Polycomb group (PcG) complexes repress developmental regulator genes by modifying their chromatin. However, how PcG proteins assemble into complexes and are recruited to their target genes is poorly understood. Here, Alfieri et al. report the crystal structure of the core of the PcG complex PhoRC, which contains the DNA-binding protein Pho and corepressor Sfmbt. The authors show that tethering of Sfmbt by Pho to Polycomb response elements is essential for Polycomb repression of developmental regulator genes in Drosophila. This study thus reveals the molecular basis for PcG protein complex assembly at specific genomic sites.
Keywords: Polycomb, Polycomb response element, PhoRC, Pho, Sfmbt, YY1
Abstract
Polycomb group (PcG) protein complexes repress developmental regulator genes by modifying their chromatin. How different PcG proteins assemble into complexes and are recruited to their target genes is poorly understood. Here, we report the crystal structure of the core of the Drosophila PcG protein complex Pleiohomeotic (Pho)-repressive complex (PhoRC), which contains the Polycomb response element (PRE)-binding protein Pho and Sfmbt. The spacer region of Pho, separated from the DNA-binding domain by a long flexible linker, forms a tight complex with the four malignant brain tumor (4MBT) domain of Sfmbt. The highly conserved spacer region of the human Pho ortholog YY1 binds three of the four human 4MBT domain proteins in an analogous manner but with lower affinity. Comparison of the Drosophila Pho:Sfmbt and human YY1:MBTD1 complex structures provides a molecular explanation for the lower affinity of YY1 for human 4MBT domain proteins. Structure-guided mutations that disrupt the interaction between Pho and Sfmbt abolish formation of a ternary Sfmbt:Pho:DNA complex in vitro and repression of developmental regulator genes in Drosophila. PRE tethering of Sfmbt by Pho is therefore essential for Polycomb repression in Drosophila. Our results support a model where DNA tethering of Sfmbt by Pho and multivalent interactions of Sfmbt with histone modifications and other PcG proteins create a hub for PcG protein complex assembly at PREs.
Polycomb group (PcG) proteins were originally identified as transcriptional repressors of HOX and other developmental regulator genes in Drosophila (Lewis 1978; Busturia and Morata 1988; Dura and Ingham 1988; Kennison 1995). In mammals, PcG proteins repress a similar, conserved set of developmental regulator genes in stem cells and during embryonic development (Boyer et al. 2006; Lee et al. 2006). In addition, PcG proteins are also required for processes such as X-chromosome inactivation (Brockdorff 2011) and are implicated in cancer progression (Mills 2010).
Biochemical studies in Drosophila revealed that PcG proteins exist in four main types of protein complexes. These protein assemblies are Polycomb-repressive complex 1 (PRC1)-type complexes, including PRC1 itself and dRAF; PRC2; Pleiohomeotic (Pho)-repressive complex (PhoRC); and Polycomb-repressive deubiquitinase (PR-DUB) (Shao et al. 1999; Muller et al. 2002; Klymenko et al. 2006; Lagarou et al. 2008; Scheuermann et al. 2010). Complexes related to PRC1, PRC2, and PR-DUB have also been characterized in mammals (Levine et al. 2002; Kuzmichev et al. 2004; Cao et al. 2005; Gearhart et al. 2006; Machida et al. 2009; Sowa et al. 2009; Yu et al. 2010; Gao et al. 2012). However, because mammals contain multiple paralogs for most complex subunits and because some of these subunits have evolved to bind additional proteins, the mammalian Polycomb machinery is generally more complex than in Drosophila. In both Drosophila and mammals, Polycomb protein complexes contain three principal histone-modifying activities: PRC2 is a histone methyltransferase that methylates histone H3 at Lys27 (H3-K27me), PRC1-type complexes possess E3 ligase activity for the monoubiquitination of histone H2A (H2Aub), and PR-DUB is a deubiquitinase for H2Aub (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002; Wang et al. 2004; Lagarou et al. 2008; Scheuermann et al. 2010). PRC1 also modifies chromatin by a noncovalent mechanism; it inhibits nucleosome remodeling by SWI/SNF complexes and compacts nucleosome arrays in vitro (Shao et al. 1999; Francis et al. 2001, 2004). Finally, PhoRC has no known chromatin-modifying activity but contains sequence-specific DNA-binding activity through Pho, the Drosophila ortholog of the mammalian transcription factor YY1 (Brown et al. 1998; Klymenko et al. 2006). In Drosophila, PcG protein complex binding at target genes is highly enriched at short cis-regulatory sequences called Polycomb response elements (PREs) (Simon et al. 1993; Chan et al. 1994; Negre et al. 2006; Schwartz et al. 2006; Tolhuis et al. 2006; Oktaba et al. 2008; for review, see Muller and Kassis 2006; Ringrose 2007). PREs frequently contain binding motifs for the PhoRC subunit Pho (Mihaly et al. 1998; Oktaba et al. 2008; Schuettengruber et al. 2009). PhoRC has been proposed to act as a tethering platform for the assembly of other PcG protein complexes at PREs (Klymenko et al. 2006). First, mutation of Pho protein-binding sites in PREs in reporter genes (Fritsch et al. 1999; Shimell et al. 2000; Busturia et al. 2001; Mishra et al. 2001) or in their native location in the genome (Kozma et al. 2008) abolishes Polycomb repression. Second, mutation of Pho-binding sites in PREs abolishes not only binding of PhoRC but also binding of other PcG proteins at PREs both in vitro and in vivo (Mohd-Sarip et al. 2005; Klymenko et al. 2006). Third, animals lacking Pho and its paralog, Pho-like (Phol), show reduction of binding of other PcG protein complexes at some PREs (Brown et al. 2003; Wang et al. 2004). Even though Pho and Phol are the only known PRE-binding proteins that are essential for repression of HOX genes, it is important to note that PRC1 and PRC2 subunits remain bound at many genomic locations in mutants lacking Pho and/or Phol (Brown et al. 2003; Wang et al. 2004). Pho and Phol are thus not the only PcG complex recruiters, and other yet unknown mechanisms must exist that help anchor PcG protein complexes at target gene chromatin.
PhoRC was initially characterized as a two-subunit complex containing Pho and Sfmbt (Klymenko et al. 2006). Pho and Sfmbt proteins form a stable dimer that can be reconstituted with recombinant proteins in vitro (Klymenko et al. 2006). Genome-wide profiling studies showed that the two proteins colocalize at a large number of PREs in Drosophila embryos and larvae (Oktaba et al. 2008). Previous structural studies of the C-terminal zinc (Zn) finger domain of the human Pho homolog YY1 revealed how it recognizes its cognate DNA-binding site (Houbaviy et al. 1996). The YY1 residues contacting DNA bases and backbone are 100% conserved in Pho (Brown et al. 1998). The YY1:DNA cocrystal structure therefore serves as an excellent model for how Pho recognizes the Pho/YY1-binding motif GCCAT. Sfmbt is a member of the malignant brain tumor (MBT) repeat family of proteins. In Drosophila, this family includes three proteins: Sfmbt, containing four MBT repeats; l(3)mbt, containing three MBT repeats; and the PRC1 subunit Scm, containing two MBT repeats. Previous biophysical and structural studies characterized the four MBT (4MBT) domain of Drosophila Sfmbt and showed that the fourth MBT repeat binds with low micromolar affinity to a variety of mono- and dimethylated lysines in the context of histone tail peptides (Klymenko et al. 2006; Grimm et al. 2009).
Here, we characterize the Pho:Sfmbt interaction using structural, biophysical, and genetic approaches. Atomic resolution structures reveal the architecture of this interaction and show that the human YY1 protein binds human Sfmbt orthologs in a similar manner. We further show that the identified Pho:Sfmbt interaction is critical for Polycomb repression of target genes in Drosophila. Together, these data reveal the molecular basis for how a DNA-binding PcG protein complex assembles at specific DNA elements in target genes.
Results
The Pho spacer region forms a stable complex with the Sfmbt 4MBT domain
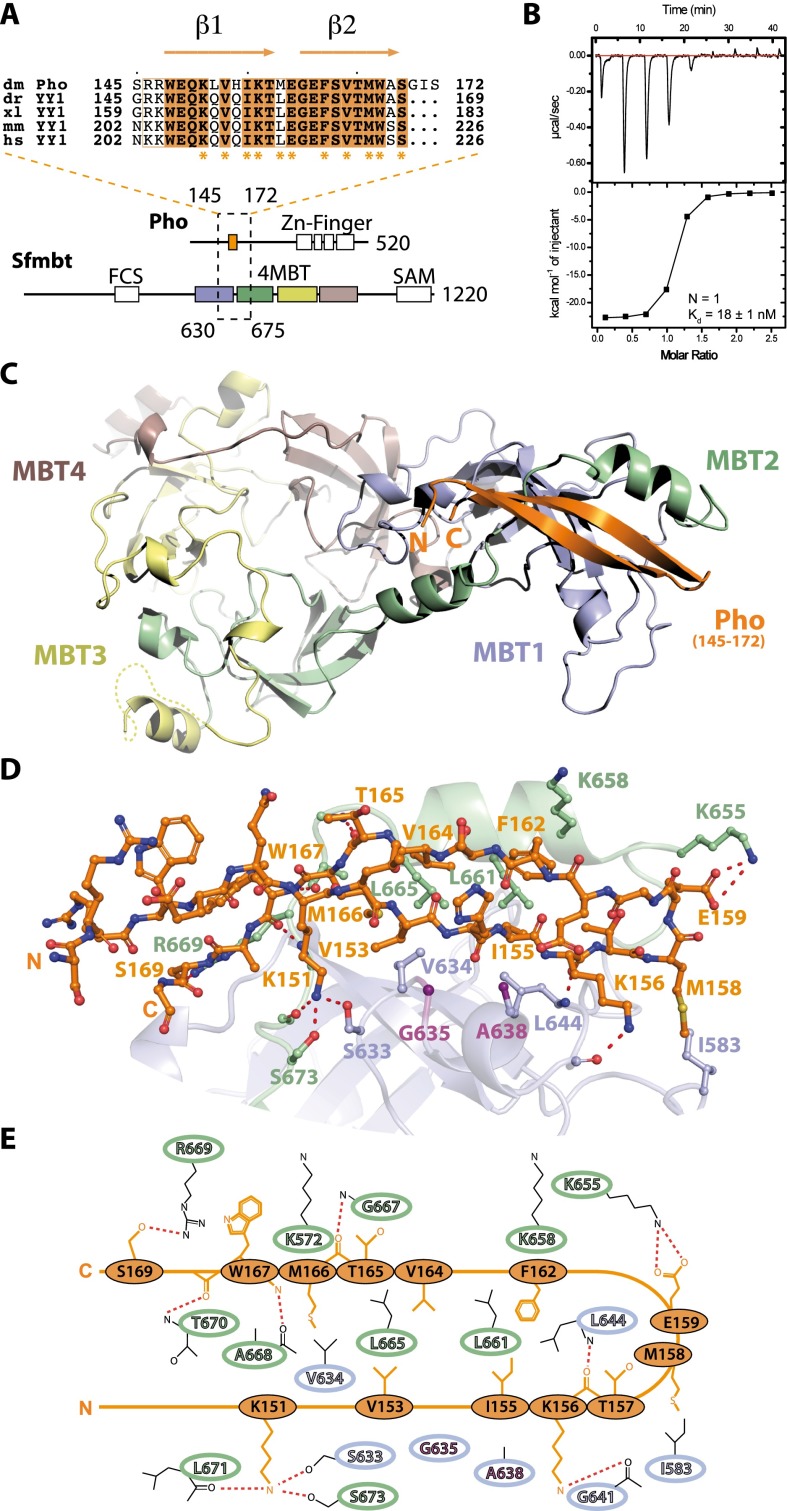
We mapped the minimal domains required for interaction between Sfmbt and Pho to the 4MBT domain of Sfmbt (Sfmbt531–980) and to a highly conserved region of ∼30 residues that has been previously named the Pho spacer (Pho145–172) (Fig. 1A; Supplemental Fig. S1A; Brown et al. 1998). In YY1, the corresponding region is also known as the REPO domain (Wilkinson et al. 2006). Isothermal titration calorimetry (ITC) showed that the Pho spacer binds with nanomolar affinity to the 4MBT domain of Sfmbt (Fig. 1B). After coexpression of these two domains in Escherichia coli, a stable minimal complex (hereafter called miniPhoRC) was obtained (Supplemental Fig. S1B,C). Using ITC, we found that miniPhoRC binds a H4K20me1 peptide with an affinity comparable with that of the Sfmbt 4MBT domain alone (Supplemental Fig. S2A). Furthermore, miniPhoRC stability was not affected by the SfmbtD917E mutation that impairs the histone Kme1/2-binding pocket (Supplemental Fig. S1B; Grimm et al. 2009). These results implied that the Pho spacer peptide contacted the 4MBT domain at a novel site, distinct from the previously characterized histone Kme1/2-binding pocket, and that this interaction was compatible with histone Kme1/2 binding.
Figure 1.
Biophysical and structural characterization of the Pho spacer:Sfmbt 4MBT interaction. (A, top) Sequence alignment of the Drosophila melanogaster Pho spacer region (dm, Q8ST83, orange) with the YY1 orthologs from Danio rerio (dr, Q7T1S3), Xenopus laevis (xl, Q6DDI1), mice (mm, Q00899), and humans (hs, P25490). Residues involved in the interaction with the Sfmbt 4MBT domain are indicated with asterisks. (Bottom) Pho and Sfmbt domain architecture. Pho spacer:Sfmbt 4MBT-interacting regions are enclosed by a dashed rectangle, and the first and last residue of the interacting regions are given. Sfmbt MBT repeats 1–4 are colored. (B) ITC data of the Pho spacer:Sfmbt 4MBT interaction. (C) Overview of the miniPhoRC complex crystal structure as a ribbon diagram presentation. (D) Close-up view of the Pho spacer:Sfmbt 4MBT interaction. Interacting residues of the Pho spacer and the Sfmbt 4MBT domain are depicted. Gly635 and Ala638 in the Sfmbt clamping helix are highlighted (purple). (E) Schematic representation of the Pho spacer:4MBT domain interaction.
Structure of the miniPhoRC
To identify the Pho spacer binding surface on the 4MBT domain and define the Pho spacer:4MBT interaction at atomic resolution, we performed crystallization trials of the purified miniPhoRC. We obtained crystals of different miniPhoRC constructs in three distinct crystal forms using the vapor diffusion method and could solve the corresponding structures at 1.95, 2.10, and 3.20 Å resolution (Supplemental Table S1). The structures were solved by molecular replacement using the 4MBT:H4K20me1 complex structure (Protein Data Bank [PDB] ID: 3h6z) (Grimm et al. 2009) as the search model. Strikingly, in all three initial σA-weighted electron density maps, we were able to detect clear additional densities corresponding to the Pho spacer peptide (Supplemental Fig. S3). In crystal form P3121, a longer Pho construct comprising 131 amino acid residues was used for cocrystallization (Supplemental Table 1), but only electron density corresponding to the Pho spacer peptide was visible. Despite three different crystal lattices, the spatial arrangement of the Pho peptide and the overall structure of the miniPhoRC are very similar (Supplemental Fig. 3). The Pho spacer peptide folds into two anti-parallel β strands connected by a β-hairpin loop, burying a total of 1900 Å2 of accessible surface upon binding. The 4MBT interaction surface is a conserved hydrophobic groove created by the end of the first MBT repeat (residues 630–650) and the beginning of the second MBT repeat (residues 651–675) (Fig. 1C). Several highly conserved residues of the Pho spacer (Val153, Ile155, Met158, Phe162, Val164, Met166, and Trp167) are engaged in hydrophobic contacts with the hydrophobic pocket of 4MBT (Fig. 1D,E). In addition, the conserved Lys151 of the Pho spacer forms hydrogen bonds with the hydroxyl groups of Sfmbt residues Ser633 and Ser673 and the carbonyl of Leu671 of the Sfmbt backbone. Furthermore, a salt bridge connects Pho Glu159 with Sfmbt Lys655, and hydrogen bonds are established between Pho Lys156 and the Sfmbt protein backbone and Pho Ser169 and Sfmbt Arg669 (Fig. 1D,E).
The hydrophobic floor of the Pho-binding pocket of Sfmbt is mainly formed by the conserved tetrad of residues Val634, Leu644, Leu661, and Leu665 that tightly pack against the Pho spacer peptide. In addition, helix 2 of MBT repeat 1 (hereafter called the clamping helix), which contains the conserved sequence motif GWCA, buttresses the Pho spacer peptide from one side against the hydrophobic side of helix 1 of MBT repeat 2 (Fig. 1C–E).
Superimposition of the miniPhoRC structure with the Sfmbt 4MBT:H4K20me1 complex structure results in a very low deviation (RMSDCα413 = 0.78 Å), indicating that Pho binding does not induce any major conformational changes in the 4MBT domain of Sfmbt (Supplemental Fig. S4). In accordance with the ITC data (Supplemental Fig. S2), the methyl-lysine-binding pocket is not significantly changed, while small changes can be observed in the Pho-binding pocket in order to accommodate the Pho peptide (Supplemental Fig. S4). Different from the Sfmbt 4MBT:H4K20me1 structure, in the miniPhoRC structure loop, residues 573–596 (M1L) in MBT repeat 1 are well structured in the two highest-resolution structures (Supplemental Figs. S3, S4). Loop residue Asp579 points toward an aromatic cage formed by Tyr612, His620, and Phe615, thereby completing a potential, additional methyl-lysine-binding pocket in MBT repeat 1 (Supplemental Fig. S4).
The Sfmbt clamping helix is required for Pho:Sfmbt complex stability
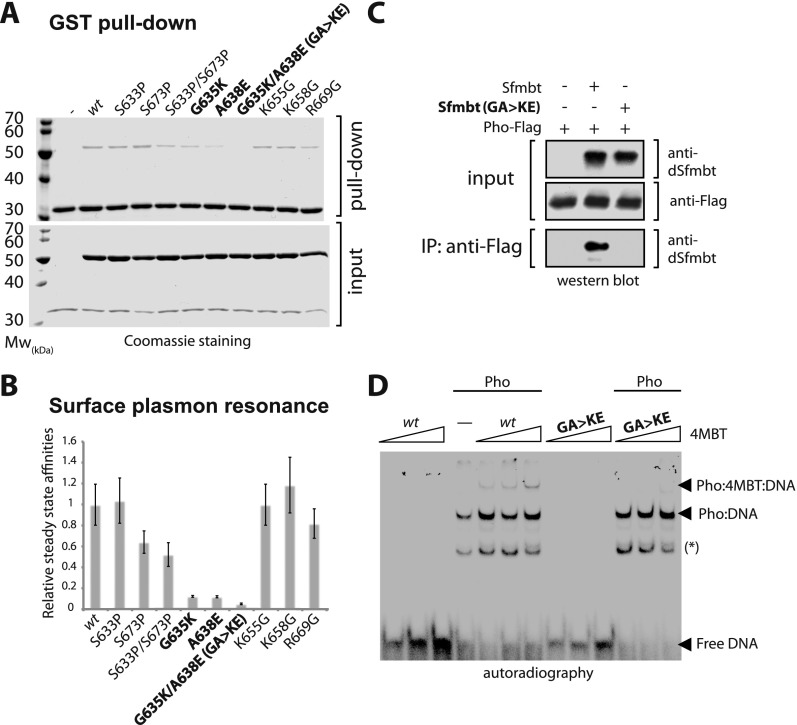
We next sought to identify Sfmbt residues that are critical for binding to the Pho spacer. A first clue came from comparing the MBT domain structures of Sfmbt and Scm, as the Pho-binding pocket of Sfmbt and the equivalent hydrophobic groove in Scm show considerable structural similarity (Supplemental Fig. S5A,B; Grimm et al. 2007, 2009), but Pho fails to form a stable complex with Scm both in vivo (Klymenko et al. 2006) and in vitro (Supplemental Fig. S5C). A more detailed structural comparison explains the inability of Pho to bind to Scm. In particular, one glutamate residue (Glu264) present in Scm protrudes into the hydrophobic groove and would clash with the Pho peptide. Indeed, when we substituted Ala638 in the clamping helix of Sfmbt with a glutamate (A638E) to mimic Glu264 present at this position in Scm (Supplemental Fig. S5), the interaction between the Pho spacer and the Sfmbt-binding pocket was strongly reduced in GST pull-down and surface plasmon resonance (SPR) experiments (Fig. 2A,B). Moreover, combining the A638E mutation with a second mutation, G635K, almost completely abolishes binding of Sfmbt to the Pho spacer (Figs. 2A,B). Using SPR, we consistently obtained lower absolute binding affinities than by using ITC, presumably because peptides needed to be immobilized at the surface during SPR experiments. However, consistent relative affinity values for wild-type and mutant proteins were obtained with both techniques. Importantly, the Sfmbt 4MBTG635K/A638E mutant protein is still able to bind the H4K20me1 peptide (Supplemental Fig. S2A,B), suggesting that the structural integrity of the MBT fold is unaffected. In contrast, disruption of the hydrogen bond network around Pho residue Lys151 by mutating Sfmbt residues Ser633 and Ser673 into proline residues only causes a twofold decrease in the binding affinity (Fig. 2A,B). Similarly, mutating Sfmbt residues Lys655, Lys658, and Arg669 into glycine residues had almost no effect on Pho spacer binding (Fig. 2A,B). The latter results are in agreement with high-temperature factors of Pho residues 155–162 that interact with these Sfmbt residues (Supplemental Fig. S6) and reflect greater structural flexibility and presumably less tight binding in this region. Consistent with our data, both complexes—miniPhoRC (data not shown) and PhoRC (Klymenko et al. 2006)—are stable at high salt concentrations, suggesting that hydrophobic interactions rather than electrostatic interactions are important for the integrity of the Pho:Sfmbt complex.
Figure 2.
In vitro mutagenesis analysis of the Pho:Sfmbt interaction. (A) GST pull-down of recombinant GST-Pho spacer and untagged Sfmbt 4MBT wild-type and structure-based mutant proteins. (B) SPR measurements of biotin-labeled Pho and Sfmbt 4MBT wild-type or mutant proteins. Results are shown as affinities relative to the Pho spacer:Sfmbt 4MBT wild-type affinity. (C) Anti-Flag affinity purifications of full-length Pho-Flag:Sfmbt wild-type or mutant complexes detected by Western blot. Antibodies used for the detection are indicated at right. (D) EMSA experiments of full-length Pho or full-length Pho:Sfmbt 4MBT wild-type and mutant complexes using a 32P end-labeled double-stranded Pho DNA-binding site. Arrows indicate full-length Pho:DNA and supershifted Pho:Sfmbt 4MBT:DNA complexes. Lanes containing only DNA and the Sfmbt 4MBT domain were used as control. The asterisk indicates the Pho DNA-binding domain/DNA complex resulting from the degradation of full-length Pho protein as confirmed by mass spectrometry (MS) (data not shown). Binding reactions were performed with 5 ng of DNA probe and 50 ng of Pho full-length protein; 50-fold, 100-fold, and 500-fold molar excess of Sfmbt 4MBT wild-type or mutant protein was added to fixed amounts of Pho protein.
In a next step, we analyzed the effects of the SfmbtG635K/A638E mutations on complex formation between the full-length Pho and Sfmbt proteins. Indeed, the SfmbtG635K/A638E protein completely failed to interact with Flag-tagged Pho in a coimmunoprecipitation experiment (Fig. 2C). This confirms the importance of the Sfmbt 4MBT domain:Pho spacer interaction and excludes that regions outside of the Sfmbt 4MBT domain and the Pho spacer would be critical for complex formation. Finally, we tested the ability of the Sfmbt 4MBT protein to form a trimeric Sfmbt:Pho:DNA complex using electrophoretic mobility shift assays (EMSA) (Fig. 2D). Full-length Pho protein induces a mobility shift of a DNA probe containing a Pho-binding site, and titrating in the Sfmbt 4MBT domain induces an additional supershift of the DNA:Pho complex, while addition of the Sfmbt 4MBTG635K/A638E mutant protein failed to produce this supershift (Fig. 2D). The Sfmbt 4MBT domain alone fails to stably associate with DNA (Fig. 2D). We conclude that interaction of the Pho spacer with the 4MBT domain is critical for DNA tethering of Sfmbt by Pho in vitro.
Pho:Sfmbt interaction is important for the recruitment and function of Sfmbt at PREs
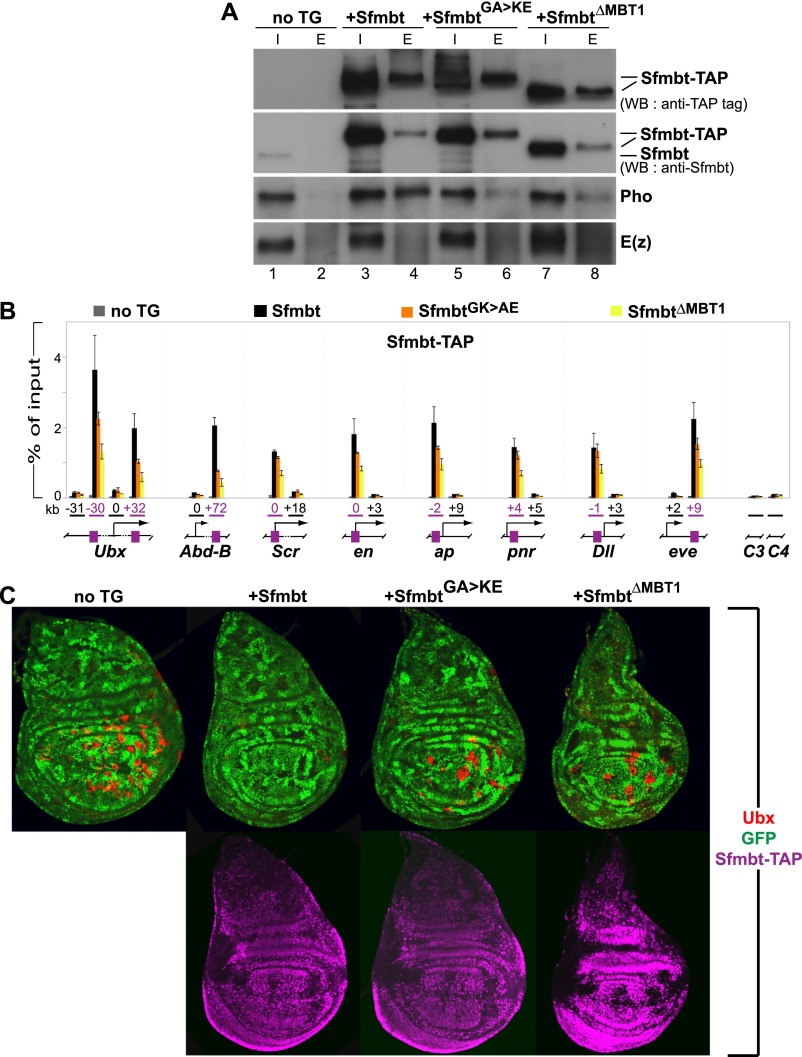
We next tested the effect of mutating the Pho-binding pocket in Sfmbt in vivo. To this end, we expressed wild-type Sfmbt, SfmbtG635K/A638E, or SfmbtΔMBT1 proteins in developing Drosophila under the control of upstream activating sequences (UASGal4) and an appropriate Gal4 driver. The transgene-encoded proteins contained a tandem affinity purification (TAP) tag at their C terminus (CTAP) to distinguish them from the endogenous Sfmbt protein. As expected, in nuclear extracts prepared from embryos expressing wild-type Sfmbt-CTAP protein, the endogenous Pho protein was robustly coimmunoprecipitated with the Sfmbt-CTAP protein (Fig. 3A, lanes 1–4). In contrast, the SfmbtG635K/A638E-CTAP and SfmbtΔMBT1-CTAP proteins only poorly coimmunoprecipitated the endogenous Pho protein (Fig. 3A, lanes 5–8).
Figure 3.
Pho:Sfmbt interaction is critical for Sfmbt function during Drosophila development. (A) Western blot analyses with the indicated antibodies of input (“I”; 2.5% of total) and eluted (“E”; 100% of total) material of IgG sepharose pull-downs of Sfmbt-CTAP proteins from nuclear extracts of 0- to 12-h-old embryos expressing the indicated Sfmbt-CTAP protein or from nontransgenic animals (“not TG”). The tagged proteins were expressed from UASGal4:Sfmbt-CTAP transgenes under the control of the daughterless:Gal4 driver. Blots were first probed with anti-Sfmbt, and then stripped and reprobed with anti-peroxidase antibody to specifically detect the Sfmbt-CTAP fusion proteins. Wild-type Sfmbt-CTAP protein coimmunoprecipitates Pho but not the PRC2-subunit E(z). Significantly lower levels of Pho are coimmunoprecipitated with the SfmbtG635K/A638E-CTAP and SfmbtΔMBT1-CTAP proteins (cf. lanes 6,8 and 4). (B) ChIP analysis monitoring binding of the indicated Sfmbt-CTAP proteins at PcG target genes in chromatin of wild-type larvae that express Sfmbt-TAP proteins under the control of the ubiquitous daughterless:Gal4 driver. Graphs show results from three independent immunoprecipitation reactions with anti-peroxidase antibody that binds to the protein A moiety of the TAP tag. ChIP signals, quantified by quantitative PCR, are presented as the mean percentage of input chromatin precipitated at each region; error bars indicate ±SD. Locations of PREs (purple boxes) and other regions relative to the transcription start sites are indicated in kilobases; control regions C3 in euchromatin and C4 in heterochromatin are located remotely from PcG target genes. Sfmbt-CTAP is specifically enriched at PREs. Levels of SfmbtG635K/A638E-CTAP protein binding are reduced at several PREs, whereas binding of SfmbtΔMBT1-CTAP is reduced twofold to fivefold at all analyzed PREs. (C) Wing imaginal discs from third instar larvae stained with antibody against the HOX protein Ubx (red, top row) and anti-peroxidase antibody to detect the Sfmbt-CTAP proteins (purple, bottom row). Clones of Sfmbt1 homozygous cells are marked by the absence of GFP (green) and were induced in animals lacking a transgene (“no TG”) or expressing the indicated Sfmbt-CTAP proteins under the control of the 69B:Gal4 driver. For unknown reasons, the SfmbtΔMBT1-CTAP is expressed at higher levels (see Supplemental Fig. S8) than the Sfmbt-CTAP and SfmbtG635K/A638E-CTAP proteins. Note that only Sfmbt1 homozygous cell clones (“no TG”) in the wing pouch but not in the notum or hinge show strong misexpression of Ubx, as described previously (Klymenko et al. 2006). To evaluate the capacity of the transgene-encoded Sfmbt proteins to repress Ubx in Sfmbt1 mutant cell clones, we therefore only analyzed mutant clones in the wing pouch area for the presence of Ubx protein. For each genotype, multiple wing imaginal discs were analyzed, and in animals expressing a CTAP fusion protein, only clones in which the fusion protein was detected by immunofluorescence labeling (shown in the bottom panel) were scored. In the “no TG” animals, 94% of Sfmbt1 homozygous clones (n = 98 clones) show misexpression of Ubx. In Sfmbt-CTAP animals, repression of Ubx is rescued in most Sfmbt1 homozygous clones, and only 4% of the clones (n = 78 clones) show misexpression of Ubx. In SfmbtG635K/A638E-CTAP animals, 81% of Sfmbt1 homozygous clones (n = 97 clones) show misexpression of Ubx. In SfmbtΔMBT1-CTAP animals, 87% of the clones (n = 31 clones) show misexpression of Ubx. The large proportion of Ubx-expressing Sfmbt1 mutant clones in SfmbtG635K/A638E-CTAP and SfmbtΔMBT1-CTAP animals suggests that these two proteins are largely nonfunctional in Polycomb repression.
Using chromatin immunoprecipitation (ChIP) assays, we next compared recruitment of the tagged Sfmbt, SfmbtG635K/A638E, and SfmbtΔMBT1 proteins to PREs of well-characterized PcG target genes in larval tissues. Like the endogenous Sfmbt protein (Supplemental Fig. S7), the Sfmbt-CTAP protein was specifically bound at the PREs but not at other analyzed regions in each of these genes (Fig. 3B). In contrast, binding of the SfmbtG635K/A638E-CTAP protein was significantly reduced at the PREs of the HOX genes Ultrabithorax (Ubx) and Abdominal-B (Abd-B) and, to a lesser extent, also at the PREs of other genes (Fig. 3B). A more drastic effect was observed in the case of the SfmbtΔMBT1-CTAP protein, for which binding was significantly reduced at all analyzed PREs (Fig. 3B). This reduced binding of SfmbtΔMBT1-CTAP was observed even though the levels of this mutant protein in larval cells were more than threefold higher compared with those of the Sfmbt-CTAP and SfmbtG635K/A638E-CTAP proteins (Supplemental Fig. S8).
We then used a genetic rescue assay to investigate whether the different Sfmbt-CTAP proteins could replace endogenous Sfmbt in target gene repression. Clones of imaginal disc cells that are homozygous for the Sfmbt1-null mutation fail to maintain PcG repression, and the HOX gene Ubx is strongly misexpressed in the mutant cells (Fig. 3C; Klymenko et al. 2006). Expression of the wild-type Sfmbt-CTAP protein rescues repression of Ubx in Sfmbt1 mutant clones (Fig. 3C). In contrast, the SfmbtG635K/A638E-CTAP and SfmbtΔMBT1-CTAP proteins both had severely compromised repressor activity, and Ubx was strongly misexpressed in the Sfmbt1 mutant clones (Fig. 3C). Thus, even though SfmbtG635K/A638E-CTAP and SfmbtΔMBT1-CTAP ChIP signals at Ubx PREs were reduced only twofold to threefold, the capacity of the mutant proteins to maintain PcG repression was strongly reduced. Taken together, these experiments demonstrate that efficient recruitment of the Sfmbt repressor protein by Pho is essential to maintain gene silencing.
Drosophila Sfmbt and human L3MBTL2 assemblies are related
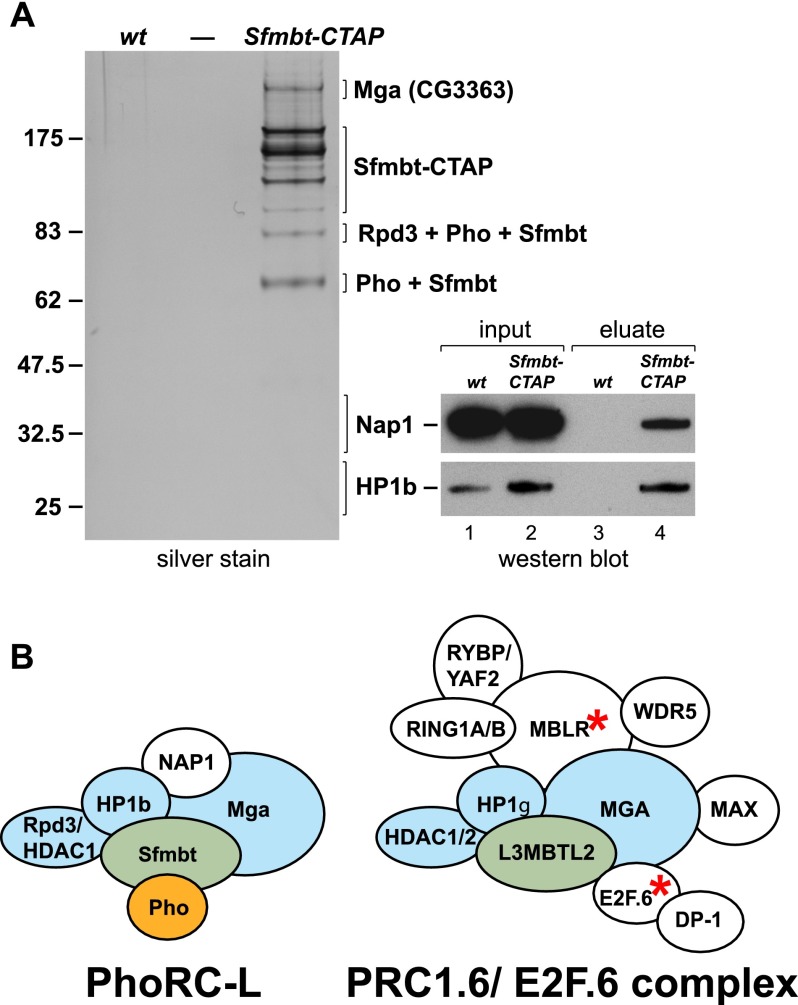
We previously isolated PhoRC through purification of a Pho-CTAP protein from Drosophila embryonic nuclear extracts (Klymenko et al. 2006). Here, we performed TAP from nuclear extracts that we generated from Sfmbt-CTAP transgenic embryos (see the Materials and Methods). Three independent purifications were analyzed by SDS-PAGE, and proteins copurifying with Sfmbt-CTAP were identified by tandem mass spectrometry (MS/MS) and liquid chromatography (LC)-MS/MS (Fig. 4; Supplemental Table S2). In addition to the Sfmbt-CTAP bait protein and Pho, all three purifications contained Rpd3/Hdac1, HP1b, Nap1, and the ortholog of the human Max gene-associated protein Mga (CG3363) (Fig. 4A). This confirms that in Drosophila, a substantial fraction of Sfmbt is associated with Pho. Further analyses will be needed to investigate whether the other copurifying proteins are all subunits of a single PhoRC protein assembly or represent different Sfmbt complexes. Using ChIP assays, we found that Rpd3/Hdac1 colocalizes with Pho and Sfmbt at all analyzed PREs (Supplemental Fig. S7), suggesting that at least Rpd3/Hdac1 may be part of a larger PhoRC assembly at PcG target genes. Interestingly, previous studies reported that the human orthologs of Rpd3/Hdac1, HP1b, and Mga copurify with human L3MBTL2, one of four human Sfmbt orthologs (Fig. 4B; Ogawa et al. 2002; Trojer et al. 2011; Gao et al. 2012). Drosophila Sfmbt and human L3MBTL2 are thus components of a conserved protein interaction network. Human L3MBTL2 assemblies, also called PRC1.6, also contain additional proteins, among which MBLR and the DNA-binding protein E2F.6 are vertebrate-specific (Fig. 4B). Intriguingly, however, the human Pho ortholog YY1 has never been identified in any of the L3MBTL2 purifications (Ogawa et al. 2002; Trojer et al. 2011; Gao et al. 2012).
Figure 4.
TAP of Sfmbt protein complexes from Drosophila embryonic nuclear extracts identifies a larger PhoRC assembly that resembles mammalian PRC1.6/E2F.6 complexes. (A) Sfmbt complexes isolated by TAP from wild-type (wt) or α-tubulin-Sfmbt-CTAP transgenic embryos. (Left) Input material for purification was normalized by protein concentration, and equivalent amounts of eluate from calmodulin affinity resin were separated on a 4%–12% polyacrylamide gel and visualized by silver staining; the molecular weight marker is indicated on the left. Sfmbt bait protein containing the calmodulin-binding tag (Sfmbt-CBP), its degradation products, and bands representing Mga (CG3363), Hdac1/Rpd3, Pho, Nap1, and HP1b were identified by MS (Supplemental Table S2). (Right) The Nap1 and HP1b proteins were undetectable as bands on silver-stained gels, and their presence was verified by Western blot analysis: total embryonic nuclear extract input material from wild-type (wt) and Sfmbt-CTAP transgenic embryos (lanes 1,2) and material eluted from the calmodulin affinity resin after purification (lanes 3,4), probed with the indicated antibodies. The Nap1 and HP1b panels come from the same batches of input and eluate material, and the same ratio of input versus eluate was loaded in both cases. (B) The Drosophila Sfmbt assembly resembles human PRC1.6/E2F.6 assemblies. Graphic representation of the larger Drosophila Sfmbt–Pho assembly with the additional proteins identified in A and therefore called PhoRC-L and the PRC1.6 assembly described in Gao et al. (2012). Note that the PRC1.6 assembly is identical to the E2F.6 assembly described in Ogawa et al. (2002) but was reported to also contain HDAC1/2 and WDR5. Drosophila Sfmbt and human L3MBTL2 proteins are labeled in green, the orthologous subunits identified in both Drosophila and human assemblies are labeled in blue, and Pho is labeled in orange. The Drosophila genome does not encode orthologs of E2F.6 and MBLR (asterisks), and this might explain why Drosophila PhoRC-L assemblies do not contain the RING1A/B ortholog Sce, the RYBP/YAF2 ortholog Rybp, and the DP-1 ortholog Dp.
Human YY1 binds 4MBT domain proteins in vitro
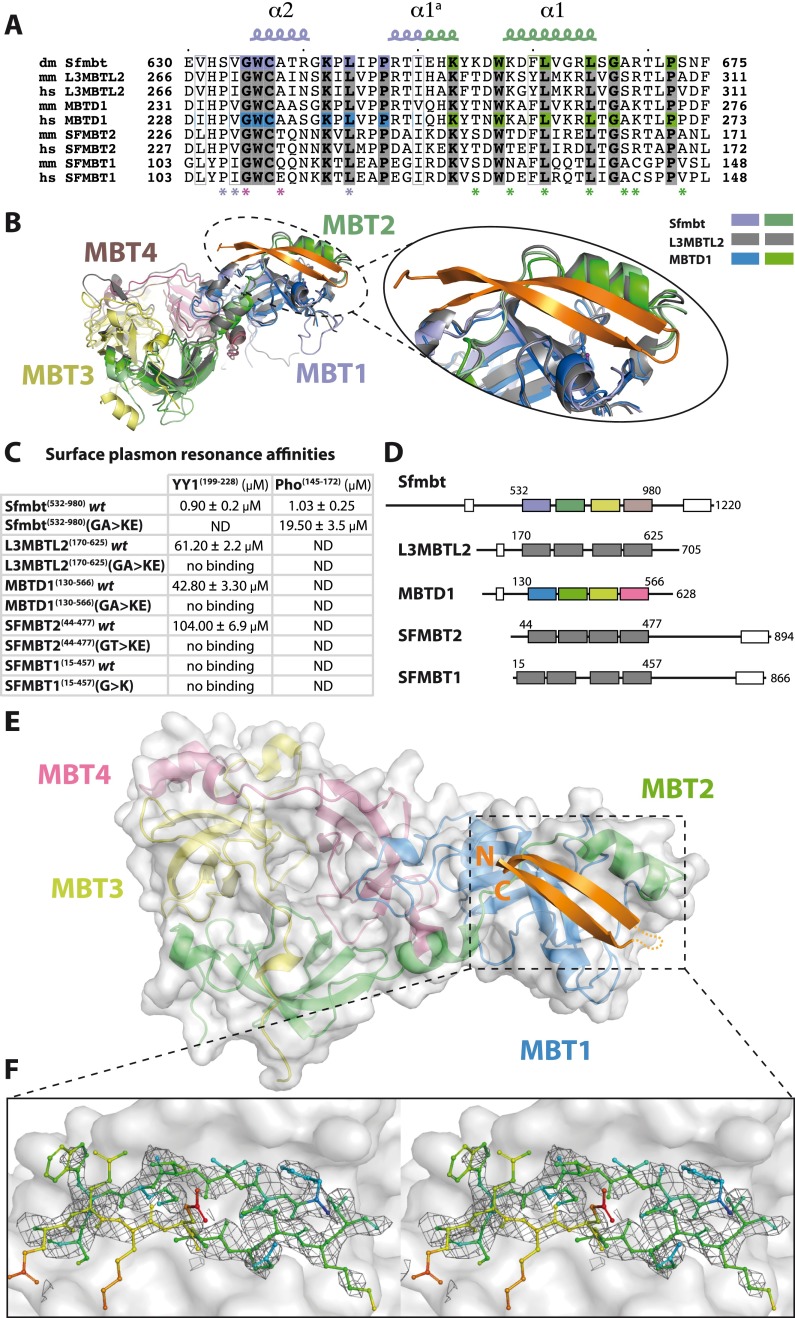
The Pho spacer is highly conserved in YY1 (Fig. 1A), and, indeed, YY1 and the Pho spacer both bind to the Drosophila Sfmbt 4MBT domain with similar affinities (Fig. 5; Supplemental Fig. S9). Similarly, the Pho-binding surface of Sfmbt is also conserved in the previously reported structures of the human L3MBTL2 and MBTD1 proteins (Fig. 5A,B). We therefore tested whether the Pho spacer:Sfmbt interaction is conserved in humans and whether the YY1 spacer would be able to bind to L3MBTL2 and the other orthologs. Indeed, the YY1 spacer specifically interacts with the 4MBT domains of L3MBTL2, MBTD1, and SFMBT2 in SPR experiments, although the affinity of these interactions is much weaker (∼50-fold) compared with the Drosophila complex (Fig. 5C). Importantly, these weak interactions were abolished by mutating MBT domain residues that were found to be critical for binding of Drosophila Sfmbt to Pho (cf. Figs. 5C and 2A,B). Moreover, the YY1 spacer failed to bind to wild-type SFMBT1 (Fig. 5C), likely because its 4MBT domain already contains a glutamate (Glu111) and thus a GWCE rather than a GWCA motif in the clamping helix (Fig. 5A). Similarly, human SFMBT2 carries a threonine instead of an alanine at position 235 in the clamping helix (Fig. 5A), possibly explaining why SFMBT2 binds YY1 with about twofold lower affinity compared with L3MBTL2 and MBTD1, both of which contain the conserved GWCA motif (Fig. 5A).
Figure 5.
Pho:Sfmbt interaction is structurally and biochemically conserved in humans. (A) Sequence alignment of the Pho spacer-binding pocket from Sfmbt (dm, Q9VK33) and mouse and human L3MBTL2 (mm, P59178; hs, Q969R5), MBTD1 (mm, Q6P5G3; hs, Q05BQ5), SFMBT2 (mm, Q5DTW2; hs, Q5VUG0), and SFMBT1 (mm, Q9JMD1; hs, Q9UHJ3). Sfmbt MBT repeats are colored as above. The same color code is used for MBTD1, where we solved the crystal structure bound to YY1. The remaining human 4MBT proteins are depicted in gray. Pho spacer:Sfmbt 4MBT-interacting residues are marked with asterisks. Glycine and alanine residues in the Sfmbt clamping helix are highlighted (purple). (B) Structural superposition of the Drosophila Pho spacer-binding pocket with the corresponding regions in the human L3MBTL2 and MBTD1 4MBT domains (PDB ID: 3f70 and 3feo). (C) Dissociation constants of YY1 or Pho spacers for D. melanogaster or human 4MBT wild-type and mutant proteins. Note that KD values measured by SPR were consistently lower than those measured by ITC, presumably due to the immobilization of the spacer peptides required for SPR. (D) Domain architecture of human 4MBT proteins. The first and last residues of the 4MBT domain constructs used in the experiments in C are indicated. The N-terminal FCS Zn finger and the C-terminal SAM domains are represented as white boxes. (E) Crystal structure of the YY1 spacer:MBTD1 4MBT complex. Color scheme of the MBT repeats according to D with the YY1 spacer depicted in orange. (F) Stereo view of the YY1 spacer σA-weighted simulated annealing omit electron density map contoured at 0.7 σ. YY1 spacer residues are depicted and colored according to temperature factors (increasing from blue to red).
Structure of the YY1:MBTD1 complex
To understand the chemical nature and the lower affinity of the interaction between YY1 spacer and human 4MBT domains, we attempted cocrystallization of the YY1 spacer with different human 4MBT proteins. We successfully crystallized and solved the structure of the MBTD1 4MBT domain bound to the YY1 spacer (Fig. 5D–F; Supplemental Table S1). The structure was solved by molecular replacement using the apo MBTD1 crystal structure (PDB ID: 3feo) as the search model, and as expected, in the initial σA-weighted electron density map, we identified additional density of the YY1 spacer peptide in the binding pocket of MBTD1 that corresponds to the Pho-binding pocket in Drosophila Sfmbt (Fig. 5E). The temperature factor of the YY1 spacer peptide in the YY1:MBTD1 structure is high (B-factorYY1 spacer = 64.1 Å2) compared with the overall B-factor (B-factoroverall = 33.8 Å2), suggesting a high flexibility of the YY1 peptide consistent with its low binding affinity for MBTD1 (Fig. 5C). Comparison of the Sfmbt- and MBTD1 spacer-binding pockets suggests that substitutions of several residues contribute to the lower binding affinity of MBTD1 for YY1. In particular, Sfmbt residues Ser633 and Ser673 that interact with the conserved Lys151 of Pho and are important for binding (Fig. 2A,B) are both changed to prolines (Pro231 and Pro271) in MBTD1 (Supplemental Fig. 10A,B). Accordingly, in the YY1 spacer, the Lys208 residue corresponding to Lys151 of Pho is disordered and not visible in the electron density of the YY1:MBTD1 complex structure (Fig. 5E,F). Additionally, the Sfmbt hydrophobic cavity formed by residues Val634, Leu665, Ala668, and Lys572 that accommodates Pho Met166 has a more hydrophilic character in MBTD1 due to the presence of Arg177 in MBTD1 instead of Lys572 in Sfmbt (Supplemental Fig. S10A,B). As a result, the thiol group of Met223 in the YY1 peptide is less well ordered (Fig. 5F). In conclusion, our YY1:MBTD1 minimal complex structure confirms that the recognition of the YY1 spacer by MBTD1 involves the same binding interface as in Sfmbt but also explains the lower affinity of YY1 for the human 4MBT proteins compared with that of Drosophila Pho for Sfmbt.
Discussion
Atomic-level information on how PcG protein complexes assemble at their target genes is essential for understanding how these key regulators repress transcription to control cell fate decisions. Progress in understanding the mechanism of PcG protein complex targeting to specific DNA sequences has come from studies in Drosophila, where these complexes assemble at PREs. In this study, we present the structural basis of how the PRE-binding PcG protein Pho tethers its partner, Sfmbt, to DNA and show that this interaction is essential for gene repression in vivo. In addition, we present structural and biochemical evidence that this interaction is likely conserved between the human Sfmbt orthologs L3MBTL2, MBTD1, and SFMBT2 and the human Pho ortholog YY1.
Molecular mode of Pho/YY1 spacer binding to MBT domain proteins
Like many other transcriptional regulators, Pho/YY1 has a modular structure: Pho/YY1 contains a Zn finger-type DNA-binding domain at the C terminus and a flexible N-terminal domain that is to a large extent intrinsically disordered. The regions most conserved between Pho and YY1 comprise the C-terminal Zn finger DNA-binding domain and the N-terminal highly conserved spacer region (also known as the REPO domain), while two acidic domains and a glycine–alanine-rich domain are present only in YY1 (Brown et al. 1998). The miniPhoRC crystal structure identifies the Pho spacer as a novel interacting motif formed by two anti-parallel β strands connected by a short β hairpin that tightly binds to a previously uncharacterized hydrophobic cavity in the Sfmbt 4MBT domain. Only a few motifs that mediate the interaction between transcriptional regulators and coactivators or corepressors have been structurally characterized. In particular, the hydrophobic face of an amphipathic helix is often used to contact other components, while so far a two-stranded β sheet has not been observed (for review, see Mapp and Ansari 2007). Our biophysical measurements show that the two proteins tightly bind to each other (Fig. 1B). The structural and mutagenesis analyses indicate that the highly conserved GWCA motif in the clamping helix of the first MBT repeat of Sfmbt is critical for PhoRC formation. Binding of the Pho spacer also induces the formation of a second potential methyl-lysine-binding pocket in MBT repeat 1 of Sfmbt. Preliminary binding studies using a small set of mono-, di-, and trimethyl-lysine-containing histone tail peptides did not reveal significant binding of any of the candidate peptides to this pocket (data not shown). A broader screen is currently in progress to determine whether this new pocket recognizes a yet uncharacterized ligand. The crystal structure of the human YY1:MBTD1 complex shows that the molecular basis of the interaction between the YY1 spacer and human 4MBT domain proteins is, in principle, conserved, while lower affinity of binding between the human proteins is correlated with the loss of polar and hydrophobic interactions of the 4MBT domains with conserved residues of the YY1 spacer peptide (Fig. 5). These findings might explain why biochemical purifications of L3MBTL2 and SFMBT1 complexes from mammalian cells have failed to recover YY1 (Trojer et al. 2011; Gao et al. 2012; Zhang et al. 2013). It should be noted that, to date, no MBTD1 purifications or genome-wide binding profiles have been reported.
The currently available evidence suggests that human L3MBTL2 may be the closest functional ortholog of Drosophila Sfmbt. First, L3MBTL2 and Sfmbt exist in related protein assemblies (Fig. 4; Ogawa et al. 2002; Trojer et al. 2011; Gao et al. 2012). Second, among the human 4MBT proteins, L3MBTL2 has the highest sequence homology with Drosophila Sfmbt and contains the critical GWCA motif in the clamping helix needed for YY1 binding (Fig. 5). Third, in both mammalian L3MBTL2 and fly Sfmbt, deletion of the first MBT repeat, which is required for the interaction with the YY1/Pho spacer, causes loss of functionality of these proteins in vivo (Fig. 3; Qin et al. 2012). Despite these remarkable similarities, genome-wide binding studies have provided little support for a general cobinding of YY1 and PcG proteins at genomic sites in murine embryonic stem (ES) cells (Vella et al. 2012). It therefore appears that, in mammals, tethering of L3MBTL2 to the majority of genomic sites is directed by other DNA-binding proteins (for example, E2F6) (Fig. 4; Ogawa et al. 2002; Trojer et al. 2011) and that YY1 may only recruit L3MBTL2 to a subset of sites.
Recruitment of Sfmbt to PREs
In vitro, mutation of the Gly and Ala residues in the GWCA motif of Sfmbt abolishes formation of a ternary Sfmbt MBT:Pho:DNA complex (Fig. 2D). Unexpectedly, our ChIP analyses showed that recruitment of the SfmbtG635K/A638E-CTAP and SfmbtΔMBT1-CTAP proteins to PREs in vivo is only partially disrupted (Fig. 3B). Considering that the SfmbtΔMBT1 protein lacks much of the Pho interaction surface, it is unlikely that tethering of the SfmbtΔMBT1-CTAP protein occurs by interaction with Pho. How are these two mutant proteins tethered to PREs? It is important to keep in mind that ChIP experiments with these mutant proteins could be performed only in wild-type larvae because Sfmbt-null mutant animals die at earlier developmental stages. Due to the presence of intact endogenous Sfmbt protein in these experiments, PcG complexes are therefore expected to still assemble properly at PREs. The mutant SfmbtG635K/A638E and SfmbtΔMBT1 proteins may therefore associate with PREs through interactions with other PcG proteins that themselves had been recruited to PREs by the native PhoRC. Such indirect interactions between Pho and Sfmbt could also explain the residual amounts of Pho protein that are coimmunoprecipitated with the mutant Sfmbt-CTAP proteins (Fig. 3A, lanes 6,8). In contrast, in the genetic rescue experiments (Fig. 3C), the endogenous Sfmbt protein is absent, and only the SfmbtG635K/A638E-CTAP or SfmbtΔMBT1-CTAP proteins are present. In this situation, tethering of these mutant Sfmbt proteins to PREs may be more drastically diminished, explaining why repression of the target gene Ubx by the SfmbtG635K/A638E-CTAP and SfmbtΔMBT1-CTAP proteins was so severely impaired (Fig. 3C). This genetic rescue assay thus demonstrates the crucial need for the direct Sfmbt:Pho interaction.
PRE-tethered Sfmbt as a hub for PcG protein complex assembly
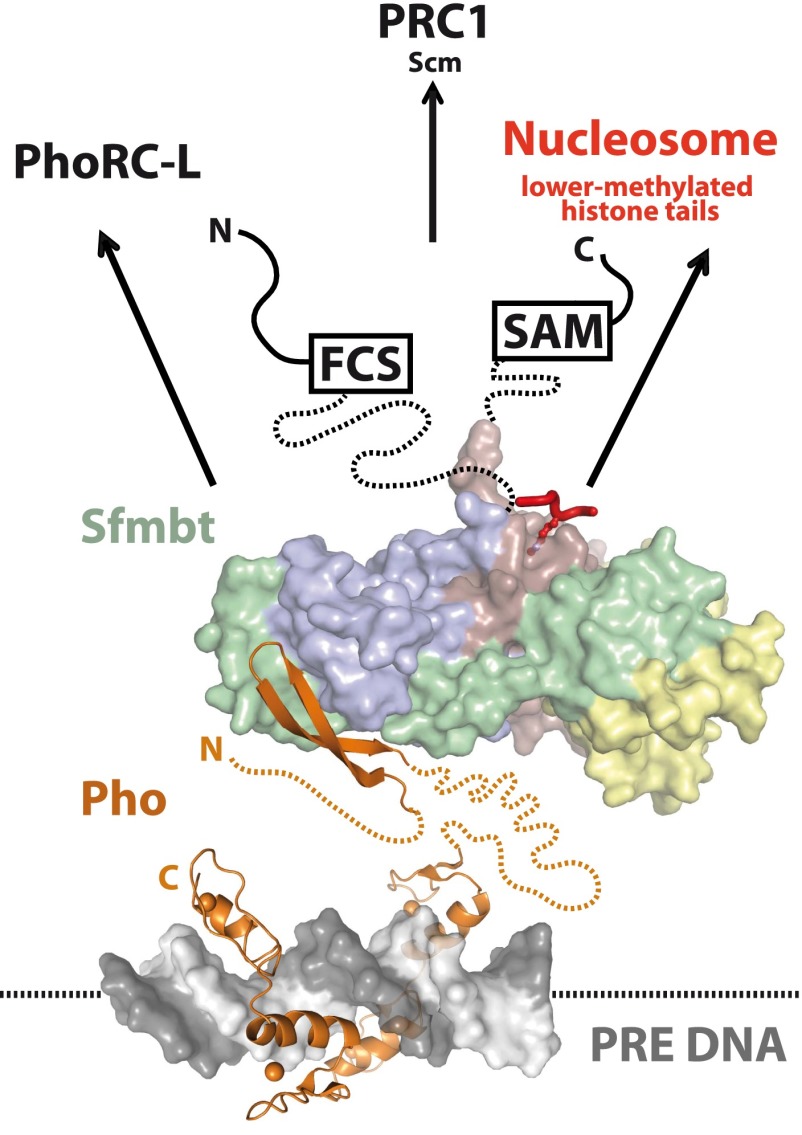
What is the function of Pho-tethered Sfmbt at PREs? A straightforward scenario is that PhoRC functions as a platform for the recruitment of other PcG complexes and the interaction with chromatin (Fig. 6). Unlike in the case of the Pho:Sfmbt interaction, it has not been possible to reconstitute stable Pho:PRC1 or Pho:PRC2 complexes with recombinant proteins (Mohd-Sarip et al. 2005, 2006; Klymenko et al. 2006). Previous studies nevertheless found both Pho and Sfmbt in purifications of the Polycomb protein from Drosophila embryos (Strubbe et al. 2011). Pho itself has been also reported to interact physically with subunits of both PRC1 (Mohd-Sarip et al. 2002, 2005, 2006) and PRC2 (Wang et al. 2004). In addition, recombinant Sfmbt and the PRC1-associated Scm protein can be reconstituted into a stable complex. This interaction thus represents a direct molecular link between PhoRC and PRC1 (Grimm et al. 2009). The C-terminal SAM domain of Sfmbt is not required for Scm binding and may thus engage in interactions with other ligands (Grimm et al. 2009). In addition, Sfmbt and Scm both bind to lower-methylated lysine residues in various histone tails (Grimm et al. 2007, 2009), and Sfmbt can also bind methylated histone tails while bound to Pho (cf. Supplemental Fig. S2). Taken together, this supports the view that PhoRC functions as a platform for the recruitment of various interactors. While Pho and Sfmbt bind each other strongly, their affinities for other binding partners such as individual PcG proteins or nucleosomes might be weaker and more transient. However, the modular architecture and the multivalency of PhoRC interactions create the hub that is necessary for the stable assembly of different PcG complexes at PREs.
Figure 6.
The PhoRC complex is a hub for multiple interactions. The Pho:Sfmbt interaction is required for the PcG-repressive function on HOX genes. The Pho/YY1 protein (orange) is recognizing the Pho-binding sites (gray and white) in a PRE through its DNA-binding domain (PDB ID: 1ubd) (Houbaviy et al. 1996) and recruits the Sfmbt 4MBT domain through its spacer region (miniPhoRC crystal structure) (this study). Pho regions flanking the spacer and connecting it to the DNA-binding domain are predicted to be disordered (dotted line). The Sfmbt MBT repeats are colored as above. Sfmbt also interacts with Scm and thereby tethers PRC1. Interaction of the fourth MBT repeat of Sfmbt with mono- or dimethylated lysines in histone tails (red) (PDB ID: 3h6z) (Grimm et al. 2009) links the PhoRC complex with nucleosomes.
Materials and methods
Protein expression and purification
The crystallization constructs containing the Pho spacer region and the Sfmbt 4MBT domain were cloned in a pETM11 vector in-frame with an N-terminal TEV-cleavable 6xHis tag and in a pCDF vector, respectively, using standard restriction cloning methods. The resulting miniPhoRC protein complex was coexpressed in the BL21(DE3) pRARE E. coli strain. The construct used for mutagenesis analysis containing the wild-type 4MBT domain was cloned in a pETM11 vector in-frame with an N-terminal TEV-cleavable 6xHis tag. All of the constructs containing the 4MBT domain mutants were generated using the Stratagene mutagenesis kit. Protein expression and purification conditions were performed as described in Grimm et al. (2009).
Crystallization and X-ray structure determination
Crystals in space group P21 were obtained by mixing equal volumes of protein solution—concentrated at 30 mg/mL in 10 mM Tris-HCl (pH 8), 150 mM NaCl, and 5 mM DTT—with reservoir solution containing 0.2 M ammonium formate (pH 6.6), and 12.5%–20% PEG-3350 in sitting drop trays. P6122 and P3121 crystals were obtained by mixing equal volumes of protein solution—concentrated at 30 mg/mL in 10 mM Tris-HCl (pH 8), 150 mM NaCl, and 5 mM DTT—with either 0.1 bicine and 20% PEG 6K or 0.1 M Bis-Tris and 3M NaCl, respectively. Crystals were cryo-cooled at 100 K using 25% glycerol as cryo-protectant. The P21 crystals diffracted to 1.95 Å resolution, and data were collected at the European Synchrotron Radiation Facility (ESRF) synchrotron and processed with the program XDS (Kabsch 2010). The crystal structure was solved with the program Phaser (McCoy et al. 2007) by molecular replacement using the Sfmbt 4MBT domain structure (PDB ID: 3h6z) (Grimm et al. 2009) as the search model. Refinement was performed with the program Phenix (Adams et al. 2010). The resulting model was used as a search model for solving the structures in crystal forms P6122 and P3121. Refinement and TLS refinement of the resulting structures were performed with Phenix (Supplemental Table S1). The YY1199–228:MBTD1 complex was reconstituted by mixing the MBTD1 4MBT domain with fivefold molar excess of YY1 spacer peptide at 4°C. The final concentration of the MBTD1 4MBT domain was 10 mg/mL. The resulting complex was cocrystallized by mixing 1 vol of the protein solution with 2 vol of a reservoir solution containing 0.25 M lithium sulfate and 20% PEG-3350. The YY1199–228/MBTD1 complex structure was solved by molecular replacement using the MBTD1 4MBT structure as the search model (PDB ID: 3feo) (Eryilmaz et al. 2009).
GST pull-down experiments
The GST-Pho spacer construct was cloned in pETM30, expressed in the BL21(DE3) pRARE E. coli strain, and purified with a GST-prep FF 16/10 column followed by gel filtration using an S200 column (GE Healthcare). Proteins were incubated with beads for 2 h at 4°C in a buffer containing 50 mM Tris-HCl, 150 mM NaCl, and 1 mM DTT. Protein-bound beads were washed three times and heated to 100°C in Laemmli sample buffer.
Flag affinity immunoprecipitation of full-length PhoRC complexes
Full-length Pho-Flag and Sfmbt wild-type and mutant proteins were expressed and purified using the baculovirus system as described (Grimm et al. 2009). Cell lysis was performed with three cycles of sonication (30 bursts, 30 sec on ice each).
EMSA experiments
Single-stranded radiolabeled oligonucleotides containing the sequence 5′-CTCCGTCGCCATAACTGTCG-3′ were labeled with ATPγP32 using T4 PNK polymerase, gel-purified, precipitated in 100% EtOH, and annealed. The EMSA experiment was performed as in Fritsch et al. (1999). Protein–DNA complexes were resolved for 50 min at 100 V in a 7% Tris-glycine acrylamide minigel.
ITC and SPR measurements
ITC was performed with a VP-ITC Microcal calorimeter (Microcal). To measure the Sfmbt 4MBT/H4K20me1 interaction, protein samples were dialyzed extensively against ITC buffer (20 mM Tris-HCl at pH 8, 150 mM NaCl, 2 mM β-mercaptoethanol), and lyophilized synthetic peptides were resuspended in the same buffer. Protein concentration in the cell was 20 μM, and the peptide concentration in the injection syringe was 400 μM. For the Sfmbt4MBT/spacer interactions, the lyophilized Pho and YY1 spacer peptides were resuspended in water and buffer containing 10 mM Tris-HCl (pH 8.5), respectively, and then dialyzed against ITC buffer (10 mM Tris-HCl at pH 8, 150 mM NaCl, 1 mM β-mercaptoethanol). Sfmbt wild-type and mutant proteins were also dialyzed against the same buffer. Peptide concentration in the cell was 5 μM, and protein concentration in the syringe was 70 μM. SPR experiments were performed with a Biacore T-200, either biotin-Pho or biotin-YY1 spacer peptides were dissolved in running buffer (10 mM Tris-HCl at pH 8, 150 mM NaCl, 0.05% Tween 20, 1 mM DTT) and immobilized in a Series S Sensor Chip SA streptavidin chip, and 4MBT wild-type and mutant proteins were injected with concentrations according to their binding affinity to the biotinylated peptide.
Generation of Sfmbt-CTAP transgenic flies
Transgenes encoding wild-type Sfmbt, double point mutant SfmbtG635K/A638E, and the truncation SfmbtΔMBT1 were cloned into a modified version of pUASTattB (Bischof et al. 2007) to create C-terminal TAP tag fusion proteins. These transgenes were then integrated into the VK00033 site (Venken et al. 2006; Bischof et al. 2007) by germline transformation, as described in Bischof et al. (2007).
Small-scale Sfmbt-CTAP pull-downs from embryo nuclear extracts
daughterless-Gal4 virgins were crossed to males heterozygous for the Sfmbt[1] allele and homozygous for each Sfmbt-CTAP version or that did not have a Sfmbt-CTAP transgene. The resulting 0- to 12-h embryos were dechorionated in bleach, washed, and dounce-homogenized in buffer NU1 (15 mM HEPES at pH 8, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA at pH 8, 0.5 mM EGTA at pH 8, 350 mM sucrose, 1 mM DTT, 1× complete protease inhibitor cocktail [Roche], 1 mM AEBSF). Nuclei were pelleted at 1500g for 10 min at 4°C, washed with low-salt buffer (15 mM HEPES at pH 8, 20% glycerol, 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA at pH 8, 1 mM DTT, 1× complete protease inhibitor cocktail [Roche], 1 mM AEBSF), and lysed by dounce homogenization in high-salt buffer (same composition as low-salt buffer, except with 400 mM KCl). Debris was pelleted by centrifuging at 20,000g for 5 min at 4°C, and the supernatant was diluted 12.5-fold to a total volume of 500 μL in immunoprecipitation buffer (15 mM HEPES at pH 8, 20% glycerol, 1.5 mM MgCl2, 200 mM KCl, 0.2 mM EDTA at pH 8, 0.1 mM DTT, 0.4% NP-40, 1× complete protease inhibitor cocktail [Roche], 1 mM AEBSF). Extracts were incubated with 60 μL of washed IgG sepharose 6 fast flow beads (GE Healthcare) for 4 h at 4°C. IgG sepharose beads were washed five times for 10 min each with 1 mL of immunoprecipitation buffer followed by two quick changes of PBS. Enriched proteins were eluted with 100 mM glycine (pH 3) and then concentrated by TCA precipitation in the presence of 0.3% sodium deoxycholate as carrier. The TCA-precipitated protein pellet was finally resuspended in 1× LDS sample buffer (Invitrogen).
Functional analysis of Sfmbt mutants in imaginal discs
Sfmbt[1] mutant clones were induced in animals expressing separate UAS-Sfmbt versions or no Sfmbt transgene under the control of the 69B Gal4 driver. Staining of larval imaginal discs 72 h after clone induction was performed following standard protocols (Beuchle et al. 2001) using anti-Ubx (clone FP3.38) and peroxidase rabbit anti-peroxidase (Sigma) to detect TAP-tagged proteins via their protein A moiety. Cy3 anti-mouse F(ab′)2 fragment (Jackson ImmunoResearch) and Cy5 anti-rabbit (Jackson ImmunoResearch) were used as secondaries. Pictures were taken on an LSM 780 confocal microscope (Zeiss).
ChIP experiments
Chromatin was prepared from imaginal discs and brains of third instar larvae as previously described (Papp and Muller 2006). These larvae were the progeny of daughterless-Gal4 virgins crossed to males that were heterozygous for the Sfmbt1 allele and homozygous for the respective UAS-Sfmbt transgene or that did not carry any UAS-Sfmbt transgene. These larvae therefore expressed the Sfmbt-CTAP versions in the ubiquitous pattern directed by daughterless-Gal4. ChIPs were performed in triplicates from three independent chromatin preparations of each genotype, as previously described (Gambetta et al. 2009). Peroxidase anti-peroxidase (Sigma) was used to specifically immunoprecipitate the TAP-tagged proteins via their protein A moiety.
Quantitative PCR (qPCR) to determine binding at specific chromosomal locations
qPCR analysis was performed as previously described (Papp and Muller 2006) using the primers listed in Supplemental Table S3.
Western blot analysis of Sfmbt-CTAP protein levels in larvae
daughterless-Gal4 virgins were crossed to males heterozygous for the Sfmbt[1] allele and homozygous for each Sfmbt-CTAP version or that did not have a Sfmbt-CTAP transgene. Inverted larval carcasses from third instar larvae were cleared of fat body, digestive track, and salivary glands to leave only brain and imaginal discs attached. Protein extracts were prepared by sonicated carcasses in 1× LDS sample buffer with a Bioruptor sonicator water bath (Diagenode). Western blots were probed with peroxidase anti-peroxidase (Sigma) to detect TAP-tagged proteins via their protein A motif or with anti-α-tubulin (Sigma) and developed using Cy5-labeled secondaries on a Typhoon FLA 7000 (GE Healthcare).
TAP of Sfmbt complexes
Previously described vectors (Rigaut et al. 1999; Klymenko et al. 2006) were used to generate an α-tubulin-Sfmbt-CTAP transgene in the Drosophila transformation vector CaSpeR with the following sequences: a 2.6-kb fragment of the α-tubulin 1 gene, including promoter and 5′ untranslated region sequences (Struhl and Basler 1993), linked to a Sfmbt cDNA fragment that contained the complete Sfmbt1–1220 ORF and fused in-frame to the C-terminal TAP tag (plasmid maps are available on request). This α-tubulin-Sfmbt-CTAP transgene failed to rescue Sfmbt1 homozygotes or Sfmbt1/Df(2L)BSC30 transheterozygotes into viable adult flies but rescued repression of Ubx in clones of Sfmbt1 homozygous cells as in the case of the UAS-Sfmbt-CTAP transgene shown in Figure 4. TAP was performed from embryonic nuclear extracts as previously described (Klymenko et al. 2006).
MS
A detailed list of peptide sequences obtained from MS analyses of purified Sfmbt complexes is shown in Supplemental Table S2.
Coordinates
The atomic coordinates and structure factors of the Drosophila Pho:Sfmbt 4MBT complexes determined at 1.9, 2.1, and 3.2 Å have been deposited under the PDB accession codes 4C5E, 4C5G, and 4C5H, respectively. Atomic coordinates and structure factors of the human YY1:MBTD1 4MBT complex have been deposited under the PDB accession code 4C5I.
Acknowledgments
We thank Sandra Müller for help with generating transgenic Drosophila. We acknowledge support by the EMBL Proteomics Core Facility and the EMBL Heidelberg Crystallization Platform as well as the EMBL/ESRF Joint Structural Biology Group for access and support at the ESRF beamlines. This project was supported by the European Commission 7th Framework Program 4DCellFate grant number 277899 to C.W.M. and J.M. and the Marie Curie Initial Training Network Nucleosome4D to C.A. J.M. acknowledges support by the Max-Planck Gesellschaft.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.226621.113.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuchle D, Struhl G, Muller J 2001. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128: 993–1004 [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Brockdorff N 2011. Chromosome silencing mechanisms in X-chromosome inactivation: Unknown unknowns. Development 138: 5057–5065 [DOI] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell 1: 1057–1064 [DOI] [PubMed] [Google Scholar]
- Brown JL, Fritsch C, Mueller J, Kassis JA 2003. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130: 285–294 [DOI] [PubMed] [Google Scholar]
- Busturia A, Morata G 1988. Ectopic expression of homeotic genes caused by the elimination of the Polycomb gene in Drosophila imaginal epidermis. Development 104: 713–720 [DOI] [PubMed] [Google Scholar]
- Busturia A, Lloyd A, Bejarano F, Zavortink M, Xin H, Sakonju S 2001. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development 128: 2163–2173 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y 2005. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 20: 845–854 [DOI] [PubMed] [Google Scholar]
- Chan CS, Rastelli L, Pirrotta V 1994. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J 13: 2553–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Dura JM, Ingham P 1988. Tissue- and stage-specific control of homeotic and segmentation gene expression in Drosophila embryos by the polyhomeotic gene. Development 103: 733–741 [DOI] [PubMed] [Google Scholar]
- Eryilmaz J, Pan P, Amaya MF, Allali-Hassani A, Dong A, Adams-Cioaba MA, Mackenzie F, Vedadi M, Min J 2009. Structural studies of a four-MBT repeat protein MBTD1. PLoS ONE 4: e7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE 2001. Reconstitution of a functional core polycomb repressive complex. Mol Cell 8: 545–556 [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL 2004. Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Fritsch C, Brown JL, Kassis JA, Muller J 1999. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 126: 3905–3913 [DOI] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Muller J 2009. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325: 93–96 [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45: 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ 2006. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol 26: 6880–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, de Ayala Alonso AG, Rybin V, Steuerwald U, Ly-Hartig N, Fischle W, Muller J, Muller CW 2007. Structural and functional analyses of methyl-lysine binding by the malignant brain tumour repeat protein Sex comb on midleg. EMBO Rep 8: 1031–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Matos R, Ly-Hartig N, Steuerwald U, Lindner D, Rybin V, Muller J, Muller CW 2009. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J 28: 1965–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Usheva A, Shenk T, Burley SK 1996. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc Natl Acad Sci 93: 13577–13582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W 2010. Xds. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA 1995. The Polycomb and trithorax group proteins of Drosophila: Trans-regulators of homeotic gene function. Annu Rev Genet 29: 289–303 [DOI] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J 2006. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev 20: 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma G, Bender W, Sipos L 2008. Replacement of a Drosophila Polycomb response element core, and in situ analysis of its DNA motifs. Mol Genet Genomics 279: 595–603 [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D 2004. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell 14: 183–193 [DOI] [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP 2008. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 22: 2799–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE 2002. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol 22: 6070–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A 2009. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem 284: 34179–34188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapp AK, Ansari AZ 2007. A TAD further: Exogenous control of gene activation. ACS Chem Biol 2: 62–75 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J, Mishra RK, Karch F 1998. A conserved sequence motif in Polycomb-response elements. Mol Cell 1: 1065–1066 [DOI] [PubMed] [Google Scholar]
- Mills AA 2010. Throwing the cancer switch: Reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer 10: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P 2001. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol Cell Biol 21: 1311–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP 2002. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol Cell Biol 22: 7473–7483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP 2005. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev 19: 1755–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP 2006. Architecture of a polycomb nucleoprotein complex. Mol Cell 24: 91–100 [DOI] [PubMed] [Google Scholar]
- Muller J, Kassis JA 2006. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr Opin Genet Dev 16: 476–484 [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G 2006. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol 4: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, Muller J 2008. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell 15: 877–889 [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J 2006. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev 20: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Whyte WA, Anderssen E, Apostolou E, Chen HH, Akbarian S, Bronson RT, Hochedlinger K, Ramaswamy S, Young RA, et al. 2012. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell 11: 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Ringrose L 2007. Polycomb comes of age: Genome-wide profiling of target sites. Curr Opin Cell Biol 19: 290–297 [DOI] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J 2010. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465: 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G 2009. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46 [DOI] [PubMed] [Google Scholar]
- Shimell MJ, Peterson AJ, Burr J, Simon JA, O'Connor MB 2000. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev Biol 218: 38–52 [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W, Shimell MJ, O'Connor M 1993. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol 158: 131–144 [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strubbe G, Popp C, Schmidt A, Pauli A, Ringrose L, Beisel C, Paro R 2011. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc Natl Acad Sci 108: 5572–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Basler K 1993. Organizing activity of wingless protein in Drosophila. Cell 72: 527–540 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M 2006. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38: 694–699 [DOI] [PubMed] [Google Scholar]
- Trojer P, Cao AR, Gao Z, Li Y, Zhang J, Xu X, Li G, Losson R, Erdjument-Bromage H, Tempst P, et al. 2011. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol Cell 42: 438–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella P, Barozzi I, Cuomo A, Bonaldi T, Pasini D 2012. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res 40: 3403–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ 2006. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS 2004. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 14: 637–646 [DOI] [PubMed] [Google Scholar]
- Wilkinson FH, Park K, Atchison ML 2006. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci 103: 19296–19301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, Hart GW, Rauscher FJ III, Drobetsky E, Milot E, et al. 2010. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol 30: 5071–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bonasio R, Strino F, Kluger Y, Holloway JK, Modzelewski AJ, Cohen PE, Reinberg D 2013. SFMBT1 functions with LSD1 to regulate expression of canonical histone genes and chromatin-related factors. Genes Dev 27: 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]