Heart formation and regeneration require cardiomyocyte commitment. Cai et al. show that the dual Nodal/BMP antagonist Cerberus-1 (Cer1) directs the SWI/SNF chromatin remodeling complex to cardiomyogenic loci in multipotent progenitors. Blocking Nodal and BMP induces Baf60c and lineage-specific transcription factors that interact with Baf60c. Knockdown of Cer1, Baf60c, or the catalytic SWI/SNF subunit Brg1 prevented cardiomyocyte differentiation. These results demonstrate how external signals from the progenitor cell environment can direct lineage-specific chromatin remodeling in order to commit cell fate.
Keywords: embryonic stem cells, cardiogenesis, Flk1/KDR, Nkx2-5, chromatin remodeling, Baf60c
Abstract
A critical but molecularly uncharacterized step in heart formation and regeneration is the process that commits progenitor cells to differentiate into cardiomyocytes. Here, we show that the endoderm-derived dual Nodal/bone morphogenetic protein (BMP) antagonist Cerberus-1 (Cer1) in embryonic stem cell cultures orchestrates two signaling pathways that direct the SWI/SNF chromatin remodeling complex to cardiomyogenic loci in multipotent (KDR/Flk1+) progenitors, activating lineage-specific transcription. Transient inhibition of Nodal by Cer1 induces Brahma-associated factor 60c (Baf60c), one of three Baf60 variants (a, b, and c) that are mutually exclusively assembled into SWI/SNF. Blocking Nodal and BMP also induces lineage-specific transcription factors Gata4 and Tbx5, which interact with Baf60c. siRNA to Cer1, Baf60c, or the catalytic SWI/SNF subunit Brg1 prevented the developmental opening of chromatin surrounding the Nkx2.5 early cardiac enhancer and cardiomyocyte differentiation. Overexpression of Baf60c fully rescued these deficits, positioning Baf60c and SWI/SNF function downstream from Cer1. Thus, antagonism of Nodal and BMP coordinates induction of the myogenic Baf60c variant and interacting transcription factors to program the developmental opening of cardiomyocyte-specific loci in chromatin. This is the first demonstration that cues from the progenitor cell environment direct the subunit variant composition of SWI/SNF to remodel the transcriptional landscape for lineage-specific differentiation.
As early as the 1920s, it was realized that uncommitted mesodermal progenitors can be induced to form cardiac cells (Bacon 1945 and references therein), and subsequent embryological and stem cell studies led to the identification of numerous paracrine signals that promote cardiogenic differentiation (Noseda et al. 2011). However, despite the intense interest in regenerating cardiomyocytes from pluripotent stem or adult progenitor cells, the extracellular signals and nuclear events that define cardiac commitment have remained obscure.
Molecular studies have identified several extracellular signaling proteins that control the multistep progression from pluripotent stem cells to terminally differentiated cardiac cells. Key among these are Wnts, Nodal, and bone morphogenetic proteins (BMPs), which exert biphasic and even triphasic effects (Mercola et al. 2011; Noseda et al. 2011). Nodal has a graded and time-dependent influence such that intermediate doses in an early developmental window induce cardiogenic progenitors (for instance, see Brennan et al. 2001). BMP and Wnt induce early mesodermal progenitors but, like Nodal, antagonize cardiac differentiation subsequently, and at least BMP is needed later for terminal differentiation (for instance, Yuasa et al. 2005; Naito et al. 2006; Kwon et al. 2007). These examples illustrate the importance of concentration and timing and support the concept that cardiogenesis depends on the tight spatiotemporal control of secreted factors (Mercola et al. 2011; Noseda et al. 2011).
An understanding of how paracrine signaling proteins and their cardiogenic inhibitors control gene expression should bridge external to genome-level signaling and possibly reveal the mechanism that commits multipotent progenitors to form cardiomyocytes and smooth muscle and vascular endothelial cells. We discovered that endoderm-derived Cerberus-1 (Cer1), by antagonizing both Nodal and BMP (Biben et al. 1998; Piccolo et al. 1999), orchestrates the expression of key SWI/SNF subunits and lineage-specific transcription factors in cardiovascular progenitors that interact to direct the developmental opening of cardiomyogenic loci in the genome. The SWI/SNF complex consists of a core catalytic subunit (either Brahma [Brm] or Brg1) and structural Brm-associated factors (BAFs). Multiple variants of certain BAFs exist (e.g., Baf60a, Baf60b, or Baf60c) and are assembled in a mutually exclusive manner into SWI/SNF, and recent studies have found that particular variants influence the physical interaction of SWI/SNF with transcription factors and lineage-specific loci in chromatin (Lessard et al. 2007; Forcales et al. 2011). The logical connection between chromatin remodeling and differentiation suggests that it might be regulated by external cues from the progenitor cell niche, yet no paracrine signals have been shown to dictate SWI/SNF composition in any developmental or regenerative context (Wu et al. 2009; Puri and Mercola 2012). Thus, our findings linking Cer1 to the Baf60 variant composition and chromatin selectivity of SWI/SNF provide a paradigm of how external signals from the progenitor cell environment can direct lineage-specific chromatin remodeling in order to commit cell fate.
Results
Cer1 is essential for stem cell cardiomyogenesis
Nodal and BMP can induce multipotent KDR/Flk1+, Mesp1+ cardiovascular progenitors, yet each must be inhibited subsequently for progenitors to differentiate into cardiomyocytes (Noseda et al. 2011). Given that stage-specific inhibition of these signals is important for cardiogenesis, we questioned whether these signals might commit progenitors to a cardiomyocyte fate. Examining transcript profiles in embryos for Cer1 (encoding a dual BMP/Nodal inhibitor) and other Nodal or BMP inhibitors—e.g., Noggin (encoding a BMP inhibitor) and Lefty1 and Lefty2 (encoding Nodal inhibitors)—revealed that each is present as Mesp1+ cardiovascular progenitors arise at embryonic day 6.5 (E6.5) but declines in abundance over the next 2 d as the progenitors make the transition to committed cardiac precursors marked by Isl1+, Mef2c+, and Nkx2-5+ (Fig. 1A). In murine embryonic stem cell (mESC) cultures, Cer1 is up-regulated during days 3–6 of cardiogenic differentiation preceding the transition from uncommitted Mesp1+ progenitors to committed precursors marked by Nkx2-5 (Fig. 1B) as well as Isl1 and Mef2c (data not shown). Unlike the profile in embryos, Lefty1 and Lefty2 are both down-regulated in mESCs such that Cer1 becomes the primary Nodal inhibitor after day 4. Transcripts for the BMP inhibitor Noggin are also present during commitment but with a slight delay at day 4 and day 5 (Fig. 1B). Thus, the temporal expression of Cer1 makes it a candidate to initiate cardiac cell commitment in ESC differentiation.
Figure 1.
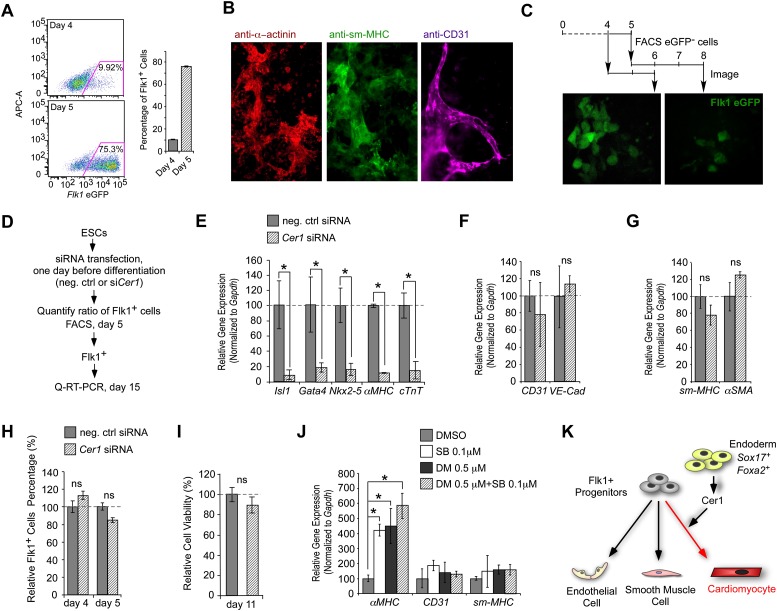
Cer1 directs cardiomyogenic differentiation by inhibiting Nodal and BMP. (A) qRT–PCR profiles showing Cer1, Noggin, and Lefty1,2 transcripts are expressed in mouse embryos at E6.5, coinciding with Mesp1 (marking uncommitted progenitors), and then decline over E7.5–E8.5 with the appearance of Isl1, Nkx2-5, and Mef2c (marking cardiac commitment). (B) During mESC differentiation, Cer1 and Mesp1 expression coincides temporally and precedes the appearance of the cardiogenic transcription factors Nkx2-5 (shown) as well as cardiogenic transcription factors Isl1 and Mef2c (not shown). Lefty1,2 mRNAs decline prior to the Cer1 peak. Arrowheads below indicate the initiation of Cer1 RNAi studies in C–F and Supplemental Figure S2 using either lentivirally transduced shRNA (before initiation of differentiation [day 0]; open triangle] or siRNA transfection (at 0 or 3 d after initiation of differentiation; solid triangles). (C–F) Cer1 controls cardiomyocyte differentiation. Cer1 siRNA-1 (siCer1) blocked αMHC induction, whereas transient overexpression of Cer1 increased αMHC levels above vector control assayed by qRT–PCR at day 8 (C). Similarly, Cer1 siRNA-1 decreased the incidence of beating colonies (D) and the yield of cardiomyocytes (day 15 in E and day 9 in F) by flow cytometry without an appreciable effect on endothelial (day 9) and smooth muscle (day 15) lineages (F). Similar results were seen under serum-containing (C,D,F) or serum-free (E) conditions. siRNAs (100 nM) or 3 μg of Cer1 expression plasmid were transfected at 1 d before differentiation (see the Supplemental Material). See Supplemental Figure S1 for validation of the siRNA and shRNA constructs and Supplemental Figure S2 for additional si/shRNA constructs and additional marker analyses. (G) Cer1 is needed for markers of lineage commitment. Cer1 shRNA did not alter markers of ectoderm (Pax6), endoderm (Foxa2, Hnf1b, and Hex), mesendoderm (Bra/T), or cardiogenic mesoderm markers (Flk1 and Mesp1) but reduced subsequent expression of cardiac markers (Nkx2-5, Mef2c, Isl1, Tbx1, Tbx5, and Gata4). Quantification of Bra/T, Foxa2, Flk1, and Mesp1 was at 4 d after the onset of differentiation, and quantification of other markers was at 8 d by qRT–PCR. (H,I) Nodal and BMP inhibition synergizes to mimic Cer1 function. (H) Heat map representation of cardiomyocyte differentiation (integrated intensity of αMHC-eGFP fluorescence by quantitative automated microscopy) (see the Supplemental Material) in ESC cultures transfected with either negative control (scrambled sequence) siRNA or siCer1 (day −1) followed by dispersal and treatment with graded doses of the Alk4,5,7 inhibitor SB-431542 (SB) and the Alk2,3,6 inhibitor Dorsomorphin (DM) between days 3 and 6 of culture. (I) Representative images of αMHC-eGFP fluorescence in cultures. Note the synergistic effect of DM and SB treatment, with dose optimum at 0.5 μM DM and 0.1 μM SB. (J–L) Optimal window for the cardiomyogenic response reveals that the synergy of SB and DM is between 3 and 6 d of differentiation (shown in J). Embryoid bodies (EBs) were treated as in H except that the dispersal and treatment window were incremented as indicated. Note that treatment increased cell number, seen as area of αMHC-eGFP fluorescence (K) and frequency distribution of fluorescence intensity by flow cytometry (L) shown relative to DMSO vehicle alone. (M) Cer1 specifies cardiomyogenic fate through inhibition of Nodal and BMP. (*) P < 0.05, unpaired Student's t-test. Error bars indicate the SEM; n = 3 in A, C, D, F, and G; n = 5 in E; n = 6 in J. See also Supplemental Figures S1 and S2.
We next tested whether modulating Cer1 would affect cardiomyocyte differentiation. Different siRNAs (siRNA-1, siRNA-2, and siRNA-3) each achieved ∼60% and 90% reduction of endogenous and recombinant Cer1, respectively (Supplemental Fig. S1A–C). Each siRNA blocked cardiogenesis when transfected into R1 mESCs 1 d before differentiation, as quantified by αMHC expression at day 8 (∼60% inhibition relative to negative control [scrambled] siRNA) (Fig. 1C). Cer1 shRNAs gave identical results (Supplemental Fig. S2A). For comparison, siRNA knockdown of the cardiogenic transcription factor Mef2c caused a comparable level of inhibition (Supplemental Fig. S2B). Probing timing, siRNA transfected at day 3 or 4 was equally effective as on day −1, suggesting that Cer1 functions between days 3–4 and day 6, when Cer1 mRNA declines (Fig. 1B; Supplemental Fig. S2B,C). Conversely, recombinant Cer1 expressed from a transfected plasmid increased αMHC transcripts greater than twofold at day 8 over vector control (Fig. 1C). Identical results were observed in CGR8 mESCs (data not shown). Cer1 siRNA-1 also decreased the incidence of beating colonies (at day 18) by 60% relative to negative control siRNA (Fig. 1D) and cardiomyocyte yield >50% by flow cytometry based on αMHC-eGFP reporter fluorescence at day 15 (17.55% ± 2.72% [negative control scrambled sequence siRNA] to 4.97% ± 0.75% [siRNA-1], P = 0.037, n = 3) in CGR8 mESCs (Fig. 1E,F). Examining other cardiopoietic lineages, we found that Cer1 knockdown did not affect differentiation of CD31+ (endothelial) or αSMA+ (smooth muscle) cells (CD31+: 1.65% ± 0.44% vs. 2.2% ± 0.43%; αSMA+: 18.57% ± 5.84% vs. 24.5% ± 8.75% [siRNA-1 vs. control siRNA, respectively]) (Fig. 1F). Thus, marker and cytometric analyses indicate that Cer1 selectively promotes cardiomyocyte differentiation.
Cer1 is required for cardiomyogenic differentiation of Flk1+ and Mesp1+ progenitors
To pinpoint the developmental stage when Cer1 acts, we profiled mesendodermal and cardiac markers in differentiating mESCs after Cer1 knockdown. Lentiviral transduction of specific shRNA against Cer1 (shCer1) decreased production of endogenous or recombinant Cer1 by up to 50%–90% (Supplemental Fig. S1D–G) and inhibited induction of αMHC in R1 mESCs by up to 50% (Supplemental Fig. S2A). Notably, Cer1 knockdown did not affect the formation of mesendoderm (Bra/T+), endoderm (Foxa2+, Hnf1b+, and Hex+), or ectoderm (Pax6+) or the appearance of Flk1+ and Mesp1+ cardiac progenitors at differentiation day 4 (Fig. 1G). Similarly, overexpression of Cer1 did not affect the formation of endoderm and cardiac progenitors at day 4 (Supplemental Fig. S2D,E). However, shCer1 decreased all subsequent markers of committed cardiac precursors (e.g., Nkx2-5, Mef2c, Isl1, Tbx1, Tbx5, and Gata4), indicating that Cer1 is needed for progression past the Flk1+ and Mesp1+ state.
Synergistic action of inhibiting Nodal and BMP
Mammalian Cer1 antagonizes both Nodal and BMP but, unlike its Xenopus counterpart, does not inhibit Wnt. To determine whether both activities are required and whether they have distinct versus redundant functions, we tested whether small molecule inhibitors selective for Nodal or BMP signaling would rescue cardiogenesis in CGR8 mESCs transfected with Cer1 siRNA-1 in a 384-well format. The selective Alk4,5,7 (type I Nodal/TGFβ receptor) inhibitor SB-431542 (SB) and Alk2,3,6 (type I BMP receptor) inhibitor Dorsomorphin (DM) were added during the time of maximal Cer1 expression (days 3–6 of differentiation, as shown in Fig. 1B). High-content microscopy was used to quantify cardiomyocyte differentiation by measuring the integrated intensity of eGFP fluorescence (area × average intensity) (see the Materials and Methods). The heat map representation (Fig. 1H; Supplemental Fig. S2F) and representative images (Fig. 1l) show that 0.25–1 μM DM significantly increased cardiogenesis in mESCs transfected with 0.01–0.5 μM Cer1 siRNA-1. SB had only a minimal effect on its own but increased cardiogenesis twofold to threefold when combined with DM, indicating synergy (Fig. 1H,J). Synergistic action of DM and SB also enhanced cardiogenesis in control ESCs transfected with inert negative control siRNA, even over the higher basal level of differentiation (Fig. 1H; Supplemental Fig. S2F). Treatment with the optimal combination (0.5 μM DM and 0.1 μM SB) for overlapping 3-d time frames defined the cardiogenic window for SB and DM between days 3 and 6 of differentiation (Fig. 1J), corresponding to the period of endogenous Cer1 expression (Fig. 1B). The inductive effect was due to increased cardiomyocyte cell number, as verified by both increased area occupied by αMHC-GFP+ cells (Fig. 1K) and increased αMHC-GFP+ cell count by flow cytometry (2.42% ± 0.61% [DMSO] to 6.59% ± 0.46% [DM + SB]; P = 0.019; n = 4) (Fig. 1L). Together, these data indicate that transient inhibition of Nodal and BMP synergizes to increase cardiomyocyte differentiation, as summarized in Figure 1M.
Endoderm-derived Cer1 is critical for cardiomyocyte differentiation of Flk1+ progenitors
Since ESC cultures are heterogeneous, progenitor populations were sorted to define the cell population that produces and responds to Cer1 (Fig. 2A,B). Cxcr4 is uniquely associated with mesendoderm formation by bioinformatics analysis (Nelson et al. 2008) and was reported as a definitive endoderm marker (Morrison et al. 2008), whereas a subset of emerging primitive streak mesoderm is Flk1+ (Kattman et al. 2006). FACS distinguished four populations in day 5 differentiating R1 ESCs: Cxcr4+, Flk1−; Cxcr4+, Flk1+; Cxcr4−, Flk1+; and Cxcr4−, Flk1− (Fig. 2A). As shown in Figure 2B, cardiac lineage markers Mef2c, Isl1, and Mesp1 were mostly in the Cxcr4+, Flk1+ population; CD31 and Mef2c were enriched in the Cxcr4−, Flk1+ population, consistent with enrichment of hemangioblasts in CD31+, Flk1+ cells (Iida et al. 2005). Endodermal cells characterized by Sox17 and Foxa2 expression were present only in the Cxcr4+, Flk1− population (Fig. 2B). Cer1, Nodal, and Bmp transcripts and their receptors were analyzed in the four populations to determine potential signaling response relationships (Fig. 2B; Supplemental Fig. 3A,B). Bmp2, Bmp4, and Noggin predominated in the Cxcr4+, Flk1+ population enriched for markers of cardiac precursors and in the Cxcr4−, Flk1+ (CD31-enriched) population, whereas Nodal transcripts were present in the Cxcr4+, Flk1− population (endoderm). Each of the four populations expressed Nodal (Alk4) and BMP (Alk3, Alk6, and Bmpr2) receptor genes. However, Cer1 was uniquely present in the Cxcr4+, Flk1− (endoderm) population (Fig. 2B), defining Cer1 as an endodermal signal (Fig. 2C).
Figure 2.
Cer1 is produced by endodermal cells. (A) Representative FACS profile of Flk1+ and Cxcr4+ precursors at day 5 of differentiation. (B) Marker analysis by qRT–PCR defines four populations: Flk1+, Cxcr4+ (cardiac precursors); Flk1+, Cxcr4− (CD31+, endothelial-enriched cells); Flk1−, Cxcr4+ (endodermal cells); and Flk1−, Cxcr4− cells. Note expression of Cer1 in the Flk1−, Cxcr4+ population (endoderm). Data are plotted as the natural log (logn) of fold change in gene expression. See Supplemental Figure S3 for additional ligand and receptor profiles; n = 6. (C) Summary showing Cer1 signaling from endoderm to regulate differentiation of cardiogenic progenitors. (*) P < 0.05, unpaired Student's t-test. Error bars indicate the SEM; n = 3. See also Supplemental Figure S3.
FACS-isolated cardiogenic progenitors were treated with selective small molecule inhibitors to test whether BMP and/or Nodal inhibition affects either progenitor number or the state of commitment. Multipotent Flk1+ progenitors emerged at day 4 (9.92%) and day 5 (75.3%) (Fig. 3A,B), as noted previously (Yang et al. 2008; Blin et al. 2010). Since many day 4 Flk1− cells become Flk1+ over time (Fig. 3C), we focused attention on day 5 Flk1+ cells. Transfection of Cer1 siRNA-1 1 d prior to differentiation caused near-total inhibition of cardiomyocyte differentiation in the day 5 Flk1+ population, marked by >85% reduction in cardiac precursor marker Isl1, Gata4, and Nkx2-5 and cardiomyocyte marker αMHC and cTnT mRNAs (relative to levels with negative control siRNA) (Fig. 3D,E). In contrast, vascular endothelial (CD31 and VE-Cad) and smooth muscle (sm-MHC and sm22) markers were not affected significantly (Fig. 3F,G), similar to results in bulk, unsorted mESC cultures (Supplemental Fig. S4B,C). The attenuation of cardiomyocyte markers did not reflect a change in the proportion of Flk1+ (Fig. 3H) or Flk1+/Cxcr4+ cells (Supplemental Fig. S4A) or increased cell death (Fig. 3I).
Figure 3.
Cer1 inhibition of Nodal/BMP signaling promotes cardiomyogenic differentiation of Flk1+ progenitors. (A–C) Cardiogenic potency of Flk1+ cells. The proportion of Flk1-eGFP+ cells increases between days 4 and 5 of differentiation (A) and gives rise to cardiomyocytes (α-actinin+), vascular smooth muscle cells (sm-MHC+), and endothelial cells (CD31+) (B). (C) Flk1-eGFP- cells isolated at day 4 or 5 express eGFP after 2 or 3 d of additional culture, respectively. R1 Flk1-eGFP reporter mESCs were used for analysis (see the Materials and Methods). (D–I) Cer1 siRNA-1 decreases cardiomyocyte differentiation. (D) Schematic of protocol for E–I. Cer1 siRNA-1 selectively decreased markers of cardiac precursors (Nkx2-5) and cardiomyocytes (αMHC and cTnT) (E) without affecting endothelial (CD31 and VE-Cad) (F) and smooth muscle (sm-MHC and sm22) (G) markers, by qRT–PCR analyzed at day 16. Cer1 siRNA-1 did not affect the fraction of Flk1+ progenitor cells at days 4 and 5 (H) or cell viability sampled at day 11 (I). Flk1+ cells were FACS-isolated from CGR8 mESCs using an anti-Flk1 antibody (see the Materials and Methods). (J) Small molecules treated with 0.1 μM SB, 0.5 μM DM, or both to block Nodal and/or BMP signaling increased αMHC without affecting CD31 or sm-MHC differentiation. Flk1+ cells were isolated at day 4 of differentiation, compounds were added between days 5 and 9, and differentiation markers were assessed at day 15. (K) Summary model indicating that Cer1 selectively promotes cardiomyogenic differentiation of Flk1+ progenitors. Results similar to J and K were observed in bulk culture (Supplemental Fig. S4B,C). (*) P < 0.05, unpaired Student's t-test. Error bars indicate the SEM; n = 3 in E–I; n = 4 in B and C. See also Supplemental Figure S4.
Since Cer1 siRNA-1 (siCer1) blocked differentiation, we tested whether SB and/or DM would enhance cardiomyogenesis. Flk1+ cells were FACS-isolated at day 4 and treated with SB or DM alone and SB + DM (0.5 μM DM and 0.1 μM SB for 4 d after FACS isolation) (Fig. 1H–J). Although day 4 Flk1+ cells would be expected to have received some inductive signaling based on the endogenous expression of Cer1, SB and DM alone each gave a striking increase in cardiomyocyte marker expression, and together gave the highest induction (Fig. 3J). SB and DM did not affect endothelial and smooth muscle markers. Thus, we conclude that blocking Nodal and BMP signaling in Flk1+ progenitors selectively activates a cardiomyocyte program of differentiation (Fig. 3K).
Nodal and BMP inhibition induce the SWI/SNF component Baf60c and cardiomyogenic transcription factors
We next sought to define the transcriptional mechanism that activated the cardiomyogenic gene program in response to BMP/Nodal inhibition by Cer1. Previous work demonstrated that the SWI/SNF subunit BAF60c is critical for both skeletal and cardiac myogenesis (Lickert et al. 2004; Takeuchi and Bruneau 2009; Forcales et al. 2011). Three Baf60 variants (Baf60a, Baf60b, and Baf60c) are incorporated into the SWI/SNF complex in a mutually exclusive manner that has been proposed to influence the selectivity of the complex for distinct sites in chromatin (Wu et al. 2009; Puri and Mercola 2012). The upstream signaling that directs the relative expression of the Baf60 variants and their activity during cardiogenesis is unknown. Quantitative analysis of the three Baf60 variants in differentiating mESCs revealed a correlation between the temporal profile of Baf60c expression and Cer1 activity (Fig. 4A). Strikingly, Baf60c expression peaked from day 6 when Baf60a and Baf60b were down-regulated, correlating to the Mesp1+/Flk1+-to-Nkx2-5+ transition (Fig. 1B). shRNA knockdown of Cer1 decreased Baf60c mRNA and protein levels as early as day 4 (Fig. 4B,C) without affecting Flk1 or Mesp1 expression (Fig. 1G) or the number of Flk1+ progenitors (Fig. 3H,I), suggesting that Cer1 induces Baf60c. Baf250a, Baf250b, and Baf180, previously implicated in myocardial development (Wang et al. 2004; Lei et al. 2012), were unaffected (Fig. 4B).
Figure 4.
Cer1 coordinately inhibits Nodal and BMP to induce Baf60c, Gata4, and Tbx5. (A) qRT–PCR profiling shows dynamic expression of Tbx5, Gata4, Baf60a, Baf60b, and Baf60c during ESC differentiation. Note that Baf60c peaks at day 6, coinciding with down-regulation of Baf60a and Baf60b, immediately following maximal expression of Cer1. (B) shRNA knockdown of Cer1 (see the Supplemental Material) down-regulated Baf60c, Tbx5, and Gata4 mRNA assayed by qRT–PCR at day 4 but not Baf250a, Baf250b, and Baf180. (C) Western blot showing decrease in Baf60c protein by Cer1 shRNA, assayed at day 6 of differentiation. (D) Profiling of gene expression in Flk1+, Cxcr4+ (cardiac precursor); Flk1+, Cxcr4− (CD31+, endothelial-enriched); and Flk1−, Cxcr4+ (endodermal) populations at day 4. Note enrichment of Baf60c, Gata4, and Tbx5 in the Flk1+, Cxcr4+ cardiac progenitor-enriched population. Data are plotted as the natural log (logn) of fold enrichment of gene expression. (E) Baf60c immunostaining in nuclei of Fkl1+ cells. Flk1-eGFP+ cells were FACS-isolated from differentiating R1 mESCs at day 5 of differentiation and allowed to resume differentiation for 4 d. (F) Cer1 siRNA-1 decreased the expression of Baf60c, Gata4, and Tbx5 but not the general mesodermal marker Mixl1 in the Flk1+, Cxcr4+ (cardiac progenitor-enriched) population. Gata4 also decreased in the Flk1+, Cxcr4− populations but not in the Flk1−, Cxcr4+ population. Data are plotted relative to levels following transfection with negative control (neg. ctrl.; scrambled sequence) siRNA. (G) Complementary effects of inhibiting Nodal and BMP on Flk1+ progenitors. The BMP inhibitor DM and recombinant BMP4 increased and decreased, respectively, the Tbx5 levels, while SB and recombinant Activin-A had no effect. Conversely, SB and Activin-A increased and decreased, respectively, the Baf60c levels, while DM and BMP4 had no effect. Both signaling pathways regulated Gata4 and αMHC. Flk1+ cells were FACS-isolated from CGR8 mESCs at day 4 of differentiation (see the Supplemental Material). Data were normalized to Gapdh levels and plotted as the natural log (logn) of fold changes in gene expression relative to untreated samples. (H,I) Similar effects of inhibiting Nodal were observed on hESCs cardiogenesis. (H) SB increased BAF60C and NKX2-5 gene expression, assayed by qRT–PCR at day 6. (I) SB (1 μM) increased the number of cardiomyocytes at day 18, represented by the αMHC-mCherry reporter fluorescence (see Supplemental Fig. S6B for flow cytometry profiles) and summarized from the results of three experiments. See the Supplemental Material and Supplemental Figure S6A for the experimental protocol. (J) BAF60C overexpression up-regulated mRNA expression of NKX2-5, αMHC, and ACTN2 in day 14 hESCs, assayed by qRT–PCR. (K–N) Baf60c overexpression rescued deficient cardiogenesis after Cer1 knockdown. (K) Schematic of protocol for L–N. Analysis by qRT–PCR at day 10 (L) and αMHC-eGFP reporter fluorescence imaging at day 15 (M,N) (see the Materials and Methods). (*) P < 0.05, unpaired Student's t-test. Error bars indicate the SEM; n = 3 independent replicates for all experiments. See also Supplemental Figures S5–S7.
Since Cer1 shRNA decreased Tbx5 and Gata4 transcript levels concomitantly (Fig. 4B), we hypothesized a coordinate regulation of the transcriptional machinery that activates cardiogenesis. Consistent with this possibility, Baf60c, Tbx5, and Gata4 were coenriched in Flk1+, Cxcr4+ (cardiogenic) precursors (Fig. 4D), and Baf60c protein was evident in the nuclei of differentiating Flk1+ cells isolated by FACS from day 5 embryoid bodies (EBs) (Fig. 4E). More importantly, siRNA knockdown of Cer1 diminished transcript levels for each of these genes in the double-positive (Flk1+, Cxcr4+) cells (Fig. 4F) without affecting the formation of the double-positive cells themselves (Supplemental Fig. S4A). Baf60c, Tbx5, and Gata4 are not regulated identically, however. Treatment with SB (Fig. 4G) or knocking down Smad2 (Supplemental Fig. S5A,B) selectively up-regulated Baf60c, whereas DM selectively up-regulated Tbx5, and both increased Gata4 in Flk1+ cells (Fig. 4G) and unsorted mESCs (both R1 and CGR8) (data not shown), consistent with the synergistic action of SB and DM.
Wnt signaling, which also participates in cardiogenesis at this developmental stage, did not affect Baf60c expression (Supplemental Fig. S5A,C,D). As for mESCs, SB also induced the expression of BAF60C in human ESCs (hESCs) (Fig. 4H) and the number of cardiomyocytes more than twofold (Fig. 4I; Supplemental Fig. S6B) at the developmental stage when uncommitted KDR+ progenitors are present (Kattman et al. 2011; Willems et al. 2011). Furthermore, lentiviral overexpression of BAF60C enhanced induction of cardiomyocyte gene expression in hESCs (Fig. 4J). Thus, Nodal inhibition plays a conserved role in induction of Baf60c.
To establish a functional relationship between endogenous Nodal/BMP inhibition and Baf60c, we tested whether Baf60c overexpression could rescue the deficit in cardiogenesis caused by siCer1 (Fig. 4K–N). Baf60c completely restored mRNA levels for Nkx2-5, αMHC, and cTnT (Fig. 4L) and levels of αMHC-eGFP reporter fluorescence (Fig. 4M,N), substantiating the idea that Baf60c mediates Cer1 function in cardiogenesis. BAF60c overexpression did not affect markers of endothelial or smooth muscle differentiation (Supplemental Fig. S7).
Cer1 mediates cardiomyogenesis through Baf60c-dependent chromatin remodeling
The preceding results suggest that Cer1 might control commitment to cardiomyocyte differentiation by modulating Baf60c and SWI/SNF function. We tested this idea directly by knocking down Baf60c (Fig. 5A–F). RNAi knockdown at the progenitor stage decreased the level of αMHC mRNA by ∼50% and the yield of cardiomyocytes in both hESCs and mESCs by ∼50%–75% (Fig. 5B; Supplemental Fig. S8A,B). The knockdown effect is identical to siRNA knockdown of Cer1, Tbx5, and Gata4 by quantitative RT–PCR (qRT–PCR) (Fig. 5B). Other cardiac markers (Nkx2-5, βMHC, MLC2v, MLC2a, cTnT, and ANF) were similarly repressed (Fig. 5C), as was α-actinin immunostaining (Fig. 5D,D′). Baf60c knockdown also blocked differentiation of FACS-isolated Flk1+ progenitors (Supplemental Fig. S8C,D), suggesting cell autonomy. In contrast, Baf60c shRNA only minimally affected marker gene expression in terminally differentiated mESC-derived cardiomyocytes (Fig. 5E–F), suggesting that its activity is necessary during cardiac commitment but is dispensable in the maintenance of the differentiated phenotype. Furthermore, inhibition or activation of Nodal signaling does not affect Baf60c expression in purified cardiomyocytes (data not shown).
Figure 5.
Cer1 activation of Baf60c-mediated chromatin remodeling is essential for the transition from Flk1+ progenitors to Nkx2-5+ cardiac precursors. (A) Schematic of experimental timeline for B–F examining Baf60c's effect in progenitors versus terminally differentiated cardiomyocytes. (B–D) After knockdown at the progenitor stage, qRT–PCR profiling of marker expression shows that siRNAs against Baf60c, Cer1, Gata4, and Tbx5 each decreased αMHC transcripts (shown in B). Similarly, siRNA against Baf60c decreased transcript levels of all cardiac markers examined (C) as well as α-actinin protein by immunostaining (D,D′). Cells were transfected with siRNAs at day 3 (Cer1, Gata4, and Tbx5) or day 5 (Baf60c), corresponding to onset of expression (Fig. 4A), and analyzed at day 10. (B,C,F) qRT–PCR data were normalized to Gapdh and plotted relative to negative control (neg. ctrl.) siRNA for each gene (set to 100%). (E–F) Effects of Baf60c shRNA knockdown in purified cardiomyocytes (see the Supplemental Material) on α-actinin staining (E,E′) and contractile protein mRNAs (F). (F) Baf60c shRNA showed no effect on most contractile protein mRNAs, except for modest decreases of αMHC and MLC2v; t-test significantly. (G,H) Epistasis experiment positions Baf60c downstream from Cer1. (G) Nkx2-5 transcript levels increased in response to Cer1 overexpression and were restored by Baf60c siRNA. (H) Conversely, Cer1 siRNA decreased Nkx2-5 expression, and this effect was completely reversed by overexpression of Baf60c. pmCer1 and siCer1 were transfected 1 d before differentiation; Baf60c siRNA and pBaf60c were transfected at day 3; Nkx2-5 expression was quantified by qRT–PCR at day 7. (I) Baf60c immunostaining colocalizes with Nkx2-5-eGFP reporter expression in differentiating mESCs. White arrowheads indicate examples of nuclear Baf60c in cells with cytoplasmic Nkx2-5-eGFP reporter fluorescence (61% ± 11% of Nkx2-5-eGFP+ cells are also Baf60c+). (J) Schematic of the ChIP-qPCR and the chromatin accessibility experiments (left) and diagram of the Nkx2-5 promoter showing the relationship of the −9-kb and −2.8-kb regions that are essential to direct expression in the heart versus stomach, pharmyx, and thyroid, respectively (right) (Lien et al. 1999; Reecy et al. 1999). Arrows indicate PCR primer pairs flanking MboII restriction endonuclease sites within the tissue-specific enhancers, which were used in the ChIP-qPCR (K–M) and endonuclease accessibility (O–Q) assays. (K–M) ChIP-qPCR assessing Baf60c occupancy (K) and H3K4me1 (L) and H3K27ac (M) histone modifications of the Nkx2-5 −9-kb and −2.8-kb enhancers and a −15-kb nonenhancer region in undifferentiated and day 6 differentiating mESCs. Fold enrichment was normalized to the IgG control. (N) Diagram of the chromatin accessibility assay. (O) Relative abundance of PCR amplicons corresponding to the −9-kb and −2.8-kb enhancer elements. Greater amplicon abundance of the −9-kb enhancer was detected in undifferentiated cells (reflecting closed chromatin) than at day 6 of differentiation, when the gene becomes active (reflecting open chromatin). siRNAs against Baf60c, Cer1, and Brg1 increased amplicon abundance, indicating decreased accessibility. In contrast, amplicons corresponding to the −2.8-kb enhancer were comparably inaccessible to the undifferentiated state under all conditions. (P) qRT–PCR profile of Nkx2-5 transcript levels (day 8) for the experiment in K normalized to scrambled sequence (negative control [neg. ctrl.]) siRNA. Note that siRNAs to Cer1, Baf60c, Brg1, Brm, Tbx5, and Gata4 all reduced Nkx2-5 transcript levels. (Q) Baf60c overexpression rescued deficient chromatin accessibility on the −9-kb cardiac enhancer after Cer1 knockdown. Cer1 siRNA-1 and pBaf60c were cotransfected at day 3, and chromatin accessibility was analyzed at day 6. (*) P < 0.05; (#) P < 0.1; unpaired Student's t-test. Error bars indicate the SEM; n = 3 independent biological replicates for all experiments. See also Supplemental Figure S8.
Probing epistasis, siRNA to Baf60c completely blocked the ability of overexpressed Cer1 to enhance cardiogenesis (Fig. 5G). Conversely, overexpressed Baf60c completely restored the reduction in cardiogenesis caused by siRNA to Cer1 (Fig. 5H). Together, these data indicate that Baf60c functions downstream from Cer1.
To pursue this model further, we asked whether Baf60c becomes localized to a native cardiac enhancer within chromatin during cardiomyocyte lineage commitment. The Nkx2-5 gene is ideally suited for this experiment since it is activated in cells that express Baf60c (61% ± 11% of cells with cytoplasmic Nkx2-5-eGFP expression are Baf60c+) (Fig. 5I, white arrowheads) and harbors two well-defined enhancers: an early cardiac-specific enhancer at −9 kb from the start site of transcription and a stomach, pharynx, and thyroid-specific enhancer at −2.8 kb that are individually capable of directing tissue-appropriate expression in transgenic mice (Lien et al. 1999; Reecy et al. 1999). The cardiomyocyte enhancer recapitulates the expression pattern of the endogenous gene in cardiogenic precursors from the onset of lineage specification to looping heart tube, after which more distally located enhancers are needed to direct expression in the heart (Lien et al. 1999; Reecy et al. 1999; Chi et al. 2005). The signals that make the −9-kb enhancer accessible during lineage commitment are unknown, although once open, its activity depends on Gata4 (Lien et al. 1999; He et al. 2011).
We performed chromatin immunoprecipitation (ChIP) followed by qPCR (ChIP-qPCR) on differentiating mESCs at day 6 when Nkx2-5 is actively transcribed. The mESCs had been transfected with either Baf60c or negative control siRNAs at day 3 (Fig. 5J). Baf60c localized to the −9-kb early cardiac enhancer but not to the −2.8-kb noncardiac enhancer or a −15-kb nonenhancer region. Occupancy of the −9-kb region was abolished by transfection of Baf60c siRNA (Fig. 5K). ChIP-qPCR using antibodies recognizing two histone modifications (H3K4me1 and H3K27ac) associated with active enhancers (Xiao et al. 2012) showed a pattern identical to Baf60c on the −9-kb cardiac enhancer but no enrichment at the noncardiac enhancer and nonenhancer region (Fig. 5K,L). Thus, Baf60c localizes to the Nkx2-5 cardiac enhancer at the time of transcriptional activation.
To determine whether Cer1 remodels chromatin via Baf60c, mESCs were transfected with either Cer1 or negative control siRNA at day 3, and cell nuclei were isolated at day 6 and assayed for chromatin accessibility by sensitivity to MboII endonuclease activity (Fig. 5J,N,O). Greater amplicon abundance upon qPCR reflects less accessible (closed) chromatin, such as from undifferentiated mESCs (Fig. 5O, black bars). Significantly fewer −9-kb amplicons were obtained from day 6 differentiating mESCs, indicative of open chromatin (Fig. 5O, gray bar). In contrast to scrambled sequence siRNA (Fig. 5O, neg. ctrl.), specific siRNAs to Cer1 increased abundance of the −9-kb cardiac enhancer amplicon to levels in undifferentiated cells, indicating that down-regulating Cer1 prevented the developmental opening of the Nkx2-5 cardiac enhancer. Similarly, siRNA to Baf60c (Fig. 5O, yellow bars) also conferred resistance to MboII, indicating that Cer1 and Baf60c are essential to make the −9-kb cardiac enhancer accessible to transcriptional machinery during cardiogenic development. siRNAs to Tbx5 and Gata4 did not prevent chromatin accessibility to MboII (Fig. 5O). Furthermore, the −2.8-kb enhancer remained in an inaccessible state at 6 d of differentiation and was unaffected by any of the manipulations, reflecting fidelity of the system (Fig. 5O).
Evaluating the two catalytic SWI/SNF subunits, siRNA to Brg1 (Fig. 5O, purple bar) but not to Brm (Fig. 5O, orange bar) also conferred resistance to MboII (Fig. 5O), suggesting that Brg1 provides the essential ATPase component of the Baf60c-based SWI/SNF complex that remodels chromatin of the Nkx2-5 cardiac enhancer.
We confirmed that greater accessibility to the −9-kb enhancer correlated with Nkx2-5 expression. Baf60c, Brg1, and Cer1 siRNAs individually reduced Nkx2-5 transcript levels (Fig. 5P). Interestingly, siRNA against Gata4 and Tbx5 also decreased the Nkx2-5 transcripts (Fig. 5P) despite lacking a significant effect on chromatin accessibility, but this is consistent with the importance of these factors for active Nkx2-5 transcription (Lien et al. 1999; He et al. 2011). Also, siRNA to Brm slightly decreased the Nkx2-5 mRNA level (Fig. 5P) even though it does not appear essential to open chromatin (Fig. 5O), possibly implying a partial redundancy with Brg1.
Finally, we used the chromatin accessibility assay to evaluate whether Baf60c is the critical mediator of Cer1's effect on chromatin. As above, siRNA to Cer1 prevented the developmental opening of the −9-kb Nkx2-5 enhancer, resulting in high amplicon abundance (Fig. 5Q, red bars). Cotransfection with a plasmid to overexpress Baf60c completely restored accessibility, resulting in low amplicon abundance (Fig. 5Q, yellow bars). Cer1 siRNA with or without Baf60c overexpression did not affect the −2.8-kb enhancer, again indicating fidelity of the system. In summary, we conclude that inhibition of Nodal and BMP signaling induces Baf60c, thereby localizing SWI/SNF to the −9-kb early cardiac enhancer and activating cardiomyogenic transcription.
Discussion
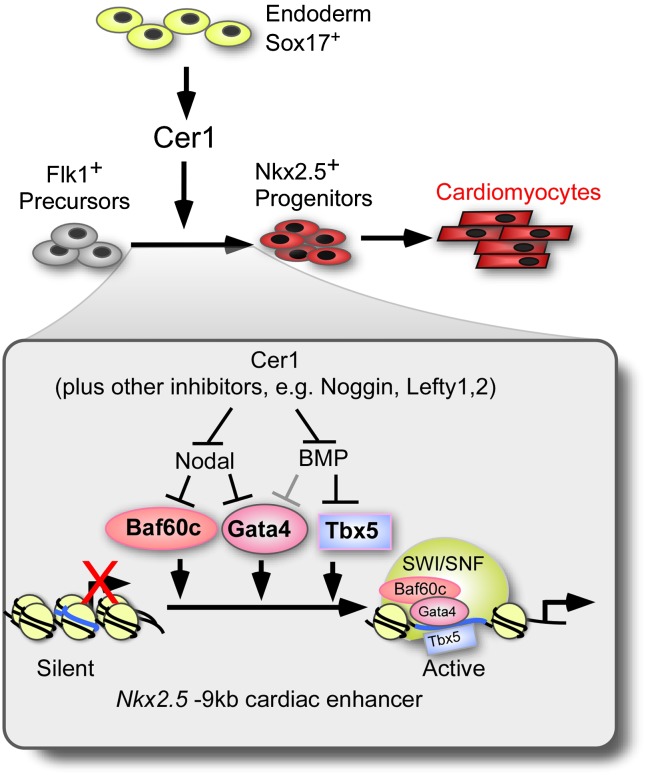
Our data support a model for cardiomyocyte commitment of multipotent KDR/Flk1+ progenitors that links extracellular signaling from endoderm to the control of chromatin remodeling. We found that inhibition of Nodal by endodermal-derived Cer1 or, experimentally, by small molecule inhibition induces the noncatalytic Baf60c subunit of SWI/SNF and the cardiogenic transcription factor Gata4. Simultaneous inhibition of BMP—also by Cer1 or small molecules—induces Tbx5 as well as Gata4, which together recruit the SWI/SNF complex to physically occupy and open cardiogenic loci in chromatin. Further siRNA and epistasis experiments show that the developmental opening of the Nkx2-5 early cardiac enhancer by Cer1 depends on Baf60c and the function of the SWI/SNF catalytic subunit Brg1. The alternate Baf60 variants (a and b), which are not selective for myogenic lineages, decline at this time. Thus, we identified a lineage control mechanism in which Nodal and BMP antagonists in the progenitor cell environment signal directly to the progenitor cell to induce Baf60c and interacting cardiogenic transcription factors, thereby altering the subunit composition of SWI/SNF and causing its selective recruitment to cardiogenic loci in chromatin, establishing a transcriptional landscape for the cardiomyocyte commitment (Fig. 6).
Figure 6.
Environmental programming of SWI/SNF to determine cardiomyocyte cell fate. Cer1 inhibition of Nodal and BMP regulates Baf60c, Tbx5, and Gata4, programming SWI/SNF to open chromatin of cardiogenic loci such as Nkx2-5, allowing access to polymerase II transcriptional machinery, and activating a cardiomyocyte program of differentiation. The model accounts for the cardiomyogenic function of endodermal cells in ESC cultures, which produce Cer1 as well as other Nodal and BMP inhibitors.
Cer1 as an inducer of ESC cardiomyocyte differentiation
The necessity of Cer1 for mESC cardiomyogenesis is consistent with its dual role as a Nodal and BMP inhibitor but was unanticipated, since cardiac induction proceeds normally in the Cer1 knockout mouse (Belo et al. 2000; Stanley et al. 2000). In mouse embryos, however, Cer1 is coexpressed with other Nodal inhibitors Lefty1 and Lefty2 and the BMP inhibitors Noggin and Chordin during the period when the heart field is specified (Bachiller et al. 2000; Perea-Gomez et al. 2002). In ESCs, Cer1 is the predominant inhibitor (Fig. 1B), while Lefty1 in particular is underrepresented in ESCs at the time of cardiomyocyte commitment (Fig. 1B). In this regard, ESC differentiation in vitro does not completely recapitulate developmental induction in vivo. Lefty1 and Cer1 have been shown to be functionally redundant in embryos, at least for primitive streak patterning (Perea-Gomez et al. 2002), and its absence might explain why our data using mESCs revealed a critical role for Cer1. We showed that Xenopus Cerberus is required for cardiogenesis (Foley et al. 2007), but XCerberus shares only modest (26%) sequence positional identity with Cer1 and is also a potent Wnt inhibitor (Bouwmeester et al. 1996; Biben et al. 1998; Piccolo et al. 1999); thus, the cardiogenic significance of Cer1 in mammals or ESCs has never been established. More importantly, the salient conclusion from our study is that the transient inhibition of Nodal and BMP dictates whether multipotent progenitors transit to the Nkx2-5+ state. This model might explain classical observations that the heart field in the embryo forms only in mesoderm adjacent to anterior endoderm (where Nodal and BMP antagonists are expressed), although cardiac competence extends throughout the early mesoderm (Nascone and Mercola 1995; Arai et al. 1997; Schultheiss et al. 1997).
A chromatin remodeling model for cardiomyogenic commitment
Our data show that Brg1 is essential for the developmental opening of the Nkx2-5 early cardiac enhancer (Fig. 5O,P). Brg1 is one of two core DNA-dependent ATPases (the other is Brm) that have been evolutionarily conserved from yeast to humans and are alternately assembled into SWI/SNF (Albini and Puri 2010; Lessard and Crabtree 2010). The developmental requirement for Brg1 to increase accessibility of the early Nkx2-5 cardiac enhancer and transcription is consistent with the idea that SWI/SNF-dependent chromatin remodeling occurs at key positions to make lineage-specific loci accessible to polymerase II transcription complexes, allowing for coordinate activation of genes involved in a specific (e.g., cardiac) differentiation program (Paige et al. 2012; Wamstad et al. 2012).
Despite its importance to lineage selection, how the catalytic subunits of SWI/SNF become localized to specific sites in chromatin remains poorly understood. In addition to the Brg1 or Brm catalytic proteins, SWI/SNF consists of 10–16 noncatalytic Brg1/Bafs in mammals, including the 60-kDa Baf60a, Baf60b, and Baf60c proteins (Albini and Puri 2010; Lessard and Crabtree 2010; Euskirchen et al. 2012). An emerging concept is that lineage selectivity is based on the mutually exclusive, combinatorial assembly of variants of the Baf structural subunits into SWI/SNF (Wu et al. 2009; Puri and Mercola 2012). The variant composition is thought to promote interactions with specific transcription factors that cause SWI/SNF to recognize discrete, lineage-appropriate sites in chromatin. For instance, Baf45b, Baf45c, and Baf53b, replacing other subunits of Bafs in SWI/SNF-like complexes, mediate the transition from proliferating neural progenitor cells to post-mitotic neurons (Lessard et al. 2007). Similarly, during commitment of myogenic stem cells to skeletal muscle differentiation, Baf60c, but not Baf60a and Baf60b, physically associates with MyoD and targets SWI/SNF to myogenic loci in chromatin, priming the chromatin at muscle loci for transcriptional activation in response to myogenic cues such as P38 signaling (Simone et al. 2004; Forcales et al. 2011). Of particular relevance to cardiomyocyte differentiation, Takeuchi and Bruneau (2009) demonstrated that Baf60c and Gata4 (augmented by Tbx5) are sufficient to induce ectopic cardiomyogenic differentiation upon overexpression in mouse embryos. Conversely, a requirement for Baf60c-mediated cardiomyogenic commitment is consistent with myocardial hypoplasia and defective morphogenesis caused by RNAi knockdown of Baf60c in mouse embryos, although cardiomyocytes did form, suggesting that additional mechanisms might compensate in embryos (Lickert et al. 2004).
The relevance of niche factors for Baf60 variant selection
Since SWI/SNF-dependent chromatin remodeling is essential for differentiation of multiple lineages, the mutually exclusive assembly of BAF subunit variants into SWI/SNF might act as a “code” to direct interactions with lineage-specific loci (Wu et al. 2009; Puri and Mercola 2012). However, the precise signals in the progenitor cell environment that control subunit variant composition were previously unknown in any context (Puri and Mercola 2012). We found that Baf60c became expressed only with differentiation and selectively within the Flk1+, Cxcr4+ (cardiogenic) precursors, peaking at the time when cardiogenic progenitors (Mesp1+/Flk1+) form cardiac progenitors (Nkx2-5+) and corresponding to a dip in Baf60a and Baf60b variant expression (Fig. 4A,H,I). Moreover, our data show that the developmental induction of Baf60c, Gata4, and Tbx5 occurs in direct response to endogenous Nodal and BMP inhibitors, implicating paracrine Nodal/BMP antagonists as the trigger for cardiomyogenic chromatin remodeling. Such a model might apply to other settings in which Baf60 variants are selectively expressed or even more broadly when SWI/SNF becomes selectively associated with lineage-specific sites in chromatin. For instance, niche factors secreted in the environment of skeletal muscle stem cells might control Baf60 variant expression and dictate whether these cells differentiate along pathological fibro-adipogenic versus regenerative myoblast lineages (S Albini, L Giordani, M Mercola, and PL Puri, unpubl.).
In summary, we propose a model that bridges extracellular signaling to chromatin remodeling in which paracrine BMP/Nodal inhibition programs the SWI/SNF complex in cardiac progenitor cells to control the transition to a cardiomyogenic precursor. It is tempting to consider the possibility that the same process might influence stem cell renewal in the adult heart. Recent studies support the conclusion that stem or progenitor-based cardiomyocyte turnover occurs in the adult mammalian heart and that the process is enhanced following injury (e.g., Hsieh et al. 2007). Since opening chromatin at cardiomyogenic loci is critical for gene activation, differentiation of adult cardiac stem cells into new cardiomyocytes after injury might also require the transient inhibition of Smad signaling to program SWI/SNF composition and create a conducive transcriptional landscape. If this were the case, it is possible that the elevated TGFβ signaling associated with fibrosis and ventricular remodeling that occurs in the injured heart might inherently antagonize the heart's regenerative potential. Therefore, it will be important to determine whether localized inhibition of TGFβ can enhance the regenerative potential of endogenous cardiac progenitor cells in the adult heart.
Materials and methods
Additional details and methods can be found in the Supplemental Material.
ESC culture
mESCs were cultured and differentiated as EBs under serum or serum-free conditions as described previously (Cai et al. 2012; Willems et al. 2012) and in the Supplemental Material.
H9 human ESCs were maintained and differentiated as described previously (Willems et al. 2011) and in the Supplemental Material. To induce cardiac differentiation, EBs were treated with BMP4, hrActivin A, and hrbFGF at the indicated stages (color blocks) as depicted in Supplemental Figure S6A. For monolayer differentiation, day 4 differentiated EBs were dissociated gently to single cells and then differentiated in StemPro 34 with 5 ng/mL hrbFGF, IWR, and SB.
qRT–PCR
The primer sequences for qRT–PCR are listed in Supplemental Table 1.
ChIP-qPCR and endonuclease accessibility assays
For ChIP, chromatin extracts were immunoprecipitated with protein A Dynabeads (Invitrogen) and specific antibodies, including anti-BAF60c (Albini et al. 2013), anti-H3K4me1 (Active Motif), anti-H3K27ac (Active Motif), and normal IgG as control. DNA was extracted followed by qPCR for amplification of specific sequences.
Endonuclease accessibility was as performed on isolated nuclei using 100 U of restriction endonuclease MBOII to digest chromatin. Reactions were stopped, and genomic DNA was extracted and subjected to qPCR. The primer sequences are listed in the Supplemental Material.
Statistics
Each experiment was repeated at least two times using a minimum of three biological replicates per condition. Statistical analysis was performed with an unpaired Student's t-test. The asterisk in the figures represents P < 0.05. Error bars indicate the SEM.
Acknowledgments
We are grateful to Dr. Yu Liu (University of Houston), Dr. Michael Schneider (Imperial College, London), Dr. Benoit Bruneau (University of California at San Francisco), Dr. Sean M. Wu (Stanford University), and Dr. Eileen D. Adamson (Stanford-Burnham Medical Research Institute) for reagents and cell lines. We are indebted to Yoav Altman and Amy Cortez for flow cytometry, Wesley L. McKeithan for help with Wnt inhibition experiments, and Herman Davidovics for general support. We thank Gabriella Minchiotti (Institute of Genetics and Biophysics, Naples) and Michael Schneider for helpful discussions. This research was supported by the Fondation Leducq Shapeheart Network and the NIH (R37HL050502, R01HL108176, and R33HL088266 to M.M.; R01AR056712 and R01AR052779 to P.L.P.; and shared service grants P30AR061303 and P30CA030199 for flow cytometry).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.225144.113.
References
- Albini S, Puri PL 2010. SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: It's time to exchange! Exp Cell Res 316: 3073–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini S, Coutinho P, Malecova B, Giordani L, Savchenko A, Forcales SV, Puri PL 2013. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep 3: 661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai A, Yamamoto K, Toyama J 1997. Murine cardiac progenitor cells require visceral embryonic endoderm and primitive streak for terminal differentiation. Dev Dyn 210: 344–353 [DOI] [PubMed] [Google Scholar]
- Bachiller D, Klingensmith J, Kemp C, Belo JA, Anderson RM, May SR, McMahon JA, McMahon AP, Harland RM, Rossant J, et al. 2000. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature 403: 658–661 [DOI] [PubMed] [Google Scholar]
- Bacon RL 1945. Self-differentiation and induction in the heart of Amblystoma. J Exp Zool 98: 87–125 [Google Scholar]
- Belo JA, Bachiller D, Agius E, Kemp C, Borges AC, Marques S, Piccolo S, De Robertis EM 2000. Cerberus-like is a secreted BMP and nodal antagonist not essential for mouse development. Genesis 26: 265–270 [PubMed] [Google Scholar]
- Biben C, Stanley E, Fabri L, Kotecha S, Rhinn M, Drinkwater C, Lah M, Wang C-C, Nash A, Hilton D, et al. 1998. Murine cerberus homolgue mCer-1: A candidate anterior patterning molecule. Dev Biol 194: 135–151 [DOI] [PubMed] [Google Scholar]
- Blin G, Nury D, Stefanovic S, Neri T, Guillevic O, Brinon B, Bellamy V, Rucker-Martin C, Barbry P, Bel A, et al. 2010. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J Clin Invest 120: 1125–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester T, Kim S-H, Sasai Y, Lu B, De Robertis EM 1996. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann's organizer. Nature 382: 595–601 [DOI] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ 2001. Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411: 965–969 [DOI] [PubMed] [Google Scholar]
- Cai W, Guzzo RM, Wei K, Willems E, Davidovics H, Mercola M 2012. A Nodal-to-TGFβ cascade exerts biphasic control over cardiopoiesis. Circ Res 111: 876–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Chatterjee PK, Wilson W III, Zhang SX, Demayo FJ, Schwartz RJ 2005. Complex cardiac Nkx2-5 gene expression activated by noggin-sensitive enhancers followed by chamber-specific modules. Proc Natl Acad Sci 102: 13490–13495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G, Auerbach RK, Snyder M 2012. SWI/SNF chromatin-remodeling factors: Multiscale analyses and diverse functions. J Biol Chem 287: 30897–30905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley AC, Korol O, Timmer AM, Mercola M 2007. Multiple functions of Cerberus cooperate to induce heart downstream of Nodal. Dev Biol 303: 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcales SV, Albini S, Giordani L, Malecova B, Cignolo L, Chernov A, Coutinho P, Saccone V, Consalvi S, Williams R, et al. 2011. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J 31: 301–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT 2011. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci 108: 5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh PC, Segers VF, Davis ME, MacGillivray C, Gannon J, Molkentin JD, Robbins J, Lee RT 2007. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med 13: 970–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida M, Heike T, Yoshimoto M, Baba S, Doi H, Nakahata T 2005. Identification of cardiac stem cells with FLK1, CD31, and VE-cadherin expression during embryonic stem cell differentiation. FASEB J 19: 371–378 [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM 2006. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell 11: 723–732 [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G 2011. Stage-specific optimization of Activin/Nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8: 228–240 [DOI] [PubMed] [Google Scholar]
- Kwon C, Arnold J, Hsiao EC, Taketo MM, Conklin BR, Srivastava D 2007. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc Natl Acad Sci 104: 10894–10899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei I, Gao X, Sham MH, Wang Z 2012. SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J Biol Chem 287: 24255–24262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard JA, Crabtree GR 2010. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol 26: 503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR 2007. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55: 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG 2004. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432: 107–112 [DOI] [PubMed] [Google Scholar]
- Lien CL, Wu C, Mercer B, Webb R, Richardson JA, Olson EN 1999. Control of early cardiac-specific transcription of Nkx2-5 by a GATA-dependent enhancer. Development 126: 75–84 [DOI] [PubMed] [Google Scholar]
- Mercola M, Ruiz-Lozano P, Schneider MD 2011. Cardiac muscle regeneration: Lessons from development. Genes Dev 25: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison GM, Oikonomopoulou I, Migueles RP, Soneji S, Livigni A, Enver T, Brickman JM 2008. Anterior definitive endoderm from ESCs reveals a role for FGF signaling. Cell Stem Cell 3: 402–415 [DOI] [PubMed] [Google Scholar]
- Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I 2006. Developmental stage-specific biphasic roles of Wnt/β-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci 103: 19812–19817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascone N, Mercola M 1995. An inductive role for the endoderm in Xenopus cardiogenesis. Development 121: 515–523 [DOI] [PubMed] [Google Scholar]
- Nelson TJ, Faustino RS, Chiriac A, Crespo-Diaz R, Behfar A, Terzic A 2008. CXCR4+/FLK-1+ biomarkers select a cardiopoietic lineage from embryonic stem cells. Stem Cells 26: 1464–1473 [DOI] [PubMed] [Google Scholar]
- Noseda M, Peterkin T, Simoes FC, Patient R, Schneider MD 2011. Cardiopoietic factors: Extracellular signals for cardiac lineage commitment. Circ Res 108: 129–152 [DOI] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. 2012. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell 151: 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Gomez A, Vella FD, Shawlot W, Oulad-Abdelghani M, Chazaud C, Meno C, Pfister V, Chen L, Robertson E, Hamada H, et al. 2002. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell 3: 745–756 [DOI] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Leyns L, Bhattacharyya S, Grunz H, Bouwmeester T, De Robertis EM 1999. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature 397: 707–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Mercola M 2012. BAF60 A, B, and Cs of muscle determination and renewal. Genes Dev 26: 2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reecy JM, Li X, Yamada M, DeMayo FJ, Newman CS, Harvey RP, Schwartz RJ 1999. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development 126: 839–849 [DOI] [PubMed] [Google Scholar]
- Schultheiss TM, Burch JB, Lassar AB 1997. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev 11: 451–462 [DOI] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL 2004. p38 pathway targets SWI–SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 36: 738–743 [DOI] [PubMed] [Google Scholar]
- Stanley EG, Biben C, Allison J, Hartley L, Wicks IP, Campbell IK, McKinley M, Barnett L, Koentgen F, Robb L, et al. 2000. Targeted insertion of a lacZ reporter gene into the mouse Cer1 locus reveals complex and dynamic expression during embryogenesis. Genesis 26: 259–264 [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG 2009. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459: 708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. 2012. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151: 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, Kang C, Skarnes WC, Tjian R 2004. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev 18: 3106–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, Cashman J, Mercola M 2011. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res 109: 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E, Cabral-Teixeira J, Schade D, Cai W, Reeves P, Bushway PJ, Lanier M, Walsh C, Kirchhausen T, Izpisua Belmonte JC, et al. 2012. Small molecule-mediated TGF-β type II receptor degradation promotes cardiomyogenesis in embryonic stem cells. Cell Stem Cell 11: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR 2009. Understanding the words of chromatin regulation. Cell 136: 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Xie D, Cao X, Yu P, Xing X, Chen CC, Musselman M, Xie M, West FD, Lewin HA, et al. 2012. Comparative epigenomic annotation of regulatory DNA. Cell 149: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. 2008. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453: 524–528 [DOI] [PubMed] [Google Scholar]
- Yuasa S, Itabashi Y, Koshimizu U, Tanaka T, Sugimura K, Kinoshita M, Hattori F, Fukami S-I, Shimazaki T, Okano H, et al. 2005. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat Biotechnol 23: 607–611 [DOI] [PubMed] [Google Scholar]