Summary
Dispersed genetic elements, such as retrotransposons and Pol-III-transcribed genes, including tRNA and 5S rRNA, cluster and associate with centromeres in fission yeast through the function of condensin. However, the dynamics of these condensin-mediated genomic associations remains unknown. We have examined the 3D motions of genomic loci including the centromere, telomere, rDNA repeat locus, and the loci carrying Pol-III-transcribed genes or long-terminal repeat (LTR) retrotransposons in live cells at as short as 1.5-second intervals. Treatment with carbendazim (CBZ), a microtubule-destabilizing agent, not only prevents centromeric motion, but also reduces the mobility of the other genomic loci during interphase. Further analyses demonstrate that condensin-mediated associations between centromeres and the genomic loci are clonal, infrequent and transient. However, when associated, centromeres and the genomic loci migrate together in a coordinated fashion. In addition, a condensin mutation that disrupts associations between centromeres and the genomic loci results in a concomitant decrease in the mobility of the loci. Our study suggests that highly mobile centromeres pulled by microtubules in cytoplasm serve as ‘genome mobility elements’ by facilitating physical relocations of associating genomic regions.
Key words: Live-cell imaging, Genome dynamics, Nuclear organization, Condensin, Fission yeast
Introduction
Genome organizations have been characterized in terms of several aspects, including chromosomal territories, nuclear bodies and genomic associations involving transcriptional regulatory elements and their target genes (Cremer and Cremer, 2010; Sexton et al., 2009; Tanizawa and Noma, 2012; Williams et al., 2010; Zhao et al., 2009). It is becoming clear that three-dimensional (3D) genome organization is connected to various nuclear processes, such as transcription, DNA replication and repair, and chromatin domain formation (Cook, 1999; Labrador and Corces, 2002; Misteli, 2007). For example, enhancer–promoter association has been demonstrated to function in gene activation (Deng et al., 2012; Mahmoudi et al., 2002; Müeller-Storm et al., 1989). Using the latest genomics method, referred to as Hi-C, which combines chromosome conformation capture (3C) and next-generation sequencing, genome-wide associations have been comprehensively captured in several organisms (Duan et al., 2010; Hou et al., 2012a; Lieberman-Aiden et al., 2009; Sexton et al., 2012; Tanizawa et al., 2010). These studies demonstrate that eukaryotic genomes are highly organized into functional structures. Although genomic associations have been comprehensively mapped for several genomes, these genomic data cannot directly represent the stability of genomic associations. For example, stable genomic associations in a few cells cannot be distinguished from a transient association in many cells. Therefore, an important problem in the current chromatin/genome biology fields is to determine the dynamics of genomic associations using cell biological approaches (Baker, 2011).
We have recently developed the ELP (Enrichment of Ligation Products) genomic approach, which is similar to Hi-C, and identified genome-wide associations in fission yeast (Tanizawa et al., 2010; Tanizawa and Noma, 2012). This genomic analysis revealed the existence of chromosomal territories and significant associations among highly transcribed genes, co-regulated genes and functionally related genes, respectively. Moreover, our microscopic studies, employing a fluorescent in situ hybridization (FISH) assay, showed that dispersed genetic elements, such as retrotransposons and RNA-polymerase-III-transcribed genes, including tRNA and 5S rRNA, referred to as Pol III genes in this article, cluster and associate with centromeres (Iwasaki et al., 2010; Tanaka et al., 2012). We also showed that condensin functions as a molecular connector among chromatin fibers and is responsible for centromeric clustering of those dispersed genetic elements. Together, these studies demonstrated that the fission yeast genome is organized into a highly elaborate functional structure through genome-wide associations. Concurrent with the rising problem in the chromatin/genome biology fields, the dynamics of genomic associations in fission yeast remains unexplored.
In this study, we have investigated the spatiotemporal associations of genomic loci in live fission yeast cells. We begin by elucidating the 3D motion of the centromeres, the telomere, the rDNA repeat locus, several genomic loci carrying Pol III genes or retrotransposons, and a control locus. Our analysis reveals that treatment with carbendazim (CBZ), a microtubule-destabilizing agent, not only strongly affects centromeric motion but also impedes movement of the other genomic regions. Our studies also demonstrate that condensin-mediated associations between centromeres and genomic loci carrying Pol III genes and retrotransposons are transient and infrequent. However, once they associate, highly mobile centromeres and the genomic regions migrate together. In addition, we show that the condensin mutation that disrupts those associations reduces the mobility of the genomic regions. Given that Pol III genes and retrotransposons are dispersed across the genome, centromeric motion coupled to microtubule polymerization can impact upon the mobility of non-centromeric genomic regions through condensin-mediated associations. Our studies might have identified a new role of centromeres and cytoplasmic microtubules in genome-wide mobility.
Results
Non-random positioning of fission yeast genomic loci
For the initial step into the examination of the dynamics of genome organization, we made several fission yeast strains carrying lacO repeats (supplementary material Fig. S1A,B). We employed strains carrying lacO or tetO repeats at the centromeres (cen1, cen2 and cen3), the telomere (tel2R), the rDNA locus, the genomic loci consisting of Pol III genes (c417 and c10H11) and retrotransposons (c947), and a control locus (c887). There are no retrotransposons nor Pol III-transcribed genes within 80 kb of the c887 control locus, and it also does not have condensin-binding sites, as determined by the ChIP-seq analysis (Tanaka et al., 2012). The FISH analysis indicated that the insertions of lacO repeats into the c417 Pol III gene locus and the c887 control locus did not affect the positioning of those genomic loci in relation to the nuclear periphery, suggesting that a lacO insertion is unlikely to drastically affect endogenous positioning of genomic regions (supplementary material Fig. S1C). We also observed that the centromeric insertions of lacO and/or tetO repeats did not cause a chromosome segregation defect, suggesting that the lacO and/or tetO insertions probably do not affect centromeric function (supplementary material Fig. S1D).
It has previously been shown that several genomic loci in budding yeast tend to localize within respective nuclear domains rather than randomly distribute throughout the nucleus (Berger et al., 2008). The Berger et al. study elegantly examined the frequent positioning of genomic loci in relation to the nuclear membrane and the nucleolus used as nuclear landmarks. The Nucloc software used in that study processed hundreds of 3D images and calculated the tendency of intra-nuclear positioning of a genomic locus in cell populations as a probabilistic density. We have employed the same approach to study spatial localization of genomic regions in fission yeast. We visualized genomic loci carrying lacO repeats in addition to nuclear landmarks, such as the nuclear membrane and the nucleolus. Probability density was calculated for the centromeres (cen1 and cen2), the telomere (tel2R), the rDNA locus, the Pol III gene locus (c417), the long-terminal repeat (LTR) retrotransposons locus (c947) and the control locus (c887) (supplementary material Fig. S2). The probability density maps revealed that the genomic regions we examined tended to localize within limited subnuclear compartments. For example, the centromeres (cen1 and cen2) were frequently positioned in proximity to the nuclear membrane but were distant from the nucleolus. The telomere (tel2R) was present near the nuclear periphery but often located away from the centromere-occupied domain. These observations suggest that the fission yeast genome might be organized into a Rabl-like chromosome conformation, which was predicted for budding yeast (Berger et al., 2008; Bystricky et al., 2005; Spector, 2003). The locus adjoining the rDNA tandem array tends to localize near the nucleolus. Consistently, rDNA repeats themselves were shown to be present in the fission yeast nucleolus (Uzawa and Yanagida, 1992). Moreover, the c417 Pol III gene locus, as well as the c947 LTR retrotransposon locus and the c887 control locus, localized within the interior domain of the nucleus, typically between the subnuclear domains occupied by the rDNA-flanking locus and the peripheral loci (cen1, cen2 and tel2R). The c887 control locus was distributed more evenly across the nucleus compared to the retrotransposon locus. We observed that the Pol III gene locus and the retrotransposon locus were present near the centromere-occupied nuclear domain in the limited cell population, whereas this positioning was not detected for the control locus, suggesting that the Pol III gene locus and the retrotransposon locus associate with centromeres in some populations (Iwasaki et al., 2010; Tanaka et al., 2012) (supplementary material Fig. S2B). These data demonstrate that the respective genomic loci tend to localize within specific subnuclear domains in fission yeast.
Movement of genomic loci in live fission yeast cells
We visualized several genomic loci and the nuclear landmarks, such as the nuclear membrane and nucleolus, in live fission yeast cells at as short as 1.5-second intervals for 5 minutes. At each time point, the position of the genomic loci was plotted in the 3D nuclear space, determined by using the nuclear landmarks. It is important to note that effects from cell migration and movement of the nucleus within the cytoplasm were filtered out using the nuclear landmarks, allowing us to examine only intra-nuclear migration of genomic loci. We observed that the centromeres (cen1 and cen2) and the telomere (tel2R) were typically located between 1.0 and 1.2 µm from the nuclear center (supplementary material Fig. S3A). The Nup61–mCherry nuclear pore complex (NPC) signals were also located at the same distance from the nuclear center, suggesting that centromeres and telomeres were both located at the nuclear periphery. In clear contrast, the rDNA-flanking locus resided between 0.2 and 0.6 µm from the nuclear center and just outside of the nucleolus. These data are consistent with the probability density maps (supplementary material Fig. S2B). Moreover, the Pol III gene loci (c417 and c10H11) and the LTR retrotransposons locus (c947) typically migrated between 0.8 and 1.0 µm from the nuclear center (supplementary material Fig. S3A). This positioning, which is relatively close to the nuclear membrane, might reflect potential associations between these genomic loci and centromeres present at the nuclear periphery. Consistently, we observed that the c887 control locus, which does not associate with centromeres, was typically positioned between 0.6 and 0.8 µm from the nuclear center and a little further away from the nuclear periphery.
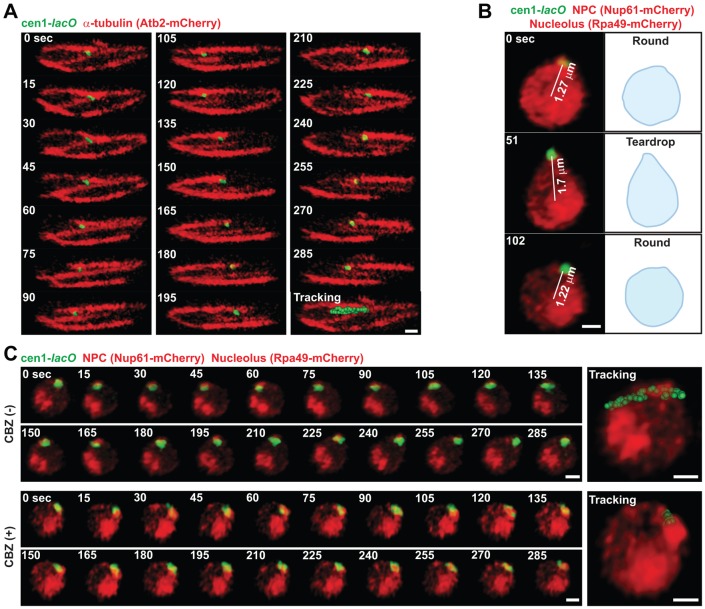
Interestingly, we observed that the cen1 was sometimes positioned more than 1.2 µm away from the nuclear center (supplementary material Fig. S3A). To further examine centromeric behavior, we next visualized the centromere, cen1, with microtubules in live cells. 3D imaging in live cells demonstrated that the centromere dynamically moves along microtubules (Fig. 1A). The centromere was typically located near the tip of the microtubule fiber and shifted position in the direction of microtubule polymerization. Live-cell imaging also showed that the centromere moves along the nuclear periphery and that the nuclear morphology was sometimes transformed from the normal round to the ‘teardrop’ shape in association with this movement (Fig. 1B). Treatment with CBZ interrupted the dynamic motion of the centromere (Fig. 1C). Together, these results suggest a model that cytoplasmic microtubules pull or move centromeres along the nuclear membrane, and that nuclear morphology can be deformed to accommodate the physical force when microtubules attempt to pull centromeres beyond the nuclear space.
Fig. 1.
Centromeric motion coupled to microtubule polymerization in cytoplasm. (A) Selected frames from a time-lapse series of the live fission yeast cell showing microtubules (red) and the centromere (cen1-lacO, green). Microtubules were visualized by mCherry fused to Atb2 (α-tubulin). Tracking of the cen1 locus is shown at the end of the time-lapse sequence. (B) Nuclear morphology and centromeric position. The centromere (cen1-lacO, green), the NPC (Nup61–mCherry, red) and the nucleolus (Rpa49–mCherry, red) were visualized in live cells. Selected frames from a time-lapse series and 3D distances between the nuclear centers and centromeric foci are shown (left). A schematic of the nuclear morphology is depicted next to the microscopic images. (C) CBZ treatment affects centromeric mobility. Cells were treated with 50 µg/ml CBZ in EMM liquid medium for 15 minutes and applied to microscopic slides with a mounting medium containing CBZ. Images were captured in 3D at 3.0-second intervals for 5 minutes. Scale bars: 1 µm.
We also investigated the movement velocities of the respective genomic loci based on 3D time-lapse imaging. We did not find a clear difference among velocities estimated from the 3D movement of the centromere (cen1), the telomere (tel2R), the Pol III gene locus, the LTR retrotransposon locus and the control locus when we estimated migration velocities at 1.5-second intervals (supplementary material Fig. S3B). Velocities of those loci were ∼0.1 µm/second. This is probably because these velocities mainly reflect random motion of the genomic loci, which has been referred to as Brownian-like or diffusional motion and has been detected from budding yeast to man (Chubb et al., 2002; Heun et al., 2001; Marshall et al., 1997). Therefore, we predict that fission yeast genomic loci are also subjected to Brownian-like motion. We next investigated velocities of the respective loci at prolonged intervals ranging from 3 to 36 seconds. Average velocities were reduced from 0.06 to 0.01 µm/second with the longer intervals, suggesting that the genomic loci typically migrate without directional movement. Interestingly, we observed that the velocity of the cen1 was enhanced to 0.02–0.03 µm/second at 24- and 36-second long intervals in some populations, whereas typical velocities for other genomic loci were ∼0.01 µm/second (supplementary material Fig. S3B). Considering that centromeres are pulled by microtubules and move along the nuclear periphery (Fig. 1), this directional motion of centromeres is mediated by the microtubules at an approximate velocity of 0.02–0.03 µm/second.
CBZ treatment reduces 3D motion of centromeres and other genomic loci
Our analyses demonstrated that the fission yeast genomic loci were mobile, yet typically restricted within distinct subnuclear domains. We next examined tracking of the respective genomic loci in terms of 3D nuclear volume using a new approach with our custom algorithm (Fig. 2A; see Materials and Methods). This approach allowed us to estimate 3D nuclear domains, referred to here as moving volumes, in which the different genomic loci migrate, for 5 minutes. We observed that the moving volumes of the centromeres (cen1 and cen2) were around 0.3 µm3 (Fig. 2B,C). Given that the nuclear volume excluding the nucleolus is ∼6.8 µm3, the centromere movement is confined to 4.4% of the nuclear volume. The average nuclear volume was estimated from 3D images visualizing the nuclear landmarks in 800 cells (supplementary material Fig. S2). The other genomic loci also migrated within relatively similar nuclear volumes (∼0.3 µm3), whereas the telomere (tel2R) was seemingly restricted to a slightly smaller nuclear volume (0.2 µm3), which is estimated as 2.9% of the entire nuclear volume (Fig. 2C).
Fig. 2.
Moving volume analysis for the genomic loci in several culturing conditions. (A) Estimation of the moving volume of the genomic locus. This schematic illustrates how occupancy of the genomic locus was estimated based on 3D time-lapse images. The moving volume of the genomic locus was defined as an accumulated number of cubes. See the Materials and Methods for details. (B) Moving volume analysis for the centromere (cen1). Cells were treated with 50 µg/ml CBZ, 40 mM CCCP or 15 mM NaN3 in liquid EMM for 15 minutes and applied to microscopic slides. The centromere (cen1-lacO, green), the NPC (Nup61–mCherry, red) and the nucleolus (Rpa49–mCherry, red) were co-visualized in live cells, and tracking of the centromere is shown (left). Images were captured in 3D at 3.0-second intervals for 5 minutes. The position of the centromere was normalized to the center of the nucleus. The moving volume of the centromere was estimated by counting the cubes (middle). The moving volume of the centromere was analyzed in five cells and data are represented as mean ± s.d. (right). (C) Moving volume analyses for the centromere (cen2), the Pol III gene loci (c417 and c10H11), the LTR retrotransposon locus (c947), the tel2R telomere locus and the control locus (c887). Typical tracking images are shown as insets. Scale bars: 1 µm.
It has been shown that inhibition of metabolic processes by ATP depletion affects rapid movement of budding yeast genomic loci (Heun et al., 2001). We analyzed whether ATP depletion, by treatment with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) and sodium azide (NaN3), has an effect on the moving volumes of the fission yeast genomic loci. We observed that ATP depletion severely reduced the moving volume of every genomic locus we examined. For example, the CCCP treatment reduced the moving volumes of the respective genomic loci to 0.05 µm3, which is estimated as only 0.74% of the nuclear volume and 16.7% of the original moving volumes. These data suggest that energy-dependent cellular activities promote motion of the interphase genomic loci (Fig. 2). To relate this phenomenon to a specific cellular process, we next focused on microtubule polymerization, an important cellular activity utilizing the energy of GTP hydrolysis (Desai and Mitchison, 1997). Remarkably, the CBZ treatment affected the moving volumes of not only the centromere, but also all other genomic loci we investigated (Fig. 2). The centromeric moving volume was reduced from 0.3 to 0.15 µm3, and the moving volumes of the other genomic loci were also decreased to 0.2 µm3, which is estimated as ∼66% of the original moving volumes. Because our analyses only reflect intra-nuclear migration of the genomic loci, the data suggest that microtubule polymerization in the cytoplasm promotes the mobility of centromeres and the other genomic loci.
To compare these results with previous studies in the other organisms, we also analyzed the mean squared change in distance (<Δd2>) based on the 3D time-lapse data (supplementary material Fig. S4) (Chubb et al., 2002; Heun et al., 2001; Marshall et al., 1997). A <Δd2> value serves as an index for diffusional motion of a genomic locus. The <Δd2> plots for the respective genomic loci showed that the <Δd2> values reached a range of 0.08 and 0.15 µm2 after 50–75 seconds, and the slopes of the <Δd2> plots typically decayed for the longer time points. These results suggest that genomic loci are mobile but tend to position within restricted subnuclear volumes, which agrees with the probability density maps (supplementary material Fig. S2B). This constrained motion of genomic loci has also been observed in other eukaryotes (Chubb et al., 2002; Heun et al., 2001; Marshall et al., 1997), suggesting that similar characteristics of the dynamic motion of genomic regions are probably conserved among eukaryotes. Moreover, we observed that ATP depletion severely decreased the <Δd2> values and that CBZ treatment also affected the <Δd2> values to a lesser extent.
Coordinated motion of centromeres and their associated loci
To begin to address the dynamics of condensin-mediated genomic associations, we decided to examine associations between centromeres and the three representative genomic loci carrying Pol III genes and LTR retrotransposons in live cells. We performed a FISH analysis and confirmed that the lacO insertion into the c417 Pol III gene locus did not affect the frequency of association between centromeres and the loci (P>0.05, Mann–Whitney U test; supplementary material Fig. S5A). We next investigated the cen2–cen3 association as a positive control for genomic associations, as fission yeast centromeres are known to stably interact with one another during interphase (Funabiki et al., 1993). The live-cell imaging data showed that the 3D distances between cen2 and cen3 foci were almost always between 0.2 and 0.6 µm during interphase (supplementary material Fig. S5B,C). Moreover, the relative positioning of cen1 and Mis12 (kinetochore) foci was similar to that of cen2 and cen3 (supplementary material Fig. S5D). Therefore, we accepted the potential technical limitation of this imaging approach by which genomic associations cannot be detected as perfect colocalization of two foci. From this point, we decided to follow the criterion that two foci positioned within 0.6 µm reflect potential associations.
We also visualized centromeres and the c417 Pol III gene locus in live cells and monitored the 3D distances between two foci continuously for 5 minutes. We observed that two foci were temporarily positioned in proximity (Fig. 3A,B). On the basis of the above criterion, centromeres potentially associate with the Pol III gene locus for ∼20 seconds at around the 156-second time point. In clear contrast, the c887 control locus was constantly separated from the centromeric foci. We observed that the control locus was rarely positioned within 0.9 µm of centromeres, whereas the c417 Pol III gene locus was frequently detected within 0.9 µm of centromeres (Fig. 3A–C). Moreover, another Pol III gene locus (c10H11) and the retrotransposon locus (c947) showed essentially the same results as the c417 Pol III gene locus (Fig. 3C). Therefore, we speculated that the genomic loci positioned within 0.9 µm of centromeres might also reflect association with centromeres. In this case, centromeres and their associating genomic loci would migrate in a coordinated fashion. Indeed, we observed that the c417 Pol III gene locus positioned between 0.6 and 0.9 µm from centromeres, with which we believe the locus does not directly associate according to the above criterion, was found to still shift along the x and y axes as centromeres moved (supplementary material Fig. S5E). The coordinated motion was also observed in cells where the c417 locus migrated within 0.6 µm of centromeres (Fig. 3D). However, the coordinated motion was not observed in cells where centromeres and the c417 locus were continuously separated by more than 0.9 µm (supplementary material Fig. S5F). To evaluate the coordinated movement between centromeres and the c417 locus, we calculated the mean correlation coefficient for each cell. This value represents similarity between the movements of centromeres and the locus. The same approach was previously employed for telomere associations in budding yeast (Bystricky et al., 2005). We found that the mean correlation coefficient for cells with centromeres and the locus always separated by more than 0.9 µm was near zero (Fig. 3E). However, the mean correlation coefficient was increased in the cases where the c417 Pol III gene locus migrated within 0.6–0.9 µm of centromeres at least once during the 5-minute observation (Fig. 3E). These results suggest that the locus positioned between 0.6 and 0.9 µm from centromeres moves in synchrony with centromeres to some extent. Our current hypothesis is that the flanking region of the c417 Pol III gene locus probably associates with centromeres and, as a result, the locus and centromeres move in a coordinated fashion. We suggest that the coordinated movement of two foci with an intervening distance of 0.6–0.9 µm reflects an indirect association. The coordinated motion reflecting direct associations was further promoted in cells where the c417 locus migrated within 0.6 µm from centromeres (Fig. 3D,E). Moreover, the mean correlation coefficient for cen2–cen3 was around 0.61, probably reflecting continuous association between the centromeres.
Fig. 3.
Coordinated migration between centromeres and their associating genomic loci. (A) The genomic locus (green), either the Pol III gene locus (c417, top) or the control locus (c887, bottom), was co-visualized with centromeres (Mis12–mCherry, red). Images were captured in 3D at 3.0-second intervals for 5 minutes and selected frames are shown. (B) Distances between centromeres and the genomic loci (c417 and c887) were measured in 3D time-lapse images used in A and plotted against time. Distances below 0.6 µm and between 0.6 and 0.9 µm are highlighted with different colors. (C) Distances between centromeres and the genomic loci, such as the Pol III gene locus (c417 and c10H11), the LTR retrotransposon locus (c947) and the control locus (c887) were measured in 3D time-lapse images. Images were captured at 3.0-second intervals for 5 minutes in more than 35 cells, and 10 examples for each locus are shown in a graph. (D) The c417 Pol III gene locus and centromeres show the coordinated movement in the case where two foci transiently migrate within 0.6 µm during the 5-minute investigation. Distances between the Pol III gene locus and centromeres are plotted against time (left). Tracking of the Pol III gene locus (green) and centromeres (red) along the x and y axes is shown (right). Coordinates of the c417 Pol III gene locus and centromeres in the x and y axes were used to calculate Δx and Δy at 3.0-seconds interval. The Δx and Δy values were plotted against time and used to calculate Pearson's correlations (bottom). (E) The coordinated motion between the c417 Pol III gene locus and centromeres. Distances between centromeres and the c417 Pol III gene locus were measured in 3D time-lapse images (n = 42). On the basis of the following criteria, cells were classified into three groups. In 7 and 13 cells, the Pol III gene locus migrated within 0.6 µm and between 0.6 and 0.9 µm from centromeres, respectively, at least once during the 5-minute investigation. In 22 cells, the two foci were always separated more than 0.9 µm. The mean correlation coefficient is calculated for each cell as the mean of Pearson's correlations in the x and y directions. The averages of the mean correlation coefficients in the three cell populations are shown. Scale bars: 1 µm.
Genomic associations in live fission yeast cells
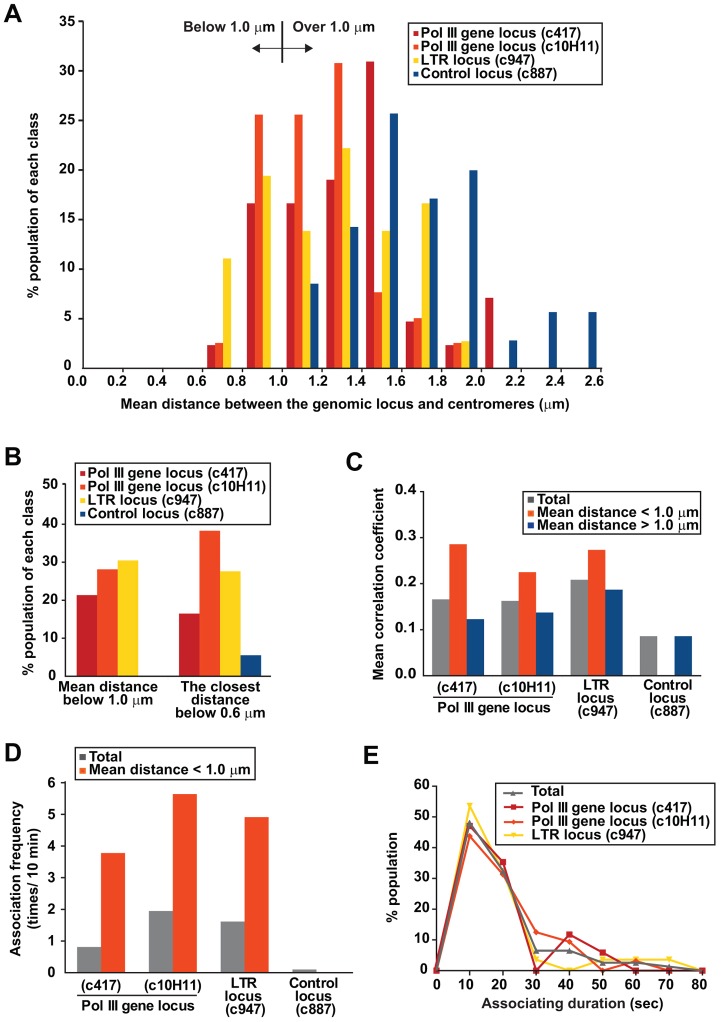
We analyzed 3D live-cell movies (∼5 minutes) co-visualizing centromeres and various genomic loci, such as the Pol III gene loci (c417 and c10H11), the LTR retrotransposon locus (c947) and the control locus. We examined more than 35 independent live cells for each locus and calculated the mean distance between centromeres and the genomic locus in each cell. Distributions of mean distances between centromeres and the respective genomic loci showed that the Pol III gene loci and the retrotransposon locus were positioned in proximity to centromeres in some cell populations, which was significantly different from the distribution with the c887 control locus (P<0.05, Mann–Whitney U test; Fig. 4A). It is noteworthy that a significant difference in distributions of mean distances was not observed when distances between centromeres and the respective genomic loci were measured in 2D images (P>0.05, Mann–Whitney U test; supplementary material Fig. S6A), suggesting that genomic associations are preferably examined in 3D.
Fig. 4.
Estimation of the association dynamics between centromeres and the genomic loci. (A) Distributions of mean distances between centromeres and the indicated genomic loci in the cell population. Distances between centromeres and the genomic loci (c417, c10H11, c947, and c887) were measured at 3.0-second intervals for 5 minutes (n = 42, 39, 36 and 35, respectively). The mean distance was calculated for each cell. Distributions of mean distances are plotted in the histogram. (B) Estimation of association frequencies in the cell population. The data in A were used to calculate the percentage of cells in each population in which mean distances between the centromeres and the indicated genomic loci were below 1.0 µm (left) and the genomic loci migrated within 0.6 µm from centromeres at least once during the 5-minute investigation (right). (C) The coordinated motion between centromeres and the genomic loci is enhanced when mean distances between two foci are below 1.0 µm. On the basis of the mean distances shown in A, cells were classified into two groups, in which mean distances between centromeres and the genomic loci were below or above 1.0 µm. The mean correlation coefficient was calculated for every cell, as described in Fig. 3E, and the averages of the mean correlation coefficients in the respective groups are shown. (D) Association frequencies between centromeres and the genomic loci. The number of times that 3D distances between centromeres and the genomic loci fell below 0.6 µm was counted to estimate their potential associations in the entire cell population and in cells where the mean distances between centromeres and the genomic loci were below 1.0 µm. (E) Durations of association between centromeres and the genomic loci were estimated for the potential associations predicted in D, and the distributions of the durations of association are plotted.
We noticed that mean distances between centromeres and the control locus were always more than 1.0 µm, implying that for loci with mean distances below 1.0 µm, the genomic loci might associate with centromeres. In ∼20–30% of cells, mean distances between centromeres and the other loci were below 1.0 µm (Fig. 4B). The coordinated motion between centromeres and the genomic loci was higher in these cells compared to the remaining cell population, which had mean distances above 1.0 µm (Fig. 4C). Moreover, we observed that the Pol III gene loci and retrotransposon locus migrated within 0.6 µm of centromeres in a similar percentage of cells (Fig. 4B). Therefore, these results suggest that these genomic associations occur in ∼20–30% of cells.
Next, we estimated association frequency and duration between centromeres and the genomic loci (Fig. 4D,E). The Pol III gene loci and LTR retrotransposon locus potentially associated with centromeres at frequencies of 1–2 times for every 10 minutes when the entire cell population was considered (Fig. 4D). Given that associations only occurs in 20–30% of cells (Fig. 4B), associations in those cells were re-estimated to occur at frequencies of 4–6 times in a 10-minute interval. Association duration was typically less than 30 seconds, although the genomic associations were sustained for 30–80 seconds in the limited cell population (Fig. 4E). These results collectively suggest that associations between centromeres and the genomic loci are clonal, infrequent and transient, but sufficient for their coordinated migration once they associate.
Role of condensin in tethering the genomic loci to centromeres
We also investigated the effects of the condensin mutations on the association between centromeres and the c417 Pol III gene locus in live cells. We observed that the mean distances between centromeres and the Pol III gene locus were significantly affected by the cut3-477 and cut14-208 condensin mutations (P<0.05, Mann–Whitney U test), whereas the c887 control locus was not affected (P>0.05, Mann–Whitney U test; Fig. 5A). Interestingly, positioning of the Pol III gene locus and retrotransposon locus in relation to the nuclear periphery was affected by the cut3-477 condensin mutation, whereas cen1 remained at the periphery (Fig. 5B). Moreover, the probability density maps suggested that the positioning of cen1 was not affected by the mutation of condensin (supplementary material Fig. S2B). Therefore, these data suggest that condensin mediates associations between centromeres and the genomic loci and, once associations are disrupted, the loci shift towards the interior domain of the nucleus.
Fig. 5.
Condensin is involved in the mobility of the genomic loci. (A) Distances between centromeres and the genomic loci, such as the c417 Pol III gene locus and the c887 control locus, were measured in cut3-477 and cut14-208 condensin mutant cells (n>35). Wild-type and cut3-477 cells were cultured at the restrictive (36°C) temperature for 2 hours and subjected to live-cell imaging analysis within a temperature-controlled chamber. The cut14-208 cells were cultured at 36°C for 1 hour. Most cells used for the analyses were in interphase. Images were captured in 3D at 3.0-second intervals for 3 minutes. Distributions of the mean distances are summarized in a graph. (B) The c417 Pol III gene locus and c947 retrotransposon locus tend to localize in the interior domain of the nucleus in the cut3-477 condensin mutant. The genomic loci (lacO, green), the NPC (Nup61–mCherry, red), and the nucleolus (Rpa49–mCherry, red) were co-visualized in wild-type and cut3-477 cells. Images were captured in 3D at 6.0-second intervals for 5 minutes. The distance from the nuclear center was divided into three zones based on the criteria depicted on the left. The mean distance between the nuclear center and the indicated genomic loci was assigned into one of these zones. (C) The moving volumes of the indicated genomic loci were estimated for the wild-type and cut3-477 condensin mutant as described in Fig. 2A. The moving volumes of the respective loci were analyzed in more than 10 cells, and data are represented as mean ± s.d.
On the basis of the moving volume data (Fig. 2), we hypothesized that centromeric motion influences genome-wide mobility through associations among centromeres and other genomic regions. To test this hypothesis, we investigated moving volumes of the genomic loci in the cut3-477 condensin mutant and observed that the Pol III gene loci (c417 and c10H11) and the retrotransposon locus (c947) became more static compared to the wild-type cells, whereas centromeres (cen1 and Mis12) were not affected (Fig. 5C). The moving volumes of the genomic loci, such as the Pol III gene loci (c417 and c10H11) and the retrotransposon locus (c947), were decreased from 0.4–0.5 µm3 to 0.25–0.35 µm3 in condensin mutant cells, which is estimated as 60–70% of the original moving volumes. Therefore, these results suggest that condensin tethers the genomic loci to centromeres, thereby allowing centromeric motion to impact upon the mobility of the associated loci.
Centromeric motion and the mobility of other genomic loci
If centromeric motion influences the mobility of other genomic loci, we should observe that genomic loci associating with centromeres migrate with centromeres, while genomic loci might become more static and independent of centromeric movement when they do not associate with centromeres. We analyzed the live-cell data (Fig. 4A) and observed a clear correlation in the moving volumes of centromeres and the genomic loci in cells with mean distances below 1.0 µm (Fig. 6A). In contrast, the moving volumes of centromeres and the genomic loci were not well correlated in cells with mean distances above 1.0 µm. Interestingly, 30 of 46 cells (∼65%) with mean distances above 1.0 µm showed a moving volume of less than 0.4 µm3 for the genomic loci, while centromeres moved independently (Fig. 6A). Moreover, we observed that, in cells with mean distances below 1.0 µm, the c417 Pol III gene locus was positioned in proximity to centromeres and they migrated in a coordinated fashion (Fig. 6B). In cells with mean distances above 1.0 µm, the Pol III gene locus became relatively more static and did not move with centromeres. All together, these studies suggest that highly mobile centromeres, pulled by microtubules, contribute to the mobility of other genomic loci through condensin-mediated associations (Fig. 6C).
Fig. 6.
The potential role of centromeres in genome-wide mobility. (A) Comparison between the moving volumes of centromeres and the genomic loci. On the basis of the mean distances shown in Fig. 4A, cells were classified into two groups in which mean distances between centromeres and the genomic loci were below or above 1.0 µm. In individual cells, the moving volumes of centromeres and the genomic loci were estimated as described in Fig. 2A. The moving volumes of centromeres and the genomic loci at the last time-point (5 minutes) are plotted. The dotted circle represents cells with a moving volume of less than 0.4 µm3 for the genomic loci. (B) Representative data showing a 5-minute tracking of centromeres (Mis12–mCherry, red) and the c417 Pol III gene locus (green) in cells with mean distances below (left) and above 1.0 µm (right). Scale bar: 1 µm. (C) A model for the role of centromeric motion in the mobility of other genomic regions. Microtubule polymerization in cytoplasm actively pushes centromeres in nucleoplasm. Centromeres transiently associate with genomic loci carrying Pol III genes and retrotransposons. These associations are mediated by condensin. Centromeres and their associating genomic loci migrate in a coordinated fashion. Because Pol IIl genes and retrotransposons are dispersed throughout the genome, centromeric motion regulated by cytoplasmic microtubules can potentially impact genome-wide mobility.
Intra-nuclear positioning of Pol III genes and their transcription
It has previously been shown that Pol III genes, which are dispersed across the fission yeast genome, cluster and associate with centromeres, and that this genome organization is coupled to the transcriptional repression of Pol III genes (Iwasaki et al., 2010). We examined whether the mobility of the Pol III gene locus affects Pol III transcription. Fission yeast cells were subjected to the CBZ treatment, and the mean distances between centromeres and the genomic loci, such as the c417 Pol III gene locus and the c887 control locus, were calculated in more than 20 cells. Distributions of the mean distances demonstrated that the CBZ treatment did not affect the positioning of the Pol III gene locus nor the control locus in relation to centromeres (P>0.05, Mann–Whitney U test), although it reduced the mobility of centromeres and the genomic loci (Fig. 2; supplementary material Fig. S6B). We also observed that tRNA expression was not affected by the CBZ treatment (supplementary material Fig. S6C). Therefore, these results suggest that the mobility of the Pol III gene locus is unlikely to be involved in the transcriptional regulation of Pol III genes.
The ChIP results indicated that the Cut14 condensin subunit was still localized at the Pol III gene locus (c417) and the centromere (cnt1) in cut3-477 mutant cells, suggesting that the condensin complex or a subset of the complex can bind to the Pol III gene locus and centromeres in cut3-477 condensin mutant cells (supplementary material Fig. S6D). However, associations between centromeres and the Pol III gene locus were significantly compromised by the cut3-477 mutation, and Pol III transcription is enhanced (Fig. 5A) (Iwasaki et al., 2010). Therefore, our current hypothesis is that condensin binding might not be sufficient to regulate Pol III genes, but their transcriptional regulation probably entails condensin-mediated positioning of the genes. Together, these results suggest that centromeric localization of Pol III genes, but not their mobility, contributes to their transcriptional regulation. It is possible that a subnuclear environment around centromeres, densely occupied by heterochromatin components, is unfavorable for optimal Pol III transcription (Kniola et al., 2001).
Discussion
Dynamics of condensin-mediated genomic associations in fission yeast
We have previously shown that centromeres associate with Pol III genes and LTR retrotransposons through condensin function (Iwasaki et al., 2010; Tanaka et al., 2012). We decided to elucidate the dynamics of the condensin-mediated associations between centromeres and the genomic loci carrying Pol III genes and retrotransposons using live-cell imaging tools developed in our laboratory. For this purpose, we first needed to define genomic associations in live cells and developed the criteria that positioning of two foci within 0.6 µm, and in a range between 0.6 and 0.9 µm, reflects direct and indirect genomic associations, respectively. These initial observations in fission yeast offer the standards to examine the dynamics of genomic associations in other organisms.
On the basis of these criteria, we estimated association frequency and duration between centromeres and the genomic loci carrying Pol III genes and retrotransposons. Our data suggest that associations between centromeres and the genomic loci investigated, potentially occur at frequencies of 1–2 times per 10-minute interval, when the entire cell population is considered. In approximately 20–30% of cells, mean distances between centromeres and the genomic loci are below 1.0 µm. In these cells, association frequencies are enhanced to 4–6 times in a 10-minute interval, indicating that there is a clonal variation for genomic associations. These estimations are probably reliable, because the control locus hardly associates with centromeres according to the same criteria. Moreover, our data also suggest that the duration of the association between centromeres and the genomic loci is typically less than 30 seconds, suggesting that condensin-mediated genomic associations are dynamic. However, prolonged associations were also observed in the limited cell population. Our previous study demonstrated that the Pol III gene locus tightly associates with centromeres during mitosis, thereby resulting in the proper assembly of mitotic chromosomes (Iwasaki et al., 2010). Therefore, the dynamics of condensin-mediated genomic associations are probably regulated during the cell cycle. Namely, condensin-mediated associations are typically unstable during interphase, as observed in this study, but probably become more static during mitosis. It is intriguing to understand the mechanism that regulates the association stability.
Protein complexes consisting of structural maintenance of chromosomes (SMC) proteins are essential for the faithful segregation of chromosomes. Two of the best-studied SMC complexes are condensin and cohesin, which are required for mitotic chromosome assembly and for holding sister chromatids together, respectively (Hagstrom and Meyer, 2003; Hirano, 2000; Koshland and Strunnikov, 1996; Laemmli et al., 1992; Nasmyth and Haering, 2005; Yanagida, 1998). These SMC complexes function as connectors between two chromatin fibers (Hirano, 2006). Recent studies have revealed that cohesin mediates functionally distinct genomic associations, including interactions between enhancers and promoters (Hadjur et al., 2009; Hou et al., 2010; Kagey et al., 2010; Mishiro et al., 2009; Nativio et al., 2009). Despite their functional importance, the dynamic nature of SMC-mediated genomic associations had remained uncharacterized. This study has examined SMC-mediated genomic associations in live cells for the first time and demonstrated that those associations are transient in the fission yeast model organism. It is possible that SMC-mediated associations in higher eukaryotes might be dynamic, as observed in fission yeast.
Role of centromeres and cytoplasmic microtubules in genome-wide mobility
Among the genomic loci we examined, centromeres most widely migrate along the nuclear membrane. In fission yeast, the interphase centromeres are known to localize adjacent to the spindle pole body (SPB), which is attached by microtubules (Ding et al., 1997; Funabiki et al., 1993). Microtubule-mediated movement of the SPB was previously observed in fission yeast (King et al., 2008). There are intricate interactions among the SPB, present at the cytoplasmic side of the nuclear surface, the SUN-KASH complex, which is integrated in the nuclear membrane, and the Csi1 protein, which connects cytoplasmic microtubules to centromeres (Chikashige et al., 2010; Hou et al., 2012b). We find that centromeric foci present near the tip of the microtubule fiber migrate along the nuclear membrane as a result of microtubule polymerization (Fig. 1A). CBZ treatment severely inhibits centromeric motion. The migration velocity of centromeres is around 0.02–0.03 µm/second. This coincides with the rate of microtubule polymerization in fission yeast (0.024 µm/second) (Tran et al., 2001). Therefore, it is likely that microtubule polymerization is a major contributor to centromeric motion. We also demonstrate that centromeric movement sometimes alters nuclear morphology into the ‘teardrop’-shape. This suggests that cytoplasmic microtubules pull centromeres along the nuclear membrane, and this pulling force is sufficient to deform the nuclear shape when centromeres are pulled beyond the nuclear volume. It has also been shown that an overproduction of the separase C-terminal fragment causes a nuclear morphological defect (Nakamura et al., 2002). These studies support the idea that the nuclear membrane is deformable. By contrast, interphase microtubules also function in the proper positioning of the nucleus in cytoplasm, revealing some stiffness of the nuclear membrane (Sawin and Tran, 2006; Tran et al., 2001). Therefore, the rigidity or flexibility of the nuclear membrane is tuned to accommodate centromeric motion, nuclear deformation, and central positioning of the nucleus in cytoplasm.
Hundreds of Pol III genes and retrotransposons, which are distributed across the fission yeast genome, mediate global genome organization by associating with centromeres (Bowen et al., 2003; Iwasaki et al., 2010; Tanaka et al., 2012). Our results indicate that centromeres and their associating genomic loci move together. Therefore, centromeric motion can facilitate the mobility of other genomic regions. In other words, once centromeric motion is inhibited, it would be expected that the mobility of many other genomic loci would reduce. Indeed, we find that CBZ treatment prevents centromeric motion and impedes movement of the other genomic loci (Fig. 2). We also find that disruption of the genomic associations between centromeres and the genomic loci by the mutation of condensin reduces the moving volumes of the loci. Therefore, we propose that centromeres serve as ‘genome mobility elements’ by connecting highly mobile centromeres to dispersed genomic regions. Our current model is that centromeric motion, driven by microtubule polymerization, can promote the mobility of other genomic regions through condensin-mediated associations (Fig. 6C). Given that microtubule polymerization is coordinated with cell growth, cell cycle and the stress response (Chang and Martin, 2009; Robertson and Hagan, 2008), this study might have identified a new mechanism by which cytoplasmic signals reflecting important cellular activities are transmitted to the nucleus and moderate the mobility of interphase genomic regions through centromeric motion coupled to microtubule polymerization. For instance, genetic materials maintain mobility while cells grow and microtubules are actively polymerized. The high mobility of genetic materials potentially facilitates associations between the transcriptional regulatory elements and their target genes, as well as DNA interactions related to recombination. Therefore, our studies shed light on condensin-mediated coordination between cellular/cytoplasmic activities and genome-wide mobility that can potentially contribute to various nuclear processes.
Materials and Methods
Strain construction and culture conditions
Strains carrying the insertions of lacO repeats were generated using a cloning-free, PCR-based method (Rohner et al., 2008). The lacO repeats were inserted into the two Pol III gene loci (c417 and c10H11), the LTR retrotransposons locus (c947), the rDNA locus, the tel2R telomere locus, and the control locus (c887). Strains carrying lacO repeats at the different genomic loci were subjected to genetic crosses to introduce LacI-GFP expressed from the his7+ locus under control of the dis1+ promoter. Nup61 and Rpa49 proteins were tagged with mCherry at the C termini of their proteins using a PCR-based module method (Bähler et al., 1998). Other strain constructions were performed using conventional genetic crosses. Yeast cells were cultured in Edinburgh Minimal Medium (EMM) at 26°C, unless otherwise noted.
Live-cell imaging
The genomic loci were visualized by the insertions of lacO and tetO repeats bound by LacI–GFP and TetR–tdTomato. The nuclear membrane, nucleolus, microtubules and centromeres were visualized by mCherry fused to Nup61 (NPC), Rpa49 (RNA polymerase III), Atb2 (α-tubulin) and Mis12 (kinetochore protein), respectively. Exponentially growing cells in liquid EMM were mounted on an agar pad that contained EMM plus 2% electrophoresis-grade agarose (Tran et al., 2004). Cells were maintained at 26°C in an environmental chamber during image acquisition. For the CBZ treatment, cells were cultured for 15 minutes in liquid EMM containing 50 µg/ml CBZ and applied to the agar pad with the same concentration of CBZ. Images were captured using a Leica TCS SP5 II AOBS spectral laser scanning confocal microscope (Leica Microsystems, Inc.) mounted on a DMI6000 inverted microscope. The system includes a 405 nm diode, multi-line Argon, and 561 nm DPSS lasers with three standard PMTs and two Leica HyD detectors. Images were captured at 1400 Hz with a 63× HCX PL APO CS oil immersion objective (NA = 1.4). Multi-channel image stacks were acquired at each time point. Time-lapse images were typically taken continuously, with a stack interval of 1.5 or 3.0 seconds, for a total of 5 minutes. Construction of 3D images and analyses on 3D data were carried out using Volocity 6.1.1 software (PerkinElmer).
Estimation of the moving volume of the genomic locus
The genomic locus (lacO, green) was co-visualized with the nuclear landmarks such as NPC (Nup61–mCherry) and the nucleolus (Rpa49–mCherry). Time-lapse images were captured at 3.0-second intervals for 5 minutes. Using the NPC foci, the center of the nucleus was estimated and the position of the genomic locus was normalized according to the nuclear center. The tracking line of the genomic locus was built by spline interpolation, resulting in a 3D tracking line at 0.3-second intervals. The nucleus, excluding the nucleolus, was divided into ∼850 cubes (0.2 µm of each side, 0.008 µm3 in volume for each cube) with a lattice arrangement. The tracking line of the genomic locus was used to assign the time-lapse occupancy of the genomic locus to the cubes. The moving volume (MV) was defined as the accumulated number of cubes occupied by the genomic locus {i.e. the number of cubes occupied by the genomic locus×volume of a cube (0.008 µm3); the percentage the nucleus is represented by: MV/[volume of the nucleus (6.8 µm3)×100]}.
Supplementary Material
Acknowledgments
We would like to thank Phong Tran for guidance on live-cell imaging; Yoshinori Watanabe and the Yeast Genetic Resource Center (YGRC) for fission yeast strains; Mitsuhiro Yanagida and Susan Gasser for plasmids; the Sanger Institute for cosmid clones; the Institut Pasteur for software; and the Wistar Institute Imaging Facility for microscopic analysis. We also thank Louise and Michael Showe for critically reading the manuscript and Mea Fuller for editorial assistance.
Footnotes
Author contributions
K.K. and K.N. designed the experiments, interpreted the data, and wrote the manuscript. K.K., O.I., C.J.C, J.R.C., and J.E.H. performed the experiments. H.T. designed the custom software and analyzed the data.
Funding
This work was supported by the National Institutes of Health (NIH) [grant number CA010815]; the NIH Director's New Innovator Award Program [number DP2-OD004348]; the G. Harold and Leila Y. Mathers Foundation; the V Foundation; the Edward Mallinckrodt Jr. Foundation; and the Wistar Pilot Project Funds (all to K.N.). Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.133678/-/DC1
References
- Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- Baker M. (2011). Genomics: Genomes in three dimensions. Nature 470, 289–294 10.1038/470289a [DOI] [PubMed] [Google Scholar]
- Berger A. B., Cabal G. G., Fabre E., Duong T., Buc H., Nehrbass U., Olivo-Marin J. C., Gadal O., Zimmer C. (2008). High-resolution statistical mapping reveals gene territories in live yeast. Nat. Methods 5, 1031–1037 10.1038/nmeth.1266 [DOI] [PubMed] [Google Scholar]
- Bowen N. J., Jordan I. K., Epstein J. A., Wood V., Levin H. L. (2003). Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 13, 1984–1997 10.1101/gr.1191603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Laroche T., van Houwe G., Blaszczyk M., Gasser S. M. (2005). Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 168, 375–387 10.1083/jcb.200409091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Martin S. G. (2009). Shaping fission yeast with microtubules. Cold Spring Harb. Perspect. Biol. 1, a001347 10.1101/cshperspect.a001347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Haraguchi T., Hiraoka Y. (2010). Nuclear envelope attachment is not necessary for telomere function in fission yeast. Nucleus 1, 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb J. R., Boyle S., Perry P., Bickmore W. A. (2002). Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12, 439–445 10.1016/S0960-9822(02)00695-4 [DOI] [PubMed] [Google Scholar]
- Cook P. R. (1999). The organization of replication and transcription. Science 284, 1790–1795 10.1126/science.284.5421.1790 [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer M. (2010). Chromosome territories. Cold Spring Harb. Perspect. Biol. 2, a003889 10.1101/cshperspect.a003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Lee J., Wang H., Miller J., Reik A., Gregory P. D., Dean A., Blobel G. A. (2012). Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 10.1016/j.cell.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Mitchison T. J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- Ding R., West R. R., Morphew D. M., Oakley B. R., McIntosh J. R. (1997). The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 8, 1461–1479 10.1091/mbc.8.8.1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y. J., Lee C., Shendure J., Fields S., Blau C. A., Noble W. S. (2010). A three-dimensional model of the yeast genome. Nature 465, 363–367 10.1038/nature08973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Hagan I., Uzawa S., Yanagida M. (1993). Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121, 961–976 10.1083/jcb.121.5.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S., Williams L. M., Ryan N. K., Cobb B. S., Sexton T., Fraser P., Fisher A. G., Merkenschlager M. (2009). Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K. A., Meyer B. J. (2003). Condensin and cohesin: more than chromosome compactor and glue. Nat. Rev. Genet. 4, 520–534 10.1038/nrg1110 [DOI] [PubMed] [Google Scholar]
- Heun P., Laroche T., Shimada K., Furrer P., Gasser S. M. (2001). Chromosome dynamics in the yeast interphase nucleus. Science 294, 2181–2186 10.1126/science.1065366 [DOI] [PubMed] [Google Scholar]
- Hirano T. (2000). Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 69, 115–144 10.1146/annurev.biochem.69.1.115 [DOI] [PubMed] [Google Scholar]
- Hirano T. (2006). At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7, 311–322 10.1038/nrm1909 [DOI] [PubMed] [Google Scholar]
- Hou C., Dale R., Dean A. (2010). Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. USA 107, 3651–3656 10.1073/pnas.0912087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C., Li L., Qin Z. S., Corces V. G. (2012a). Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell 48, 471–484 10.1016/j.molcel.2012.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H., Zhou Z., Wang Y., Wang J., Kallgren S. P., Kurchuk T., Miller E. A., Chang F., Jia S. (2012b). Csi1 links centromeres to the nuclear envelope for centromere clustering. J. Cell Biol. 199, 735–744 10.1083/jcb.201208001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki O., Tanaka A., Tanizawa H., Grewal S. I., Noma K. (2010). Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell 21, 254–265 10.1091/mbc.E09-09-0790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., van Berkum N. L., Ebmeier C. C., Goossens J., Rahl P. B., Levine S. S. et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. C., Drivas T. G., Blobel G. (2008). A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell 134, 427–438 10.1016/j.cell.2008.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniola B., O'Toole E., McIntosh J. R., Mellone B., Allshire R., Mengarelli S., Hultenby K., Ekwall K. (2001). The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell 12, 2767–2775 10.1091/mbc.12.9.2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Strunnikov A. (1996). Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol. 12, 305–333 10.1146/annurev.cellbio.12.1.305 [DOI] [PubMed] [Google Scholar]
- Labrador M., Corces V. G. (2002). Setting the boundaries of chromatin domains and nuclear organization. Cell 111, 151–154 10.1016/S0092-8674(02)01004-8 [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Käs E., Poljak L., Adachi Y. (1992). Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr. Opin. Genet. Dev. 2, 275–285 10.1016/S0959-437X(05)80285-0 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O. et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T., Katsani K. R., Verrijzer C. P. (2002). GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 21, 1775–1781 10.1093/emboj/21.7.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F., Straight A., Marko J. F., Swedlow J., Dernburg A., Belmont A., Murray A. W., Agard D. A., Sedat J. W. (1997). Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7, 930–939 10.1016/S0960-9822(06)00412-X [DOI] [PubMed] [Google Scholar]
- Mishiro T., Ishihara K., Hino S., Tsutsumi S., Aburatani H., Shirahige K., Kinoshita Y., Nakao M. (2009). Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 28, 1234–1245 10.1038/emboj.2009.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. (2007). Beyond the sequence: cellular organization of genome function. Cell 128, 787–800 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- Mizukami T., Chang W. I., Garkavtsev I., Kaplan N., Lombardi D., Matsumoto T., Niwa O., Kounosu A., Yanagida M., Marr T. G. et al. (1993). A 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell 73, 121–132 10.1016/0092-8674(93)90165-M [DOI] [PubMed] [Google Scholar]
- Müeller-Storm H. P., Sogo J. M., Schaffner W. (1989). An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell 58, 767–777 10.1016/0092-8674(89)90110-4 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Nagao K., Nakaseko Y., Yanagida M. (2002). Cut1/separase C-terminus affects spindle pole body positioning in interphase of fission yeast: pointed nuclear formation. Genes Cells 7, 1113–1124 10.1046/j.1365-2443.2002.00586.x [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C. H. (2005). The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74, 595–648 10.1146/annurev.biochem.74.082803.133219 [DOI] [PubMed] [Google Scholar]
- Nativio R., Wendt K. S., Ito Y., Huddleston J. E., Uribe-Lewis S., Woodfine K., Krueger C., Reik W., Peters J. M., Murrell A. (2009). Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 5, e1000739 10.1371/journal.pgen.1000739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K., Allis C. D., Grewal S. I. (2001). Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293, 1150–1155 10.1126/science.1064150 [DOI] [PubMed] [Google Scholar]
- Robertson A. M., Hagan I. M. (2008). Stress-regulated kinase pathways in the recovery of tip growth and microtubule dynamics following osmotic stress in S. pombe. J. Cell Sci. 121, 4055–4068 10.1242/jcs.034488 [DOI] [PubMed] [Google Scholar]
- Rohner S., Gasser S. M., Meister P. (2008). Modules for cloning-free chromatin tagging in Saccharomyces cerevisae. Yeast 25, 235–239 10.1002/yea.1580 [DOI] [PubMed] [Google Scholar]
- Sadaie M., Naito T., Ishikawa F. (2003). Stable inheritance of telomere chromatin structure and function in the absence of telomeric repeats. Genes Dev. 17, 2271–2282 10.1101/gad.1112103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K. E., Tran P. T. (2006). Cytoplasmic microtubule organization in fission yeast. Yeast 23, 1001–1014 10.1002/yea.1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T., Bantignies F., Cavalli G. (2009). Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin. Cell Dev. Biol. 20, 849–855 10.1016/j.semcdb.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G. (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Spector D. L. (2003). The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72, 573–608 10.1146/annurev.biochem.72.121801.161724 [DOI] [PubMed] [Google Scholar]
- Tanaka A., Tanizawa H., Sriswasdi S., Iwasaki O., Chatterjee A. G., Speicher D. W., Levin H. L., Noguchi E., Noma K. (2012). Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol. Cell 48, 532–546 10.1016/j.molcel.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H., Noma K. (2012). Unravelling global genome organization by 3C-seq. Semin. Cell Dev. Biol. 23, 213–221 10.1016/j.semcdb.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H., Iwasaki O., Tanaka A., Capizzi J. R., Wickramasinghe P., Lee M., Fu Z., Noma K. (2010). Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 38, 8164–8177 10.1093/nar/gkq955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. T., Marsh L., Doye V., Inoué S., Chang F. (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153, 397–411 10.1083/jcb.153.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. T., Paoletti A., Chang F. (2004). Imaging green fluorescent protein fusions in living fission yeast cells. Methods 33, 220–225 10.1016/j.ymeth.2003.11.017 [DOI] [PubMed] [Google Scholar]
- Uzawa S., Yanagida M. (1992). Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci. 101, 267–275 [DOI] [PubMed] [Google Scholar]
- Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., Martienssen R. A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- Williams A., Spilianakis C. G., Flavell R. A. (2010). Interchromosomal association and gene regulation in trans. Trends Genet. 26, 188–197 10.1016/j.tig.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M. (1998). Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol. 8, 144–149 10.1016/S0962-8924(98)01236-7 [DOI] [PubMed] [Google Scholar]
- Zhao R., Bodnar M. S., Spector D. L. (2009). Nuclear neighborhoods and gene expression. Curr. Opin. Genet. Dev. 19, 172–179 10.1016/j.gde.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.