Abstract
Objective
Early metastasis is a major biological feature of pancreatic cancer. The current study examined whether silencing Slc38a1, a gene involved in energy metabolism, using short hairpin RNA (shRNA) could inhibit the growth, migration, and invasiveness of pancreatic cancer cells.
Methods
A series of Slc38a1 shRNAs were designed and cloned into the pGPU6/GFP/Neo vectors. An shRNA with the most efficacious inhibitory action on SCL38A1 expression (65% inhibition) upon screening in DH5α bacteria was used to transfect SW1990 human pancreatic cancer cells. Cell growth, migration, and invasiveness were examined using cell counting kit-8, Boyden chamber without and with Matrigel, respectively.
Results
Transfection of SW1990 cells with the SLCs38A1 shRNA significantly decreased the proliferation (P<0.0001) and migratory potential (by 46.7%, P=0.0399) of the cancer cells. Invasiveness, however, was not affected.
Conclusions
Inhibiting Slc38a1 using shRNA technology could decrease the growth and migration of representative pancreatic cancer cells. However, the fact that invasiveness was not affected suggested that SLC38A1 is unlikely to be responsible for early metastasis.
Keywords: Slc38a1, shRNA, pancreatic cancer
Introduction
Transportation of small neutral amino acids and N-methyl amino acids by Type A amino acid transporters (ATA) plays an important role in cell physiology. Three subtypes of ATA (ATA1, ATA2 and ATA3) have been identified. ATA1 is also referred to as SNAT1 or SLC38A1. SLC38A1 is over-expressed in hilar cholangiocarcinoma (1) and primary hepatic carcinoma (2), and thought to promote cancinogenesis and cancer metastasis.
In the current study, we constructed a eukaryotic vector that carried a siRNA to inhibit the expression of Slc38a1 gene and transfected SW1990 cells (a representative human pancreatic adenocarcinoma cell line). The results indicated that selective inhibition of Slc38a1 gene could inhibit the growth and migration, but not the invasiveness of SW1990 cells.
Materials and methods
Materials
The eukaryotic expression vector pGPU6/GFP/Neo was from Zimmer Company (Shanghai, China). DH5α Escherichia coli (E. coli) were from the Cancer Research Institute, China Medical University (Shenyang, China). The restriction endonucleases Bbs I and BamH I, T4 ligase, and Taq enzyme were from TaKaRa (Dalian, China). RPMI-1640 culture medium, fetal bovine serum (FBS), trypsin, lipofectamine 2000 and Trizol reagent were from Gibico (Invitrogen, Carlsbad, CA, USA). Plasmid DNA extraction kit was from Qiagen (Shanghai, China). Reverse transcription kit was from Promega (Fitchburg, WI, USA). All oligonucleotides, including PCR primers, were synthesized by Shanghai Ying Jun Bio-technology. Cell counting kit-8 (CCK-8) was from Dojindo Laboratories (Kumamoto, Japan). Boyden chamber for migration assay was from Corning Costar (Rochester, NY, USA). Matrigel (0.3 mg/mL) was from BD Biosciences (Bedford, MA, USA).
Cell culture
SW1990 cells were obtained from Shanghai Cell Institute, Chinese Academy of Sciences, and expressed a high level of SLC38A1. Cells were cultured in RPMI-1640 medium supplemented with 10% FBS, 100 µg/mL streptomycin, 100 U/mL penicillin in a humidified atmosphere containing 5% CO2 at 37 °C.
shRNA synthesis and vector construction/verification
The SCL38A1 siRNAs, and a negative control (with scrambled sequence with no match to any known gene) were selected based on the full-length cDNA of human Slc38a1 mRNA (gene bank number: NM_001077484.1) using an siRNA design software by Ambion (Table 1). An siRNA against GAPDH was included as a positive control (shPC) to verify transfection reliability, RNA extraction and gene expression quantification. To avoid premature termination, TTCAAGAGA was used in the loop of all constructs. The PCR products were annealed at the following condition: 95 °C 5 min, 85 °C 5 min, 75 °C 5 min, 70 °C 5 min, 4 °C preservation. The annealing process yielded 10 µmol/L shRNA template, which was diluted to 20 nmol/L for ligation.
Table 1. The target sites and target sequences of Slc38a1 shRNA plasmids.
| Plasmid | Target sequence |
|---|---|
| sh1 | 5'-CACCGGATGATAACATTAGCAATTTCAAGAGAATTGCTAATGTTATCATCCTTTTTTG-3' |
| 5'-GATCCAAAAAAGGATGATAACATTAGCAATTCTCTTGAAATTGCTAATGTTATCATCC-3' | |
| sh2 | 5'-CACCGCAATGACTCCAATGATTTCATTCAAGAGATGAAATCATTGGAGTCATTGCTTTTTTG-3' |
| 5'-GATCCAAAAAAGCAATGACTCCAATGATTTCATCTCTTGAATGAAATCATTGGAGTCATTGC-3' | |
| sh3 | 5'-CACCGCATTGTTCCAGAGCTAAATTTCAAGAGAATTTAGCTCTGGAACAATGCTTTTTTG-3' |
| 5'-GATCCAAAAAAGCATTGTTCCAGAGCTAAATTCTCTTGAAATTTAGCTCTGGAACAATGC-3' | |
| sh4 | 5'-CACCGCATACTCTTGGTTGTTATCATTCAAGAGATGATAACAACCAAGAGTATGCTTTTTTG-3' |
| 5'-GATCCAAAAAAGCATACTCTTGGTTGTTATCATCTCTTGAATGATAACAACCAAGAGTATGC-3' | |
| shNC | 5'-CACC GTTCTCCGAACGTGTCACGT CAAGAGATT ACGTGACACGTTCGGAGAA TTTTTT G-3' |
| 5'-GATC CAAAAAA TTCTCCGAACGTGTCACGT AATCTCTTG ACGTGACACGTTCGGAGAAC-3' | |
| shPC | 5'-CACC GTATGACAACAGCCTCAAG TTCAAGAGA CTTGAGGCTGTTGTCATAC TTTTTT G-3' |
| 5'-GATC CAAAAAA GTATGACAACAGCCTCAAG TCTCTTGAA CTTGAGGCTGTTGTCATAC-3' |
After agarose electrophoresis purification, shRNA was inserted to the pGPU6/GFP/Neo vector at the BamH I and Bbs sites. The resulting vector was inoculated to competent DH5α E. coli, selected using kanamycin resistance, and verified with sequencing by Shanghai Ying Jun Bio-technology. A total of four constructs, referred to as pGPU6/GFP/Neo-shRNA-1-4 (sh1-4), were tested in cultured SW1990 cells. The experiment also included pGPU6/GFP/Neo-shNC (shNC) as a negative control, and pGPU6/GFP/Neo-shPC (shPC) as a positive control.
Experiments in SW1990 cells
One day after the seeding in 6-well plates, the same amount of SW1990 cells (2×105/well) were transfected with sh1-4, shNC, or shPC using Lipofectamine 2000. A mock transfection (transfection reagents only) was also included. After incubation at 37 °C in a CO2 incubator for 48 h, Slc38a1 mRNA was examined using RT-PCR. Briefly, total RNA was extracted for RT-PCR amplification of Slc38a1 cDNA. The PCR products were separated using a 1.5% agarose gel, and analyzed using an imaging system from BioRad (Hercules, CA, USA).
Cell number/growth was examined by counting the cell number at 48, 72 and 96 h after the transfection using a colorimetric CCK-8 assay at 450 nm. Migration was examined using a modified Boyden chamber containing a gelatin-coated polycarbonate membrane filter (8-µm pore size). Briefly, RPMI-1640 containing 10% FBS was placed in the lower chamber. SW1990 (at a final concentration of 5×104 cells/mL) was suspended in serum-free RPMI-1640 in the upper chamber, and incubated at 37 °C under 5% CO2 for 8 h. Cells migration was quantified by counting the cells that migrated to the lower chamber using crystal violet staining and an optical microscope, and expressed as the mean number of cells per field (average of 10 random fields). The invasion was examined after 24 h culture in Boyden chamber, with the upper surface of the filter coated with 20 µL Matrigel. All experiments were conducted in triplicate.
Statistical analysis
All data are expressed as x̄±s, and analyzed using an one-way analysis of variance (ANOVA) with the SPSS 13.0 software (SPSS Inc., Chicago, IL, USA). P<0.05 was considered statistically significant.
Results
Verification of shRNA vector
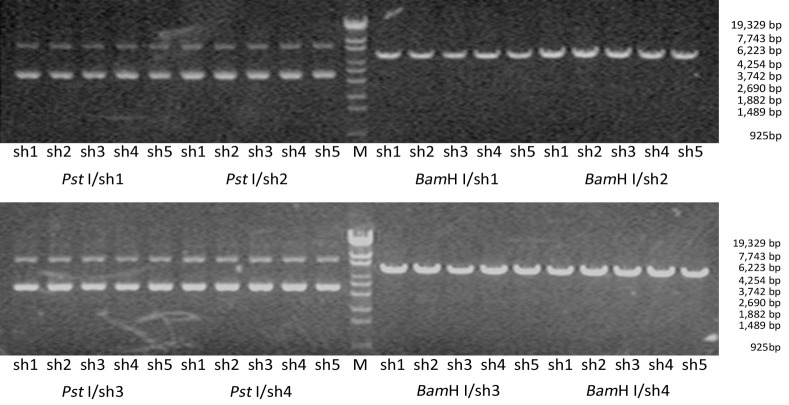
Electrophoresis after digestion with Pst I and BamH I confirmed that the shRNA size conformed to the design (Figure 1). Two clones for each construct were further verified with sequencing. The sequence analysis conformed to the design in all cases.
Figure 1.
Identification of the recombinant vectors. BamH I and Pst I were used to digest and identify the extraction plasmid. Positive recombinant vector can be cut apart by BamH I but not Pst I. M, Lambda/Eco130I.
Expression of the pGPU6/GFP/Neo vector that included a cassette of green fluorescent protein revealed >70% transfection efficiency (Figure 2).
Figure 2.
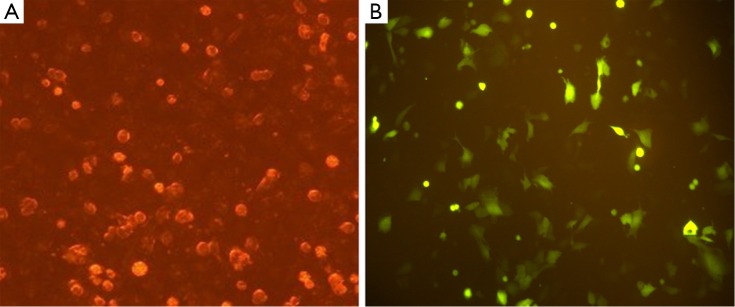
Transfection of SW1990 cells with sh4. (A) Cells transfected with recombinant plasmid sh4 under a light microscope, 200×; (B) Cells transfected with recombinant plasmid sh4 containing a GFP gene under a fluorescent microscope, 200×.
Effects on Slc38a1 expression
At the 48th hour after the transfection, Slc38a1 mRNA was decreased by all four shRNAs (P<0.05 vs. the negative control; Figure 3). The effect of sh4 on Slc38a1 expression was strongest among the four shRNAs (at 65.28%±3.54%), and was hence used for further experiments.
Figure 3.
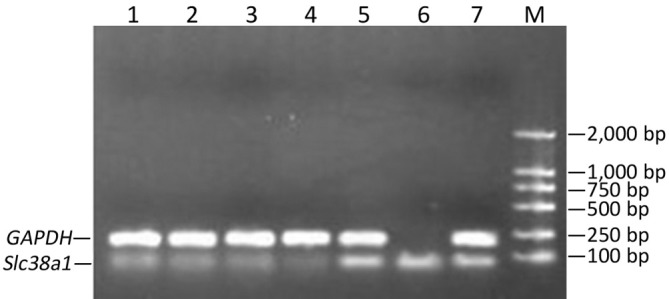
Slc38a1 mRNA in SW1990 cells after transfection of Slc38a1 shRNAs. Lanes l-4, cells transfected with sh1-4; lane 5, cells transfected with shNC; lane 6, cells transfected with shPC; lane 7, mock transfection; M, molecular ladder.
Effects on proliferation, migration, and invasion of SW1990 cells
Sh4 decreased the proliferation (P<0.0001, Figure 4), migration (by 46.7%, P=0.0399; Figure 5), but not invasiveness (Figure 6) of SW1990 cells as reflected by Matrigel assay.
Figure 4.
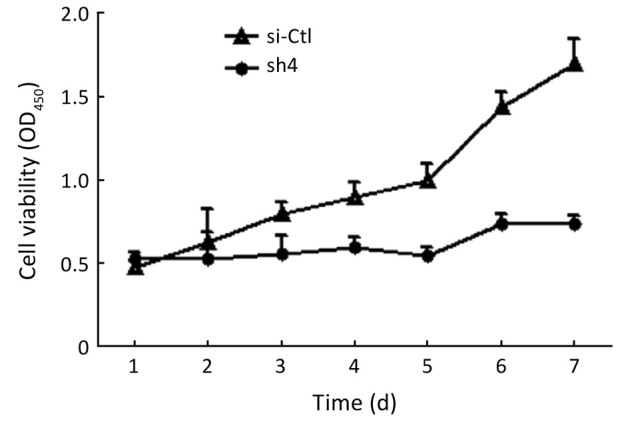
sh4 significantly inhibited the growth of SW1990 cells.
Figure 5.
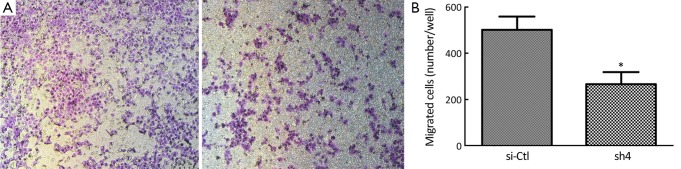
Migration assay of SW1990 cells. (A) sh4 decreased the migration of SW1990 cells in transwell chambers; (B) The number of migrated cells. *, P<0.01 vs. scrambled control.
Figure 6.
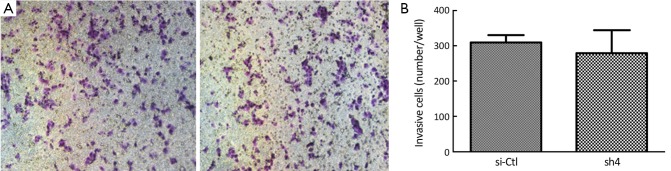
Invasion assay of SW1990 cells. (A) sh4 could not influence the invasiveness of SW1990 cells; (B) The number of invasive cells through the Matrigel.
Discussion
Three types (L, A and B) of amino acid carriers were proposed in 1960s (3). In addition to a critical role in transporting amino acids to organs with significant barrier between the blood and tissue, much more data implicated amino acid transporters involved malignant transformation of mammalian cells or its invasion. Among the various amino acid transporters, LAT-1 of type L system is up-regulated in a variety of tumors, including carcinoma of the urinary bladder (4), adenocarcinoma of the esophagus (5), oral squamous cell carcinoma (6), and particularly in breast cancer (7), choriocarcinoma BeWo cells (8) and hepatic cancer (9). SLC38A1 is a subtype of type A system which is Na+-dependent and transports amino acids from extracellular space into the cells (10). Enhanced SLC38A1 expression has been observed in several other types of malignancies, including liver cancer (2), hilar cholangiocarcinoma (1) and C6 glioma (11). SLC38A1 over-expression is thought to promote cancer cell migration (2).
RNA interference (12-13) technology is widely used to investigate the function of a specific gene (14-15). shRNAs are typically small hairpin RNAs with stem loop structure, and could inhibit the expression of target genes (16). In the present study, we synthesized a series of shRNAs against Slc38a1 using the BLAST analysis (www.ncbi.nlm.nih.gov/BLAST). Upon transfection with the resulting vector, the shRNAs significantly decreased the expression of the target gene in SW1990 pancreatic cancer cells. Selective inhibiting the expression of Slc38a1 profoundly reduced cell proliferation of pancreatic cancer SW1990 cells, as well as the migration. However, invasiveness (Boyden chamber experiments with Matrigel) was not affected, suggesting that SLC38A1 may promote carcinogenesis and migration of pancreatic cancer, but establishment of metastatic lesions requires additional processes/molecules.
The success in building the Slc38a1 shRNA expression vector in the current study also provided important experimental basis for further function study.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- 1.Yu WL, Cong WM, Zhang Y, et al. Overexpression of ATA1/SLC38A1 predicts future recurrence and death in Chinese patients with hilar cholangiocarcinoma. J Surg Res 2011;171:663-8 [DOI] [PubMed] [Google Scholar]
- 2.Kondoh N, Imazeki N, Arai M, et al. Activation of a system A amino acid transporter, ATA1/SLC38A1, in human hepatocellular carcinoma and preneoplastic liver tissues. Int J Oncol 2007;31:81-7 [PubMed] [Google Scholar]
- 3.Christensen HN. Role of amino acid transport and countertransport in nutrition and metabolism. Physiol Rev 1990;70:43-77 [DOI] [PubMed] [Google Scholar]
- 4.Kim DK, Kanai Y, Choi HW, et al. Characterization of the system L amino acid transporter in T24 human bladder carcinoma cells. Biochim Biophys Acta 2002;1565:112-21 [DOI] [PubMed] [Google Scholar]
- 5.Lin J, Raoof DA, Thomas DG, et al. L-type amino acid transporter-1 overexpression and melphalan sensitivity in Barrett's adenocarcinoma. Neoplasia 2004;6:74-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim DK, Ahn SG, Park JC, et al. Expression of L-type amino acid transporter 1 (LAT1) and 4F2 heavy chain (4F2hc) in oral squamous cell carcinoma and its precusor lesions. Anticancer Res 2004;24:1671-5 [PubMed] [Google Scholar]
- 7.Bhat HK, Vadgama JV. Role of estrogen receptor in the regulation of estrogen induced amino acid transport of System A in breast cancer and other receptor positive tumor cells. Int J Mol Med 2002;9:271-9 [PubMed] [Google Scholar]
- 8.Novak D, Quiggle F, Haafiz A.Impact of forskolin and amino acid depletion upon System A activity and SNAT expression in BeWo cells. Biochimie 2006;88:39-44 [DOI] [PubMed] [Google Scholar]
- 9.Campbell WA, Thompson NL. Overexpression of LAT1/CD98 light chain is sufficient to increase system L-amino acid transport activity in mouse hepatocytes but not fibroblasts. J Biol Chem 2001;276:16877-84 [DOI] [PubMed] [Google Scholar]
- 10.Seow HF, Broer S, Broer A, et al. Hartnup disorder is caused by mutations in the gene encoding the neutral amino acid transporter SLC6A19. Nat Genet 2004;36:1003-7 [DOI] [PubMed] [Google Scholar]
- 11.Ogura M, Takarada T, Yoneda Y, et al. Exacerbated vulnerability to oxidative stress in astrocytic C6 glioma cells with stable overexpression of the glutamine transporter slc38a1. Neurochem Int 2011;58:504-11 [DOI] [PubMed] [Google Scholar]
- 12.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806-11 [DOI] [PubMed] [Google Scholar]
- 13.Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411:494-8 [DOI] [PubMed] [Google Scholar]
- 14.Chakraborty C.Potentiality of small interfering RNAs (siRNA) as recent therapeutic targets for gene-silencing. Curr Drug Targets 2007;8:469-82 [DOI] [PubMed] [Google Scholar]
- 15.Rubinson DA, Dillon CP, Kwiatkowski AV, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 2003;33:401-6 [DOI] [PubMed] [Google Scholar]
- 16.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002;296:550-3 [DOI] [PubMed] [Google Scholar]