphic among 90 different populations of D. melanogaster analyzed by in situ hybridization on polytene chromosomes of salivary glands [21].
Transposition events were identified by molecular analysis in Drosophila laboratory stocks showing genomic instability [22] and in the progeny of a single female collected in the wild [23]. Recently, Bari1 transposition has been observed in a genetic background in which the piRNA pathway, which suppresses transposable elements activity, was deregulated [24].
Several mechanisms regulating transposition rate are well characterized. They are either based on repressors produced by the transposon [25] or are evolved by the host to protect its genome from excessive insertional mutation events [26] [27] [28].
Several DNA transposons can be used as vectors for moving exogenous DNA sequences into chromosomes by mimicking the natural process of horizontal gene transfer under laboratory conditions; these include the plant transposons Ac/Ds and Spm systems [29] and transposons from animals including SB [10] and piggyBac [30]. Engineered transposons are useful tools for biotechnology [31], medicine [32] [33] and genetics [34] [35].
To test the essential biological features of the Bari family of mobile elements, we utilized the Bari1 element as model. Here we show that Bari1 encodes a transposase that compartmentalizes to the nucleus both in insect and mammalian cells, and that it can bind the TIRs of the Bari1 transposon. Although we were unable to demonstrate its ability to catalyze the transposition reaction, transcriptional analyses identified unexpected transposase transcripts expressed under different experimental conditions. We discuss the possible causes of the low transposition activity of the Bari1 element in vivo and in vitro, and propose that, in addition to already known regulatory circuits, a post-transcriptional regulation mechanism may also control the transposition of the Bari elements. Due to the relationship of Bari1 to other well-known mobile elements, these results could be of importance in the field of the transposon biology. Their importance as source of variability in the eukaryotic genome as well as substrates in the development of novel integration tools requires a deep biological knowledge of their regulation repertoire.
Results
In silico Analysis of the Bari1 Transposase
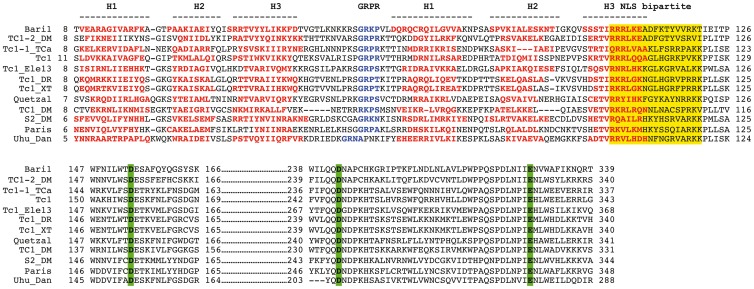
To obtain preliminary information on the functional domains of the Bari1 transposase we compared it with several transposases encoded by Tc1-like elements. Multiple alignment analysis, coupled with secondary structure prediction, identified several functional domains typical of the Tc1/mariner transposases (figure 1). Several protein motifs, such as the DNA binding domain, the NLS (Nuclear Localization Signal) and the catalytic domain, have been shown to be of great importance for the transposase function (see [36] for a review), and they can also be recognized in the Bari1 transposase. A bipartite DNA binding domain thought to be responsible for recognition of the transposon termini can be easily detected at the N-terminus of the protein. This domain is divergent in sequences among the transposases, but the predicted alpha helices of both HTH motifs rely in similar position with respect to each other, indicating functional, rather than sequence conservation.
Figure 1. Partial multiple alignment of Tc1-like transposase sequences.
Residues of the DNA binding domain (consisting of the H1-H3 alpha helices) are red boldfaced, the GRPR domain is blue boldfaced. NLSs are highlighted in yellow and the acidic triads of the catalytic domains (DDE) are highlighted in green.
A slightly divergent GRPR motif (GRKP) motif characteristic of the homeo-domain proteins [37] is also present at position 59 of the Bari1 transposase and between the two HTH motifs. This domain, followed by an additional HTH region (i.e. the Homeo-like domain), is present in all the transposases aligned. The multiple alignment also highlights the presence of a putative bipartite NLS rich in basic aminoacids. The catalytic domain, characterized by the typical DDE motif, is also recognizable in the primary sequence of Bari1 transposase.
Notwithstanding these similarities, the PredictProtein prediction tool (http://www.predictprotein.org/) did not predict the NLS signal, while the PSORT prediction tools give ambiguous results (not shown). For these reasons we aimed at experimentally testing nuclear import of the Bari1 transposase.
In vivo Analysis of the Subcellular Localization of the Bari1 Transposase
To investigate the nuclear import signals in the Bari1 transposase we performed a series of immuno-detection tests after overexpression of the transposase in two model cellular systems, the Drosophila S2R+ and the human HepG2 cells. After expression of either full length (ASE1) or truncated versions (ASE1/Δ 159–339 and ASE1/Δ 1–158) of the Bari1 transposase fused to the V5 tag in the two cell types, the localization of the fusion proteins was detected using a monoclonal anti-V5 antibody.
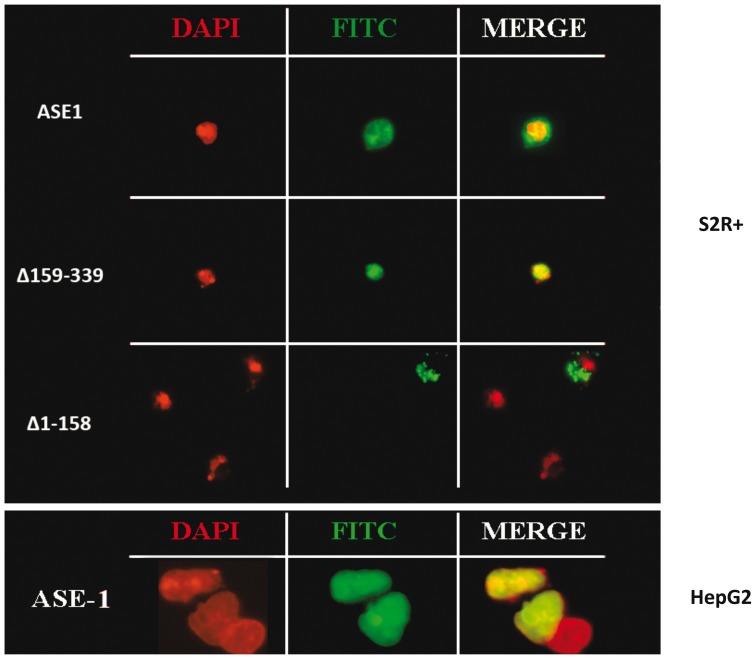
The results obtained are summarized in figure 2 (upper panel, first row), and clearly show that the full-length Bari1 transposase localizes to the nucleus in both cell types, thus indicating that a nuclear import signal in the protein is functional in both insect and mammalian cells. Furthermore, to precisely map the NLS, we tested the subcellular localization of two truncated forms of Bari1 transposase in Drosophila cells. As shown in figure 2 (upper panel, second and third row), the ASE1/Δ 159–339 amino-terminal portion of Bari1 transposase (aminoacids 1–158) retains its nuclear localization, while the Δ1–158 carboxyl-terminal part (aminoacids 159–339) does not. These results clearly indicate that the first 158 aminoacids of the Bari1 transposase contain the NLS, which probably maps to the K/R rich amino acidic sequence, a motif conserved in different Tc1 like transposases (see figure 1).
Figure 2. Sub-cellular localization of the Bari1 transposase.
Upper Panel. Localization of the full length (ASE1), the N-terminal (Δ159–339) and the carboxyl terminal (Δ1–158) portion of the Bari1 transposase in S2R+ cells. Lower Panel. Localization of the full length Bari1 transposase in HepG2 cells. The left column shows the DAPI fluorescence signal (nuclear DNA), the middle column shows the FITC fluorescence signal (indicating fusion protein localization), and the right column shows the merged fluorescence signal.
In vitro Assay of the Binding of the Bari1 Transposase to the Bari1 TIRs
In the nucleus, a Tc1-like transposase must bind specifically to the recognition sequence in the transposon DNA (i.e. its terminal inverted repeats). The DNA-binding domains of several Tc1-like transposase, e.g. Tc1 [38] and SB [10], maps to the N-terminal regions, and can be easily predicted using the available secondary structure prediction tools in combination with a multiple alignments (figure 1).
The Bari1 terminal inverted repeat structure contains three DRs, which are the putative binding sites for the transposase [16]. Here we report the results of extensive in vitro and in vivo assays of the transposase/transposon interaction.
Expression of the full-length Bari1 transposase was induced in E. coli and the expected 43 kD 6XHis-transposase fusion protein (hereafter T16) was purified by exchange chromatography. Similarly, a 27 kD polypeptide corresponding to the 198 C-terminal aminoacids of the transposase fused to a His-tag (hereafter C9) was expressed and purified (see Figure S1 for detailed figures related to protein expression and purification).
The purified proteins obtained were used to assess interaction of the transposase with the Bari1 terminal sequences in EMSA experiments. As shown in figure 3, DNA fragments containing combinations of the three DRs identified within the Bari1 left IR [16], were used as target for the binding.
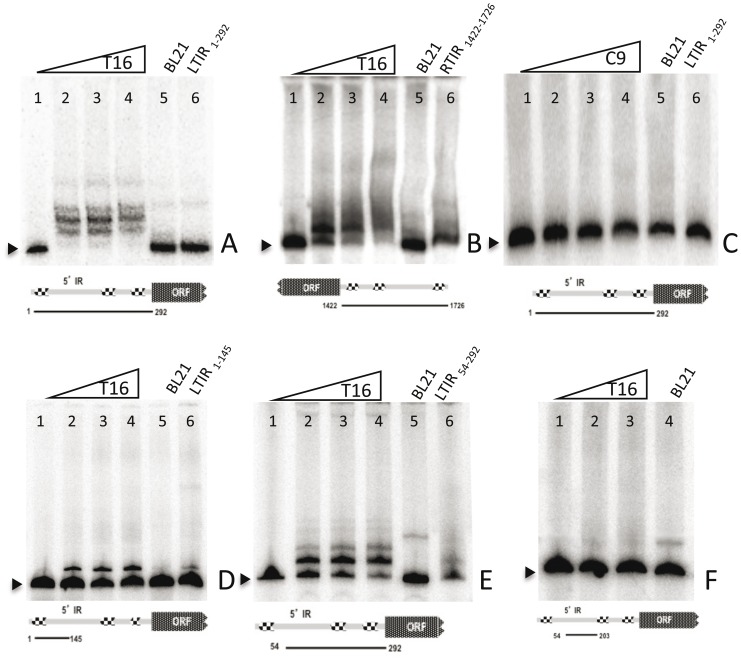
Figure 3. EMSA experiments.
Panel A. The radiolabeled fragment corresponding to the full length left TIR containing 3(lane 1) migrates faster than the same fragment incubated with increasing amounts of Bari1 transposase (T16) (lanes 2, 3, 4). No retardation is observed when the same fragment is incubated with non-induced bacterial extracts (lane 5). The binding is specifically disrupted by a 50X excess of the unlabeled fragment (lane 6). Panel B. Same as panel A, except that the right IR was tested. Panel C. Same as panel A, except that the carboxyl terminal part of the transposase was tested. Panels D, E, F. demonstration of the interaction of the full-length transposase (T16) with left TIR sub-fragments containing one (panel D), two (panel E) or none (panel F) DRs.
Nucleoprotein complexes were analyzed by EMSA after incubation of the T16 protein with radiolabeled DNA fragments corresponding either to the left or the right IRs of Bari1, comparing the patterns observed to those obtained with protein extracts from control (non-induced) bacterial cultures. For both the left and the right IR, slow migrating bands can be observed in the electrophoretic pattern (figure 3, panel A and B, lanes 2–4) compared to the pattern of the free fragment (figure 3, panel A and B, lanes 1), or to the pattern of fragments incubated with protein extracts lacking T16 protein (figure 3, panel A and B, lanes 5). Especially for the left IR, several protein/DNA complexes were visible in the gel, indicating the presence of multiple binding sites for the transposase in the IR, or binding of multiple transposase molecules per IR (figure 3 panel A, lanes 2–4).
Unlabeled IR fragment added in excess as specific competitor DNA inhibited the DNA-protein interaction (figure 3, panels A–E, lanes 6), whereas excess of lambda DNA did not appreciably disrupt the binding (not shown). These results clearly show that the Bari1 transposase specifically binds both Bari1 IRs, and so must contain a functional DNA binding domain. Similarly, it can be concluded that the Bari1 termini contain multiple binding sites for the transposase.
Similar experiments were performed with a purified protein containing the 198 C-terminal aminoacids of the transposase lacking the putative DNA binding domain. As can be observed in figure 3 (panel C) this protein completely lacks the DNA binding properties, as shown by the absence of the slow migrating bands in the electrophoretic pattern. Our results indicate that the transposase domain involved in DNA binding is contained in the first 197 aminoacids of the protein, presumably in the region containing the predicted HTH domains (see figure 1).
Finally, to test the binding properties of the transposase to each of the three DRs of the Bari1 terminal repeats we performed mobility shift assays using truncated versions of the left terminal repeat containing respectively one (figure 3, panel D), two (figure 3, panel E) or none (figure 3, panel F) of the DRs. The results indicate that the Bari1 IRs contain multiple binding sites for the transposase, whereas no shifted bands can be observed using a fragment not containing any DR sequence (figure 3, panel F).
In vivo Assay of the Binding of the Bari1 Transposase to the Bari1 TIRs
To further demonstrate the DRs-mediated interaction observed in the EMSA experiments, we performed a biased One Hybrid assay in yeast. We constructed yeast strains carrying three copies of either the outer (3×Lo), the middle (3×Lm), or the inner (3×Li) DR, in each case integrated upstream of a LacZ reporter cassette. The three copies of the DRs serve as “bait” for the binding of the transposase “prey” protein expressed from the yeast plasmid pACT2-ASE1 (see Materials and Methods). The LacZ reporter activation is expected only upon bait/prey interaction. Figure 4 shows the results of the β-galactosidase assay on integrant yeast strains transformed with a plasmid expressing either the full length (figure 4, panels A, C, E, G) or the carboxyl terminal part (figure 4, panels B, D, F, H) of the transposase. As can be observed in figure 4A, the full-length transposase is able to turn on the LacZ reporter as the expected result of its interaction with the complete left Bari1 TIR, thus confirming the results of the EMSA experiments (figure 3). Similarly, the LacZ reporter system is also activated in strains transiently expressing the full-length transposase and containing three copies of either the Lo or the Li DR fragments (figure 4, C and G respectively), again confirming that the transposase interacts specifically with the DRs of the terminal inverted repeats. The interaction of the transposase with three copies of the middle DR (Lm) appears to be very weak, as indicated by the very faint blue staining compared to those obtained with the Lo and Li targets. By contrast, and as expected, the carboxyl terminal part of the transposase (aminoacids 142–339), which lacks the DNA binding domain, is completely unable to activate the LacZ reporter when either the entire TIR or the DRs were used as bait (figure 4, B, D, F, H).
Figure 4. Yeast One hybrid Assay.
Left column: LacZ reporter system activation in yeast by full-length transposase expressed due to the interaction with the entire 5′TIR (A), the outer DR (three repeats) (C), the middle DR (three repeats) (E) or the inner DR (three repeats) (G). Right column: Bari1 transposase lacking the DNA binding domain completely fails to interact with the Bari1 5′ TIR or with its DRs (B, D, F, H).
Taken together, these results clearly shows that the Bari1 transposase is able to bind in vitro and in vivo the DRs contained in the transposon termini, a crucial initial step of the transposition process.
Excision and Integration are Rate-limiting Steps of Bari1 Transposition
We have tested the transposition activity of Bari1 using classic transposition assay. This is a simple assay aimed to demonstrate the activity of both isolated and reconstructed transposons; the assay is usually performed in heterologous cellular systems in order to bypass repressive circuitry acting on the original cellular environment from which the element has been isolated. On the other hand, testing transposition in homologous cellular systems ensures that necessary and species-specific co-factors are present. For these reasons, we have tested Bari1 transposition in cultured human HepG2 or Drosophila S2R+ cells after co-transfection of a plasmid carrying a marked transposon containing a neomycin or blasticidin antibiotic resistance gene plus a helper plasmid expressing the Bari1 transposase. As shown in figure 5, there was no increase in resistant foci with respect to the controls, indicating lack of transposition in either system assayed.
Figure 5. Transposition assay.
Resistant colonies resulting from the transfection of Drosophila S2R+ cells (top) and human HepG2 cells (bottom) with a mixture of donor plasmid and helper plasmid or with the donor plasmid alone.
Similarly, a transposon excision assay based on PCR amplification of the “empty” donor plasmid molecules in cultured cells transfected as described above failed to detect transposon excision (data not shown).
Analysis of Bari1 transcripts in the hsp83scratch mutant
Thereafter, we asked if Bari1 transposition could be affected by mechanisms acting at the transcriptional or post-transcriptional level. To overcome any possible repressive epigenetic mechanism acting on Bari1 [24] [39] [40], we analyzed transcripts in homozygous hsp83scratch D. melanogaster mutants.
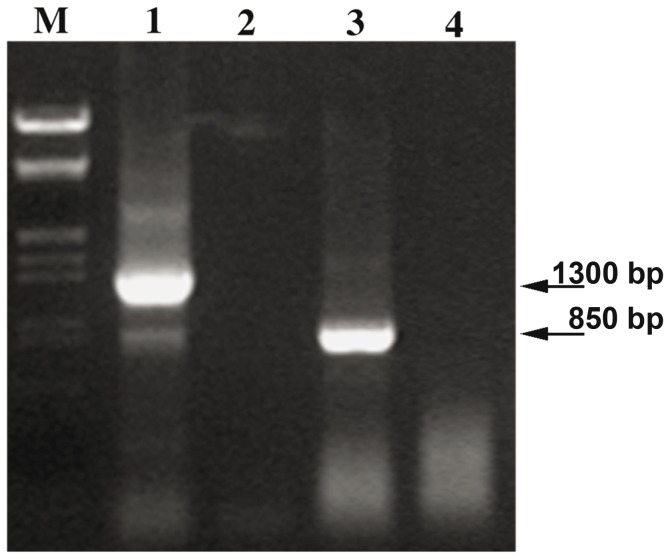
As previously reported, in the hsp83scratch mutant there is a strong germ line deregulation of several mobile genetic elements, including Bari1 [24]. We analyzed by RT-PCR the Bari1 transcripts from hsp83scratch ovaries and testes, tissues where instability of the mobile genetic elements is known to occur. In contrast to the ovaries, where only a full-length 1,396 bp long cDNA was found, in the testes an 882 bp long cDNA was present (figure 6, lanes 1 and 3). No transcripts were detected in the somatic tissues (carcasses) of the hsp83scratch strain (VS and MPB personal communication and unpublished) indicating transcriptional repression in somatic tissues in vivo.
Figure 6. Bari1 transcriptional analyses in the Drosophila hsp83scratch strain.
Results of RT-PCR performed on ovaries (lanes 1 and 2) and testes (lanes 3 and 4) of homozygous hsp83scratch mutant (lanes 1 and 3) or wild type flies (lanes 2 and 4). M: λ/Eco-Hind molecular weight marker. Arrows indicates the approximate molecular weight of the amplified cDNAs.
The shorter transcript harbors a 514 bp internal deletion with respect to the full-length transcript (figure 7, panel A, right, and figure S2). Interestingly, this male specific mRNA potentially encodes a 194 aa protein entirely lacking the catalytic domain of the transposase (figure 7, panel C, right). On the other hand, no Bari1 transcripts were detected in the gonads of a wild type D. melanogaster strain (figure 6, lanes 2 and 4) confirming that Bari1 is transcriptionally repressed in wild type flies.
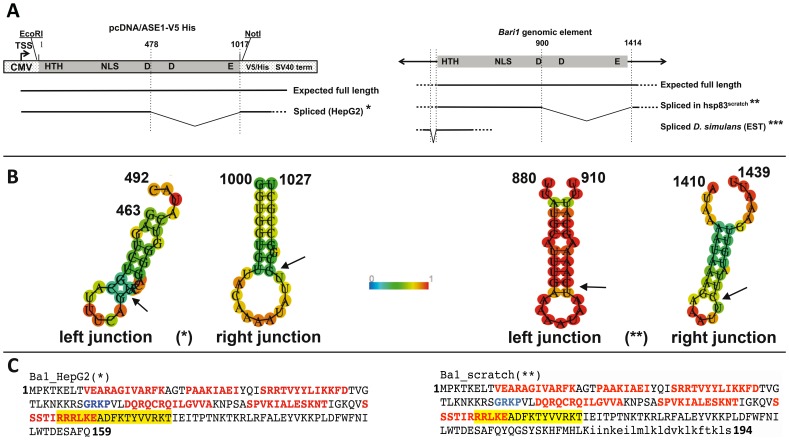
Figure 7. Structural analysis of the transcripts.
Panel A. Structural analyses of the transcripts isolated in HepG2 cells (*), in the testes of hsp83scratch homozygous males (**) or identified in silico by D. simulans ESTs database (***). TSS: transcriptional start site. Positions are relative to the pcDNA/ASE1 plasmid sequence (left) assuming as position 1 the first nucleotide of the cloned fragment, or to the full-length element sequence (right). Panel B. Secondary structure prediction of the RNA sequences across the splicing sites of the HepG2-specific transcript marked with an asterisk (left) and the hsp83scratch –specific transcript marked with two asterisks (right). Arrows indicate the splicing sites. Colored scale indicates base-pairing (unpairing) probabilities. Panel C. Putative protein products of the transcripts isolated in HepG2 cells (left) and in the testes of hsp83scratch homozygous males (right). Color code and protein residues numbering are match those showed in figure 1. New amino acids added upon RNA processing are shown in italic.
To exclude the possibility that an aberrant genomic copy carrying the same deletion could produce the observed testis-specific transcript, we have performed a PCR analysis on genomic DNA extracted from hsp83scratch homozygous mutants using the same primers used for the RT PCR analysis. The results are compatible with the absence of defective copies of Bari1 in the mutant genome hsp83scratch (figure S3).
Analysis of Bari1 transcripts in HepG2 and S2R+ cells
To further support the above results we have analyzed the transcripts arising upon transient overexpression of the Bari1 transposase in HepG2 cells. Although these cells are of somatic origin, this experimental system mimics in part the hsp83scratch mutant and, therefore, no piRNA-mediated epigenetic regulation is expected. Sequencing of cDNA clones revealed two transcripts of different size: one corresponding to the expected 1,2 Kb full-length Bari1 transcript, the second to an unexpected shorter transcript. The shorter cDNA harbors a 540 bp deletion extending from position 478 (relative to the full-length coding region) to the NotI cloning site present in the pcDNA/ASE1 plasmid expressing the Bari1 transposase (see figure 7, panel A, left, and figure S2). Also this mRNA isolated in HepG2 cells potentially encodes a 159 aa protein entirely lacking the catalytic domain of the transposase (figure 7, panel C, left).
A similar analysis was performed in S2R+ cells transfected with the pAC5/ASE1-V5His plasmid. In this case we failed to detect any processed cDNA, being able to detect a single cDNA, corresponding in size to the full-length transcript, even after additional cycles of nested amplification (data not shown).
The shorter transcripts reported above are unlikely to originate from spliceosomal-mediated splicing events, because canonical donor and acceptor consensus sites are absent in the corresponding positions of the full-length transcript (see figure S2). Also, no potential splicing sites are identified in silico by the NNSPLICE tool [41] (not shown). However, a careful analysis of the full-length Bari1 transcript by the RNAfold program [42] revealed the presence of a stem-loop secondary structures at the regions encompassing the sequences removed in both shorter transcripts described above. Figure 7B shows these structures as predicted in the “unspliced” transcripts. It is worth noting that the left and the right junctions share the respective cleavage positions, the left cleavage occurring at the first bond in the stem and the cleavage at the right junction occurring at the first bond in the loop (see arrows in figure 7B).
Analysis of Bari1 transcripts in related Drosophila species
We have mined the main ESTs databases in search of Bari1 transcripts carrying deletions similar to those observed in the above described cDNA clones. No similar defective transcripts can be found in the FlyBase and NCBI ESTs databases, except for a D. simulans cDNA clone (GenBank: DK347895.1; cloneID = dsif17g09), which differs from the full-length transcript by a 66 bp deletion in the UTR region, but it is probably the product of a canonical splicing event (figure 7, panel A left and figure S2, second pairwise alignment). No Bari1 genomic copy carrying this 66 bp deletion was detected by a BLAST search of the D. simulans genomic databases, thus excluding that it is the transcriptional product of a defective copy of the transposon.
On the basis of these results, we suggest that Bari1 transcripts may undergo processing, thereby resulting in catalytically inactive transposase polypeptides.
Discussion
The importance of transponsable elements as source of variability in the eukaryotic genome and their use in the development of novel integration tools (e.g. for gene therapy and functional genomics) dictates a better understanding of their biology. The Tc1/mariner superfamily of transposable elements has been extensively studied, particularly in order to explain the wide diffusion of its members in a variety of animal populations. At present, much detailed information is available about the key structural and functional features of many mobile elements of this superfamily, e.g. about the tempo and mode of their invasion of host species [43] [36], about the mechanisms underlying the specificity of the “cut and paste” reaction [44], about the host strategies that limit the harmful effect of their mobility within the host genome [45], and about their value as vectors in transgenesis experiments [46]. The Bari transposons, belonging to this remarkably family, are present in almost every species of Drosophila so far analyzed [17] [16]. With the aim of identifying and characterizing in depth the structural and functional features underlying the intriguingly successful diffusion of many members of the Tc1/mariner family throughout a wide range of host species, we started a detailed analysis of the Bari1 transposition process. In this paper, we focus on three informative aspects of this process, i.e: 1) the cellular localization of the transposase; 2) the transposon-transposase physical interaction; 3) the possibility that a novel regulative post-transcriptional mechanism based on alternative splicing may, at least in some cell types and genetic backgrounds, repress the transposition of the Bari1 elements.
The Bari1 Transposase Possesses a Functional NLS and is Capable of Binding the Terminal Ends of the Transposon
In order to achieve correct subcellular localization nuclear proteins contain NLSs [47], and this is also the case for the transposases encoded by highly active elements such as Activator and Mu of maize [48] [49], Tag1 of Arabidopsis [50], mariner of Drosophila [51] and BmTc1 of the silkworm [52].
The nuclear compartmentalization of the transposase expressed by Bari1, supposed to act on the chromosomal copies of the transponson, was never characterized before. In order to address this issue we transiently overexpressed Bari1 transposase and performed immuno-localization experiments both in S2R+ and HepG2 cells. Our finding that Bari1 transposase localized to the nucleus of both systems strongly suggested the presence of a functional NLS signal. Subsequent deletion analysis suggested that the NLS signal should be included within the first 158 amino acids of the protein, and multiple alignment analysis allowed us to locate this signal at positions 106–121 of the transposase (see figure 1).
Purified Bari1 transposase specifically binds to the left and right TIR of the transposon. Our in vitro results show that the binding occurs at the three DR sequences occurring at the transposon termini [16]. However, the binding to the right transposon end appears to be significantly less efficient as compared to the left one (figure 3, panel A and B). This could be due either to the (quite low) sequence divergence between the DRs of the right and left termini, or to the much higher divergence of the intervening sequences that separate the DRs. Since recognition of both ends is the initial and crucial step in the transposition mechanism [53], the decreased binding affinity of one of the terminal sequences could significantly affect transposition efficiency.
In vivo experiments performed in yeast highlighted a strong binding of the transposase to two out the three DRs: the transposase/DR interaction resulted strong for the Lo and Li DRs but very weak for Lm. Similar data have been previously reported for the Sleeping Beauty transposon [13], whose middle (half) DR enhances, but is not essential for transposition, suggesting that the Lm repeat contributes to the protein-DNA interactions without a direct involvement.
Is Bari1 Transposition Controlled by Additional Post-transcripional Regulation Mechanisms?
Despite the correct sub-cellular localization of the transposase and its ability to bind both TIRs of the element, no transposition or excision of Bari1 elements was detectable by classic assays performed on the cultured cell lines we utilized. Several explanations might be possible. First, as previously suggested [23], a poor catalytic activity of the transposase could lead to genomic mobility of the element only in rare instances. In this case, specific amino acid substitutions in the region encompassing the catalytic domain of the transposase could significantly improve the efficiency of the transposition process. Essentially, this was the strategy that allowed the birth of the engineered Sleeping Beauty element [10].
As mentioned above, Bari1 has been shown to be an active transposon in D. melanogaster, albeit with a limited mobility [21] [23], suggesting the existence of control mechanisms that maintain a very low frequency of transposition.
Controlling mechanisms are not uncommon: the mobility of several transposable elements is regulated by host loci (e.g. gypsy-flamenco [54] [55], ZAM-COM [56]) or by self-encoded repressors (e.g. P element [53]). Unstable strains, in which an allelic form of a controlling locus allows a higher rate of transposition, are a further demonstration of the existence of control systems regulating genomic transposon mobility. An unstable strain, in which several transposable elements, including Bari1, are deregulated, has been recently described [24].
Previous studies have highlighted that Bari1 is subjected to tight germline post-transcriptional regulation by the piRNA pathway [24] [39] [40]. Similarly, a regulation system based on the siRNA post-transcriptional gene silencing protects somatic cells against Bari1 transposition in Drosophila [57]. In addition, the chromatin state of Bari1 genomic copies is directly linked to the piRNA pathway in germline cells [39] and possibly to the siRNA pathway [58].
The results presented in this work suggest that at least one additional level of control may exist on Bari1. This evidence comes from transcript analysis of overexpressed Bari1 transposase in HepG2 cells, which should lack both piRNA- and siRNA-based regulation on Bari1 as well as epigenetic control at the chromatin level, being Bari1 normally absent in the human genome. We have observed that, in this particular experimental condition, unusually processed Bari1 transcripts can be detected, opening a suggestive scenario to a hitherto unreported mechanism relying on post-transcriptional modifications of Bari1 transcripts. Very similar results were found after Bari1 transcript analysis in the testes of a Drosophila unstable mutant strain in which the piRNA pathway has been disrupted and transcription of Bari1 has been previously demonstrated. We hypothesize that the predicted stem-loop secondary structures at the splicing sites of Bari1 full-length transcript (see Figure 7 panel B), which are in part similar to the secondary structures of natural targets of the so-called “non-conventional” splicing system.
It has been postulated before that secondary structure can be important in pre-mRNA processing, and this has been in some cases experimentally documented [59] [60].
Unconventional splicing is a special class of splicing events, which does not involve spliceosomes and occurs in the cytoplasm. The unconventional splicing system recognizes mRNA secondary structures at the boundaries of the intervening intron sequence and plays a key role in the Unfolded Protein Response (UPR) [61], as it leads to the production of spliced mRNAs which can be translated into functional transcription factors as the transcription factors XBIP [62] and HAC1 [63]. These transcription factors activate UPR in mammals and in yeast respectively. Dmel\Xbp1 has been identified as the D. melanogaster counterpart of the mammalian XBP1 protein, and its transcript is subject to unconventional splicing in the salivary glands of third instar larvae [64]. Upon accumulation of unfolded protein in the endoplasmic reticulum, XBP1 mRNA is processed to an active form by the endonuclease inositol-requiring enzyme 1, IRE1. The resulting loss of 26 nt (23 nucleotides in D. melanogaster [64]) from the spliced mRNA causes a frame-shift and produces the XBP1(S) isoform, which is the functionally active transcription factor.
An attractive speculation is that similar mechanism could be involved in the regulation of transposon mobility, although at present the IRE1 protein has not been shown to target transposon transcripts, and XBP1 (HAC1 in yeast) is the only known transcript processed by IRE1. However, little it is known about the endonucleolytic activity of IRE1 on other cellular mRNAs including transcripts of transposon origin.
The short unexpected transcripts that we have detected upon transient overexpression of the transposase gene might be simply explained as a side effect of IRE1 activity induced by the activation of the UPR pathway. In the testes of hsp83scratch mutants this side effect could be enhanced, as a consequence of the concomitant deregulation of multiple families of transposable element [24]. Similarly, in the HepG2 human cells the Bari1 transcript could be “intercepted” by IRE1 upon transient overexpression of the transposase gene, which has been tested under the transcriptional control of a strong promoter. We hypothesize that the protein products potentially encoded by the unusually spliced transcripts (see figure 7 panel C) could play a role in transposition suppression, acting as a dominant negative form of the transposase. In fact although these putative proteins lack the catalytic domains they could be still able to bind the transposon termini (i.e. contain the DNA binding domain) and, possibly, to form heterodimers with wild type transposase molecules.
It is worth noting that we have not detected processed transcripts in transfected S2R+ cell, and in the ovary of the hsp83scratch mutant (figure 6), suggesting that we are facing a complicate, and multi-level, regulation issue. Interestingly, the finding that Bari1 fails to transpose in S2R+ cells indicates that the inhibition occurs even in absence of transcript processing suggesting that other general mechanisms control Bari1 activity (i.e. folding or the poor catalytic activity).
In conclusion, we have performed a functional analysis of the Bari1 transposon and observed that, despite the nuclear localization of the transposase and its ability to bind the transposon terminal inverted repeats, Bari1 has no detectable transposition activity. We speculate that post-transcriptional processing of mRNA could interfere with the transposition of Bari1 elements, particularly in absence of primary (i.e. piRNA-mediated) host defense mechanisms. This regulatory system could involve canonical (spliceosomal-mediated) or unconventional (i.e. IRE1-mediated) splicing and cooperate with other already known or yet to be discovered regulatory circuits in controlling the genomic mobility of the elements of the Bari1 transposon family, and possibly the mobility of elements of other families as well. Further studies are needed to demonstrate our hypothesis and to assess the existence of additional levels of regulation of a member of the Tc1-mariner superfamily.
Materials and Methods
Drosophila Stocks and Cell Cultures
Fly stocks were maintained on standard cornmeal-agar medium at 25°C.
The hsp83scratch strain has been described by Specchia and co-authors [24].
S2R+ cells (DGRC, Bloomington, USA) were cultured in Schneider’s insect medium supplemented with 10% FBS, 1% penicillin/streptomycin, at 26°C.
HepG2 cells (ATTC, Manassas, USA) were grown in Dulbecco’s minimum essential medium supplemented with 10% FBS, 200 mM glutamine, 1% penicillin/streptomycin, and maintained at 37°C with 5% CO2.
For transposition and excision assays either blasticidin (25 µg/ml) or G418 (1 mg/ml) were added to the medium as selective agents depending on the cell type used.
Transfection and Immuno-detection of Recombinant Proteins
One day prior to transfection cells were seeded and let grow into 6-wells plates containing sterile glass coverslips. Respectively 1×106 and 5×105 S2R+ and HepG2 cells were transfected with 1 µg of purified plasmids DNA using TransIt LT1 (Mirus).
For immunofluorescence staining, the cells attached to slides were washed with phosphate-buffered saline and fixed with 4% formaldehyde for 10 minutes at room temperature followed by three washes in PBS. Blocking was performed with a solution containing 10% fetal bovine serum and 0,5% of Triton X-100 for 30 minutes followed by two washes in PBS for 2 minutes each.
Cells were incubated with a dilution 1∶500 of V5 antibody (Invitrogen) conjugated with FITC fluorochrome for 2 hours. After three washes in PBS, the cells were stained with DAPI (4',6-diamidino-2-phenylindole) and mounted with anti-fade (DABCO).
Slides were imaged under an Olympus epifluorescence microscope equipped with a cooled CCD camera. At least 100 positive cells per slide were observed. Grey-scale images, obtained by separately recording FITC and DAPI fluorescence, were pseudo-colored and merged to obtain the final image using Adobe Photoshop software.
Expression Plasmids Construction
A PCR-based strategy was used to clone the transposase ORF, and the derivatives amino and carboxyl terminal fragments into the pAC5.1/V5-His vector (Invitrogen).
Bari1_UP/Bari1_Low, Bari1_UP/Bari1_N-Ter Low, Bari1_C-Ter Up/Bari1_Low respectively were used to amplify DNA sequences encoding the full length Bari1 transposase, its NH-terminal and its COOH-terminal fragments.
The PCR products were digested with EcoRI and NotI restriction enzymes and cloned into the pAc5.1 V5-His C vector in-frame with and upstream the V5-His tag coding sequence of the plasmid. The fusion constructs were sub-cloned in pcDNA3.1 (Invitrogen) using EcoRI and BamHI. All plasmids were sequence-verified.
Cloning in the expression plasmid pET100/D-TOPO was performed using Ba381-topo-U/Ba1398L and Ba802U/Ba1398L to obtain the pET/T16 and pET/C9 respectively.
Recombinant Protein Expression and Purification
Induction of the T16 protein was obtained in E. coli strain BL21(DE3) (Novagen) by the addition of 1 mM IPTG at 0.5 OD600 and continued for 2.5 hr at 37°C. Cells were sonicated in 25 mM HEPES (pH 7.5), 1 M NaCl, 15% glycerol, 0.25% Tween 20, 2 mM b-mercaptoethanol, 1 mM PMSF, and 10 mM imidazole (pH 8.0) was added to the soluble fraction before it was mixed with Ni-NTA resin (Qiagen) according to the recommendations of the manufacturer. The resin was washed with sonication buffer containing 30% glycerol and 50 mM imidazole; bounded proteins were eluted with sonication buffer containing 300 mM imidazole and dialyzed overnight against sonication buffer without imidazole.
Electrophoretic Mobility Shift Assay (EMSA)
The fragments used in EMSA assays were obtained by amplification using the following primer combinations.
Ba_EW4/Ba_292L to obtain the full-length left TIR (figure 3 panel A, C). Ba_1422U/Ba_EW5 to obtain the full-length right TIR fragment (figure 3 panel B). Ba_A/Ba_292L to obtain the fragment depicted in figure 3 panel E. Ba_EW4/Ba_F to obtain the fragment depicted in figure 3 panel D. Ba_A/IR_Ba204 to obtain the fragment depicted in figure 3 panel F.
Fragments were cloned in the pGEMT-easy vector (Promega) and subsequently released by double digestion to obtain fragments with the protruding ends necessary for end labeling.
The fragments tested were end-labeled by mean of a filling-in reaction, using [α 32P]dATP and the Klenow fragment. Nucleoprotein complexes were formed in 25 mM Hepes pH 7.6, 1 mM EDTA pH 8, 50 mM NaCl, 1 mM DTT, 0.1 mg BSA, 2.5 mM Spermidin, 10% Glycerol, 0.1 mg poly [dI][dC] in a total volume of 20 µl. Reactions contained 1 ng labeled probe, and 1.5 ng of purified transposase. After 20 min incubation on ice, 5 µl of loading dye containing 50% glycerol and bromophenol blue was added and the samples loaded onto a 4% polyacrylamide gel in 0.25X TBE buffer.
Yeast Methods
The YM4271 yeast strain was grown in SD medium supplemented with aminoacid, which complement auxotrophies.
Yeast transformation was performed using the TRAFO methods described in [65].
One-hybrid experiments were essentially carried out following the MatchMaker One Hybrid System manual (Clontech). Briefly, Synthetic oligonucleotides (see table S1) containing three tandem repeats of the outer, inner, or middle DRs were designed in order to create double stranded oligonucleotides with protruding ends compatible with the EcoRI and SalI sites of the pLacZi plasmid. Annealing was performed for the following oligonucleotides couples: Z3Lo+/Z3Lo−; Z3Lm+/Z3Lm−; Z3Li+/Z3Li− to obtain the pLaczi-3Lo pLaczi-3Lm pLaczi-3Lm plasmids respectively. Annealed oligonucleotides were cloned into the pLacZi plasmid vector. Similarly, the entire left TIR of Bari1 was amplified with the Ba_EW4/Ba_292L oligonucleotides and cloned into pLacZi. These plasmids were linearized and independently transformed into the S. cerevisiae YM4271 strain in order to obtain integration at the URA locus. Integrants were selected on SD agar plates lacking uracil.
The background expression level of the reporter system was determined by a standard β-galactosidase assay. Colonies were transferred to Whatman filter paper discs and lysed with liquid nitrogen. Filters were then exposed to Z-buffer (Na2HPO4.7H2O 60 mM, NaH2PO4.H2O 40 mM, KCl 10 mM, MgSO4 1 mM, β–mercaptoethanol 50 mM, pH 7) containing X-gal (5-bromo-4-chloro-indolyl-β-D-galactopyranoside 0,33 mg/ml). Only clones without LacZ basal expression in 8 hours were selected for further analyses.
The selected positive colonies were then transformed with a plasmid expressing the prey protein, obtained by cloning the sequence encoding either the full-length transposase or the C terminal portion into the pACT2 vector into the EcoRI and XhoI sites and in frame with the GAL4-AD. β-galactosidase activity was assayed for 6 hours.
Oligonucleotides used for the transposase gene amplification were Ba381U_pACT2/Ba1381L_pACT2 and Ba805U_pACT2/Ba1381L_pACT2 for the full-length and the carboxyl terminal portion of the transposase respectively.
All recombinant plasmids obtained were sequence-verified.
Transcriptional Analysis
RNA was extracted with TRIZOL. Cultured cells were directly processed after two washes in PBS 1X. Quantitation and estimation of RNA purity were performed using a NanoDrop spectrophotometer.
1 µg RNA was converted to cDNA using the QIAQuick reverse transcription kit and following the manufacturer’s instruction. RNA samples from transfected HepG2 cells were amplified with the Bari1 UP/V5 Rev primer primers, while RNA samples from hsp83scratch were amplified with the BaintB_UP/BaintB_low primers.
Transposition and Excision Assays
Transposition assays were performed as described in [10].
Donor plasmids were constructed starting from the p28/47D plasmid clone containing a Bari1 element with the flanking sequences from the 47D region of the polytene chromosomes. Either the blasticidin resistance cassette or the neomycin resistance cassette were amplified from the pCMV-beta and from the pCoBlast plasmids respectively and inserted into the KpnI site of Bari1. Helper plasmids were the same used for the subcellular localization of the transposase.
Cells were co-transfected with a donor plasmid and a helper plasmid expressing the full-length transposase. Two days after the transfection the medium was supplemented with the selective agent (blasticidin or G418) and cultured for two to three weeks.
For the excision assay, cells were co-transfected as described above and two to five days after transfection plasmid DNA was extracted from the transfected cells using the modified Hirt method [66]. Excision_FOR/excision_REV and M13for/M13rev oligonucleotides were used to detect “empty” donor plasmid molecules.
In silico Methods
Pairwise alignments were performed using either the NCBI online tools or the LALIGN tool (http://embnet.vital-it.ch/software/LALIGN_form.html).
Multiple alignments were performed using the Multalin tool (http://multalin.toulouse.inra.fr/) [67]. Protein secondary structures predictions were performed using the PhD secondary structure prediction method (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_phd.html) [68]. Sequences used for construction of the multiple alignment in figure 1 were retrieved from the Repbase database (www.girinst.org) [69].
NLS predictions were performed using the PSORT program (http://psort.hgc.jp/) [70].
RNA secondary structures were predicted using the RNAfold web service tool (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
Supporting Information
Purification of the Bari1 transposase protein. Panel A. PAGE of several protein extract from different purification steps of the T16 protein. BL21: total BL21 cell lysate not induced; T16: total cell lysate from transformed BL21 cells without IPTG induction; T16*: total cell lysate from transformed BL21 cells after induction with 1 mM IPTG 37°C; T16*S: soluble fraction from T16*. The red arrow indicates the induced protein; T16*P: insoluble fraction from T16*; M: molecular weight marker. The 50 and 40 KD bands of the marker are indicated. Panel B. Western blotting with the anti 6HIS/GLY antibody specifically recognizes the induced T16 protein. Panel C. PAGE of several protein extract from different purification steps of the C9 protein. M: molecular weight marker (the 30, 25 and 20 KD bands are indicated); BL21-NI: total BL21 cell lysate not induced; BL21-I: total cell lysate after induction with 1 mM IPTG 37°C; C9-NI: total cell lysate from transformed BL21 cells without IPTG induction; C9-I: total cell lysate from transformed BL21 cells after induction with 1 mM IPTG 37°C. The red arrow indicates the induced protein.
(PPTX)
Alignment of the processed transcripts identified in this study to the reference sequences.
(DOCX)
The genome of hsp83scratch mutant does not contain defective copies of Bari1 . M – λ/Eco-Hind molecular weight marker; Lane 1 - PCR product from Oregon-R DNA; Lane 2 - PCR product from hsp83scratch homozygous flies. Arrows indicates fragments of the expected size (about 1200 bp). Primers used are the same used in the RT-PCR experiments and described in the Methods section.
(TIF)
List of the oligonucleotides used in this study.
(DOCX)
Acknowledgments
We wish to thank to Prof. Paolo Barsanti and Dr. Konstantinos Lefkimmiatis for critical reading of the manuscript and useful suggestions.
Funding Statement
This work was carried out despite the strong and indiscriminate cut to Italian public research. This work was carried out using residual material from previously approved projects in part from Università degli Studi di Bari and in part from Ministero dell'Istruzione e Ricerca. VS acknowledges the grant “Futuro in ricerca 2010” no. RBFR10V8K6. RMM was formerly supported by an EMBO short term fellowship ASTF 182–2004. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yuan YW, Wessler SR (2011) The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci U S A 108: 7884–7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shao H, Tu Z (2001) Expanding the diversity of the IS630-Tc1-mariner superfamily: discovery of a unique DD37E transposon and reclassification of the DD37D and DD39D transposons. Genetics 159: 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lampe DJ, Churchill ME, Robertson HM (1996) A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J 15: 5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 4. Plasterk RH (1996) The Tc1/mariner transposon family. Curr Top Microbiol Immunol 204: 125–143. [DOI] [PubMed] [Google Scholar]
- 5. Auge-Gouillou C, Hamelin MH, Demattei MV, Periquet G, Bigot Y (2001) The ITR binding domain of the Mariner Mos-1 transposase. Mol Genet Genomics 265: 58–65. [DOI] [PubMed] [Google Scholar]
- 6. Lampe DJ, Grant TE, Robertson HM (1998) Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149: 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Merriman PJ, Grimes CD, Ambroziak J, Hackett DA, Skinner P, et al. (1995) S elements: a family of Tc1-like transposons in the genome of Drosophila melanogaster. Genetics 141: 1425–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Franz G, Savakis C (1991) Minos, a new transposable element from Drosophila hydei, is a member of the Tc1-like family of transposons. Nucleic Acids Res 19: 6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petrov DA, Schutzman JL, Hartl DL, Lozovskaya ER (1995) Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci U S A 92: 8050–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ivics Z, Hackett PB, Plasterk RH, Izsvak Z (1997) Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91: 501–510. [DOI] [PubMed] [Google Scholar]
- 11. Brillet B, Bigot Y, Auge-Gouillou C (2007) Assembly of the Tc1 and mariner transposition initiation complexes depends on the origins of their transposase DNA binding domains. Genetica 130: 105–120. [DOI] [PubMed] [Google Scholar]
- 12. Ivics Z, Izsvak Z, Minter A, Hackett PB (1996) Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci U S A 93: 5008–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Izsvak Z, Khare D, Behlke J, Heinemann U, Plasterk RH, et al. (2002) Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem 277: 34581–34588. [DOI] [PubMed] [Google Scholar]
- 14. Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB (2002) Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol 318: 1221–1235. [DOI] [PubMed] [Google Scholar]
- 15. Fischer SE, van Luenen HG, Plasterk RH (1999) Cis requirements for transposition of Tc1-like transposons in C. elegans. Mol Gen Genet 262: 268–274. [DOI] [PubMed] [Google Scholar]
- 16. Moschetti R, Chlamydas S, Marsano RM, Caizzi R (2008) Conserved motifs and dynamic aspects of the terminal inverted repeat organization within Bari-like transposons. Mol Genet Genomics 279: 451–461. [DOI] [PubMed] [Google Scholar]
- 17. Moschetti R, Caggese C, Barsanti P, Caizzi R (1998) Intra- and interspecies variation among Bari-1 elements of the melanogaster species group. Genetics 150: 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caizzi R, Caggese C, Pimpinelli S (1993) Bari-1, a new transposon-like family in Drosophila melanogaster with a unique heterochromatic organization. Genetics 133: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marsano RM, Milano R, Minervini C, Moschetti R, Caggese C, et al. (2003) Organization and possible origin of the Bari-1 cluster in the heterochromatic h39 region of Drosophila melanogaster. Genetica 117: 281–289. [DOI] [PubMed] [Google Scholar]
- 20. Marsano RM, Moschetti R, Barsanti P, Caggese C, Caizzi R (2003) A survey of the DNA sequences surrounding the Bari1 repeats in the pericentromeric h39 region of Drosophila melanogaster. Gene 307: 167–174. [DOI] [PubMed] [Google Scholar]
- 21. Caggese C, Pimpinelli S, Barsanti P, Caizzi R (1995) The distribution of the transposable element Bari-1 in the Drosophila melanogaster and Drosophila simulans genomes. Genetica 96: 269–283. [DOI] [PubMed] [Google Scholar]
- 22. Junakovic N, Di Franco C, Terrinoni A (1997) Evidence for a host role in regulating the activity of transposable elements in Drosophila melanogaster: the case of the persistent instability of Bari 1 elements in Charolles stock. Genetica 100: 149–154. [PubMed] [Google Scholar]
- 23. Marsano RM, Caizzi R, Moschetti R, Junakovic N (2005) Evidence for a functional interaction between the Bari1 transposable element and the cytochrome P450 cyp12a4 gene in Drosophila melanogaster. Gene 357: 122–128. [DOI] [PubMed] [Google Scholar]
- 24. Specchia V, Piacentini L, Tritto P, Fanti L, D'Alessandro R, et al. (2010) Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 463: 662–665. [DOI] [PubMed] [Google Scholar]
- 25. Roche SE, Schiff M, Rio DC (1995) P-element repressor autoregulation involves germ-line transcriptional repression and reduction of third intron splicing. Genes Dev 9: 1278–1288. [DOI] [PubMed] [Google Scholar]
- 26. Chiu YL, Greene WC (2008) The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol 26: 317–353. [DOI] [PubMed] [Google Scholar]
- 27. Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13: 335–340. [DOI] [PubMed] [Google Scholar]
- 28. Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285. [DOI] [PubMed] [Google Scholar]
- 29. Osborne BI, Baker B (1995) Movers and shakers: maize transposons as tools for analyzing other plant genomes. Curr Opin Cell Biol 7: 406–413. [DOI] [PubMed] [Google Scholar]
- 30. Fraser MJ, Brusca JS, Smith GE, Summers MD (1985) Transposon-mediated mutagenesis of a baculovirus. Virology 145: 356–361. [DOI] [PubMed] [Google Scholar]
- 31. Clark KJ, Geurts AM, Bell JB, Hackett PB (2004) Transposon vectors for gene-trap insertional mutagenesis in vertebrates. Genesis 39: 225–233. [DOI] [PubMed] [Google Scholar]
- 32. Ivics Z, Izsvak Z (2006) Transposons for gene therapy! Curr Gene Ther. 6: 593–607. [DOI] [PubMed] [Google Scholar]
- 33. Aronovich EL, Bell JB, Belur LR, Gunther R, Koniar B, et al. (2007) Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: implications for non-viral gene therapy of mucopolysaccharidoses. J Gene Med 9: 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawakami K (2005) Transposon tools and methods in zebrafish. Dev Dyn 234: 244–254. [DOI] [PubMed] [Google Scholar]
- 35. Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, et al. (2009) Transposon-mediated genome manipulation in vertebrates. Nat Methods 6: 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Plasterk RH, Izsvak Z, Ivics Z (1999) Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet 15: 326–332. [DOI] [PubMed] [Google Scholar]
- 37. Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, et al. (1994) Homeodomain-DNA recognition. Cell 78: 211–223. [DOI] [PubMed] [Google Scholar]
- 38. Colloms SD, van Luenen HG, Plasterk RH (1994) DNA binding activities of the Caenorhabditis elegans Tc3 transposase. Nucleic Acids Res 22: 5548–5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang SH, Elgin SC (2011) Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci U S A 108: 21164–21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, et al. (2011) Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development 138: 4039–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reese MG, Eeckman FH, Kulp D, Haussler D (1997) Improved splice site detection in Genie. J Comput Biol 4: 311–323. [DOI] [PubMed] [Google Scholar]
- 42. Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL (2008) The Vienna RNA websuite. Nucleic Acids Res 36: W70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lohe AR, Moriyama EN, Lidholm DA, Hartl DL (1995) Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol Biol Evol 12: 62–72. [DOI] [PubMed] [Google Scholar]
- 44. Claeys Bouuaert C, Chalmers R (2013) Hsmar1 transposition is sensitive to the topology of the transposon donor and the target. PLoS One 8: e53690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- 46. Ivics Z, Izsvak Z The expanding universe of transposon technologies for gene and cell engineering. Mob DNA 1: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nair R, Carter P, Rost B (2003) NLSdb: database of nuclear localization signals. Nucleic Acids Res 31: 397–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boehm U, Heinlein M, Behrens U, Kunze R (1995) One of three nuclear localization signals of maize Activator (Ac) transposase overlaps the DNA-binding domain. Plant J 7: 441–451. [DOI] [PubMed] [Google Scholar]
- 49. Ono A, Kim SH, Walbot V (2002) Subcellular localization of MURA and MURB proteins encoded by the maize MuDR transposon. Plant Mol Biol 50: 599–611. [DOI] [PubMed] [Google Scholar]
- 50. Liu D, Crawford NM (1998) Characterization of the putative transposase mRNA of Tag1, which is ubiquitously expressed in Arabidopsis and can be induced by Agrobacterium-mediated transformation with dTag1 DNA. Genetics 149: 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lohe AR, De Aguiar D, Hartl DL (1997) Mutations in the mariner transposase: the D,D(35)E consensus sequence is nonfunctional. Proc Natl Acad Sci U S A 94: 1293–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mikitani K, Sugasaki T, Shimada T, Kobayashi M, Gustafsson JA (2000) The chitinase gene of the silkworm, Bombyx mori, contains a novel Tc-like transposable element. J Biol Chem 275: 37725–37732. [DOI] [PubMed] [Google Scholar]
- 53.Craig NL (2002) Mobile DNA II. Washington, D.C.: ASM Press. xviii, 1204 p., [1232] p. of col. plates p.
- 54. Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A (1995) Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139: 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A (2007) The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175: 1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Desset S, Buchon N, Meignin C, Coiffet M, Vaury C (2008) In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS One 3: e1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kawamura Y, Saito K, Kin T, Ono Y, Asai K, et al. (2008) Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453: 793–797. [DOI] [PubMed] [Google Scholar]
- 58. Yasuhara JC, Wakimoto BT (2008) Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin-heterochromatin transition zones. PLoS Genet 4: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Graveley BR (2005) Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 123: 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Singh NN, Singh RN, Androphy EJ (2007) Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res 35: 371–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bernales S, Papa FR, Walter P (2006) Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 22: 487–508. [DOI] [PubMed] [Google Scholar]
- 62. Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891. [DOI] [PubMed] [Google Scholar]
- 63. Kawahara T, Yanagi H, Yura T, Mori K (1997) Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol Biol Cell 8: 1845–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Souid S, Lepesant JA, Yanicostas C (2007) The xbp-1 gene is essential for development in Drosophila. Dev Genes Evol 217: 159–167. [DOI] [PubMed] [Google Scholar]
- 65. Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96. [DOI] [PubMed] [Google Scholar]
- 66. Arad U (1998) Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. Biotechniques 24: 760–762. [DOI] [PubMed] [Google Scholar]
- 67. Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rost B, Sander C (1993) Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol 232: 584–599. [DOI] [PubMed] [Google Scholar]
- 69. Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, et al. (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467. [DOI] [PubMed] [Google Scholar]
- 70. Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purification of the Bari1 transposase protein. Panel A. PAGE of several protein extract from different purification steps of the T16 protein. BL21: total BL21 cell lysate not induced; T16: total cell lysate from transformed BL21 cells without IPTG induction; T16*: total cell lysate from transformed BL21 cells after induction with 1 mM IPTG 37°C; T16*S: soluble fraction from T16*. The red arrow indicates the induced protein; T16*P: insoluble fraction from T16*; M: molecular weight marker. The 50 and 40 KD bands of the marker are indicated. Panel B. Western blotting with the anti 6HIS/GLY antibody specifically recognizes the induced T16 protein. Panel C. PAGE of several protein extract from different purification steps of the C9 protein. M: molecular weight marker (the 30, 25 and 20 KD bands are indicated); BL21-NI: total BL21 cell lysate not induced; BL21-I: total cell lysate after induction with 1 mM IPTG 37°C; C9-NI: total cell lysate from transformed BL21 cells without IPTG induction; C9-I: total cell lysate from transformed BL21 cells after induction with 1 mM IPTG 37°C. The red arrow indicates the induced protein.
(PPTX)
Alignment of the processed transcripts identified in this study to the reference sequences.
(DOCX)
The genome of hsp83scratch mutant does not contain defective copies of Bari1 . M – λ/Eco-Hind molecular weight marker; Lane 1 - PCR product from Oregon-R DNA; Lane 2 - PCR product from hsp83scratch homozygous flies. Arrows indicates fragments of the expected size (about 1200 bp). Primers used are the same used in the RT-PCR experiments and described in the Methods section.
(TIF)
List of the oligonucleotides used in this study.
(DOCX)