Muenke syndrome model mice (Fgfr3P244R/+) have hearing loss associated with a supporting cell fate transformation of two Deiters' cells to two pillar cells. Mansour et al. find that the hearing loss is not rescued by reducing the FGFR3c ligand FGF8, but reducing FGF10 is sufficient to rescue cochlear form and function. The P244R mutation caused FGFR3b and FGFR3c to acquire responsiveness to FGF10. Fgf10 heterozygosity allows a gradual reversion of fate-switched cells toward the normal phenotype.
Keywords: FGFR3 P244R, pillar cell, Deiters' cell, cell fate transformation, FGF10, mouse
Abstract
The stereotyped arrangement of cochlear sensory and supporting cells is critical for auditory function. Our previous studies showed that Muenke syndrome model mice (Fgfr3P244R/+) have hearing loss associated with a supporting cell fate transformation of two Deiters' cells to two pillar cells. We investigated the developmental origins of this transformation and found that two prospective Deiters' cells switch to an outer pillar cell-like fate sequentially between embryonic day 17.5 (E17.5) and postnatal day 3 (P3). Unexpectedly, the Fgfr3P244R/+ hearing loss and supporting cell fate transformation are not rescued by genetically reducing fibroblast growth factor 8 (FGF8), the FGF receptor 3c (FGFR3c) ligand required for pillar cell differentiation. Rather, reducing FGF10, which normally activates FGFR2b or FGFR1b, is sufficient for rescue of cochlear form and function. Accordingly, we found that the P244R mutation changes the specificity of FGFR3b and FGFR3c such that both acquire responsiveness to FGF10. Moreover, Fgf10 heterozygosity does not block the Fgfr3P244R/+ supporting cell fate transformation but instead allows a gradual reversion of fate-switched cells toward the normal phenotype between P5 and at least P14. This study indicates that Deiters' and pillar cells can reversibly switch fates in an FGF-dependent manner over a prolonged period of time. This property might be exploited for the regulation of sensory cell regeneration from support cells.
The sensory epithelium of the mammalian cochlear duct comprises a stereotyped mosaic of sensory and supporting cells, both of which are derived from a prosensory progenitor pool and are critical to auditory function. From cochlear base to apex, there are four longitudinal rows of sensory hair cells situated above a basilar membrane. The row closest (medial) to the central axis of the coiled cochlea is composed of inner hair cells, and the three more lateral rows are composed of outer hair cells. These sensory cells convert sound-induced movement of the basilar membrane into electrical signals that travel through the cochlear nerve and into the brain for auditory perception. Each type of hair cell has associated supporting cells that transmit basilar membrane movement to the sensory cells, provide rigidity to the epithelium, and assist with recycling ions and neurotransmitters produced by hair cells. Viewed in cross-section (Fig. 1B,C), an inner phalangeal cell supports the inner hair cell, and a Deiters' cell supports each outer hair cell. In addition, two pillar cells separate the rows of inner and outer hair cells, forming the tunnel of Corti. Dysfunction of any of these cell types consequent to genetic mutation or environmental or age-related insults causes hearing impairment, and in most cases, sensory hair cell death ensues. In fish and birds, hair cells regenerate naturally by either direct transdifferentiation or proliferation and subsequent differentiation of remaining support cells. Mammalian hair cells, however, do not normally regenerate, and most strategies designed to promote such regeneration are based on manipulating the developmental signals controlling cochlear prosensory progenitor proliferation and differentiation (for review, see Kelly and Chen 2009; Groves 2010; Monzack and Cunningham 2013; Rubel et al. 2013; Wan et al. 2013).
Figure 1.
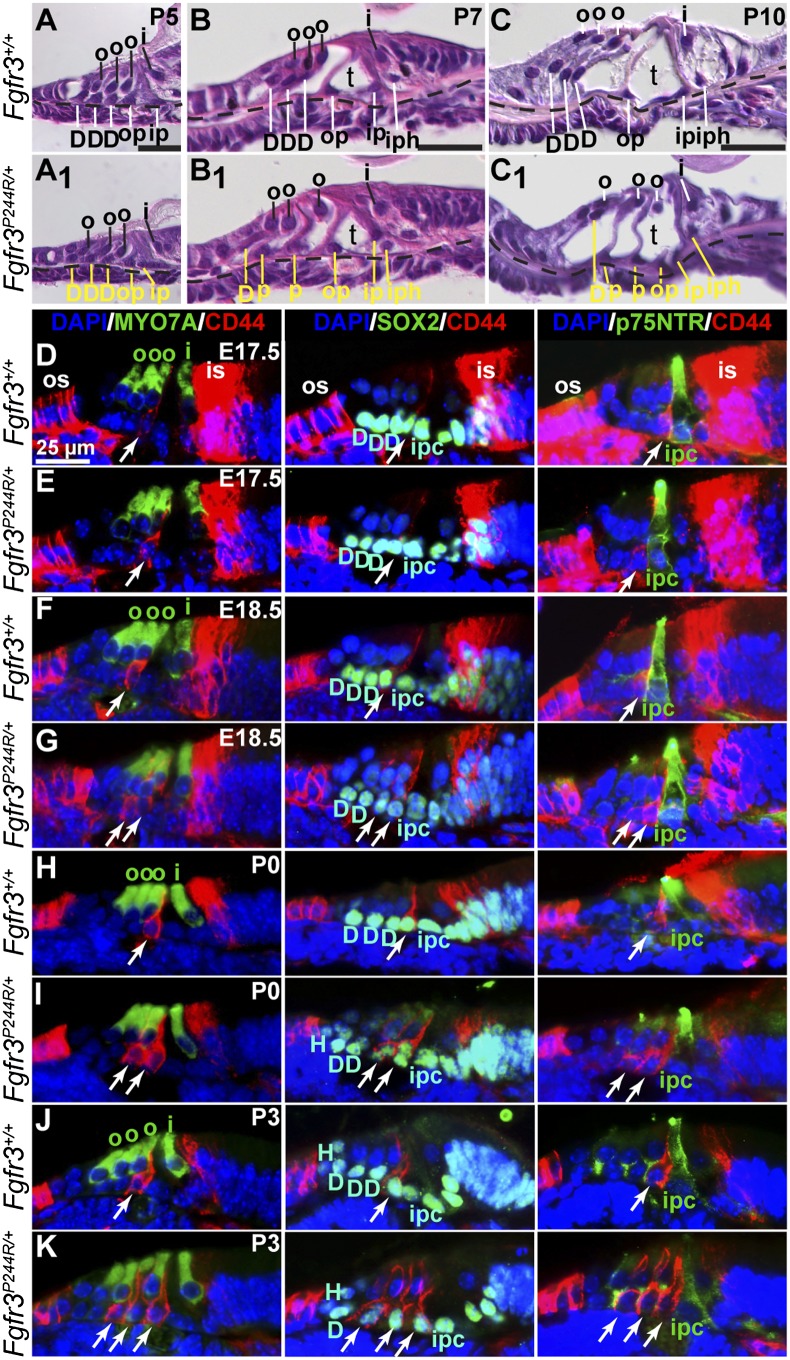
A sequential transformation of two Deiters' cells to two pillar cells occurs between E17.5 and P3 in Muenke syndrome model cochleae. Hematoxylin and eosin-stained basal cochlear cross-sections from wild-type (A–C) and Fgfr3P244R/+(A1–C1) mice. Stage is indicated at the top right of the wild-type sections, and a dotted line marks the basilar membrane. Yellow lines and text in mutant panels indicate support cell nuclei and presumptive identification based on position. Bars in the bottom right of A–C indicate 25 μm and apply to corresponding mutant panels. (D–K) Mid-base cochlear cross-sections of wild-type and Fgfr3P244R/+samples treated with primary antibodies directed against CD44 (outer pillar cells) and MYO7A (hair cells) (left column), CD44 and SOX2 (support-cell nuclei) (middle column), or CD44 and p75NTR (inner pillar cells) (right column). The color of the name of each antigen shown in the column headings indicates the color of the secondary antibodies used for detection. DAPI (blue) labels all nuclei. Genotypes are indicated at the left of each row, and stages are indicated at the top right of the first panel in each row. White arrows indicate outer pillar or pillar-like cells. The bar shown in D applies. For all panels, lateral is to the left. (D) Deiters' cells; (H) Hensen's cell; (i) inner hair cell; (ipc), inner pillar cell; (is) inner sulcus; (o) outer hair cell; (op) outer pillar; (os) outer sulcus; (t) tunnel of Corti.
Fibroblast growth factor (FGF) signaling plays critical dosage-sensitive roles in many aspects of inner ear development, from induction of the otic placode through otic vesicle morphogenesis to cochlear sensory and supporting cell fate specification (for review, see Schimmang 2007; Groves and Fekete 2012; Wu and Kelley 2012). Mammalian FGFs comprise an 18-member family of secreted ligands that signal by activating FGF receptor (FGFR) tyrosine kinases, which are encoded by four genes. Fgfr genes 1–3 are alternatively spliced such that differential inclusion of either of two exons encoding a portion of the FGF-binding domain leads to alternative production of “b” and “c” receptor isoforms. These differ with respect to FGF-binding specificity and affinity in cell culture-based activity and in vitro binding assays (for review, see Goetz and Mohammadi 2013). Mutations in ligand/receptor pairs often lead to shared loss-of-function phenotypes. For example, FGF3 and FGF10 both preferentially activate FGFR2b, and loss of any one of these genes causes abnormal morphogenesis of the otic vesicle (Pirvola et al. 2000; Pauley et al. 2003; Hatch et al. 2007). In the developing cochlear sensory epithelium, shared loss-of-function phenotypes suggest that FGF8, which is expressed specifically by the developing inner hair cell, signals through FGFR3, expressed in the laterally adjacent developing support cells, to promote differentiation of the two closest support cells as pillar cells. Thus, mice lacking Fgfr3 or humans carrying a presumed dominant-negative mutation in FGFR3 are hearing impaired, and the mice show incomplete differentiation of pillar cells (Colvin et al. 1996; Toydemir et al. 2006; Hayashi et al. 2007; Puligilla et al. 2007). Similarly, mice lacking inner ear expression of Fgf8 also show aberrant pillar cell differentiation (Jacques et al. 2007; Zelarayan et al. 2007). FGFR3c is presumed to be the relevant isoform for pillar cell specification because although Fgfr3c-null mice have not been tested for hearing loss, they show other phenotypes associated with complete loss of Fgfr3, whereas Fgfr3b-null mice have no reported abnormalities (Eswarakumar and Schlessinger 2007).
Just as a loss of FGF signaling leads to hearing impairment, so, too, does a gain of FGF signaling. Mice lacking the negative feedback regulator of FGF signaling, Sprouty2, show significant hearing loss and a cochlear supporting cell fate transformation of one Deiters' cell to one pillar cell such that they have three, instead of two, rows of pillar cells and only two, instead of three, rows of Deiters' cells. Significantly, the hearing loss and supporting cell fate transformation are rescued when one copy of Fgf8 is removed from the Spry2-null background (Shim et al. 2005). These data, together with the loss-of-function studies (Colvin et al. 1996; Hayashi et al. 2007; Jacques et al. 2007; Puligilla et al. 2007; Zelarayan et al. 2007), support the model of an FGF signaling gradient in the developing cochlear sensory epithelium in which support cell progenitors experiencing the highest levels of FGF8/FGFR3 signaling differentiate as pillar cells, and those that are farther from the FGF source differentiate as Deiters' cells (Shim et al. 2005).
A gain of FGF signaling can also be caused by FGFR missense mutations. Such dominant mutations are frequent causes of craniosynostosis in humans, and some, like that causing Muenke syndrome (Online Mendelian Inheritance in Man [OMIM] 602849; FGFR3 P250R), are also associated with hearing loss (Agochukwu et al. 2010). The Muenke syndrome mutation is located in the extracellular domain of FGFR3, affects both the “b” and “c” receptor isoforms, and increases the binding affinity of FGFR3c for certain FGFs (Ibrahimi et al. 2004b). Although Muenke syndrome hearing loss was originally thought to be variable, we found that all subjects have a mild–moderate predominantly low-frequency sensorineural (cochlear) hearing loss of ∼20–40 decibels (dB). Furthermore, mice heterozygous for the paralogous mutation (FGFR3 P244R) have 100% penetrant hearing loss that is similar in character (worse at low frequencies than at high) but more severe in degree (up to 60 dB, depending on genetic background). The Muenke syndrome model mice have a support cell fate transformation similar to that of Spry2-null mice but more severe: Two rows of Deiters' cells are converted to two rows of pillar cells such that there is a total of four rows of pillar cells and one row of Deiters' cells (Mansour et al. 2009).
Here, we explore the development of this phenotype and determine the FGF ligand responsible for the Muenke syndrome model inner ear phenotypes by assaying for rescue of auditory function and support cell fate consequent to reducing cochlear-expressed Fgfs in the Fgfr3P244R/+ background. Our results implicate an unexpected ligand driving Fgfr3P244R/+ cochlear pathogenesis, and our studies of the development and rescue of the support cell phenotype reveal an extended period of FGF-dependent plasticity in the identity of supporting cells that might ultimately prove useful in regulating sensory hair cell regeneration.
Results
The Muenke syndrome model cochlear support cell fate transformation starts between embryonic day 17.5 (E17.5) and E18.5 and is complete by postnatal day 3 (P3)
To determine when the supporting cell fate transformation occurred in the Muenke syndrome model, we first examined histologic basal cross-sections of wild-type and Fgfr3P244R/+cochleae between E15.5 and P10. In P5 (Fig. 1A) and younger (data not shown) wild-type samples, each of the presumptive outer pillar (op), inner pillar (ip), and three Deiters' (D) cell nuclei were located immediately above the basilar membrane (dashed line) and beneath the inner (i) and outer (o) hair cell nuclei. At these stages Fgfr3P244R/+ samples showed no obvious differences in cytoarchitecture (Fig. 1A1, support cell nuclei are indicated by yellow lines, and presumptive identities are based on position). By P7 in wild-type samples, the tunnel of Corti (t) opened, and while the two pillar cell nuclei remained quite close to the basilar membrane, the three Deiters' cell nuclei had moved upward with the hair cell nuclei (Fig. 1B). In contrast, P7 Fgfr3P244R/+ samples had an abnormal appearance. Although the tunnel of Corti had opened and there were five support cell nuclei found lateral to the inner hair cell, four were still located immediately above the basilar membrane, as expected for pillar cells. Only one support cell nucleus (the most lateral) had moved upward and was close to the nucleus of the most lateral outer hair cell, assuming the position expected for a Deiters' cell (Fig. 1B1). By P10, just prior to the onset of hearing in mice, the differences between the two genotypes were more striking. While wild-type sections showed normally located pillar and Deiters' cell nuclei (Fig. 1C), the Fgfr3P244R/+ sample had four pillar-like cells and only one with a Deiters'-like position (Fig. 1C1), similar to the differences reported in mature samples (Mansour et al. 2009).
To determine when molecular differences between supporting cell types were first evident as well as the time course of supporting cell fate transformation, we examined immunostained mid-base cochlear cross-sections prepared from wild-type and Fgfr3P244R/+ samples between E17.5 and P3 (Figs. 1D–K). Outer pillar cells were labeled with antibodies directed against CD44, which is also expressed by a subset of cells in the inner and outer sulci on either side of the sensory epithelium but not by inner pillar cells (Hertzano et al. 2010). Sensory hair cell bodies and support cell nuclei were identified using antibodies directed against MYO7A and SOX2, respectively. DAPI was used to label all nuclei. We found that in wild-type samples, CD44 immunoreactivity was just barely detectable at E17.5 and was localized to a single organ of Corti support cell (Fig. 1D, white arrows) adjacent to the innermost (medial) outer hair cell (Fig. 1D, o). The CD44 staining surrounded a SOX2-positive nucleus that was adjacent to three SOX2-positive Deiters' cell nuclei (Fig. 1D, D), identifying it as an outer pillar cell. The same pattern of immunoreactivity was found in the E17.5 Fgfr3P244R/+ sample (Fig. 1E), indicating that the cell fate change seen in older animals had not yet occurred. One day later, at E18.5, we found that relative to the wild-type sample (Fig. 1F), the Fgfr3P244R/+ epithelium had acquired an additional CD44-positive cell that costained for SOX2, but the number of SOX2-positive nuclei located immediately above the basilar membrane was unchanged (Fig. 1G), suggesting that one Deiters' cell had converted to an outer pillar cell-like fate. Immunostaining of P0 samples (Figs. 1H,I) was similar to those at E18.5, but by P3, we found that relative to wild-type samples (Fig. 1J), Fgfr3P244R/+ samples now had three CD44-positive cells with SOX2-positive nuclei, and there was still no change in the total number of SOX2-positive nuclei above the basilar membrane (Fig. 1K). The CD44-positive cells did not costain with antibodies directed against the inner pillar cell marker p75NTR in any sample (Fig. 1D–K, third column). Thus, the cell fate transformation of two Deiters' cells to two pillar cells, identified as outer pillar-like cells based on CD44 staining, occurred gradually starting at E18.5 and finishing by P3. Attempts to determine whether the outer pillar-like cells seen in Fgfr3P244R/+ cochleae retain any Deiters'-like character by labeling them with antibodies directed against S100A1 (Hayashi et al. 2007; Puligilla et al. 2007) were frustrated by persistent colabeling of pillar cells even in wild-type controls (data not shown).
FGF signaling expands laterally in Muenke syndrome model cochleae
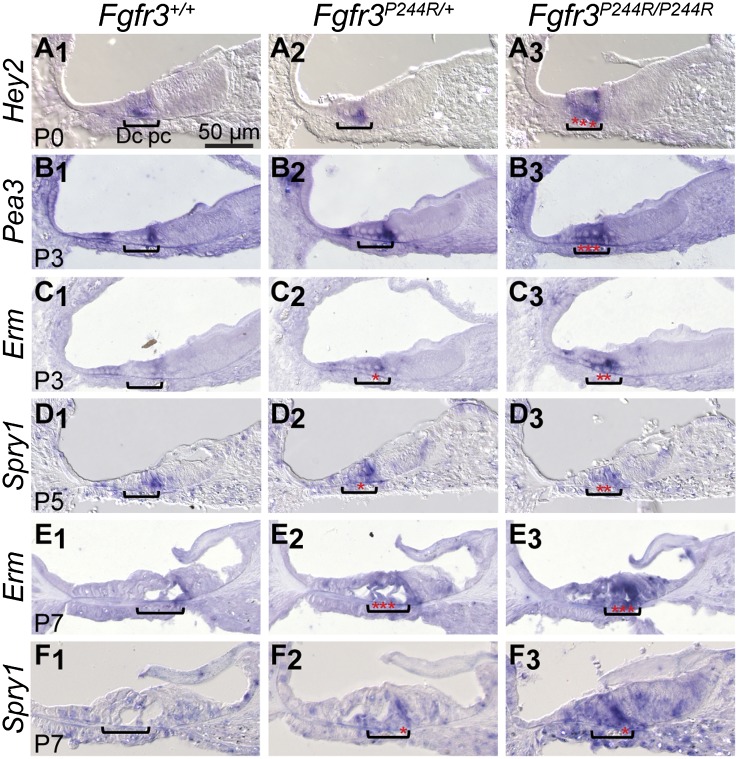
After E18.5, developing Deiters' and pillar cells are the only cochlear cell types that express Fgfr3 (see Supplemental Fig. S1; Hayashi et al. 2010). To determine where FGF signaling activity changes during the development of the Fgfr3P244R/+ cochlear phenotype, we assayed expression of known transcriptional targets of FGF signaling (Hey2, Pea3/Etv4, Erm/Etv5, and Spry1) by in situ hybridization to RNA in wild-type control, heterozygous, and homozygous mutant sections (Fig. 2). At P0, Hey2 was expressed in developing pillar cells (Fig. 2A1; Doetzlhofer et al. 2009), and although unaffected in heterozygotes, its domain expanded laterally into the Deiters' cell region in homozygotes (Fig. 2A2,A3). At P3, Pea3 and Erm were expressed strongly and weakly, respectively, in cells flanking the Deiters' cell area in wild-type samples (Fig. 2B1,C1; Hayashi et al. 2008). Pea3 expression expanded into the Deiters' cell area in homozygotes (Fig. 2B3), and Erm expression expanded in both heterozygotes and homozygotes (Fig. 2C2,C3). At P5, Spry1 was expressed in wild-type developing pillar cells (Fig. 2D1; Shim et al. 2005), and ectopic expression was detected in the Deiters' cell region of both heterozygotes and homozygotes (Fig. 2D2,D3). At P7, weak expression of Erm and Spry1 was evident in wild-type pillar cells (Fig. 2E1,F1), and ectopic expression of both genes was evident in both heterozygotes and homozygotes (Fig. 2E2,E3,F2,F3). Thus, in Fgfr3P244R/+ cochleae, FGF signaling was expanded and/or elevated in cells that would ordinarily develop as Deiters' cells but assumed an outer pillar cell-like fate. Furthermore, this expansion persisted beyond P3, the stage at which the supporting cell fate transformation was complete.
Figure 2.
FGF signaling expands into the Deiters' cell region of Fgfr3P244R/+ and/or Fgfr3P244R/P244R cochleae. In situ hybridization of mid-base cochlear cross-sections with cRNA probes for known transcriptional targets of FGF signaling is shown. Genotypes (on the 129S6 inbred background) are indicated at the top of each column. Developmental stages are shown at the bottom left of each row, and probe names are indicated at the left of each row. Brackets indicate the location of the developing Deiters' cells, and red stars highlight expansion and/or intensification of the hybridization signal into this region. For each panel, lateral is to the left, and the bar shown in A1 applies.
The hearing loss and supporting cell fate transformation characteristic of the Muenke syndrome model are rescued by reducing the copy number of Fgf10
The hearing loss and one-to-one Deiters'-to-pillar cell fate transformation characteristic of Spry2-null mice are rescued by reducing the copy number of Fgf8 (Shim et al. 2005). To determine the FGF ligand responsible for the hearing loss and supporting cell fate transformation seen in Fgfr3P244R/+ cochleae, we first reduced the copy number of Fgf8 and several other FGFs that, like FGF8, are expressed in the cochlear duct and effectively interact with FGFR3c in cell culture-based activity or in vitro binding assays (Ibrahimi et al. 2004b; Zhang et al. 2006). We then assayed auditory brainstem response (ABR) thresholds to broadband click and pure tone pip stimuli of 8, 16, and 32 kHz (representing the low-, middle-, and high-frequency ranges of mouse hearing) in 3- to 5-wk-old animals and calculated the degree of hearing loss.
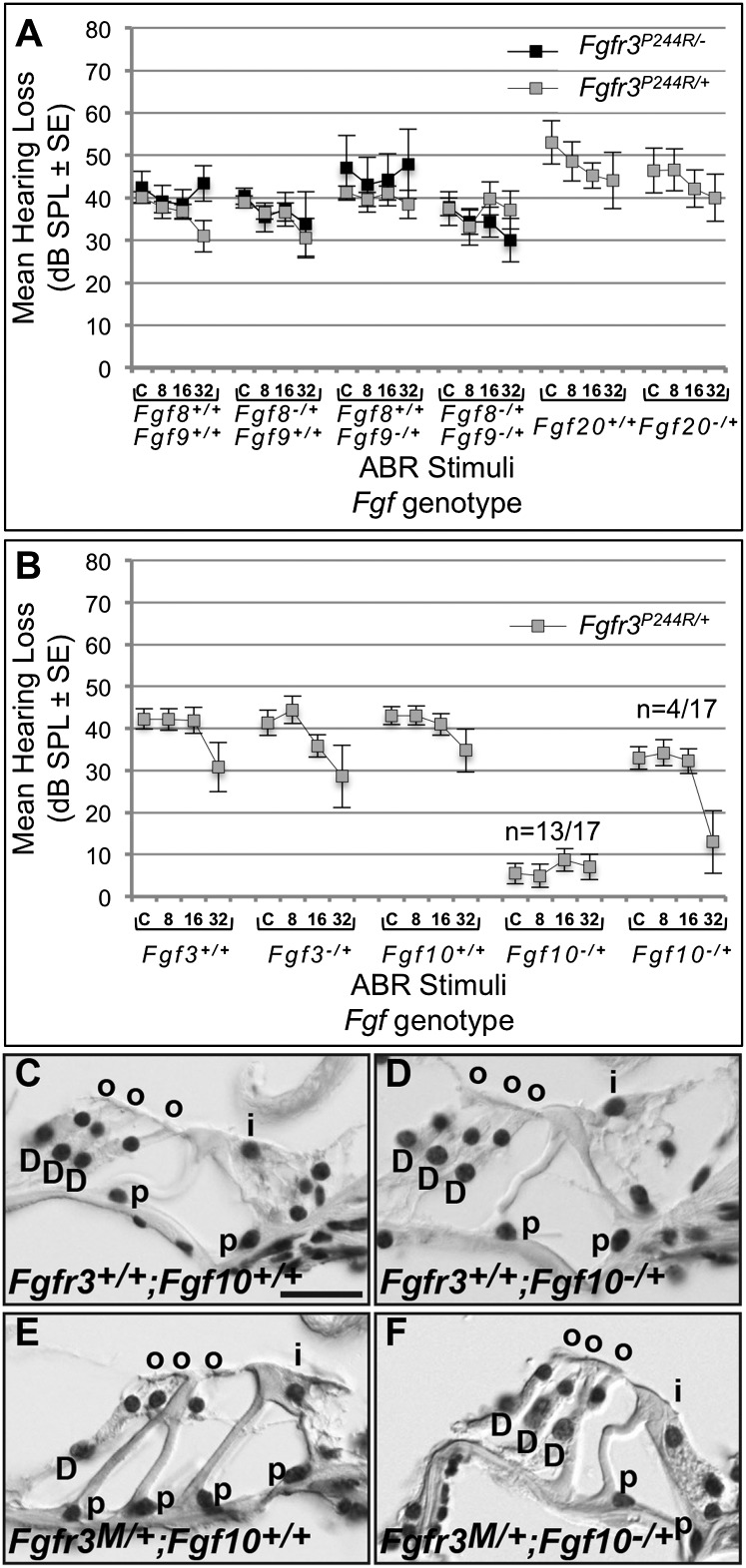
Preliminary studies of Fgfr3P244R/+ mice lacking one copy of Fgf8, one copy of Fgf9, or one copy each of Fgf8 and Fgf9 (Supplemental Fig. S1 of Mansour et al. 2009) were expanded, and we also determined ABRs of mice with those same genotypes after removing the remaining wild-type copy of Fgfr3. In addition, we tested Fgfr3P244R/+ mice in which one copy of Fgf20 was removed. Fgf20 encodes another potential FGFR3c ligand (Zhang et al. 2006) that is expressed in the developing prosensory domain and is required for outer hair cell differentiation (Hayashi et al. 2008; Huh et al. 2012). Even in a mixed genetic background, Fgfr3P244R/+;Fgf8+/+;Fgf9+/+ mice had hearing loss that was 100% penetrant, mild–moderate in degree (30–40 dB), and more severe (40 dB) at low frequencies than at high (30 dB) (Fig. 3A, left four gray boxes). None of the copy number reductions involving Fgf8, Fgf9, Fgf20, or Fgfr3 rescued Fgfr3P244R/+ hearing loss, and in some cases, they actually made the hearing loss worse (Fig. 3A).
Figure 3.
The hearing loss and cochlear support cell fate transformation characteristic of Muenke syndrome model mice are rescued by removing one copy of Fgf10. (A) Gray boxes show mean hearing loss (in decibels; SPL ± tandard error of the mean [SEM] difference) on the Y-axis plotted against click (C) and 8, 16, and 32 kHz tone pip stimuli calculated following ABR testing of Fgfr3P244R/+ and control mice heterozygous for null alleles of Fgf8, Fgf9, Fgf8, and Fgf9, and Fgf20. Black boxes show the mean hearing loss after removal of one copy of Fgfr3 and Fgf8, Fgf9, or Fgf8 and Fgf9. (B) Gray boxes show mean hearing loss of Fgfr3P244R/+ mice to the same stimuli as in A and heterozygous for either Fgf3 or Fgf10. The Fgf10 results are further split into the majority (13 of 17) showing full restoration of auditory thresholds and the minority (four of 17) in which only high-frequency hearing improved. (C–F) Hematoxylin and eosin-stained cochlear cross-sections from ABR-tested mice of the genotypes indicated at the bottom left of each panel. (D) Deiters' cell; (i) inner hair cell; (o) outer hair cell; (p) pillar cell. For C–F, lateral is to the left, and the bar shown in C (20 μm) applies to all panels.
Next, we noticed that Fgf3 and Fgf10, which encode effective ligands for FGFR2b and FGFR1b (Zhang et al. 2006) but are unable to bind in vitro to either FGFR3c or FGFR3c P250R (equivalent to mouse FGFR3c P244R) (Ibrahimi et al. 2004b), are expressed weakly and strongly, respectively, in the vicinity of the developing cochlear support cells (Supplemental Figs. S1, S2 for Fgf10; Wilkinson et al. 1989; Pirvola et al. 2000; Pauley et al. 2003). Removing one copy of Fgf3 from Fgfr3P244R/+ mice had no effect on hearing loss. Remarkably, however, removing one copy of Fgf10 was sufficient to completely restore ABRs in 13 of 17 Fgfr3P244R/+ mice and improved high-frequency responses in the remaining four such animals (Fig. 3B).
To assess cochlear support cell differentiation in mice with the double-heterozygous genotype, we prepared histologic sections of inner ears following ABR testing. As expected, wild-type and Fgf10−/+ control animals with normal hearing had cochleae that showed the normal pattern of three Deiters' cells and two pillar cells (Fig. 3C,D). Support cells from hearing-impaired Fgfr3P244R/+ mice were transformed, showing one Deiters'-like and four pillar-like cells (Fig. 3E), similar to those of inbred Fgfr3P244R/+ mice reported previously (Mansour et al. 2009) and hearing impaired Fgfr3P244R/+; Fgf8−/+ mice (data not shown). Support cells of normal hearing Fgfr3P244R/+; Fgf10−/+ mice (Fig. 3F), however, appeared similar to those of controls. Therefore, these data provide genetic evidence that FGF10 is responsible for generating the Muenke model hearing loss and cochlear supporting cell fate transformation.
Rescue of the Muenke syndrome model Deiters'-to-pillar cell fate transformation by Fgf10 reduction occurs gradually after P5
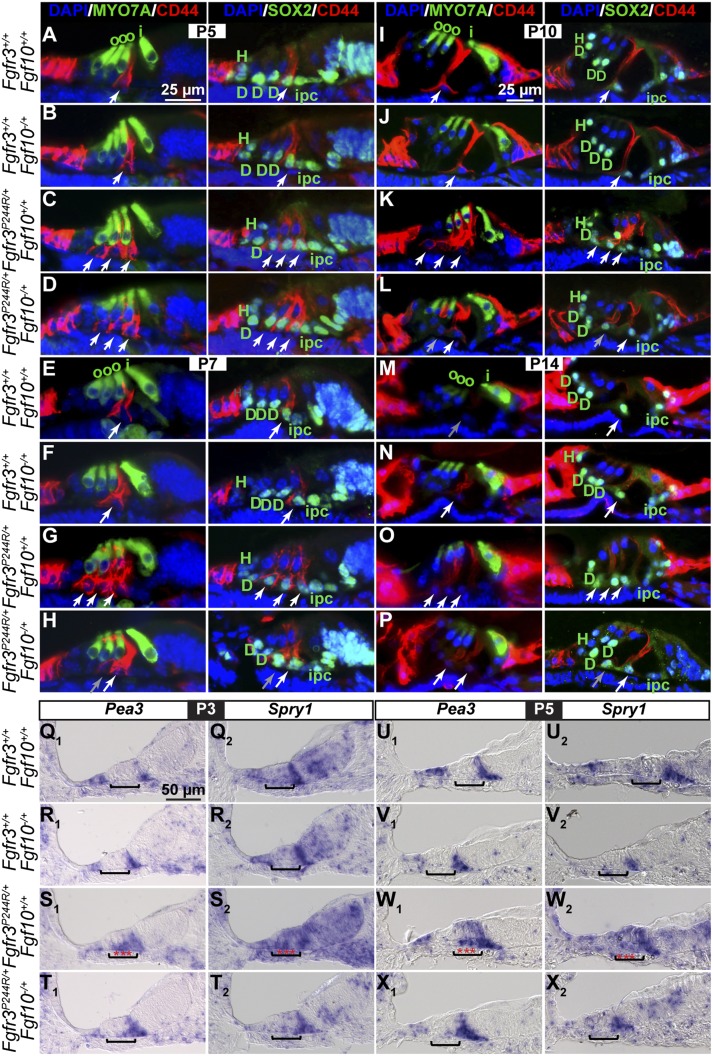
To determine when Fgf10 heterozygosity rescues the Fgfr3P244R/+ support cell fate transformation, we examined immunostained mid-base cochlear cross-sections prepared from wild-type, Fgf10−/+, Fgfr3P244R/+, and Fgfr3P244R/+;Fgf10−/+ samples between E17.5 and P14 (Fig. 4A–P). No differences in Deiters'-like versus outer pillar-like cell numbers between Fgfr3P244R/+ and Fgfr3P244R/+;Fgf10−/+ samples were apparent between E17.5 and P3 (data not shown). Even at P5, a stage when cochlear Fgf10 expression is diminishing naturally in a basal-to-apical wave (Supplemental Fig. S2), relative to the control samples with one outer pillar cell and three Deiters' cells (Fig. 4A,B) and Fgfr3P244R/+ samples with three outer pillar-like cells and one Deiters'-like cell (Fig. 4C), Fgfr3P244R/+;Fgf10−/+ samples still had a cochlear support cell phenotype similar to that of Fgfr3P244R/+ samples, with only a slight diminishment in CD44 staining of the most lateral outer pillar-like cell (Fig. 4D). It was not until P7, when Fgf10 expression in the cochlear base is almost extinguished (Supplemental Fig. S2), that we first detected a reversion of Fgfr3P244R/+;Fgf10−/+ support cells back toward a more normal phenotype. Fgfr3P244R/+;Fgf10−/+ sections showed two outer pillar-like cells (the outermost of which had weak CD44 expression) (Fig. 4H, gray arrow) and two Deiters'-like cells (Fig. 4H). A similar phenotype was seen in P10 Fgfr3P244R/+;Fgf10−/+ samples relative to control and Fgfr3P244R/+ samples (Figs. 4I–L). Even at P14, when the CD44 immunoreactivity in outer pillar cells was severely diminished even in control samples (Figs. 4M,N), Fgfr3P244R/+;Fgf10−/+ support cells were much closer to but not entirely normal, with residual CD44 expression in one more support cell than normal (Figs. 4O,P). This may suggest that either double heterozygotes are never entirely normal (although the large majority become hearing-competent) (Fig. 3B) or fate transformation is not complete until after the stages at which CD44 expression can be detected. Regardless, heterozygosity for Fgf10 does not prevent the cochlear support cell fate transformation characteristic of the Muenke syndrome model. Rather, it appears that the levels of FGF10 in a heterozygote become insufficient to maintain the transformation, and this permits a reversion to the normal phenotype after the fact.
Figure 4.
Rescue of the Fgfr3P244R/+ Deiters'-to-pillar cell fate transformation by Fgf10 heterozygosity proceeds gradually after P5 and is preceded by reversal of FGF signaling expansion into the Deiters' cell region. (A–P) Immunostaining of mid-base cochlear cross-sections prepared from wild-type and Fgf10−/+ controls and Fgfr3P244R/+ and Fgfr3P244R/+;Fgf10−/+ samples with antibodies directed against CD44 (outer pillar cells) and MYO7A (hair cells) or CD44 and SOX2 (support cell nuclei). The color of the name of each antigen shown in the column headings indicates the color of the secondary antibodies used for detection. DAPI (blue) labels all nuclei. White arrows indicate outer pillar cells. Gray arrows indicate CD44-positive cells with diminished staining. (Q–X) In situ hybridization of P3 and P5 mid-base cochlear cross-sections with cRNA probes directed against FGF signaling targets Pea3 and Spry1. Brackets indicate the location of the developing Deiters' cells, and red stars indicate expansion and/or intensification of the hybridization signal in Fgfr3P244R/+ samples. Genotypes are indicated at the left of each row, stages are indicated above each set of four genotypes, and for each panel, lateral is to the left. The bar in A applies to A–H, the bar in I applies to I–P, and the bar in Q applies to Q–X. (D) Deiters' cells; (H) Hensen's cell; (i) inner hair cell; (ipc) inner pillar cell; (o) outer hair cell.
Removing one copy of Fgf10 rescues the expansion of FGF signaling in Muenke syndrome model cochleae
To ask whether removing one copy of Fgf10 from Fgfr3P244R/+ mice affected FGF signaling in the developing Deiters' cell region, we examined Pea3 and Spry1 expression at P3 and P5. At both stages, Pea3 expression flanked developing Deiters' cells in wild-type and Fgf10−/+ control samples (Fig. 4Q1,R1,U1,V1), expanded into the Deiters' cell region in Fgfr3P244R/+;Fgf10+/+ samples (Fig. 4S1,W1), and was restored to normal in Fgfr3P244R/+;Fgf10−/+ samples (Fig. 4T1,X1). The more obvious lateral spreading of Pea3 expression in this P3 Fgfr3P244R/+ sample (Fig. 4S1) relative to that in Figure 2B2 could be a consequence of genetic background and/or in situ hybridization development times. Similarly, Spry1, which was expressed most strongly in the wild-type and Fgf10−/+ control pillar cell region (Fig. 4Q2,R2,U2,V2) and expanded into the Deiters' cell region of Fgfr3P244R/+ samples (Fig. 4S2,W2), was restored to its normal domain in Fgfr3P244R/+;Fgf10−/+ samples (Fig. 4T2,X2). Therefore, rescue of the Muenke syndrome model support cell phenotype correlates with a reduction in FGF signaling indicators in the developing Deiters' cell region.
FGF10 activates FGFR3b P244R and FGFR3c P244R but not their wild-type counterparts
Since FGF10 is not an effective ligand for FGFR3b or FGFR3c in cell culture-based assays (Zhang et al. 2006) and does not bind to human FGFR3c or FGFR3c P250R in surface plasmon resonance assays (Ibrahimi et al. 2004b), one mechanism that could explain how FGF10 mediates the Muenke syndrome model support cell phenotype would be if FGF8 activation of the mutant receptor induces expression of an FGF10 receptor, such as FGFR2b or FGFR1b, in developing Deiters' cells, making them responsive to FGF10. However, we found that not only was there no change in cochlear Fgfr2 expression in Fgfr3P244R heterozygotes and homozygotes relative to wild-type samples, there were also no changes in Fgfr1 or Fgfr3 expression, and furthermore, neither Fgf8 nor Fgf10 expression was altered (Supplemental Fig. S1).
Next, we asked whether the Muenke syndrome model cochlear phenotype might be caused by FGF10 activation of signaling through FGFR3b P244R, the human equivalent of which was not tested in the binding assay (Ibrahimi et al. 2004b). RT–PCR assays confirmed that both the Fgfr3b and Fgfr3c splice isoforms are indeed expressed in wild-type and Fgfr3P244R/+ cochlear epithelia, with the level of Fgfr3c consistently higher than that of Fgfr3b, despite extensive cycles of amplification (Supplemental Fig. S3).
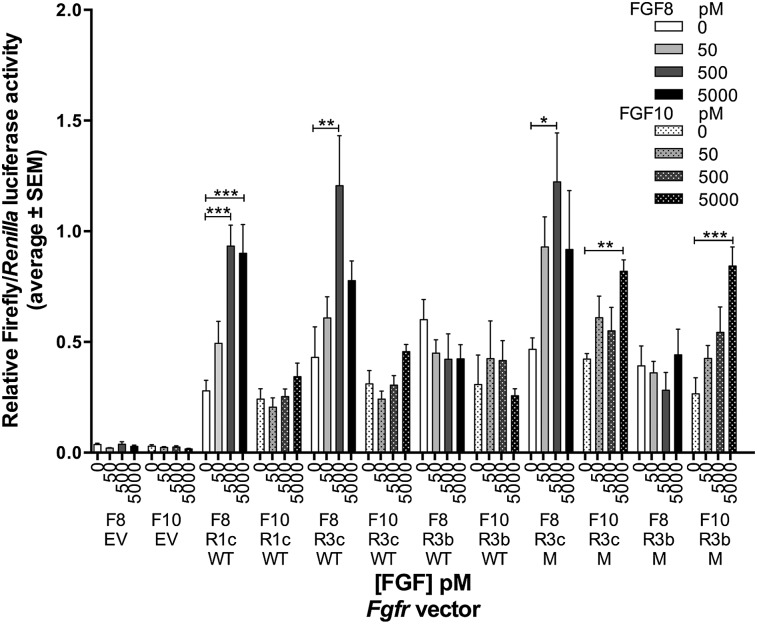
To determine the relative abilities of FGF8 and FGF10 to activate the wild-type and P244R versions of FGFR3b and FGFR3c, we transiently transfected expression vectors for each wild-type and mutant receptor isoform together with an FGF-responsive firefly luciferase reporter plasmid and a Renilla luciferase transfection control plasmid into L6 rat myoblasts, treated the transfected cells with several FGF concentrations, and determined the ratio of firefly to Renilla luciferase activity (Fig. 5). The Fgfr3 cDNAs were modified previously such that the extracellular domain encodes FGFR3b or FGFR3c, but the intracellular domain is derived from FGFR1 (Ornitz et al. 1996). Renilla luciferase activity from the transfection control vector was robust but showed no FGF responsiveness (data not shown). Neither FGF was able to induce luciferase activity in cells transfected with an expression vector lacking an Fgfr cDNA (empty vector), whereas, as expected, FGF8, but not FGF10, induced luciferase activity significantly above background levels in cells transfected with a wild-type FGFR1c expression vector. FGFR1c was also activated by FGF2 to the same extent seen by Raivio et al. (2009) (data not shown). These control conditions confirmed that there is no endogenous FGFR expression in L6 cells (Olwin and Hauschka 1989; Roghani et al. 1996) and that there is endogenous FGF expression and/or perdurant FGF from the serum used to culture the L6 cells.
Figure 5.
FGF10 activates both FGFR3b P244R and FGFR3c P244R but not their wild-type counterparts. Bar graph from one of three replicate experiments showing luciferase activity induced by FGF8 or FGF10 stimulation of FGFRs. L6 myoblasts were transfected with the indicated FGFR expression plasmid (or an empty vector [EV]), an FGF-responsive firefly luciferase plasmid, and a Renilla luciferase transfection control plasmid. Cells were treated with the indicated concentrations (in picomolar) of FGF8 (F8) or FGF10 (F10). The ratio of firefly to Renilla luciferase activity for each assay is shown by the height of each bar. Error bars represent 1 SEM. Horizontal bars topped with asterisks indicate significant induction of luciferase activity using Dunnett's multiple comparison test. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001; (WT) wild type; (M) P244R.
As expected, we found that, in all three trials, FGF8, but not FGF10, significantly induced wild-type FGFR3c (approximately threefold) (Fig. 5). FGF2 was also able to activate FGFR3c (data not shown). Also as expected, in all three trials, neither FGF8 nor FGF10 activated wild-type FGFR3b. In contrast, although FGF8 significantly activated FGFR3c P244R to an extent similar to wild-type FGFR3c in all three trials, in two of the three trials, FGF10 also significantly activated FGFR3c P244R (approximately twofold) (Fig. 5). Finally, FGF8 did not activate FGFR3b P244R, but FGF10 did so significantly in all three trials (approximately threefold) (Fig. 5). Thus, FGF8 can activate FGFR3c P244R similarly to the wild-type receptor, and FGF10 is an effective ligand for both of the Muenke syndrome FGFR3 receptor isoforms.
Discussion
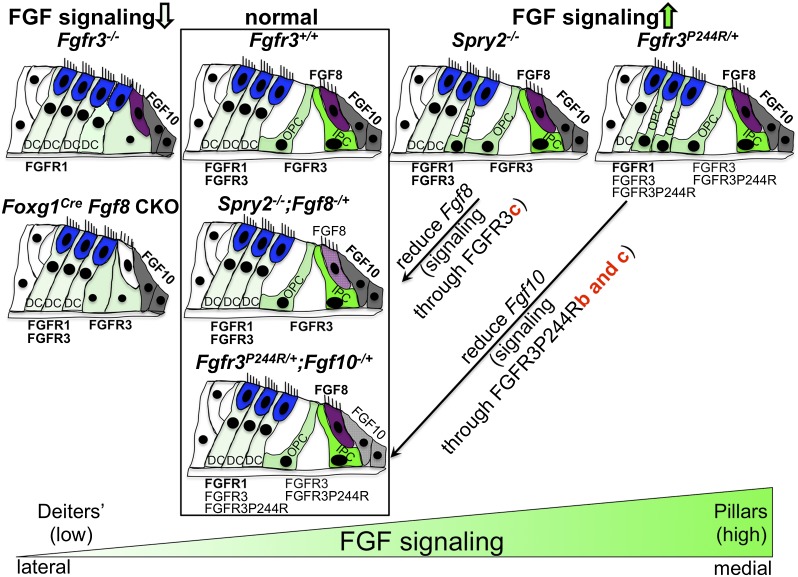
Collectively, our data show that in the mouse Fgfr3P244R/+ Muenke syndrome model cochlea, two Deiters' cells transform sequentially into two outer pillar-like cells between E17.5 and P3, and this transformation is associated with the expansion of FGF signaling into the developing Deiters' cell region. Surprisingly, the Muenke syndrome model hearing loss and supporting cell fate transformation are not rescued by reducing the copy number of FGF8, the FGFR3c ligand required for pillar cell differentiation, or reducing other cochlear FGF ligands that activate FGFR3c but rather reducing FGF10, a ligand that normally activates FGFR2b and FGFR1b. In addition, we found that the Muenke syndrome mutation changes FGFR3b and FGFR3c such that both acquire responsiveness to FGF10. These results are integrated into a diagram summarizing the effects of loss and gain of FGF signaling on cochlear support cell differentiation, in which low levels of FGFR3 signaling promote Deiters' cell differentiation, and high levels promote pillar cell differentiation (Fig. 6). Intriguingly, we also found that FGF10 heterozygosity does not prevent the supporting cell fate transformation but instead, starting between P5 and P7, when Fgf10 transcripts naturally begin to diminish in the cochlear base, appears to allow a gradual reversion toward the normal phenotype. This is associated with a reduction of FGF signaling in the developing Deiters' cell region, indicating that Deiters' and pillar cells can reversibly switch fates in an FGF-dependent manner over a prolonged period of time.
Figure 6.
Model of FGFR3 signaling levels controlling differentiation of cochlear support cells. Normal support cell differentiation of two pillar cells and three Deiters' cells is depicted in the second column (boxed). The column at the far left shows cochlear differentiation when FGFR3 signaling is reduced; i.e., when Fgfr3 is globally absent or Fgf8 is conditionally absent in the Foxg1 domain, which includes the entire cochlear duct. Both conditions lead to incomplete or abnormal differentiation of pillar cells. The third column shows cochlear differentiation when FGFR3 signaling is increased by global deletion of Spry2. These cochleae have a cell fate transformation of one Deiters' cell to one pillar cell. The column at the far right shows cochlear differentiation when FGFR3 signaling is increased in the Muenke syndrome model (Fgfr3P244R/+). These cochleae have a cell fate transformation of two Deiters' cells to two pillar cells. Rescue of the Spry2−/− phenotype by Fgf8 heterozygosity and of the Fgfr3P244R/+ phenotype by Fgf10 heterozygosity is depicted in the same column as the wild type (boxed). The genetic rescue results together with the L6 transfection assay data suggest that normal pillar cell differentiation is controlled by activation of FGFR3c by FGF8, whereas the Muenke syndrome model phenotypes arise by inappropriate activation of FGFR3b P244R and/or FGFR3c P244R by FGF10. Inner hair cells are colored in purple, outer hair cells are in blue, Fgf10-expressing cells are in gray, and support cells are in a shade of green depending on the level of FGFR3 signaling (light: low; dark: high). (DC) Deiters' cell; (IPC) inner pillar cell; (OPC) outer pillar cell.
Our previous histologic study of ABR-tested Muenke syndrome model heterozygotes and homozygotes showed that the FGFR3 P244R mutation induces two Deiters' cells to transform into two presumptive pillar cells (Mansour et al. 2009). The present study examined the development and molecular identity of support cells in the Muenke syndrome model. Inner pillar cells are the first support cells to differentiate (Thelen et al. 2009), and this occurs at E16 in rats (approximately E14.5 in mice). By E17 (approximately E15.5 in mice), these cells express p75NTR (von Bartheld et al. 1991), although the timing of specificity for inner pillar cells in mice may be somewhat later (Mueller et al. 2002; Woods et al. 2004). We found that the earliest expression of the only known outer pillar cell marker, CD44 (Hertzano et al. 2010), occurred in controls at E17.5, when inner pillar cells, identified by p75NTR staining, had clearly already differentiated. Since the number of CD44-positive cells increased in Fgfr3P244R/+ cochlear cross-sections by one at E18.5 and by two at P3 but p75NTR-positive cells did not change and neither did the number of SOX2-positive support cells, we conclude that Muenke syndrome model cochleae have a cell fate transformation from Deiters' to outer pillar-like cells that occurs over 4–5 d. This fate transformation is likely a direct effect of increased signaling by FGFR3 P244R because Fgfr3 is expressed in developing support cells, and the Deiters' cell region of Fgfr3P244R/+ cochleae shows ectopic and/or enhanced expression of several FGF target genes.
Normally, FGF8 promotes pillar cell differentiation by activating FGFR3 (Hayashi et al. 2007; Jacques et al. 2007; Puligilla et al. 2007; Zelarayan et al. 2007), so increasing FGF signaling by removing Spry2, the negative feedback regulator that is initially expressed broadly in cochlear support cells but eventually becomes restricted to Deiters' cells, causes one Deiters' cell to transform into a pillar cell by P7. This transformation is rescued by removing one copy of Fgf8 (Shim et al. 2005). The Spry2−/− cell fate transformation is complete a bit later and is weaker than that found in Fgfr3P244R/+ cochleae, suggesting that the P244R mutation leads to a higher level of FGF signaling in developing Deiters' cells than occurs in Spry2-null mice. Thus, developing cochlear support cells are exquisitely sensitive to the level of FGF signaling, and it is conceivable that even higher levels of FGF signaling could convert all three Deiters' cells to outer pillar cells. Normal Deiters' cells are presumably protected from such high levels of signaling by the inability of FGF10 to activate wild-type FGFR3 isoforms.
The failure of phenotypic rescue found for Fgfr3P244R/+ animals heterozygous for Fgf8 and/or lacking a normal copy of Fgfr3 and the success of phenotypic rescue following Fgf10 copy number reduction suggest that the P244R mutation is neomorphic. Our finding that FGFR3b P244R and FGFR3c P244R gain responsiveness to FGF10 in a cell culture assay supports this conclusion. Previous surface plasmon resonance assays of FGF10 interactions with FGFR3c P250R did not detect binding (Ibrahimi et al. 2004b); however, it is possible that our cell culture assay, which involves a hybrid receptor with an intracellular domain derived from FGFR1, provides greater sensitivity. The extent of FGF10 activation of FGFR3c P244R in cell culture, however, was less than that found with FGF10 and FGFR3b P244R and was statistically significant in only two out of three cases. Nevertheless, this interaction may well be relevant in the cochlear duct, where Fgf10 mRNA and presumably FGF10 protein are highly expressed and where both Fgfr3 splice isoforms are also found, with our results suggesting that Fgfr3c is likely more prevalent than Fgfr3b. Determining whether one or both FGFR3 isoforms are responsible for the observed phenotypes in vivo would require simultaneous knock-in of the P244R mutation and knockout of each of the “b”- and “c”-specific Fgfr3 exons. In the absence of such information, the L6 cell culture assay nevertheless provides a system that could be exploited to identify small molecules that interfere with FGF10/FGFR3b or FGF10/FGFR3c P244R interactions and have no effect on the FGF8/FGFR3c signaling required for normal development. A similar approach could also be relevant to the ligand-dependent Apert syndrome mutations that render FGFR2c sensitive to “b”-type ligands and FGFR2b sensitive to “c”-type ligands (Yu et al. 2000; Ibrahimi et al. 2004a; Hajihosseini et al. 2009). Such molecules could have therapeutic utility.
It was unexpected that Fgfr3P244R/+ mice do not frequently exhibit coronal craniosynostosis, the original defining feature of Muenke syndrome (Muenke et al. 1997; Twigg et al. 2009). Our present results suggest a possible explanation. It could be that mice do not express high enough levels of an FGF ligand, such as FGF10, capable of activating the Muenke syndrome FGFR3 in the developing coronal suture, whereas humans may do so. Thus, studies of therapies for Muenke syndrome based on the amelioration of hearing loss in the mouse model could also prove relevant to human craniosynostosis.
The rescue of Muenke syndrome model cochlear phenotypes by Fgf10 heterozygosity begs the question of the normal role of Fgf10 in the cochlear duct. Fgf10 is very strongly expressed in the developing cochlear ganglion and all of the developing inner ear prosensory patches, eventually resolving in the cochlear duct to the broad inner sulcus (Pirvola et al. 2000; Pauley et al. 2003) immediately medial to the developing inner hair cell, which expresses Fgf8. Fgf10-null mutants have severely deranged vestibular system morphogenesis and abnormalities of cochlear and vestibular innervation (Pauley et al. 2003; Ohuchi et al. 2005), but cochlear duct defects have not been reported and are currently being assessed.
The prolonged period of development of the Muenke syndrome model support cell fate transformation and the finding that Fgf10 heterozygosity does not block the phenotype but instead allows fate-transformed support cells to convert gradually back to a normal fate indicate a great deal of FGF-dependent plasticity in the identity of these cells and has implications for strategies aimed at sensory cell regeneration. Mammalian hearing loss can occur by many different genetic and/or environmental insults, but the end result is the death of sensory hair cells, which cannot naturally regenerate. However, support cells often remain and, in birds and fish, appear to be the source of replacement hair cells, whether by direct transdifferentiation or proliferation followed by differentiation (Monzack and Cunningham 2013; Rubel et al. 2013; Wan et al. 2013). Indeed, immature mouse pillar cells can be induced in cultured cochleae to convert to hair cells when both Notch and FGF signaling are blocked (Doetzlhofer et al. 2009), and following noise damage, Notch inhibition in vivo has a limited ability to convert residual cochlear support cells into functional hair cells (Mizutari et al. 2013). Furthermore, induced expression of Atoh1 in immature but not mature Deiters' cells promotes hair cell differentiation (Liu et al. 2012). Interestingly, mature Deiters' cells continue to express Fgfr3 well past the stage when cochlear FGF expression is extinguished (Hayashi et al. 2010). The exquisite sensitivity of supporting cells to FGF signaling levels suggests that it will be important to carefully calibrate the FGF signaling environment when attempting to promote hair cell generation from residual support cells. Finally, other strategies for hearing restoration involve generation of hair cells in vitro from embryonic stem cells or induced pluripotent stem cells followed by reintroduction into the damaged ear. Some of these already rely on manipulation of FGF signaling to promote otic progenitor production (Oshima et al. 2010; Chen et al. 2012; Koehler et al. 2013), but further consideration of FGF signaling levels at later stages of differentiation might be helpful in producing immature support cell progenitors that could be transplanted and then induced to differentiate in situ into hair cells.
Materials and methods
Mutant mice and genotyping
All experiments reported here complied with protocols approved by the University of Utah Institutional Animal Care and Use Committee. The mouse Fgfr3P244R allele (Fgfr3tm1.1Aomw; Mouse Genome Informatics [MGI]: 3831364) serving as a model of Muenke syndrome has been described (Twigg et al. 2009). Fgf8−/+ mice (Fgf8tm1Mrc; MGI: 2150350) were described by Moon and Capecchi (2000). Fgf9−/+ mice (Fgf9tm1Dor; MGI: 2135961) were described by Colvin et al. (2001). Fgfr3−/+ mice (Fgfr3tm1Dor; MGI: 1931521) were described by Colvin et al. (1996). Fgf20−/+ mice (Fgf20tm1.1Dor; MGI: 5425761) were described by Huh et al. (2012). The Fgf3−/+ (Fgf3tm1.1Sms; MGI: 3767558) and Fgf10−/+ (Fgf10tm1.1Sms; MGI: 3526181) strains were generated and described previously (Urness et al. 2010). Embryonic development was considered as E0.5 on the day a mating plug was observed. The day of birth was considered P0. Primer sets used for PCR-based genotyping of each allele are shown in Supplemental Table 1.
The Fgfr3+/+, Fgfr3P244R/+, and Fgfr3P244R/P244R cochleae that were studied histologically (Fig. 1) and assayed for expression of FGF signaling targets (Fig. 2) or Fgf or Fgfr gene expression (Supplemental Fig. S1) all came from an intercross of heterozygotes maintained on the 129S6 inbred background characterized previously (Mansour et al. 2009), whereas all other samples in this study were derived from mixed genetic backgrounds consequent to crossing the Fgfr3P244R allele to the various Fgf loss-of-function alleles.
Immunostaining of cochlear cryosections
For E17.5 and E18.5 samples, the whole head was bisected in the sagittal plane and fixed for 2 h at room temperature in 4% paraformaldehyde prepared in phosphate-buffered saline (PBS). For postnatal samples, the temporal bone was dissected and fixed for 2 h at room temperature or overnight at 4°C and then decalcified overnight at 4°C in a solution of 10% EDTA/PBS. Heads or temporal bones were embedded in sucrose/gelatin as described (Hurley et al. 2003) with the following modifications: 5% sucrose infiltration overnight at 4°C, 15% sucrose infiltration overnight at 4°C, and 15% sucrose/7.5% gelatin (Sigma, G2500; Bloom, 300) infiltration overnight at 37°C. Samples were cryosectioned at 14-μm thickness in the sagittal or modiolar parallel planes, respectively; collected on SuperFrost Plus slides; and stored at −20°C. Primary antibodies were diluted into PBS/5% normal serum of the secondary antibody species/0.2% Triton X-100 and applied at the following dilutions: rat anti-CD44 (1:800 for E18.5 and younger and 1:1000 for P0 and older; BD Biosciences, no. 550538), rabbit anti-MYO7A (1:1000; Proteus Biosciences, no. 25-6790), goat anti-SOX2 (1:250; Santa Cruz Biotechnology, no. sc-17320), and rabbit anti-p75NTR (1:650; Millipore, no. 07-476). Secondary antibodies were all from Invitrogen and diluted 1:400 into PBST/5% normal serum (Alexa Fluor 594 goat anti-rat [A11007], AlexaFluor 488 donkey anti-rat [A21208], Alexa Fluor 488 goat anti-rabbit [A11034], and Alexa Fluor 594 donkey anti-goat [A11058]). DAPI was included in the mounting medium (Vectashield, Vector Laboratories). For technical reasons, the secondary antibodies used to detect SOX2 with CD44 in Figures 1 and 4 were red and green, respectively. However, to provide visual consistency across the entire data set, we used Photoshop CS6 to exchange the red and green channels such that SOX2-expressing cells appear green (or pale green, when overlapped with DAPI) and CD44-expressing cells appear red.
Cochlear RNA in situ hybridization
Temporal bones were dissected from postnatal animals, fixed, embedded in paraffin, sectioned parallel to the modiolus at 10 μm, and hybridized as described (Urness et al. 2008). Digoxigenin-labeled cRNA probes were prepared from cDNA-containing plasmids or following PCR amplification of 3′ untranslated regions (UTRs) from mouse genomic DNA. Erm(Etv5), Fgf8, Fgf10, Fgfr1, and Spry1 probes were described previously (Li et al. 2007; Urness et al. 2010, 2011). Pea3(Etv4) came from IMAGE cDNA 5318783. The cDNA clone was cut with SacI and transcribed with T7 RNA polymerase to yield a 3′ UTR antisense probe of 515 b. The Fgfr2 cDNA clone was described by Hayashi et al. (2010) and provided by the investigators. Hey2 and Fgfr3 3′ UTRs were first amplified with T7 promoter-containing primers Hey2F (5′-GTAACTGATGTCGTCCATTTCGG-3′) plus Hey2R (5′- ggatcctaatacgactcactatagggagAAACCAGGGTTCACTAGAGGTATGG-3′) and Fgfr3F (5′- CTAGACTCCATGTCCAGAGTGG-3′) plus Fgfr3R (5′- ggatcctaatacgactcactatagggagTGGGACTAAACGCTATGAACCTGG-3′), respectively, and the antisense strand was transcribed using T7 RNA polymerase.
ABR measurements
All animals were tested at 3–5 wk of age for response thresholds to broadband click and 8, 16, and 32 kHz pure tone pip stimuli as described (Mansour et al. 2009). Testing was performed blind with respect to genotype, and in the mixed genetic background, it was not possible to infer genotypes based on craniofacial phenotypes except in very rare instances for the Fgfr3P244R/+ genotype. To simplify the data presentation and analysis, we calculated a mean hearing loss value by subtracting the Fgfr3+/+;FgfX−/+ mean threshold value from the corresponding Fgfr3P244R/+;FgfX−/+ (or Fgfr3P244R/−;FgfX−/+) mean value and then computed the standard error of the mean difference. Some of the data for the animals from which one copy of Fgf8, Fgf9, or both genes were removed were presented previously, with the Fgfr3+/+;FgfX−/+ and Fgfr3P244R/+;FgfX−/+ data plotted separately (Supplemental Material; Mansour et al. 2009), but these data were augmented by additional animals bred specifically for this study and are shown along with the entirely new data obtained following removal of one copy of Fgfr3, Fgf20, Fgf3, and Fgf10 so that the ABR rescue result can be better appreciated.
RT–PCR of cochlear RNA
Total RNA was isolated from four wild-type and four heterozygous P3 cochlear epithelia using Qiagen RNAeasy. One microgram of RNA was reverse-transcribed using MMLV polymerase (RETROscript, Ambion), and 1 μL of cDNA was added to a PCR mixture containing a common Fgfr3 forward primer, p1 (5′-CAAGTTTGGCAGCATCCGGCAGAC-3′), and either a “b”-specific reverse primer, p2 (5′-TCTCAGCCACGCCTATGAAATTGGTG-3′), or a “c”-specific reverse primer, p3 (5′-CACCACCAGCCACGCAGAGTGATG-3′). To detect Fgf10 expression, we used the following primers: F10-F (5′-TGTCTTCGTTCCCTGTCACCTG-3′) and F10-R (5′-CATACATTTGCCTGCCATTGTG-3′). Forty cycles of amplification were performed. The Fgfr3b-specific product is 350 base pairs (bp), the Fgfr3c-specific product is 373 bp, and the Fgf10 product is 450 bp.
Transient transfection of L6 myoblasts to assay FGFR activity
Chimeric mouse Fgfr3 cDNA clones that consist of the coding sequences for the extracellular domain of FGF3b and FGFR3c, each fused to coding sequences for the intracellular tyrosine kinase domain of FGFR1 (Ornitz et al. 1996), were mutated to the corresponding Muenke syndrome form by changing an exon 7 C to a G (coding for P244R) using the QuickChange kit (Stratagene). Mutations were confirmed by sequencing. Both the wild-type and mutated cDNAs were transferred into the pcDNA1 expression vector used by Chellaiah et al. (1994) to express other Fgfr3 constructs. A human FGFR1c expression vector (Raivio et al. 2009) was used as a control. The FGF-responsive OCFRE-luc vector was described previously (Newberry et al. 1996). The pGL4.73 Renilla luciferase expression vector (Promega) served as a transfection control. Rat L6 myoblasts were purchased from American Type Culture Collection. Purified FGFs were purchased from Peprotech and diluted and stored as recommended by the manufacturer.
The FGFR activity assays were conducted as described by Raivio et al. (2009) with the following modifications. The assays were scaled down for the 96-well format, and each FGF/FGFR combination was replicated six times. Only four FGF concentrations (0, 50, 500, and 5000 pM) were assayed. Following a personal communication from Dr. Yisrael Sidis, we used FuGeneHD (Promega) as the transfection reagent and did not starve the cells of serum prior to FGF treatment. Luciferase activities were determined using the Promega Dual-Luciferase assay system and a microplate luminometer (Veritas). Firefly luciferase units for each well were normalized by dividing by the corresponding Renilla luciferase units and averaged across the six replicates. Each set of assays was repeated three times, and a representative assay is shown.
The data were graphed and analyzed using GraphPad Prism. One-way ANOVA with Dunnett's post-hoc test for multiple comparisons identified FGF/FGFR combinations exhibiting significant induction of luciferase activity in each experimental replicate. The assay graphed in Figure 5 shows each significant induction marked with asterisks indicating the adjusted P-value.
Acknowledgments
We are grateful to the following colleagues for providing mutant mice, reagents, or expertise: Jerry Spangrude (anti-CD44), Charlie Murtaugh (anti-SOX2), Anne Moon (Fgf8−/+ mice), Nelly Pitteloud and Yisrael Sidis (FGFR1c expression clone, FGF-responsive OCFRE luciferase vector, detailed protocols for and consultation regarding the L6 assays), Mike Howard (luciferase assay advice, pGL4.73, and luminometer), Shannon Odelberg (consultations on GraphPad Prism for statistical analyses), Mark Hill and Sara Rees (assistance with in situ hybridization and/or ABR tests in the early phases of this project), and David Ornitz (Fgf9−/+, Fgfr3−/+, and Fgf20−/+ mice; wild-type Fgfr3b/1 and Fgfr3c/1 expression clones). The Fgfr3−/+ strain was sent by Olivia Bermingham-McDonogh, and the Fgf9−/+ strain was transferred from Irene Hung. We thank Gary Schoenwolf and Jan Christian for critical comments on the manuscript. Supported by a grant from the NIH/NIDCD to S.L.M. (R01DC002043, including an ARRA Administrative Supplement Providing Summer Research Experiences).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.228957.113.
References
- Agochukwu NB, Doherty ES, Muenke M 2010. Muenke syndrome. In GeneReviews (ed. Pagon RA, et al.), Seattle, WA [Google Scholar]
- Chellaiah AT, McEwen DG, Werner S, Xu J, Ornitz DM 1994. Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J Biol Chem 269: 11620–11627 [PubMed] [Google Scholar]
- Chen W, Jongkamonwiwat N, Abbas L, Eshtan SJ, Johnson SL, Kuhn S, Milo M, Thurlow JK, Andrews PW, Marcotti W, et al. 2012. Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature 490: 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM 1996. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet 12: 390–397 [DOI] [PubMed] [Google Scholar]
- Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM 2001. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell 104: 875–889 [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N 2009. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell 16: 58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Schlessinger J 2007. Skeletal overgrowth is mediated by deficiency in a specific isoform of fibroblast growth factor receptor 3. Proc Natl Acad Sci 104: 3937–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz R, Mohammadi M 2013. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol 14: 166–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK 2010. The challenge of hair cell regeneration. Exp Biol Med (Maywood) 235: 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, Fekete DM 2012. Shaping sound in space: The regulation of inner ear patterning. Development 139: 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajihosseini MK, Duarte R, Pegrum J, Donjacour A, Lana-Elola E, Rice DP, Sharpe J, Dickson C 2009. Evidence that Fgf10 contributes to the skeletal and visceral defects of an Apert syndrome mouse model. Dev Dyn 238: 376–385 [DOI] [PubMed] [Google Scholar]
- Hatch EP, Noyes CA, Wang X, Wright TJ, Mansour SL 2007. Fgf3 is required for dorsal patterning and morphogenesis of the inner ear epithelium. Development 134: 3615–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Cunningham D, Bermingham-McDonogh O 2007. Loss of Fgfr3 leads to excess hair cell development in the mouse organ of Corti. Dev Dyn 236: 525–533 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, Bermingham-McDonogh O 2008. Fgf20 is required for sensory epithelial specification in the developing cochlea. J Neurosci 28: 5991–5999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, Younkins C, Bermingham-McDonogh O 2010. Expression patterns of FGF receptors in the developing mammalian cochlea. Dev Dyn 239: 1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzano R, Puligilla C, Chan SL, Timothy C, Depireux DA, Ahmed Z, Wolf J, Eisenman DJ, Friedman TB, Riazuddin S, et al. 2010. CD44 is a marker for the outer pillar cells in the early postnatal mouse inner ear. J Assoc Res Otolaryngol 11: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SH, Jones J, Warchol ME, Ornitz DM 2012. Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biol 10: e1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley PA, Clarke M, Crook JM, Wise AK, Shepherd RK 2003. Cochlear immunochemistry–a new technique based on gelatin embedding. J Neurosci Methods 129: 81–86 [DOI] [PubMed] [Google Scholar]
- Ibrahimi OA, Zhang F, Eliseenkova AV, Itoh N, Linhardt RJ, Mohammadi M 2004a. Biochemical analysis of pathogenic ligand-dependent FGFR2 mutations suggests distinct pathophysiological mechanisms for craniofacial and limb abnormalities. Hum Mol Genet 13: 2313–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi OA, Zhang F, Eliseenkova AV, Linhardt RJ, Mohammadi M 2004b. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Hum Mol Genet 13: 69–78 [DOI] [PubMed] [Google Scholar]
- Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW 2007. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development 134: 3021–3029 [DOI] [PubMed] [Google Scholar]
- Kelly MC, Chen P 2009. Development of form and function in the mammalian cochlea. Curr Opin Neurobiol 19: 395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KR, Mikosz AM, Molosh AI, Patel D, Hashino E 2013. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 500: 217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Scott DA, Hatch E, Tian X, Mansour SL 2007. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development 134: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Dearman JA, Cox BC, Walters BJ, Zhang L, Ayrault O, Zindy F, Gan L, Roussel MF, Zuo J 2012. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters' cells to immature hair cells by Atoh1 ectopic expression. J Neurosci 32: 6600–6610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Twigg SR, Freeland RM, Wall SA, Li C, Wilkie AO 2009. Hearing loss in a mouse model of Muenke syndrome. Hum Mol Genet 18: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutari K, Fujioka M, Hosoya M, Bramhall N, Okano HJ, Okano H, Edge AS 2013. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron 77: 58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzack EL, Cunningham LL 2013. Lead roles for supporting actors: Critical functions of inner ear supporting cells. Hear Res 303: 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AM, Capecchi MR 2000. Fgf8 is required for outgrowth and patterning of the limbs. Nat Genet 26: 455–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KL, Jacques BE, Kelley MW 2002. Fibroblast growth factor signaling regulates pillar cell development in the organ of corti. J Neurosci 22: 9368–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenke M, Gripp KW, McDonald-McGinn DM, Gaudenz K, Whitaker LA, Bartlett SP, Markowitz RI, Robin NH, Nwokoro N, Mulvihill JJ, et al. 1997. A unique point mutation in the fibroblast growth factor receptor 3 gene (FGFR3) defines a new craniosynostosis syndrome. Am J Hum Genet 60: 555–564 [PMC free article] [PubMed] [Google Scholar]
- Newberry EP, Boudreaux JM, Towler DA 1996. The rat osteocalcin fibroblast growth factor (FGF)-responsive element: An okadaic acid-sensitive, FGF-selective transcriptional response motif. Mol Endocrinol 10: 1029–1040 [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Yasue A, Ono K, Sasaoka S, Tomonari S, Takagi A, Itakura M, Moriyama K, Noji S, Nohno T 2005. Identification of cis-element regulating expression of the mouse Fgf10 gene during inner ear development. Dev Dyn 233: 177–187 [DOI] [PubMed] [Google Scholar]
- Olwin BB, Hauschka SD 1989. Cell type and tissue distribution of the fibroblast growth factor receptor. J Cell Biochem 39: 443–454 [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M 1996. Receptor specificity of the fibroblast growth factor family. J Biol Chem 271: 15292–15297 [DOI] [PubMed] [Google Scholar]
- Oshima K, Shin K, Diensthuber M, Peng AW, Ricci AJ, Heller S 2010. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell 141: 704–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B 2003. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn 227: 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, Thesleff I, Fritzsch B, Dickson C, Ylikoski J 2000. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci 20: 6125–6134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puligilla C, Feng F, Ishikawa K, Bertuzzi S, Dabdoub A, Griffith AJ, Fritzsch B, Kelley MW 2007. Disruption of fibroblast growth factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Dev Dyn 236: 1905–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio T, Sidis Y, Plummer L, Chen H, Ma J, Mukherjee A, Jacobson-Dickman E, Quinton R, Van Vliet G, Lavoie H, et al. 2009. Impaired fibroblast growth factor receptor 1 signaling as a cause of normosmic idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab 94: 4380–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghani M, Mohammadi M, Schlessinger J, Moscatelli D 1996. Induction of urokinase-type plasminogen activator by fibroblast growth factor (FGF)-2 is dependent on expression of FGF receptors and does not require activation of phospholipase Cγ1. J Biol Chem 271: 31154–31159 [DOI] [PubMed] [Google Scholar]
- Rubel EW, Furrer SA, Stone JS 2013. A brief history of hair cell regeneration research and speculations on the future. Hear Res 297: 42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmang T 2007. Expression and functions of FGF ligands during early otic development. Int J Dev Biol 51: 473–481 [DOI] [PubMed] [Google Scholar]
- Shim K, Minowada G, Coling DE, Martin GR 2005. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell 8: 553–564 [DOI] [PubMed] [Google Scholar]
- Thelen N, Breuskin I, Malgrange B, Thiry M 2009. Early identification of inner pillar cells during rat cochlear development. Cell Tissue Res 337: 1–14 [DOI] [PubMed] [Google Scholar]
- Toydemir RM, Brassington AE, Bayrak-Toydemir P, Krakowiak PA, Jorde LB, Whitby FG, Longo N, Viskochil DH, Carey JC, Bamshad MJ 2006. A novel mutation in FGFR3 causes camptodactyly, tall stature, and hearing loss (CATSHL) syndrome. Am J Hum Genet 79: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SR, Healy C, Babbs C, Sharpe JA, Wood WG, Sharpe PT, Morriss-Kay GM, Wilkie AO 2009. Skeletal analysis of the Fgfr3(P244R) mouse, a genetic model for the Muenke craniosynostosis syndrome. Dev Dyn 238: 331–342 [DOI] [PubMed] [Google Scholar]
- Urness LD, Li C, Wang X, Mansour SL 2008. Expression of ERK signaling inhibitors Dusp6, Dusp7, and Dusp9 during mouse ear development. Dev Dyn 237: 163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urness LD, Paxton CN, Wang X, Schoenwolf GC, Mansour SL 2010. FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev Biol 340: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urness LD, Bleyl SB, Wright TJ, Moon AM, Mansour SL 2011. Redundant and dosage sensitive requirements for Fgf3 and Fgf10 in cardiovascular development. Dev Biol 356: 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bartheld CS, Patterson SL, Heuer JG, Wheeler EF, Bothwell M, Rubel EW 1991. Expression of nerve growth factor (NGF) receptors in the developing inner ear of chick and rat. Development 113: 455–470 [DOI] [PubMed] [Google Scholar]
- Wan G, Corfas G, Stone JS 2013. Inner ear supporting cells: Rethinking the silent majority. Semin Cell Dev Biol 24: 448–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, McMahon AP 1989. Expression pattern of the FGF-related proto-oncogene int-2 suggests multiple roles in fetal development. Development 105: 131–136 [DOI] [PubMed] [Google Scholar]
- Woods C, Montcouquiol M, Kelley MW 2004. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci 7: 1310–1318 [DOI] [PubMed] [Google Scholar]
- Wu DK, Kelley MW 2012. Molecular mechanisms of inner ear development. Cold Spring Harb Perspect Biol 4: a008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Herr AB, Waksman G, Ornitz DM 2000. Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proc Natl Acad Sci 97: 14536–14541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelarayan LC, Vendrell V, Alvarez Y, Dominguez-Frutos E, Theil T, Alonso MT, Maconochie M, Schimmang T 2007. Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev Biol 308: 379–391 [DOI] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM 2006. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem 281: 15694–15700 [DOI] [PMC free article] [PubMed] [Google Scholar]