Abstract
N-α-terminal acetylation is one of the most common, but least understood modifications of eukaryotic proteins. Although a high degree of conservation exists between the N-α-terminal acetylomes of plants and animals, very little information is available on this modification in plants. In yeast and humans, N-α-acetyltransferase complexes include a single catalytic subunit and one or two auxiliary subunits. Here, we report the positional cloning of TRANSCURVATA2 (TCU2), which encodes the auxiliary subunit of the NatB N-α-acetyltransferase complex in Arabidopsis. The phenotypes of loss-of-function tcu2 alleles indicate that NatB complex activity is required for flowering time regulation and for leaf, inflorescence, flower, fruit and embryonic development. In double mutants, tcu2 alleles synergistically interact with alleles of ARGONAUTE10, which encodes a component of the microRNA machinery. In summary, NatB-mediated N-α-terminal acetylation of proteins is pleiotropically required for Arabidopsis development and seems to be functionally related to the microRNA pathway.
Introduction
One of the most common modifications of eukaryotic proteins is N-α-acetylation of the amino terminus, which is found in 50–70% of yeast proteins and 70–90% of human proteins, but occurs only rarely in prokaryotic proteins (reviewed in [1]). N-α-acetylation occurs in the cytoplasm and is carried out by N-α-acetyltransferases, which transfer an acetyl group from acetyl-CoA to the α-amino group of the first amino acid of a protein. Unlike other protein modifications, N-α-terminal acetylation is irreversible and, although it was first described over 50 years ago [2], little is known about its function.
In yeast and mammals, N-α-terminal acetylation involves several complexes, termed NatA to NatF, which exhibit substrate specificity (reviewed in [1]). The NatA, NatB and NatC complexes contain a catalytic subunit and one or two auxiliary, non-catalytic subunits. The Nat3 (N-acetyltransferase 3) and Mdm20 (mitochondrial morphology and distribution 20) subunits of the yeast and human NatB complex cosediment with ribosomes, which suggests that they act on nascent polypeptides [3]–[5]. A recently revised nomenclature for N-terminal acetyltransferases [6] renamed the Nat3 and Mdm20 proteins as Naa20 and Naa25, respectively. The human NatB complex seems to be required for cell cycle progression [5]. The Ard1 (Naa10) catalytic subunit of the human NatA complex has been isolated in cytoplasmic and nuclear fractions, which suggests additional functions for Ard1, other than cotranslational N-α-terminal acetylation [7]. Recent work showed that the X-linked Ogden syndrome is caused by a specific missense mutation affecting the Naa10 catalytic subunit of the human NatA complex [8].
Several studies in yeast and mammals have shown that N-α-terminal acetylation has diverse molecular functions including inhibition of protein translocation to endoplasmic reticulum [9], regulation of protein degradation [10] and mediation of protein interactions [11]. It has been believed for many years that N-α-terminal acetylation protects proteins from degradation [12], [13]. In fact, proteins with N-α-terminal acetylation are more stable in vivo than non-acetylated proteins [14]. This could be due to ubiquitination, which involves direct conjugation of ubiquitin to the N-terminal amino acid and promotes the subsequent degradation of ubiquitinated proteins [15]. Thus, acetylation of the amino terminus would prevent ubiquitination and stabilize proteins [16]. This occurs, for example, with the p16 and p14/p19ARF tumor suppressor proteins in human and mouse cells [15], [17], [18]. However, an acetylated amino terminus did not affect the destabilization of a protein by ubiquitin-independent mechanisms [19]. In fact, it has been proposed that, in some cases, acetylated amino terminal sequences may act as signals for degradation [10], [20]. At least eight yeast proteins that are acetylated at their amino termini are recognized by the Doa10 ubiquitin ligase [10], [16], [21]. It has been shown recently in yeast that NatB-mediated acetylation does not alter the stability of proteins [22]. Although these observations suggest opposite roles for N-α-terminal acetylation, several mechanisms could coexist, each acting on specific groups of proteins under certain conditions.
N-α-terminal acetylation has different functions in different proteins, as shown by studies performed in yeast [16]: actin and tropomyosin activity requires NatB-mediated N-α-terminal acetylation [3], [23], [24]; Tfs1p (Twenty-five suppressor) lipid binding protein requires N-terminal acetylation by NatB to inhibit carboxypeptidase Y [25]; delivery of the Arl3p (Arf-related protein 3) and Grh1p (GRASP65 protein homologue 1) GTPases to the Golgi apparatus requires NatC-mediated acetylation [26]–[28]; association of Trm1p-II (Transcriptional regulation of methanol induction 1 protein II) with the inner nuclear membrane also requires acetylation [29]; and Sir3p (Silent information regulator 3 protein) and Orc1p (Origin recognition complex 1 protein) activities in gene silencing require acetylation by NatA [30], [31]. However, the mechanisms by which acetylation affects protein function remain unclear. For example, a study of several NatB substrates led to the conclusion that the absence of acetylation has no apparent impact on their subcellular localization [32].
There is a dearth of information on the activity of Nat complexes in plants. Mutant phenotypes caused by genes encoding components of the Nat complexes have been identified for only one gene, that encoding the MAK3 (NAA30) catalytic subunit of NatC. The atmak3-1 mutation was isolated in a search for insertional mutants with reduced performance of photosystem II and exhibits a smaller rosette, paler leaves and less thylakoidal multiprotein complexes than wild type [33]. The Mak3p (Naa30p), Mak10p (Naa35p) and Mak31p (Naa38p) components of yeast NatC are essential [34], but null alleles of the putative Arabidopsis ortholog of yeast MAK10 do not cause any visible mutant phenotype. In Arabidopsis, NatC-mediated N-α-terminal acetylation of certain chloroplast protein precursors seems to be necessary for the accumulation of mature proteins in this organelle [33]. A recent proteome-wide mass spectrometry analysis that allowed enrichment and selection of N-terminal peptides from Arabidopsis and human samples identified hundreds of N-α-acetylated proteins; the results indicated a high degree of convergence between the plant and animal kingdoms in both components and substrates of the N-α-acetylation machinery. An unknown mechanism seems to be responsible for N-α-acetylation (after removal of the transit peptide) of a high proportion of chloroplast proteins encoded by Arabidopsis nuclear genes [35].
Here, we report the positional cloning and characterization of Arabidopsis TRANSCURVATA2 (TCU2), which encodes the auxiliary subunit of the NatB complex. The pleiotropic phenotypes caused by loss of function of TCU2 indicate that NatB-mediated N-α-terminal acetylation plays a role in leaf flatness, flowering time, inflorescence structure, flower size, silique structure and size, and embryonic development. In addition, genetic interactions between TCU2 alleles and alleles of ARGONAUTE10 (AGO10) suggest an unexpected link between N-α-terminal acetylation and the microRNA pathway.
Results
Morphological and histological phenotypes of the P14 6.3 mutant line
A screen for ethylmethane sulfonate (EMS)-induced leaf mutants in a Ler background, conducted in the laboratory of J.L. Micol [36], identified two lines (P14 6.3 and P14 20.1) with indistinguishable phenotypes, vegetative leaves that bend down obliquely to the primary vein. We tested for monogenic inheritance in the F2 of their backcrosses to Ler using a chi-square test with the Yates correction for one degree of freedom. These mutant lines exhibit monogenic recessive inheritance, as shown by the 3∶1 phenotypic segregation ratios found for P14 6.3 (78 wild-type plants: 15 mutant plants; chi-square = 3.44) and P14 20.1 (79∶19; chi-square = 1.36). Since these non-complementing lines had been isolated from the same parental group (see Methods), they were considered likely to be identical, and only the P14 6.3 line (Figure 1B) was further studied.
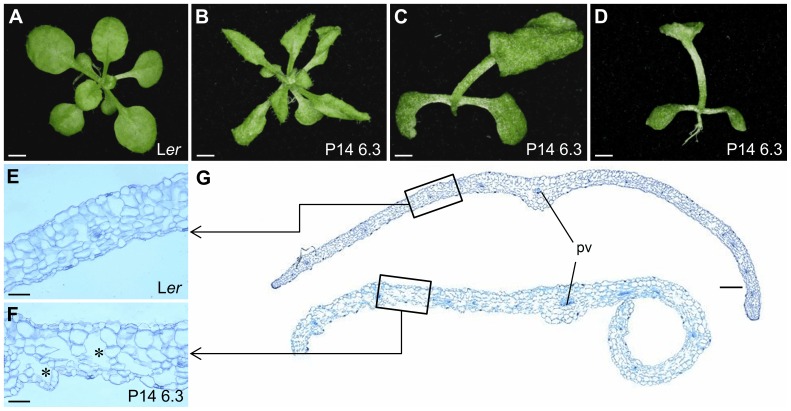
Figure 1. Leaf morphology and histology in the P14 6.3 line.
(A, B) Rosettes of (A) Ler and (B) P14 6.3 plants. (C) Top and (D) lateral views of a P14 6.3 plant showing fusion of the first pair of vegetative leaves. (E, F) Transverse sections of the central region of the lamina, midway between the leaf margin and the primary vein of (E) Ler and (F) P14 6.3 third-node leaves. Asterisks indicate large intercellular air spaces. (G) Margin to margin transverse sections of third-node leaves. pv: primary vein. Pictures were taken (A–D) 22 das and (E–G) 21 das. Scale bars: (A, B, D) 2 mm, (C) 1 mm, (E, F) 50 µm and (G) 200 µm.
The vegetative leaves of P14 6.3 are reticulated with protuberant veins that are greener than the interveinal tissues. Around 10% of P14 6.3 plants (Figure 1C, D) showed only one large leaf with a duplicated primary vein, fusion of the first two leaves, or a single, trumpet-shaped leaf apparently abaxialized; these aberrant seedlings are viable and develop into fertile plants. This phenotype, which is likely a consequence of perturbation of shoot apical meristem function, was evident in around 10% of the progeny resulting from selfing of any P14 6.3 plant, regardless of the parental phenotype. Plants with this phenotype were not examined in the phenotypic analyses described below.
Vegetative leaves are smaller in P14 6.3 than in wild type, except for the first pair, which are similar to those of Ler. To study their morphology and histology, first- and third-node P14 6.3 leaves were collected 21 days after stratification (das) and cleared with chloral hydrate. Dark-field micrographs were used to draw venation pattern diagrams (Figure S1), which were subjected to morphometric analysis. Only small differences in venation pattern were found with the wild type, except for the duplicated primary vein shown by the fused first pair of leaves occasionally seen in P14 6.3 seedlings.
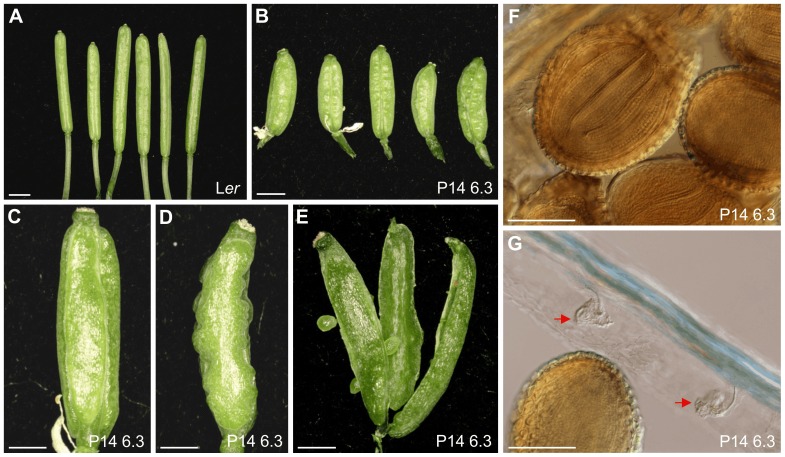
Optical microscopy of leaf cross-sections (Figure 1E-G) showed that there is more heterogeneity in mesophyll cell and airspace sizes in P14 6.3 than in wild type. No differences with the wild type were observed in root apex structure, root hair number and root length, although root hairs are longer in P14 6.3 than in Ler. In P14 6.3, the stem is shorter than wild type (Figure 2A), is fasciated and winding, and has some lateral filiform bumps and aborted flowers (Figure 2B, C). In Ler, flowers appear clustered and separate from each other when siliques begin to elongate, but in P14 6.3 they separate from the cluster before silique formation (Figure 2D). Floral organs of P14 6.3 are shorter, particularly the petals (Figure 2E-H). Mutant siliques are short, thick and rugged compared to those of Ler, contain many aborted or unfertilized ovules and almost all exhibit three or even four valves instead of the two usually seen in wild type (Figure 3).
Figure 2. Inflorescence structure and floral morphology in P14 6.3.
(A) Lateral view of Ler and P14 6.3 inflorescences. (B, C) Details of abnormal structures observed in P14 6.3 stems: (B) a filamentous bulge, and (C) an aborted lateral flower. (D) Inflorescence apex in Ler and P14 6.3 plants. The red arrow indicates an aborted lateral flower. (E, F) Flowers of the inflorescence apex from (E) Ler and (F) P14 6.3. (G, H) Different sizes of (G) flowers from the inflorescence apex and (H) petals from Ler and P14 6.3. Pictures were taken 35 das. Scale bars: (A) 2 cm, (B, C) 0.5 mm, (D–F) 2 mm and (G, H) 1 mm.
Figure 3. Silique morphology in P14 6.3.
(A, B) Siliques from (A) Ler and (B) P14 6.3 plants. (C–E) Three valves with a rough surface were shown in most P14 6.3 siliques: (C) front and (D) lateral views of closed siliques and (E) a silique with longitudinally open septa. (F) Seeds containing mature embryos and (G) aborted or unfertilized ovules (red arrows) in dissected P14 6.3 siliques. Pictures were taken 35 das. Scale bars: (A, B) 2 mm, (C–E) 1 mm and (F, G) 200 µm.
Positional cloning of the TCU2 gene
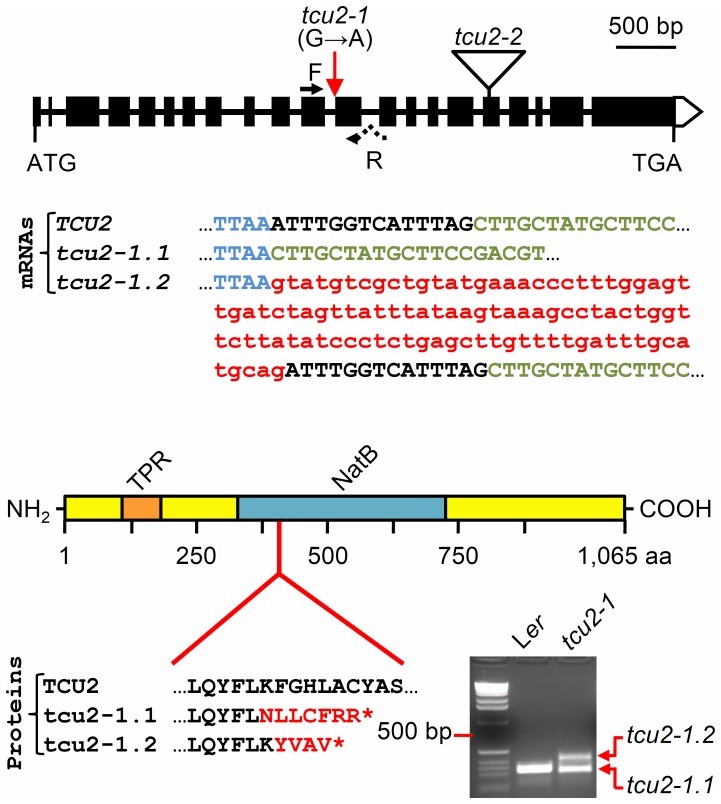
We termed the causal mutation for the phenotype of the P14 6.3 line transcurvata2-1 (tcu2-1). Low-resolution mapping of the tcu2-1 mutation had already been carried out by linkage analysis [37]. We developed new molecular markers (Table S1), most of them based on the small insertions or deletions (In/Del) described in the Monsanto database (http://www.arabidopsis.org/browse/Cereon/index.jsp) and used them for the analysis of a mapping population of 1,137 F2 plants derived from a P14 6.3×Col-0 cross. This allowed us to define a 176-kb candidate interval encompassing 52 annotated genes (Figure S2). To identify TCU2, we first obtained 81 lines carrying T-DNA insertions in genes in the candidate interval; however, none of the lines exhibited the same mutant phenotype as tcu2-1. However, sequencing of 28 of the candidate genes in the TCU2 region revealed a G→A transition in the last nucleotide of the eleventh intron of At5g58450. This mutation damages a splicing acceptor site, generating two aberrant mRNA variants, one of which (tcu2-1.2) retains intron eleven of At5g58450, while the other (tcu2-1.1) lacks the first 14 nucleotides of exon twelve. We demonstrated these alterations by sequencing PCR amplification products obtained from reverse transcribed tcu2-1 mRNA (Figure 4).
Figure 4. Structure of the TCU2 gene, its mutant alleles and their effects on the TCU2 protein.
Blue and green capital letters represent nucleotides of exons eleven and twelve, respectively, of the TCU2 gene, which are present in TCU2 wild-type mRNA and the tcu2-1.1 and tcu2-1.2 mutant mRNA variants. Red lowercase letters highlight the intron that is not removed from tcu2-1.2 during splicing; black capital letters indicate the nucleotides from the 5′ end of wild type exon twelve that are removed from tcu2-1.1 during splicing. The TCU2 protein contains several tetratricopeptide repetitions (TPR; residues 89 to 181) and a NatB domain (residues 365 to 725). The mutant proteins have 436 (tcu2-1.1) and 434 (tcu2-1.2) amino acids. A picture of a 3%-agarose gel stained with ethidium bromide is shown at the lower right corner, which shows the PCR amplification products obtained from reverse transcribed total mRNA extracted from Ler and P14 6.3 whole plants.
P14 6.3 is a tcu2-1 zll-2 double mutant
We found several publicly available lines carrying T-DNA insertions in At5g58450 in the Col-0 genetic background, but only one line (GK_819_A05) exhibited a mutant phenotype. The corresponding allele of TCU2 was termed tcu2-2 (Figure 4). As observed in P14 6.3, plants homozygous for the tcu2-2 allele showed reticulated leaves. However, the strong leaf folding that characterizes P14 6.3 plants was mild in tcu2-2 plants. Both reticulation and weak folding were similar in the leaves of tcu2-2 homozygous plants and the tcu2-1/tcu2-2 heterozygous F1 progeny of a P14 6.3×tcu2-2 cross (Figure 5A, B). We concluded that these two lines are allelic and that some phenotypic traits of tcu2-1 are influenced by a recessive modifier present in the Ler background of P14 6.3 but absent in the Col-0 background of tcu2-2.
Figure 5. Some phenotypic traits of the tcu2-2 mutant.
(A–C) Rosettes of (A) tcu2-1/tcu2-1, (B) tcu2-2/tcu2-2 and (C) tcu2-1/tcu2-2 plants. (D) Siliques from tcu2-2/tcu2-2 (left) and Col-0 (right) plants. (E) tcu2-1/tcu2-2 (left) and wild-type sibling (right) siliques. Only three-valved siliques are shown from tcu2-2/tcu2-2 and tcu2-1/tcu2-2 in D and E; the wild-type siliques shown have two valves. (F) Detail of a tcu2-2/tcu2-2 silique with four valves. Pictures were taken (A, C) 21 das, (B) 18 das and (D-F) 50 das. Scale bars: (A–E) 2 mm and (F) 1 mm.
Four phenotypic traits that the P14 6.3 and tcu2-2 lines do not share are: fusion of the first two leaves (in about 10% of P14 6.3 plants but never seen in tcu2-2); reduced silique length (only in P14 6.3); three or more valves (in almost all P14 6.3 siliques but only in some tcu2-2 siliques) and reduced plant stature (small in P14 6.3 but similar to wild type in tcu2-2). The first three of these traits can be caused in a Ler genetic background by loss-of function alleles of AGO10 (ARGONAUTE10; also called PINHEAD [PNH] and ZWILLE [ZLL]), a known component of the microRNA (miRNA) machinery [38]–[40]. AGO10 and TCU2 are linked at a distance of about 30 cM. Taken together, these observations prompted us to obtain a mutant carrying the pnh-2 loss-of-function allele of AGO10, in a Ler genetic background. The pnh-2 mutant exhibited the expected, previously published phenotype [41], [42] under our growth conditions (Figure S3): around 10% of the pnh-2 plants showed fusion of the first two leaves, and all plants exhibited small flowers, which developed into thick siliques, most of which had three valves, containing some aborted or unfertilized ovules. These phenotypic traits were less severe in pnh-2 than in P14 6.3.
Sequencing of AGO10 in the P14 6.3 and P14 20.1 mutant lines and their wild type Ler revealed a G→A transition in its exon twelve, which is predicted to cause a Gly707→Asp change in the AGO10 protein (Figure 6). Since an identical mutation has already been published and named zll-2 [43], we decided to use this symbol for the mutation found in AGO10 of P14 6.3 and P14 20.1. This result revealed that the latter lines carry two mutations: tcu2-1 in TCU2 and zll-2 in AGO10. Next-generation, whole genome sequencing of P14 6.3 DNA confirmed the presence of both tcu2-1 and zll-2 homozygous mutations, as well as the absence of any other G→A or C→T homozygous mutations (those typically caused by EMS) affecting either exons or splicing donor or acceptor signals in the genes encompassed by the 176-kb interval candidate delimited by linkage analysis (Figure S2). In addition, we obtained tcu2-2 pnh-2, a different double mutant combination of loss-of-function alleles of TCU2 and AGO10, whose phenotype was very similar to that of P14 6.3, the only exception being plant height, which was higher in tcu2-2 pnh-2 than in P14 6.3 (see below).
Figure 6. Structure of the AGO10 gene and its alleles studied in this work.

Exons and introns are shown as black rectangles and lines, respectively. White boxes represent the 5′ and 3′ untranslated regions. Arrows point to the position of the zll-2 and pnh-2 point mutations.
To study their individual phenotypic effects, we segregated the tcu2-1 and zll-2 mutations in the F2 progeny of a cross of the tcu2-1 zll-2 double mutant to Ler. The tcu2-1 single mutant exhibited leaf reticulation and mild leaf folding (Figure 5C), and only a few three-valved siliques (see below). By contrast, the zll-2 single mutant showed fusion of the first two leaves in about 10% of its seedlings and most of its siliques were three-valved. Flowering was earlier in the tcu2-1 zll-2 double mutant (22.64±0.21 das; n = 91) than in Ler (24.98±0.28 das; n = 90) (Figure S4). Early bolting is clearly caused by the tcu2-1 mutation, since flowering time was 22.8±0.57 das (n = 86) in the tcu2-1 single mutant and 24.05±0.04 das (n = 91) in the zll-2 single mutant. Because of their early flowering, adult tcu2-1 and tcu2-1 zll-2 plants had fewer vegetative leaves than wild type.
Only a few thick siliques with three or four valves (Figure 5D, E) were observed in heterozygous tcu2-1/tcu2-2, homozygous tcu2-2 (siliques with 3 or 4 valves/all siliques = 0.02±0.03; n = 5 plants; 55 das), or homozygous tcu2-1 (0.13±0.08) plants, but this phenotype was observed in an increased (ten-fold) proportion compared with their respective wild types (Col-0, 0.002±0.004; Ler, 0.01±0.01). The structure of the apical region of the inflorescence was also similar in the tcu2-1 and tcu2-2 single mutants and in the tcu2-1 zll-2 double mutant.
TCU2 encodes the auxiliary subunit of the NatB complex
The TCU2 gene is annotated to encode a protein of 1,065 aa and 12.15 kDa (Figure 4) containing several tetratricopeptide repetitions, which are assumed to confer a helical structure and to mediate binding to other proteins. TCU2 also contains a domain with similarity to Mdm20p (Naa25p), the auxiliary subunit of the N-α-terminal acetyltransferase B (NatB) complex of organisms as diverse as mammals and yeast, which participates in the N-α-terminal acetylation of proteins.
Three N-α-terminal acetyltransferase complexes, NatA, NatB and NatC, have been characterized in Saccharomyces cerevisiae, each of which includes a catalytic subunit and one or more auxiliary subunits [44]. Yeast Mdm20p is the best known auxiliary, non-catalytic subunit of the NatB complex, which N-α-terminally acetylates proteins having Met-Glu, Met-Asp or Met-Asn residues at the amino terminus [3], [44]. Like Mdm20p, TCU2 contains a TPR domain with multiple tandem tetratricopeptide repeats and a NatB_MDM20 non-catalytic domain (first identified in yeast Mdm20p [23]). TPR domains are composed of 3-16 degenerate tandem repeats of 34 amino acids that form structural domains in proteins that facilitate the assembly of large protein complexes (reviewed in [45]). The NatB_MDM20 domain, of unknown function, is very conserved and occupies half of the NatB auxiliary subunit in all organisms studied. TCU2 shows 18.70% identity and 40.30% similarity to Mdm20p. Overall similarity and identity percentages between TCU2 and several of its putative orthologues in higher plants were of 71.02 and 59.81 (castor oil plant), 72.45 and 61.04 (grapevine), 62.47 and 49.77 (rice), and 69.74 and 59.80 (poplar), respectively (Figure S5).
The two mutant mRNA variants produced by the tcu2-1 allele are translated into truncated proteins of 434 and 436 aa (Figure 4), which lack most of the NatB_MDM20 domain but still contain an intact protein-binding TPR domain. The NatB_MDM20 domain affected by the tcu2-1 mutation is highly conserved in both monocotyledoneous and dicotyledoneous plants. The tcu2-2 allele carries a T-DNA insertion in exon seventeen, disrupting the region that encodes the NatB_MDM20 domain (Figure 4).
Expression of the TCU2 gene and subcellular localization of the TCU2 protein
We determined the expression pattern of TCU2 in silico, according to the LRU Arabidopsis database Browser (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.Cgi; http://www. genevestigator.ethz.ch; [46]) (Figure S6). To experimentally assess the spatial expression pattern of TCU2, we generated a TCU2pro:GUS construct (see Methods), in which the TCU2 endogenous promoter drives expression of the β-glucuronidase (GUS; [47]) gene, and transferred this construct into plants. GUS activity was detected in all stages and tissues analyzed (Figure S7), and expression of the reporter gene was higher in the vascular bundles, hydathodes, leaf primordia and the base of the trichomes.
To assess the subcellular localization of the TCU2 protein, we generated a 35Spro:TCU2:GFP construct (see Methods) and transferred it into plants. The GFP fluorescent signal was concentrated in the cytoplasm of proliferating cells and absent from the nucleus (Figure S8).
Silencing of the TCU2 gene
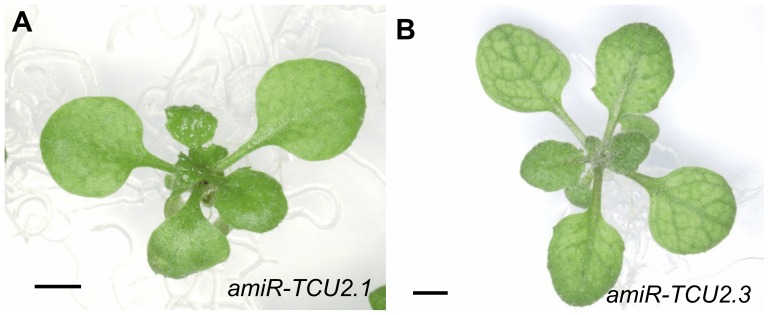
To further confirm the effects of TCU2 loss of function, we assessed the consequences of the post-transcriptional repression of this gene. A transgene designed to express amiR-TCU2, an artificial microRNA (amiRNA) targeting the At5g58450 gene, was constructed, cloned and transferred into Arabidopsis plants (see Methods). Among the 48 T1 transformants that we obtained, two showed in their T2 progeny the weak leaf folding shown by the tcu2 mutants (Figure 7). These amiRNA-producing transgenic lines also exhibited reticulated leaves and mild early flowering, similar to tcu2-1 and tcu2-2 (amiR-TCU2.1, 22.87±1.46 das; amiR-TCU2.3, 25.26±0.96; Col-0, 27.87±1.70; n = 30).
Figure 7. Rosette phenotype of amiR-TCU2 transgenic lines.
Pictures were taken 16 das. Scale bars: 2 mm.
Double mutant combinations of tcu2 alleles with nbc-1 and er-1
At1g03150 encodes the putative catalytic subunit of Arabidopsis NatB. To identify phenotypes shared by mutants affected in the non-catalytic and catalytic subunits of the NatB complex, we obtained the only available line (SAIL_323_B05) carrying a T-DNA insertional allele of At1g03150, which is in the Col-0 genetic background. We named this allele nbc-1 (for NatB catalytic subunit-1). It carries a T-DNA insertion that maps 25 bp upstream of nucleotide +1 of the transcription unit of At1g03150 and 85 bp upstream of nucleotide +1 of At1g03140, which are divergently transcribed.
Homozygous nbc-1 plants exhibited a mutant phenotype distinguishable from both tcu2-2 and Col-0. However, tcu2-2 and nbc-1 plants shared some traits, such as reticulated leaves, early flowering, increased proportion of siliques with more than two valves and aborted or unfertilized ovules in siliques (Figure S9). We also obtained tcu2-2 nbc-1 double mutant plants, which exhibited an additive overall phenotype, except that the number of siliques per plant, scored 55 das, was strongly reduced in the double mutant: 235.00±24.00 siliques in Col-0, 250.20±52.67 in tcu2-2, 255.00±96.10 in nbc-1, and 154.5±33.49 in tcu2-2 nbc-1. More than two valves were seen in 0.60±0.89 siliques in Col-0, 7.20±9.63 in tcu2-2, 5.60±5.59 in nbc-1, and 5.60±7.30 in tcu2-1 nbc-1.
Measurement of plant height revealed that tcu2-1, tcu2-2 and nbc-1 genetically interact with erecta-1 (er-1), the loss-of-function allele of the ERECTA gene carried by the Ler accession [48]. Plant height measured at 49 das was 39.10±6.18 cm in Ler (er-1), 50.28±4.89 cm in Col-0, 35.22±4.54 cm in tcu2-2, 7.05±2.23 cm in tcu2-2 er-1, 34.86±8.06 cm in nbc-1, and 15.75±7.08 cm in nbc-1 er-1 (n = 10 plants). The interaction between ER and TCU2 explains the difference in height between tcu2-2 pnh-2 and P14 6.3 (tcu2-1 zll-2 er-1). The reduction in inflorescence size caused by the interaction between tcu2 alleles and er-1 causes a reduction in silique number (239.40±64.01 siliques per plant in Ler, and 63.50±20.34 in P14 6.3; n = 5 plants; 59 das).
Discussion
TCU2 is the Arabidopsis ortholog of the human and yeast genes encoding the NatB non-catalytic subunit
Although many eukaryotic proteins are subjected to N-α-terminal acetylation, the function of this co- and post-translational modification is known only for a few proteins in a few species, Homo sapiens and Saccharomyces cerevisiae in particular. Knowledge of this process in the plant kingdom is particularly poor: only a few genes encoding components of the N-α-terminal acetylation machinery have been studied, and only one of these genes, ATMAK3, has been found to cause a visible phenotype when mutated. ATMAK3 encodes a cytoplasmic protein, the Arabidopsis ortholog of the catalytic subunit of human and yeast NatC complex; the atmak3-1 mutant exhibits slightly pale leaves and reduced photosynthesis and growth [33].
We describe here an EMS-induced Arabidopsis mutant line that we initially named P14 6.3, which was found to carry the tcu2-1 and zll-2 point, loss-of-function alleles of the linked TCU2 and AGO10 genes, respectively. We positionally cloned TCU2, which encodes a protein homologous to the non-catalytic subunit of the NatB N-α-terminal acetylase complex of yeast and mammals, and identified tcu2-2, a T-DNA insertional allele of TCU2. We found that TCU2 is broadly expressed in plants, and that TCU2 protein is present in the cytoplasm. These are the expected spatial and subcellular localization patterns for NatB-mediated N-α-terminal acetylation enzymatic activity, which is known to occur in all animal tissues in the cytoplasm (reviewed in [1]).
Loss of function mutation of TCU2 causes a pleiotropic phenotype that includes leaf reticulation and weak leaf folding, early flowering, unfertilized or aborted ovules in siliques, and an increase in the proportion of siliques with more than two valves compared to wild type. These phenotypes were shown by homozygous tcu2-1 or tcu2-2 plants, the heterozygous F1 progeny of their intercrosses, and transgenic lines expressing an amiRNA targeting TCU2. These observations suggest that the non-catalytic subunit of the NatB N-α-terminal acetylase complex is required for N-α-terminal acetylation of proteins in Arabidopsis and that NatB-mediated N-α-terminal acetylation is required in Arabidopsis for vegetative and reproductive development, including leaf development, flowering time, flower and silique development, as well as gametogenesis and/or fertilization. These observations substantially expand the list of developmental processes for which N-α-terminal acetylation in general, and that mediated by NatB in particular, is known to be required in Arabidopsis.
We also studied the nbc-1 mutant, which carries a loss-of-function allele of the gene encoding the putative catalytic subunit of NatB. The tcu2 and nbc-1 mutants showed reticulated leaves, early flowering, increased proportion of siliques with more than two valves, and aborted or unfertilized ovules in siliques. These shared traits clearly indicate that the NBC and TCU2 proteins participate in the same processes, as can be expected for the subunits of a complex. Further studies of the nbc-1 and tcu2 mutants will be required to explain the differences between their phenotypes and will provide useful insights into the N-α-terminal acetylome in plants.
TCU2 is required for vegetative and reproductive development
Several large-scale analyses of the N-α-acetylome have been recently conducted; these studies aimed to answer the many open questions on the functional role of Nat complexes in Saccharomyces cerevisiae [49], [50], Homo sapiens [35], [50]-[52], Drosophila melanogaster [53], Arabidopsis thaliana [35], Halobacterium salinarum and Natronomonas pharaonis [54], [55]. Although these studies provide a much better overview of the process at the proteome scale, gene-centric analyses are lacking, at least in the plant kingdom. Given that the TCU2 protein is highly conserved among higher plants, the tcu2 alleles that we describe here will inform analyses of NatB function.
In the tcu2 mutants, leaves are reticulated, a phenotype that is usually caused by defective mesophyll development [56]. Cross sections showed an increase in mesophyll intercellular air spaces. In previously described reticulated mutants [56]-[60], leaf interveinal tissues contain fewer and smaller mesophyll cells than wild type. Plastid number, size and/or differentiation status is also altered in reticulated mutants, suggesting a perturbation in retrograde signaling (from the chloroplasts to the cell nucleus), which in turn affects mesophyll growth and development [61]. A limited supply of essential metabolites in early stages of leaf development has also been proposed as a cause of the defective mesophyll structure seen in reticulated mutants: early differentiating tissues, as the perivascular, bundle-sheath cells, would be able to properly differentiate, but differentiation of the remaining tissues would be arrested or retarded [62]. Metabolic pathways that take place at least in part within chloroplasts are impaired in Arabidopsis reticulated mutants, such as reticulata (re), venosa3 (ven3) and ven6, and chlorophyll a/b binding protein underexpressed (cue1). CUE1 encodes a phosphoenolpyruvate (PEP)/phosphate antiporter localized at the chloroplast inner membrane [63], and VEN3 and VEN6 encode the two subunits of carbamoyl phosphate synthetase (CPS), which is required in the chloroplasts for arginine biosynthesis [60].
Because of the known relationship between the reticulated phenotype and chloroplast dysfunction, a tempting hypothesis is that some processes related to chloroplast metabolism and/or biogenesis are impaired in the tcu2 mutants. It is worth mentioning here that a recently discovered kingdom-specific property of the Arabidopsis N-α-acetylome is the high prevalence of N-α-terminal acetylation of neo N-termini of nuclear-encoded proteins imported into the chloroplast. These proteins are processed to remove their chloroplast transit peptides during translocation to the chloroplast and seem to be N-α-terminal acetylated after removal of the transit peptide. Nothing is known on the mechanism underlying such post-translational modification, which was shared by 277 peptides of a total of 1054 found to be N-α-terminal acetylated [35].
Only a few mutations have been reported to increase the number of Arabidopsis silique valves, most of them exhibiting low penetrance. Several of these mutations seem to perturb the regulation of floral meristem function through the CLAVATA (CLV)/WUSCHEL (WUS) pathway. Mutations in the CLV genes produce fruits with extra valves (reviewed in [64], [65]). Also, FILAMENTOUS FLOWER (FIL) and YABBY3 (YAB3) negatively regulate CLV3 and WUS [66], and many of the fruits of fil yab3 double mutants have three valves [67]. The CLV genes encode regulatory peptides that are known to have different modifications, one of which might be N-α-terminal acetylation. In fact, the amino terminus of CLV3 is Met-Asp, one of the canonical NatB substrates.
TCU2 alleles synergistically interact with alleles of AGO10 and ER
microRNAs (miRNAs) participate in the regulation of many aspects of plant development [68]-[70], including leaf [71], [72] and fruit [73] development. RNA-induced silencing complexes (RISC) mediate the action of miRNAs on their target mRNAs. A key RISC component in Arabidopsis is AGO1, and the closest AGO1 paralog in Arabidopsis is AGO10 [74]. AGO10 was proposed to participate in the miRNA pathway causing translational repression of miRNA targets [75] and has recently been found to regulate AGO1 by sequestering miRNAs that down-regulate members of the HOMEODOMAIN-LEUCINE ZIPPER (HD-ZIP III) family of transcription factors [40]. Reduction of mRNA levels of HD-ZIP III genes leads to defects in shoot apical meristem functions, alteration of organ polarity and abnormal formation of carpels. AGO10 participates in the repression of APETALA2 (AP2), which promotes WUS activity [38].
We segregated the tcu2-1 and zll-2 mutations from the tcu2-1 zll-2 double mutant. We also obtained tcu2-2 pnh-2, another combination of loss-of-function alleles of TCU2 and AGO10. We considered some phenotypic traits of the tcu2-1 zll-2 double mutant to be additive. For example, mutant alleles of AGO10 cause fusion of the first two leaves, silique shortening and generalized formation of more than two valves in siliques; mutant alleles of TCU2 cause leaf reticulation, early flowering and an increase in the proportion of siliques with more than two valves. As mentioned above, alteration in the number of silique valves is a rare phenotype; the observation of a perturbation in this trait in the tcu2 and ago10 mutants, though at different penetrance levels, cannot be put down to chance and strongly suggests that both TCU2 and AGO10 participate in fruit development.
Comparison of the phenotypes of the tcu2-1, tcu2-2, zll-2 and pnh-2 single mutants and their double mutant combinations clearly indicates that the TCU2 and AGO10 genes interact. The strong leaf folding shown by the double mutants is clearly a synergistic phenotype, which is absent from the single mutants. Indeed, leaves are almost normal in the pnh-2 and zll-2 mutants, and only mildly bent in the tcu2 mutants. The leaf phenotype of the double mutants carrying mutant alleles of TCU2 and AGO10 is very similar to that of single mutants affected in AGO7, another member of the AGO family [76], when grown in our standard culture conditions (data not shown). Taken together, these observations suggest a functional relationship between TCU2 and AGO10, in leaf development in particular. Two simple hypotheses may explain these observations: the AGO10 protein itself, or any of the proteins regulated by AGO10, is acetylated by NatB in the wild type, or the expression of TCU2 is directly or indirectly regulated by the miRNA pathway. However, the N-terminus of the AGO10 protein does not show any of the canonical signals of NatB substrates, which makes unlikely its N-α-terminal acetylation by the NatB complex.
The genetic interactions that we found, of er-1 with tcu2 alleles and nbc-1, also indicate a functional relationship. The ER protein is assumed to be required for the regulation of plant size and belongs to a leucine-rich receptor-like serine/threonine kinase family of plant signaling receptors [48]. Further research will be required to shed light on the nature of the interactions of the tcu2 and nbc-1 mutations with er-1. These interactions, however, clearly suggest that Ler is a better choice than other commonly used wild-type accessions for studies of mutations affecting components of Nat complexes, since it provides a sensitized genetic background that renders a much better visualization of the effects of loss of function of components of the Nat machinery.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana (L) Heynh. (hereafter, Arabidopsis) Landsberg erecta (Ler) and Columbia-0 (Col-0) wild-type accessions were obtained from the Nottingham Arabidopsis Stock Center (NASC; Nottingham, UK) and then propagated at our institute for further analysis. The P14 6.3 and P14 20.1 lines were isolated in a Ler background after EMS mutagenesis in a screen in which mutants were given protocol numbers, as PN X.Y: PN indicates the corresponding parental group, X refers to the number of the plate where the mutant was isolated and Y is an ordinal assigned to each of the mutants found in a given plate [36]. Each parental group was the pooled M2 seed progeny of about 600 M1 plants exposed to EMS. The P14 6.3 and P14 20.1 M2 mutant lines were considered likely to carry identical mutation(s) because they were phenotypically indistinguishable and had been isolated among seeds belonging to the same parental group (P14).
All other Arabidopsis seeds used in this work were provided by NASC or the Arabidopsis Biological Resource Center (ABRC; Columbus, Ohio, USA). Seed sterilization and sowing, plant culture and crosses were performed as previously described [36], [77]. In brief, seeds were sown on plates containing MS agar medium (half-strength Murashige and Skoog salts, 0.7% plant agar [Duchefa], pH 5.7, and 1% sucrose) and stratified (4°C in the dark) for 48 h and then transferred to either Conviron TC16 or TC30 growth chambers set to our standard conditions (continuous light at approximately 75 µmol·m-2·s-1, 20±1°C, 60-70% relative humidity). When required, plants were transferred into pots containing a 2∶2∶1 mixture of perlite:vermiculite:sphagnum moss and grown in walk-in growth chambers set to our standard conditions.
Plant gross morphology, histology and histochemical assays
Leaf clearing and fixation, embedding, microscopy and morphometry were performed as previously described [58], [60], [78], [79]. Venation diagrams were obtained from micrographs by hand drawing on the screen of a Wacom Cintiq 18SX Interactive pen display (http://www.wacom.com/) and using the Adobe Photoshop CS3 (http://www.adobe.com) software. Morphometric analyses of the diagrams (n≥10) were performed with ImageJ 1.36b [80] (http://rsb.info.nih.gov/ij/index.html/), Scion Image 4.0.3.2 (http://scion-image.software.informer.com/) and NIS-Elements AR 2.30 (Nikon Imaging; http://www.nis-elements.com/). GUS assays were performed as previously described [81].
Positional cloning and molecular characterization of TCU2 and its mutant alleles
Low-resolution mapping of the tcu2-1 mutation was performed as previously described [82], [83]. For the fine mapping of the TCU2 gene, SSLP, SNP and In/Del markers were developed based on the polymorphisms between Ler and Col-0 described in the Monsanto Arabidopsis Polymorphism Collection database (http://www.arabidopsis.org). Synthetic oligonucleotides were purchased from Sigma-Aldrich UK (Table S1). Genomic DNA was extracted, PCR amplified and sequenced as previously described [84]. For the sequencing of TCU2 and AGO10 alleles, PCR amplification products spanning their transcription units were obtained using wild-type and mutant genomic DNA as templates. To confirm the presence and position of T-DNA insertions, DNA was extracted and PCR amplified. Sequencing reactions, RNA extractions and qRT-PCR were performed as described in [85], using the primers shown in Table S2. The multiple alignment of protein sequences shown in Figure S6 was obtained using ClustalX 2.0 [86] and shaded with Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
Next-generation sequencing
Total DNA was purified from P14 6.3 rosettes collected 18 das using the DNeasy Plant Midi Kit (Qiagen) following the instructions of the manufacturer and sent for sequencing at BGI-Hong Kong. Whole genome sequencing was performed in an Illumina HiSeq 2000 and returned paired reads with an average length of 90 nt. A reference-guided assembly of the clean reads and a SNP report were obtained with SeqMan NGen (Lasergene 11; DNASTAR), using as a reference the genome sequence of Ler-0 obtained in the ‘19 genomes of Arabidopsis thaliana’ project, available at http://mus.well.ox.ac.uk/19genomes/.
DNA constructs
Gateway (Invitrogen) entry and destination vectors were used to obtain all the constructs used in this work. To visualize TCU2 expression, we PCR amplified a 1,349-bp segment of genomic DNA from Ler, including most of the first exon of TCU2 (At5g58440), the intergenic region between TCU2 and At5g58450 and 324 bp from the 5′ end of the transcription unit of At5g58450. To visualize the subcellular localization of the TCU2 protein, we PCR amplified a Ler genomic segment of 5.2 kb encompassing the entire TCU2 transcription unit. The amplification products were cloned into pGEM-T Easy221, sequence verified and subsequently subcloned into pMDC164 (TCU2pro:GUS) or pMDC85 (35Spro:TCU2:GFP). The constructs obtained in this way were then transferred from Escherichia coli One Shot Match1 cells to Agrobacterium tumefaciens LBA4404, and the transformants thus obtained were used to infect Col-0 and Ler plants by the floral dip method [87]. The transgenic plants were selected on MS agar medium supplemented with 15 µg·ml-1 hygromicin.
amiRNA construction
Arabidopsis transgenic lines expressing the amiR-TCU2 artificial microRNA (5′- UAGGAGAUUACUUAAGUCGAC -3′) were obtained as will be described in detail elsewhere (Jover-Gil, Paz-Ares, Micol and Ponce, unpublished). In brief, the transgene designed to express amiR-TCU2 was constructed in the backbone of the gene encoding miR319a, an endogenous Arabidopsis miRNA. The sequence of the mature miR319a miRNA was replaced by that of amiR-TCU2, as described in [88] and at http://wmd3.weigelworld.org/cgi-bin/webapp.cgi. The amiR-TCU2 construct was flanked with attB1 and attB2 sites to enable use of the Gateway technology, and inserted by means of a BP reaction into pGEM-T Easy221. The plasmid obtained in this way was used to transform Escherichia coli DH5α cells. Plasmid DNA was isolated from transformants and the insert was transferred into the pMDC32 destination vector [89] and then mobilized into Agrobacterium tumefaciens C58C1 cells. Arabidopsis plants were transformed by infection using the floral dip method [87].
Supporting Information
Leaf venation pattern in the P14 6.3 line. Diagrams are shown for first- and third-node leaves, indicating the leaf margin in orange. Duplicated primary veins are highlighted with arrows in the diagram representing a fused first pair of leaves in P14 6.3. Pictures were taken 21 das. Scale bars: 2 mm.
(PDF)
Fine mapping of the TCU2 gene. Red bars represent segments of chromosome 5, and green boxes, BAC clones corresponding to the candidate interval, which is highlighted in blue. The number of informative recombinants identified for each of the molecular markers used for linkage analysis is shown in parentheses.
(PDF)
Some phenotypic traits of the pnh-2 mutant grown under our standard culture conditions. (A) Apical region of the inflorescence. (B) Fused first pair of leaves (arrow). (C) Adult plant. (D) Flowers of the apical region of the inflorescence, some of them showing siliques beginning to elongate. (E) Mature siliques. (F-H) Detail of three-valved siliques: (F) closed and (G, H) longitudinally open through a septum. Arrows highlight a few aborted or unfertilized ovules. Pictures were taken (B) 18 das and (A, C, D-H) 50 das. Scale bars: (A, B, D, E) 2 mm, (C) 2 cm and (F-H) 1 mm.
(PDF)
Early flowering in the P14 6.3 line. (A) Lateral view of P14 6.3 and Ler rosettes. (B, C) Cotyledons and vegetative leaves of (B) P14 6.3 and Ler (C) arranged from left to right in order of appearance. Leaf reticulation is clearly visible in B. Pictures were taken (A) 25 and (B, C) 28 das. Scale bars: (A) 1 cm and (B, C) 2 mm.
(PDF)
Multiple alignment of the NatB_MDM20 domains of TCU2 and some of its putative orthologs in higher plants. At, Arabidopsis thaliana (NP_200653.2); Rc, Ricinus communis (XP_002516347.1); Vv, Vitis vinifera (XP_002273069.1); Pt, Populus balsamifera subsp. trichocarpa (XP_002319956.1); and Os, Oryza sativa (Os NP_001055256.1). A triangle indicates the conserved amino acid changed by the tcu2-1 mutation. Amino acid residues identical or similar in all five sequences are shaded black or grey, respectively. See http://pfam.sanger.ac.uk/family/PF09797?type=Family#tabview=tab1 for a list of all the unique domain organisations or architectures in which this domain is found.
(PDF)
TCU2 expression in Arabidopsis development. Expression data from the Arabidopsis Electronic Fluorescent Pictograph (eFP) Browser for At5g58450 (TCU2) expression levels throughout all Arabidopsis developmental stages.
(PDF)
Spatial expression analysis of TCU2. GUS staining of TCU2pro:GUS transgenic plants in (A) leaves, (B) a root, (C) the basal region of trichomes and (D-I) whole rosettes. Plant material was collected at the time shown in each picture (in das). Scale bars: (A) 1 mm, (B) 0.5 mm, (C) 100 µm and (D-I) 2 mm.
(PDF)
Subcellular localization of the TCU2 protein. Confocal micrographs are shown from the apex and root elongation zone of transgenic plants obtained after transformation by infection with Agrobacterium tumefaciens cells carrying the pMDC85 vector (A) without insert or (B) with the TCU2 insert. GFP emission is shown in green. The cloning site of pMDC85 is flanked by two tandem 35S promoters and the GFP coding sequence. Hence, GFP is expressed from a 35Spro:GFP transgene in the plant shown in A, and a TCU2:GFP fusion protein is expressed from a 35Spro:TCU2:GFP transgene in the plant shown in B. Nuclear exclusion of the TCU2:GFP protein is clearly visible in the dividing cells of the root tip shown in B. Pictures were taken 14 das. Scale bar: 50 µm.
(PDF)
Effects of the tcu2 and nbc-1 mutations on embryonic development. Dissected siliques are shown from selfed (A) Ler, (B) tcu2-1, (C) Col-0, (D) tcu2-2 and (E) nbc-1 plants. Arrows indicate abnormal seeds that are likely to be aborted or unfertilized ovules. Pictures were taken 59 das. Scale bar: 250 µm.
(PDF)
Oligonucleotide sets used for the fine mapping of TCU2. *Labeled with HEX (4,7,2′,4′,5′,7′-hexachloro-6-fluorescein).
(PDF)
Other oligonucleotide sets used in this work. a,bThese oligonucleotides include at their 5′ ends the aattB1 and battB2 sequences, which are shown in italics.
(PDF)
Acknowledgments
The authors thank H. Candela for useful discussions, and R. Sarmiento-Mañús, J.M. Serrano, F.M. Lozano, T. Trujillo, L. Serna, D. Navarro, A. Torregrosa and J.M. Sánchez-Larrosa for technical assistance.
Funding Statement
This research was supported by grants from the Ministerio de Economía y Competitividad of Spain (BIO2008-01900 to M.R.P.) and the Generalitat Valenciana (PROMETEO/2009/112 to M.R.P. and J.L.M.). A.F.-A. and M.M.A.-P. hold fellowships from the Ministerio de Ciencia e Innovación of Spain. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Starheim KK, Gevaert K, Arnesen T (2012) Protein N-terminal acetyltransferases: when the start matters. Trends Biochem Sci 37: 152–161. [DOI] [PubMed] [Google Scholar]
- 2. Narita K (1958) Isolation of acetylpeptide from enzymic digests of TMV-protein. Biochim Biophys Acta 28: 184–191. [DOI] [PubMed] [Google Scholar]
- 3. Polevoda B, Cardillo TS, Doyle TC, Bedi GS, Sherman F (2003) Nat3p and Mdm20p are required for function of yeast NatB Nalpha-terminal acetyltransferase and of actin and tropomyosin. J Biol Chem 278: 30686–30697. [DOI] [PubMed] [Google Scholar]
- 4. Polevoda B, Brown S, Cardillo TS, Rigby S, Sherman F (2008) Yeast N(alpha)-terminal acetyltransferases are associated with ribosomes. J Cell Biochem 103: 492–508. [DOI] [PubMed] [Google Scholar]
- 5. Starheim KK, Arnesen T, Gromyko D, Ryningen A, Varhaug JE, et al. (2008) Identification of the human N(alpha)-acetyltransferase complex B (hNatB): a complex important for cell-cycle progression. Biochem J 415: 325–331. [DOI] [PubMed] [Google Scholar]
- 6. Polevoda B, Arnesen T, Sherman F (2009) A synopsis of eukaryotic N-alpha-terminal acetyltransferases: nomenclature, subunits and substrates. BMC Proc 3 Suppl 6S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arnesen T, Anderson D, Baldersheim C, Lanotte M, Varhaug JE, et al. (2005) Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem J 386: 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rope AF, Wang K, Evjenth R, Xing J, Johnston JJ, et al. (2011) Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am J Hum Genet 89: 28–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forte GM, Pool MR, Stirling CJ (2011) N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol 9: e1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hwang CS, Shemorry A, Varshavsky A (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA (2011) N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334: 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jornvall H (1975) Acetylation of protein N-terminal amino groups structural observations on alpha-amino acetylated proteins. J Theor Biol 55: 1–12. [DOI] [PubMed] [Google Scholar]
- 13. Persson B, Flinta C, von Heijne G, Jornvall H (1985) Structures of N-terminally acetylated proteins. Eur J Biochem 152: 523–527. [DOI] [PubMed] [Google Scholar]
- 14. Hershko A, Heller H, Eytan E, Kaklij G, Rose IA (1984) Role of the alpha-amino group of protein in ubiquitin-mediated protein breakdown. Proc Natl Acad Sci USA 81: 7021–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ben-Saadon R, Fajerman I, Ziv T, Hellman U, Schwartz AL, et al. (2004) The tumor suppressor protein p16(INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system. Direct evidence for ubiquitination at the N-terminal residue. J Biol Chem 279: 41414–41421. [DOI] [PubMed] [Google Scholar]
- 16. Arnesen T (2011) Towards a functional understanding of protein N-terminal acetylation. PLoS Biol 9: e1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ciechanover A, Ben-Saadon R (2004) N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol 14: 103–106. [DOI] [PubMed] [Google Scholar]
- 18. Kuo ML, den Besten W, Bertwistle D, Roussel MF, Sherr CJ (2004) N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev 18: 1862–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pena MM, Melo SP, Xing YY, White K, Barbour KW, et al. (2009) The intrinsically disordered N-terminal domain of thymidylate synthase targets the enzyme to the ubiquitin-independent proteasomal degradation pathway. J Biol Chem 284: 31597–31607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varshavsky A (2008) Discovery of cellular regulation by protein degradation. J Biol Chem 283: 34469–34489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kreft SG, Hochstrasser M (2011) An unusual transmembrane helix in the endoplasmic reticulum ubiquitin ligase Doa10 modulates degradation of its cognate E2 enzyme. J Biol Chem 286: 20163–20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Helbig AO, Rosati S, Pijnappel PW, van Breukelen B, Timmers MH, et al. (2010) Perturbation of the yeast N-acetyltransferase NatB induces elevation of protein phosphorylation levels. BMC Genomics 11: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singer JM, Shaw JM (2003) Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proc Natl Acad Sci USA 100: 7644–7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coulton AT, East DA, Galinska-Rakoczy A, Lehman W, Mulvihill DP (2010) The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers permits the discrete regulation of specific myosins in fission yeast. J Cell Sci 123: 3235–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caesar R, Blomberg A (2004) The stress-induced Tfs1p requires NatB-mediated acetylation to inhibit carboxypeptidase Y and to regulate the protein kinase A pathway. J Biol Chem 279: 38532–38543. [DOI] [PubMed] [Google Scholar]
- 26. Behnia R, Panic B, Whyte JR, Munro S (2004) Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol 6: 405–413. [DOI] [PubMed] [Google Scholar]
- 27. Setty SR, Strochlic TI, Tong AH, Boone C, Burd CG (2004) Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat Cell Biol 6: 414–419. [DOI] [PubMed] [Google Scholar]
- 28. Behnia R, Barr FA, Flanagan JJ, Barlowe C, Munro S (2007) The yeast orthologue of GRASP65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic. J Cell Biol 176: 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murthi A, Hopper AK (2005) Genome-wide screen for inner nuclear membrane protein targeting in Saccharomyces cerevisiae: roles for N-acetylation and an integral membrane protein. Genetics 170: 1553–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang X, Connelly JJ, Wang CL, Sternglanz R (2004) Importance of the Sir3 N terminus and its acetylation for yeast transcriptional silencing. Genetics 168: 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Geissenhoner A, Weise C, Ehrenhofer-Murray AE (2004) Dependence of ORC silencing function on NatA-mediated Nalpha acetylation in Saccharomyces cerevisiae . Mol Cell Biol 24: 10300–10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Caesar R, Warringer J, Blomberg A (2006) Physiological importance and identification of novel targets for the N-terminal acetyltransferase NatB. Eukaryot Cell 5: 368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pesaresi P, Gardner NA, Masiero S, Dietzmann A, Eichacker L, et al. (2003) Cytoplasmic N-terminal protein acetylation is required for efficient photosynthesis in Arabidopsis. Plant Cell 15: 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Polevoda B, Sherman F (2000) Nalpha -terminal acetylation of eukaryotic proteins. J Biol Chem 275: 36479–36482. [DOI] [PubMed] [Google Scholar]
- 35. Bienvenut WV, Sumpton D, Martinez A, Lilla S, Espagne C, et al. (2012) Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-alpha-acetylation features. Mol Cell Proteomics 11: M111.015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berná G, Robles P, Micol JL (1999) A mutational analysis of leaf morphogenesis in Arabidopsis thaliana . Genetics 152: 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robles P, Micol JL (2001) Genome-wide linkage analysis of Arabidopsis genes required for leaf development. Mol Genet Genomics 266: 12–19. [DOI] [PubMed] [Google Scholar]
- 38. Ji L, Liu X, Yan J, Wang W, Yumul RE, et al. (2011) ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis . PLoS Genet 7: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Q, Yao X, Pi L, Wang H, Cui X, et al. (2009) The ARGONAUTE10 gene modulates shoot apical meristem maintenance and leaf polarity establishment by repressing miR165/166 in Arabidopsis. Plant J 57: 28–40. [DOI] [PubMed] [Google Scholar]
- 40. Zhu H, Hu F, Wang R, Zhou X, Sze SH, et al. (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McConnell JR, Barton MK (1995) Effect of mutations in the PINHEAD gene of Arabidopsis on the formation of shoot apical meristems. Dev Genet 16: 358–366. [Google Scholar]
- 42. Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, et al. (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481. [DOI] [PubMed] [Google Scholar]
- 43. Moussian B, Schoof H, Haecker A, Jurgens G, Laux T (1998) Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J 17: 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Polevoda B, Sherman F (2003) N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol 325: 595–622. [DOI] [PubMed] [Google Scholar]
- 45. Allan RK, Ratajczak T (2011) Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16: 353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, et al. (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perrot M, Massoni A, Boucherie H (2008) Sequence requirements for Nalpha-terminal acetylation of yeast proteins by NatA. Yeast 25: 513–527. [DOI] [PubMed] [Google Scholar]
- 50. Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, et al. (2009) Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA 106: 8157–8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Helbig AO, Gauci S, Raijmakers R, van Breukelen B, Slijper M, et al. (2010) Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol Cell Proteomics 9: 928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Starheim KK, Gromyko D, Velde R, Varhaug JE, Arnesen T (2009) Composition and biological significance of the human Nalpha-terminal acetyltransferases. BMC Proc 3 Suppl 6S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goetze S, Qeli E, Mosimann C, Staes A, Gerrits B, et al. (2009) Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster . PLoS Biol 7: e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Falb M, Aivaliotis M, Garcia-Rizo C, Bisle B, Tebbe A, et al. (2006) Archaeal N-terminal protein maturation commonly involves N-terminal acetylation: a large-scale proteomics survey. J Mol Biol 362: 915–924. [DOI] [PubMed] [Google Scholar]
- 55. Aivaliotis M, Gevaert K, Falb M, Tebbe A, Konstantinidis K, et al. (2007) Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis . J Proteome Res 6: 2195–2204. [DOI] [PubMed] [Google Scholar]
- 56. Pérez-Pérez JM, Esteve-Bruna D, González-Bayón R, Kangasjärvi S, Caldana C, et al. (2013) Functional redundancy and divergence within the Arabidopsis RETICULATA-RELATED gene family. Plant Physiol 162: 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li H, Culligan K, Dixon RA, Chory J (1995) CUE1: A mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell 7: 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. González-Bayón R, Kinsman EA, Quesada V, Vera A, Robles P, et al. (2006) Mutations in the RETICULATA gene dramatically alter internal architecture but have little effect on overall organ shape in Arabidopsis leaves. J Exp Bot 57: 3019–3031. [DOI] [PubMed] [Google Scholar]
- 59. Kinsman EA, Pyke KA (1998) Bundle sheath cells and cell-specific plastid development in Arabidopsis leaves. Development 125: 1815–1822. [DOI] [PubMed] [Google Scholar]
- 60. Mollá-Morales A, Sarmiento-Mañús R, Robles P, Quesada V, Pérez-Pérez JM, et al. (2011) Analysis of ven3 and ven6 reticulate mutants reveals the importance of arginine biosynthesis in Arabidopsis leaf development. Plant J 65: 335–345. [DOI] [PubMed] [Google Scholar]
- 61. Yu F, Fu A, Aluru M, Park S, Xu Y, et al. (2007) Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ 30: 350–365. [DOI] [PubMed] [Google Scholar]
- 62. Rosar C, Kanonenberg K, Nanda AM, Mielewczik M, Brautigam A, et al. (2012) The leaf reticulate mutant dov1 is impaired in the first step of purine metabolism. Mol Plant 5: 1227–1241. [DOI] [PubMed] [Google Scholar]
- 63. Fischer K, Kammerer B, Gutensohn M, Arbinger B, Weber A, et al. (1997) A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 9: 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Carles CC, Fletcher JC (2003) Shoot apical meristem maintenance: the art of a dynamic balance. Trends Plant Sci 8: 394–401. [DOI] [PubMed] [Google Scholar]
- 65. Ripoll JJ, Ferrándiz C, Martínez-Laborda A, Vera A (2006) PEPPER, a novel K-homology domain gene, regulates vegetative and gynoecium development in Arabidopsis . Dev Biol 289: 346–359. [DOI] [PubMed] [Google Scholar]
- 66. Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y (2008) Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 20: 1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. González-Reig S, Ripoll JJ, Vera A, Yanofsky MF, Martínez-Laborda A (2012) Antagonistic gene activities determine the formation of pattern elements along the mediolateral axis of the Arabidopsis fruit. PLoS Genet 8: e1003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jover-Gil S, Candela H, Ponce MR (2005) Plant microRNAs and development. Int J Dev Biol 49: 733–744. [DOI] [PubMed] [Google Scholar]
- 69. Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen X (2012) Small RNAs in development - insights from plants. Curr Opin Genet Dev 22: 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pulido A, Laufs P (2010) Co-ordination of developmental processes by small RNAs during leaf development. J Exp Bot 61: 1277–1291. [DOI] [PubMed] [Google Scholar]
- 72. Jover-Gil S, Candela H, Robles P, Aguilera V, Barrero JM, et al. (2012) The microRNA pathway genes AGO1, HEN1 and HYL1 participate in leaf proximal-distal, venation and stomatal patterning in Arabidopsis. Plant Cell Physiol 53: 1322–1333. [DOI] [PubMed] [Google Scholar]
- 73. Karlova R, van Haarst JC, Maliepaard C, van de Geest H, Bovy AG, et al. (2013) Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J Exp Bot 64: 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang Z, Zhang X (2012) Argonautes compete for miR165/166 to regulate shoot apical meristem development. Curr Opin Plant Biol 15: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, et al. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190. [DOI] [PubMed] [Google Scholar]
- 76. Hunter C, Sun H, Poethig RS (2003) The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol 13: 1734–1739. [DOI] [PubMed] [Google Scholar]
- 77. Ponce MR, Quesada V, Micol JL (1998) Rapid discrimination of sequences flanking and within T-DNA insertions in the Arabidopsis genome. Plant J 14: 497–501. [DOI] [PubMed] [Google Scholar]
- 78. Candela H, Martínez-Laborda A, Micol JL (1999) Venation pattern formation in Arabidopsis thaliana vegetative leaves. Dev Biol 205: 205–216. [DOI] [PubMed] [Google Scholar]
- 79. Rubio-Díaz S, Pérez-Pérez JM, González-Bayón R, Muñoz-Viana R, Borrega N, et al. (2012) Cell expansion-mediated organ growth is affected by mutations in three EXIGUA genes. PLoS One 7: e36500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42. [Google Scholar]
- 81. Robles P, Fleury D, Candela H, Cnops G, Alonso-Peral MM, et al. (2010) The RON1/FRY1/SAL1 gene is required for leaf morphogenesis and venation patterning in Arabidopsis. Plant Physiol 152: 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ponce MR, Robles P, Lozano FM, Brotons MA, Micol JL (2006) Low-resolution mapping of untagged mutations. In: Salinas J, Sánchez-Serrano JJ, editors. Arabidopsis protocols, 2nd edition: Humana Press. pp. 105–113. [DOI] [PubMed]
- 83. Ponce MR, Robles P, Micol JL (1999) High-throughput genetic mapping in Arabidopsis thaliana . Mol Gen Genet 261: 408–415. [DOI] [PubMed] [Google Scholar]
- 84. Pérez-Pérez JM, Ponce MR, Micol JL (2004) The ULTRACURVATA2 gene of Arabidopsis encodes an FK506–binding protein involved in auxin and brassinosteroid signaling. Plant Physiol 134: 101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Barrero JM, González-Bayón R, del Pozo JC, Ponce MR, Micol JL (2007) INCURVATA2 encodes the catalytic subunit of DNA polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana . Plant Cell 19: 2822–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 87. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 88. Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis . Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Leaf venation pattern in the P14 6.3 line. Diagrams are shown for first- and third-node leaves, indicating the leaf margin in orange. Duplicated primary veins are highlighted with arrows in the diagram representing a fused first pair of leaves in P14 6.3. Pictures were taken 21 das. Scale bars: 2 mm.
(PDF)
Fine mapping of the TCU2 gene. Red bars represent segments of chromosome 5, and green boxes, BAC clones corresponding to the candidate interval, which is highlighted in blue. The number of informative recombinants identified for each of the molecular markers used for linkage analysis is shown in parentheses.
(PDF)
Some phenotypic traits of the pnh-2 mutant grown under our standard culture conditions. (A) Apical region of the inflorescence. (B) Fused first pair of leaves (arrow). (C) Adult plant. (D) Flowers of the apical region of the inflorescence, some of them showing siliques beginning to elongate. (E) Mature siliques. (F-H) Detail of three-valved siliques: (F) closed and (G, H) longitudinally open through a septum. Arrows highlight a few aborted or unfertilized ovules. Pictures were taken (B) 18 das and (A, C, D-H) 50 das. Scale bars: (A, B, D, E) 2 mm, (C) 2 cm and (F-H) 1 mm.
(PDF)
Early flowering in the P14 6.3 line. (A) Lateral view of P14 6.3 and Ler rosettes. (B, C) Cotyledons and vegetative leaves of (B) P14 6.3 and Ler (C) arranged from left to right in order of appearance. Leaf reticulation is clearly visible in B. Pictures were taken (A) 25 and (B, C) 28 das. Scale bars: (A) 1 cm and (B, C) 2 mm.
(PDF)
Multiple alignment of the NatB_MDM20 domains of TCU2 and some of its putative orthologs in higher plants. At, Arabidopsis thaliana (NP_200653.2); Rc, Ricinus communis (XP_002516347.1); Vv, Vitis vinifera (XP_002273069.1); Pt, Populus balsamifera subsp. trichocarpa (XP_002319956.1); and Os, Oryza sativa (Os NP_001055256.1). A triangle indicates the conserved amino acid changed by the tcu2-1 mutation. Amino acid residues identical or similar in all five sequences are shaded black or grey, respectively. See http://pfam.sanger.ac.uk/family/PF09797?type=Family#tabview=tab1 for a list of all the unique domain organisations or architectures in which this domain is found.
(PDF)
TCU2 expression in Arabidopsis development. Expression data from the Arabidopsis Electronic Fluorescent Pictograph (eFP) Browser for At5g58450 (TCU2) expression levels throughout all Arabidopsis developmental stages.
(PDF)
Spatial expression analysis of TCU2. GUS staining of TCU2pro:GUS transgenic plants in (A) leaves, (B) a root, (C) the basal region of trichomes and (D-I) whole rosettes. Plant material was collected at the time shown in each picture (in das). Scale bars: (A) 1 mm, (B) 0.5 mm, (C) 100 µm and (D-I) 2 mm.
(PDF)
Subcellular localization of the TCU2 protein. Confocal micrographs are shown from the apex and root elongation zone of transgenic plants obtained after transformation by infection with Agrobacterium tumefaciens cells carrying the pMDC85 vector (A) without insert or (B) with the TCU2 insert. GFP emission is shown in green. The cloning site of pMDC85 is flanked by two tandem 35S promoters and the GFP coding sequence. Hence, GFP is expressed from a 35Spro:GFP transgene in the plant shown in A, and a TCU2:GFP fusion protein is expressed from a 35Spro:TCU2:GFP transgene in the plant shown in B. Nuclear exclusion of the TCU2:GFP protein is clearly visible in the dividing cells of the root tip shown in B. Pictures were taken 14 das. Scale bar: 50 µm.
(PDF)
Effects of the tcu2 and nbc-1 mutations on embryonic development. Dissected siliques are shown from selfed (A) Ler, (B) tcu2-1, (C) Col-0, (D) tcu2-2 and (E) nbc-1 plants. Arrows indicate abnormal seeds that are likely to be aborted or unfertilized ovules. Pictures were taken 59 das. Scale bar: 250 µm.
(PDF)
Oligonucleotide sets used for the fine mapping of TCU2. *Labeled with HEX (4,7,2′,4′,5′,7′-hexachloro-6-fluorescein).
(PDF)
Other oligonucleotide sets used in this work. a,bThese oligonucleotides include at their 5′ ends the aattB1 and battB2 sequences, which are shown in italics.
(PDF)