SUMMARY
Pseudomonas aeruginosa produces several phenazines including the recently described 5-methyl-phenazine-1-carboxylic acid (5MPCA), which exhibits a novel antibiotic activity towards pathogenic fungi such as Candida albicans. Here we characterize the unique antifungal mechanisms of 5MPCA using its analog phenazine methosulfate (PMS). Like 5MPCA, PMS induced fungal red pigmentation and killing. Mass spectrometry analyses demonstrated that PMS can be covalently modified by amino acids, a process that yields red derivatives. Furthermore, soluble proteins from C. albicans grown with either PMS or P. aeruginosa were also red and demonstrated absorbance and fluorescence spectra similar to that of PMS covalently linked to either amino acids or proteins in vitro, suggesting that 5MPCA modification by protein amine groups occurs in vivo. The red-pigmented C. albicans soluble proteins were reduced by NADH and spontaneously oxidized by oxygen, a reaction that likely generates reactive oxygen species (ROS). Additional evidence indicates that ROS generation precedes 5MPCA-induced fungal death. Reducing conditions greatly enhanced PMS fungal uptake and killing. Since 5MPCA demonstrated to be more toxic than other phenazines that are not modified, such as pyocyanin, we propose that the covalent binding of 5MPCA promotes its accumulation in target cells and contributes to its antifungal activity in mixed-species biofilms.
INTRODUCTION
A significant proportion of human microbial infections are biofilm-associated (Douglas, 2003; Lynch and Robertson, 2008), and many of these biofilms are formed by multiple microbial species. It has been demonstrated that in mixed-species biofilms containing bacteria and fungi, a range of different interactions can occur, including increased resistance to antimicrobials, enhanced surface colonization and interspecies antagonism (Morales and Hogan, 2010; Peleg et al., 2010; Shirtliff et al., 2009). Interestingly, the nature of these interspecies interactions can determine the fate of the microbial populations within these biofilms (Lynch and Robertson, 2008; Shirtliff et al., 2009) and thus probably the outcome of polymicrobial diseases. Some interspecies interactions may only occur in a biofilm setting due to the lability of secreted compounds or the need for high local concentrations of microbial products for activity. Analysis of the antagonistic relationship between the human opportunistic pathogens Pseudomonas aeruginosa and Candida albicans (Brand et al., 2008; Cugini et al., 2007; Gibson et al., 2009; Hogan and Kolter, 2002; Kerr, 1994), two species commonly found together in mixed species biofilm-related infections (Bauernfeind et al., 1987; Chotirmall et al., 2010; Gupta et al., 2005; Hughes and Kim, 1973), has provided insight into the many different types of interactions occurring between species coexisting in close proximity. These interactions include killing by secreted factors and signaling events that modulate virulence properties (Brand et al., 2008; Cugini et al., 2007; Gibson et al., 2009; Hogan and Kolter, 2002; Hogan et al., 2004; Kerr, 1994).
Bacterially-produced phenazines have been known for their toxicity towards other bacteria (Baron and Rowe, 1981; Hassan and Fridovich, 1980) and eukaryotes ranging from nematodes to humans (Kerr et al., 1999; Lau et al., 2004a; Mahajan-Miklos et al., 1999; Mavrodi et al., 2006; Ran et al., 2003; Thomashow et al., 1990). Phenazine production by Pseudomonads has been demonstrated to be particularly important in interactions with fungi (Anjaiah et al., 1998; Bolwerk et al., 2003; Thomashow and Weller, 1988). For example, Pseudomonas chlororaphis colonizes and forms biofilms on the phytopathogen Fusarium oxysporium, and contributes to fungal biocontrol by secreting phenazine-1-carboxamide (Bolwerk et al., 2003). In addition, P. aeruginosa secretes two phenazines, phenazine-1-carboxylate (PCA) and pyocyanin (PYO) (Fig. 1A), which also have been shown to antagonize fungal growth (Kerr et al., 1999; Thomashow et al., 1990). By using P. aeruginosa TnMariner mutants in the phenazine pathway, we have recently demonstrated that when the bacterium grows in close proximity to C. albicans, 5-methyl-phenazinium-1-carboxylate (5MPCA; see Fig. 1A for structure) induces fungal death more efficiently than the other characterized phenazines produced by P. aeruginosa (Gibson et al., 2009). Notably, this death is associated with the development of a red pigmentation and intracellular fluorescence of C. albicans cells (Gibson et al., 2009). While 5MPCA has been previously described as a putative precursor of PYO (Fig. 1A), a biological role for this phenazine has been poorly described since it has not been found to accumulate in the medium of P. aeruginosa mono-cultures (Byng et al., 1979).
Fig. 1.
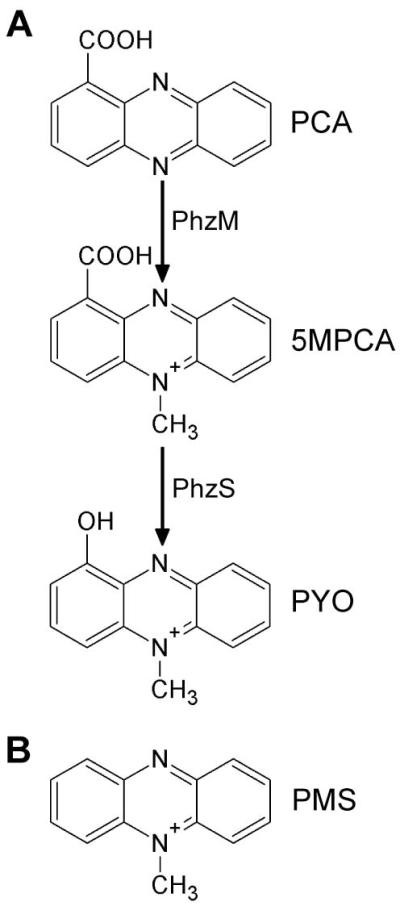
Phenazines and phenazimiums. A. Biosynthetic pathway for P. aeruginosa PYO and its immediate precursors. B. Chemical structure of PMS, a 5MPCA analog (only the 5-methyl-phenazinium ion is shown).
Here, we describe the elements that make 5MPCA a potent antifungal toxin and the properties that make this methylphenazinium particularly effective in bacterial-fungal biofilms. We found that phenazine methosulfate (PMS), a commercially available and well characterized methylphenazinium (Fig. 1B), could be used as a powerful surrogate for P. aeruginosa-produced 5MPCA, which is relatively unstable under physiological conditions (Hansford et al., 1972). We show evidence for 5MPCA and PMS uptake by C. albicans over time, and that the metabolic activity and reducing conditions within colony biofilms facilitate this uptake. PMS and 5MPCA were concentrated within the fungal cell and the accumulation of this phenazine is due to its covalent binding to cellular amines within the fungus, to yield red species. Our results show that this covalent modification does not alter methylphenaziniums redox activity, a chemical property that allows methylphenaziniums or their red derivatives to generate reactive oxygen species (ROS). In fact, we detected intracellular ROS at early time points upon exposure to methylphenaziniums, and mutants sensitive to oxidative stress were more susceptible to killing by these compounds. We found that if this chemical modification of PMS occurs outside the fungal cell, the PMS red derivative neither induced the formation of the cell-bound red pigment nor was toxic to C. albicans, although its redox activity was retained. We propose that the permanent localization of these red methylphenazinium derivatives within the cell allows for the constant generation of toxic levels of ROS that finally help to kill the fungus. Finally, we show that PYO, the well-characterized P. aeruginosa toxin, was neither modified nor accumulated within the fungal cell and that it was less toxic than 5MPCA. Together our data indicate that the unusual chemical reactivity of 5MPCA contributes to its concentration within fungal cells, where it retains toxic activity. 5MPCA therefore represents a particularly useful antifungal within mixed-species biofilms where producer and target cells are in close proximity.
RESULTS
PMS and 5MPCA induce red pigmentation and death of C. albicans
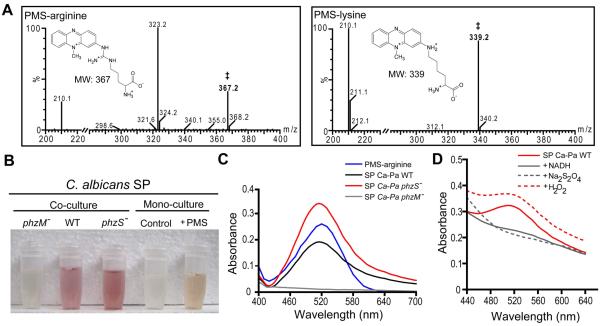
When C. albicans lawns are spot inoculated with P. aeruginosa WT, such as strain PA14, a red, phenazine-derived pigmentation is detected (Fig, 2A). Previous studies indicate that the production of 5MPCA by P. aeruginosa is both necessary and sufficient for C. albicans red coloration and death (Gibson et al., 2009). Since the availability of 5MPCA is limited due to the fact that it has not been found to accumulate in P. aeruginosa monoculture supernatants (Byng et al., 1979), and its chemical synthesis is difficult (Hansford et al., 1972), we tested PMS (Fig. 1B) as a structural analog that would allow us to study the mechanisms of 5MPCA activity towards C. albicans. Similar to what was observed in C. albicans–P. aeruginosa co-cultures (Fig. 2A and Gibson et al., 2009), C. albicans grown on agar containing PMS, which is yellow in these medium conditions, developed a red coloration that increased over time (Fig. 2B). In addition, C. albicans cells within PMS-grown colony biofilms were killed progressively, while controls cultured without PMS retained full viability (Fig. 2C).
Fig. 2.
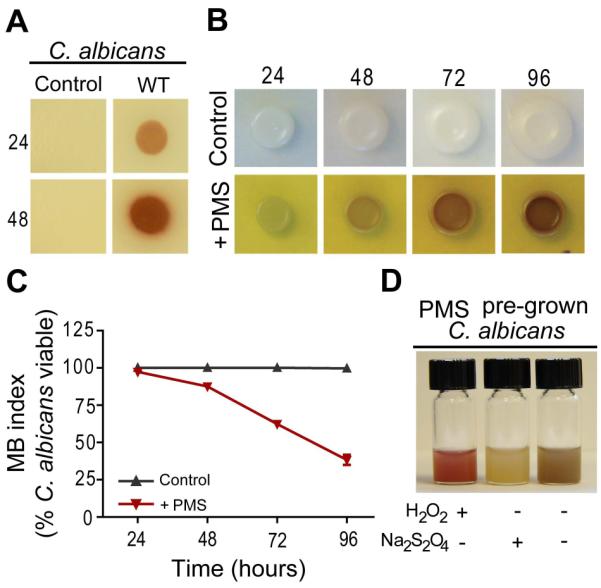
PMS and 5MPCA effects on C. albicans. A. Plate co-cultures of C. albicans lawns inoculated with P. aeruginosa PA14 (WT) or without P. aeruginosa (control) after 24 and 48 hours of incubation. B. C. albicans colonies grown at 30°C on YNBA medium with or without (control) 1 mM PMS. The PMS-supplemented medium remained yellow throughout the experiment. C. C. albicans viability was determined by methylene blue staining of samples collected at the indicated time points after growth on either control or PMS medium (n=2). D. C. albicans grown for 72 hours on 1mM PMS-containing medium was suspended in saline solution and then photographed after the addition of few crystals of Na2S2O4, 10 μl of 30% H2O2 or without treatment. Data are representative of three independent experiments.
Phenazines are well known for their different spectral properties in their oxidized and reduced states (Price-Whelan et al., 2007). Consistent with the hypothesis that the red pigmentation of C. albicans is directly derived from the redox active molecule, PMS, a suspension of cells from colonies grown on PMS-containing medium became colorless upon the addition of the strong reducing agent, sodium hydrosulfite (Na2S2O4), and restored their redness after adding the oxidizing agent, hydrogen peroxide (H2O2) (Fig. 2D). The effects of the reduction or oxidation of the cell suspension were repeatedly reversible (data not shown). Notably, these redox changes exhibited by PMS-grown C. albicans were identical to those observed for the red fungal cells from co-cultures with P. aeruginosa (Gibson et al., 2009). Taken together, these data indicate that PMS closely recapitulates the effects that 5MPCA induces in C. albicans.
Methylphenaziniums form redox active, red derivatives via covalent binding to amino acids
Since the red coloration of fungal cells induced by 5MPCA and PMS (Fig. 2A and B) remained associated with whole cells or cell lysates despite extraction with aqueous or organic solvents (Gibson et al., 2009 and data not shown), we hypothesized that these methylphenaziniums are modified within cells to form red derivatives bound to cellular constituents. Based on the early observation that PMS forms a red derivative with an amine group fused to carbon 2, namely amino-PMS (2-amino-10-methyl phenazinium methyl sulfate), upon treatment with strong ammonia in air (Kehrmann and Havas, 1913), we postulated that amino-containing compounds within the fungal cells could participate in a similar type of reaction. To test this hypothesis, we first determined if PMS could be modified in vitro by amino acids. Upon mixing the yellow PMS with excess arginine free base under alkaline conditions (pH 10.0), a strong wine red color developed over a 5 minute period of vigorous mixing in air. When we used physiological conditions (pH 7.5) to perform the reaction with arginine, PMS required reduction with Na2S2O4 prior to amino acid addition, and red color development proceeded slowly over 24 hours. Analysis of the red-colored product by mass spectrometry identified a molecular mass ion at 367.2, which corresponds to the mass of the covalent adduct of PMS with arginine (Fig. 3A). The mass of this adduct is consistent with the attack of the amine group of arginine on the phenazinium ring (Fig. 3A inset). Moreover, the red product was water soluble and had other spectral properties similar to those previously described for amino-PMS (Kehrmann and Havas, 1913), leading us to predict that this covalent modification is most likely occurring at the C-2 position as in amino-PMS (Fig. S1A). Similarly, other amino acids also covalently linked to PMS, giving rise to PMS-lysine (MW 339.2; Fig. 3A), PMS-alanine (MW 282.1) and PMS-serine (MW 298.1). Only unreacted PMS (MW 195) was detected in reactions of PMS with amino acids performed at physiological pH, but lacking Na2S2O4 (See Supporting Information for experimental details). To test if this chemical modification could occur in presence of more complex amino-containing compounds, such as proteins, we mixed reduced PMS with an excess of bovine serum albumin (BSA). A similar red product was formed which exhibited the same visible absorption spectrum as described below for the PMS-arginine derivative (data not shown) (see Supporting Information for experimental details).
Fig. 3.
Methylphenazinium modification by amino acids and cellular proteins. A. LC-MS/ES ionization analysis of the in vitro-produced red products from PMS reaction with amino acids. (A-insets) Proposed chemical structures of the corresponding (‡) adduct of PMS-arginine or PMS-lysine. B. Soluble proteins (SP) extracted from C. albicans cultured on medium with or without (control) 1 mM PMS and with P. aeruginosa PA14 (WT), phzS::TnM (phzS−) phzM::TnM (phzM−) for 96 and 48 hours, respectively. C. Absorption spectra of the in vitro-produced red compound formed by reacting PMS with arginine (PMS-arginine), compared with representative absorption spectra of the SP extracted from C. albicans co-cultured with P. aeruginosa WT (Ca-Pa WT), phzS::TnM (Ca-Pa phzS−) or phzM::TnM (Ca-Pa phzM−). To minimize artifacts caused by turbidity, these latter spectra were obtained from difference of the reduced (by adding 80 mM of Na2S2O4) minus the oxidized spectra of each extract. D. Redox changes of absorption spectra of the red SP extracted from C. albicans cocultured with P. aeruginosa WT in its oxidized form (Ca-Pa WT) and after reduction with 2 mM NADH or 80 mM Na2S2O4, and followed by reoxidation with 280 mM H2O2.
These chemical findings suggested a mechanism whereby PMS and 5MPCA could be modified within the fungus by cellular amines associated with macromolecules. To test if in vivo modification of the cellular amines by methylphenaziniums would yield red proteins within the fungus, we recovered the soluble fungal proteins from C. albicans grown with or without PMS, or co-cultured with three P. aeruginosa strains: WT, phzS::TnM (a 5MPCA overproducer) or phzM::TnM (5MPCA-deficient) (Fig. 1A). Soluble proteins from fungal cells exposed to 5MPCA-producing P. aeruginosa or PMS, but not control cultures, showed a red coloration (Fig. 3B) that remained pellet-associated after precipitation with trichloroacetic acid (TCA) (data not shown). Similarly to the PMS-arginine derivative and PMS-modified BSA, red soluble proteins from cells exposed to 5MPCA during co-culture conditions exhibited an identical intense absorption at 520 nm in the visible spectrum (Fig. 3C), suggesting that modification of cellular amines by 5MPCA occurs in vivo. The methylphenazinium-modification of proteins is likely occurring via a covalent linkage as prolonged treatment of C. albicans red soluble proteins with strong denaturants (8 M urea or 6 M guanidinium chloride) did not dissociate the red pigmentation of the protein fraction even after TCA precipitation (Macheroux et al., 1998). Likewise, PMS-modified BSA retained its red pigmentation upon treatment with urea (data not shown). Together, these data provide evidence for an intracellular covalent modification of proteins by methylphenaziniums that yield red derivatives within the fungal cells.
Whole C. albicans cells from co-cultures with P. aeruginosa (Gibson et al., 2009) or from colonies grown with PMS showed that the red pigmentation was redox-dependent (Fig. 2D). Likewise, both the PMS-arginine and red soluble proteins exhibited redox activity. PMS-arginine was reversibly reduced to a colorless compound by Na2S2O4 and re-oxidized upon aeration (Fig. S2A), and C. albicans proteins from co-cultures with P. aeruginosa WT (Fig. 3D) or phzS::TnM (Fig. S2B) strains also demonstrated changes in the absorbance profile upon reduction with Na2S2O4 and NADH, or oxidation with H2O2. Together, these data demonstrate that the modification of the methylphenazinium contributing to its localization does not affect its redox activity.
Intracellular modification of methylphenaziniums is required for fungal killing
5MPCA modification appeared to be more efficient within the fungal cells, as red derivatives were not detected in the extracellular milieu of P. aeruginosa–C. albicans co-cultutres (data not shown). We next determined whether methylphenazinium modification needed to occur within the cell, after fungal uptake, to efficiently induce red pigmentation, toxicity and fungal killing. To assess this, the in vitro synthesized red PMS-arginine derivative was added to C. albicans cultures and both pigmentation and cell viability were evaluated. Strikingly, while fungal cells grown in agar containing unmodified PMS developed red coloration and died (Fig 2B and C), C. albicans incubated with the in vitro-generated PMS-arginine compound did not develop red pigmentation and did not show a decrease in viability (Fig. S3A). C. albicans cells suspended in buffer and treated with the PMS-arginine derivative in either its oxidized form or after reduction with Na2S2O4 also remained viable during the course of the experiment and did not develop red pigmentation (Fig. S3B and data not shown). These data indicate that methylphenazinium modification must occur within the fungal cells in order to correlate with killing.
Pyocyanin does not react with amino acids and is not modified within fungal cells
While the biological activity of 5MPCA has been poorly described, its derivative, PYO is a well-known toxin and antibiotic (Lau et al., 2004b; Moustafa Hassan and Fridovich, 1980; Muller, 2009). We have previously demonstrated that a P. aeruginosa phzS::TnM mutant, which lacks the ability to produce PYO but still makes 5MPCA (Fig. 1A), kills C. albicans more efficiently than the WT strain which secretes both 5MPCA and PYO (Gibson et al., 2009). To compare the effects of PYO to the 5MPCA-analog PMS on the fungus, we assessed the viability of C. albicans grown on medium containing either of these compounds. While PMS-grown fungal cells developed a stably associated red color (Fig. 2B), cells grown on PYO did not show red pigmentation even after extended incubation times (data not shown). Furthermore, the fraction of inviable cells in the PYO-exposed C. albicans cultures (40 ± 4.1 %) was significantly lower than that of PMS-treated cells (72 ± 4.1 %) after 72 hours, indicating that under these colony biofilm conditions, PMS exhibits higher toxicity towards the fungus than PYO (Fig. S4A). Importantly, mass spectrometry experiments determined that neither oxidized nor reduced PYO were modified in vitro by arginine (Fig. S4B), lysine, alanine or serine as only the unreacted pyocyanin mass (MW 211.1) was detected. These data underscore the unusual properties of 5MPCA that enable its localization and concentration within the target fungal cells.
ROS generated by methylphenaziniums or their derivatives contribute to C. albicans killing
Other studies have shown that PMS (Maridonneau-Parini et al., 1986; Nishikimi et al., 1972; Ottaway, 1966) and other phenazines such as PYO (Hassan and Fridovich, 1980; Muller, 2002), in their reduced forms can rapidly react with oxygen to produce ROS, such as superoxide and, by dismutation of superoxide, H2O2. Since the PMS-arginine derivative synthesized in vitro (Fig. S2A), the PMS-modified BSA (data not shown) and the red-pigmented soluble proteins extracted from red cells of C. albicans (Fig. 3D and S2B) demonstrated redox properties under oxidizing and reducing conditions, we tested if fungal killing by methylphenaziniums or their derivatives was related to ROS generation. To measure intracellular accumulation of ROS at early time points, C. albicans cells cultured for 18 hours in presence or absence of PMS were treated with the ROS-detecting probe, dichlorodihydrofluorescein diacetate (DCFH-DA), and the green fluorescence indicative of intracellular ROS was measured using flow cytometry. We found that the fungal cells exposed to PMS exhibited higher fluorescence intensity than untreated control cells. A positive control of cells cultured with a sublethal concentration of H2O2 also showed increase in fluorescence (Fig. 4A). Thus, these results demonstrate that methylphenaziniums or their derivatives generate ROS in vivo. It has been reported that when C. albicans is exposed to ROS, it induces the expression of enzymes (catalase) or scavengers (glutaredoxin and thioredoxin systems) that reduce the damage caused by oxidation (Barelle et al., 2004; Enjalbert et al., 2007). To assess if there is induction of these ROS-responsive genes after exposure to PMS-generated ROS, we measured the fluorescence of C. albicans CAI-4 strains harboring the promoterless green fluorescent protein (GFP) fused to either CAT1 (catalase), TRX1 or TTR1 (thioredoxin system) promoter regions (Enjalbert et al., 2006) at early time points using flow cytometry. We found that for all ROS responsive promoters evaluated, PMS exposure induced a marked increased in the levels of green fluorescence in comparison with control untreated cells (Fig. S5A). Although a slight shift in the background fluorescence of the pGFP control strain harboring GFP under control of the basal ADH1 promoter (Barelle et al., 2004) was observed upon treatment with PMS, the fold change in the mean fluorescence intensity of PMS treated vs. untreated cells was much higher for reporter strains (CAT1, 2.83; TTR1, 4.0 and TRX1, 4.24) than for the control pGFP strain (1.86) (Fig. S5A). As described in more detail below, we speculate that the background fluorescence is due to PMS.
Fig. 4.
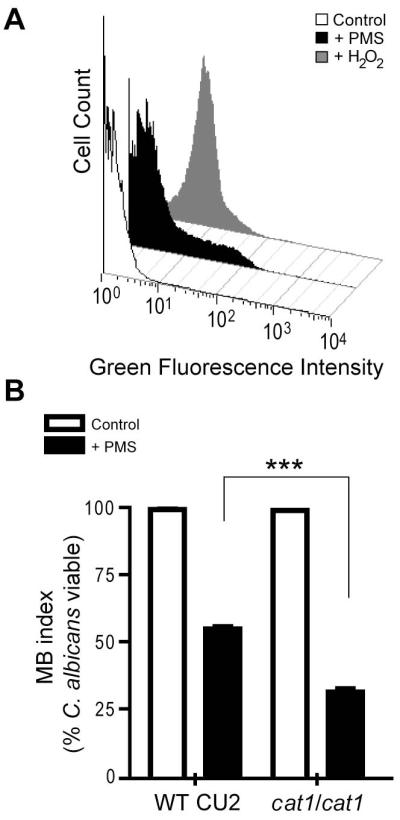
Oxidative stress and cell death in C. albicans. A. Effect of PMS on ROS generation in C. albicans cells determined by DCFH-DA staining. Cells were grown on YNBA with or without (control) 1 mM PMS, and with 10 mM H2O2 at 30°C. Fungal cells were harvested after 18 hours growth, washed and incubated with DCFH-DA for 30 minutes. Green fluorescence was evaluated by flow cytometry. B. C. albicans CAT1/CAT1 (WT CU2) or cat1/cat1 were grown on YNBA medium alone (control) and with PMS for 72 hours. Fungal survival determined by MB staining (n=3). Data are representative of at least two independent experiments ***P<0.01.
To determine if the pigments derived from PMS or 5MPCA correlated with fungal cell death via ROS generation, we tested if a catalase-deficient mutant (cat1/cat1) that is unable to efficiently detoxify H2O2, is more sensitive to 5MPCA-PMS-mediated toxicity than its parental strain. Thus, C. albicans CU2 (WT) and its cat1/cat1 mutant derivative were cultured with or without PMS. Notably, after 72 hours of growth on PMS-containing medium, C. albicans cat1/cat1 mutant developed a more intense red pigmentation in comparison to its parental strain (data not shown). Furthermore, through methylene blue viability analysis (Fig. 4B) and clonogenecity assays (Fig. S5B), we demonstrated that C. albicans CU2 was approximately 2-fold more resistant to PMS than the mutant lacking CAT1. No significant variation on proportion of dead cells was found in either of the strains cultured without PMS (Fig. 4B and S5B). Interestingly, addition of the antioxidant rutin, a flavonoid that protects against lipid peroxidation and inhibits the generation H2O2 (Shcherbachenko et al., 2007), caused a small but reproducible protection of both WT and cat1/cat1 cells against PMS-mediated red killing (data not shown). Together, these results indicate that PMS or its red derivatives induce the generation of oxygen radicals, such as H2O2, and that this process is one of the mechanisms that contributes to fungal killing.
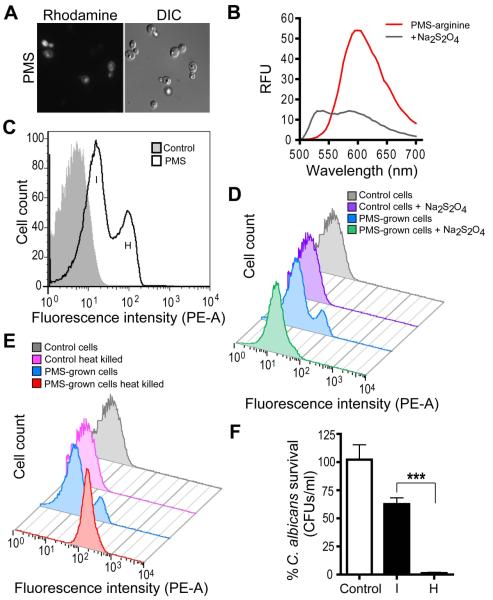
Modified methylphenaziniums exhibit fluorescence that correlates with 5MPCA-PMS uptake, modification and fungal killing
P. aeruginosa 5MPCA induced a redox active intracellular fluorescence within C. albicans that was visible by microscopy (Gibson et al., 2009) and a similar intracellular fluorescence was also observed in PMS-exposed fungal cells (Fig. 5A). Spectrofluorimetric analysis of PMS indicated that by itself it is not fluorescent (data not shown), but its derivative PMS-arginine exhibits fluorescence in the red emission wavelength range when excited at 488 nm. This fluorescence was partially reduced upon addition of Na2S2O4 (Fig. 5B). The red proteins extracted from C. albicans grown with P. aeruginosa or on PMS-medium also exhibited this characteristic red fluorescence (Fig. S2C), suggesting that the phenazinium-induced modification is responsible for the fungal intracellular fluorescence. These observations lead us to propose C. albicans intracellular fluorescence as a means for in vivo monitoring the uptake and modification of methylphenaziniums. We performed a time course analysis of the PMS-induced intracellular fluorescence of C. albicans using flow cytometry. At early time points after PMS exposure, we found a population exhibiting intermediate (I) fluorescence (Fig. S6A); this population decreased over the course of 96 hours, with a concomitant increase in a highly (H) fluorescent population (Fig. 5C and S6A for time course). C. albicans co-cultured with P. aeruginosa demonstrated similar fluorescence profiles (Fig. S6B). Since we found that methylphenaziniums modified in vitro (Fig. 5B) and in vivo (Gibson et al., 2009) exhibited a maximal fluorescence in the oxidized state, we hypothesized that the highly fluorescent population (H) correlated with changes in the intracellular redox environment upon the cessation of metabolism, and was representative of cell death. To test this, C. albicans was grown on medium with or without PMS for 24 hours and then, fluorescence of collected cells was analyzed by flow cytometry before and after treatment with a strong reducing agent. We found that reduction was sufficient to shift the fluorescence intensity of the peak corresponding to the H population closer to the peak associated with intermediate intensity (I) (Fig. 5D). Consistently, heat treatment of PMS-grown cells led to increased endogenous fluorescence, likely due to the loss of a reducing environment within the fungal cells upon death (Fig. 5E). Cell death alone was not sufficient to induce intermediate or high levels of fluorescence since C. albicans control cultures killed by heat treatment (Fig. 5E) did not exhibit changes in fluorescence compared with untreated control cultures. Furthermore, cells from I and H populations were FACS-sorted and their viability was assessed. While 60 ± 13.8 % of cells exhibiting intermediate fluorescence formed colonies, virtually none of the cells in the highly fluorescent population were colony formers (Fig. 5F). Hence, these results demonstrate that the intermediate levels of fluorescence are indicative of methylphenazinium uptake and modification, and that transformation precedes fungal death.
Fig. 5.
Intracellular fluorescence due to PMS uptake, modification and redox state. A. Microscopic view of epifluorescence (rhodamine) and DIC images of C. albicans grown for 72 hours in the presence of 1 mM PMS. B. Fluorescence emission spectra of PMS-arginine derivative. The red product obtained from the chemical reaction performed at pH 10.0 (see Supporting Information section for more details) was scanned before and after addition 80 mM Na2S2O4 using a SpectraMax M2 spectrophotometer. Excitation was at 488 nm. C. Flow cytometry analysis of C. albicans grown for 24 hours on YNBA medium (gray histogram) or medium containing 1 mM PMS (open histogram). PMS-cultured cells exhibited either intermediate (I) or high (H) fluorescence in the phycoerythrin (PE) channel. D and E. Flow cytometry analysis of C. albicans pre-grown on YNBA without (control) or with 1mM PMS for 24 hours and then treated: D. Cells assessed after treatment with 100 mM of Na2S2O4. E. C. albicans heat killed at 80°C for 30 minutes. Data are representative of two independent experiments. F. Viability analysis of C. albicans grown for 24 hours on YNBA medium alone (control) or with 1 mM PMS. Colony-forming units (CFUs) were determined for ~5×104 sorted cells from C. albicans populations exhibiting either (I) or (H) fluorescence detected by FACS. ***P<0.01 (n=3).
The microenvironment within colony biofilms promotes PMS-mediated killing
Evidence suggested that the environmental conditions within colony biofilms favor the lethal action of P. aeruginosa-derived 5MPCA or PMS. We found that during the first 24 hours of PMS exposure, prior to the appearance of visible red pigmentation, a green emerald color developed below C. albicans colonies (Fig. S7A). This color has been reported to correspond to a reduced semiquinoid form of PMS distinct from its fully reduced (colorless) or oxidized (yellow) species (Halaka et al., 1982; Zaugg, 1964). Thus, we hypothesized that PMS reduction is occurring within the colony biofilms and is required for phenazinium uptake and modification by C. albicans. To test this idea, we measured by flow cytometry the fluorescence intensity of yeast cells sampled from colonies grown on YNBA agar and resuspended in saline solution containing oxidized or reduced PMS. Reduction of PMS by the mild reducing agent ascorbic acid (AA) was performed prior to C. albicans addition (Chayet et al., 1963; Garcia-Sancho and Herreros, 1983; Konings et al., 1971). A bright green color was observed upon addition of AA to the PMS-containing solution (data not shown), confirming the production of the semiquinoid form under these mildly reducing conditions (Zaugg, 1964). Interestingly, we found that prior reduction of this methylphenazinium by AA dramatically accelerated the appearance of intermediate cellular fluorescence (Fig. 6A) and detection of red coloration of the fungal pellet (data not shown). In addition, reduced PMS (PMS+AA) triggered a five log decrease in C. albicans viability as quickly as 8 hours post exposure, while neither AA nor PMS alone had any toxic effect on fungal cells suspended in saline solution lacking any energy source to support metabolism (Fig. 6B).These data suggest that reduction of PMS or 5MPCA markedly promotes uptake of these methylphenaziniums. Therefore, the conditions within oxygen-limited biofilms may enhance the antifungal activity of these methylphenaziniums and consequently, their toxicity.
Fig. 6.
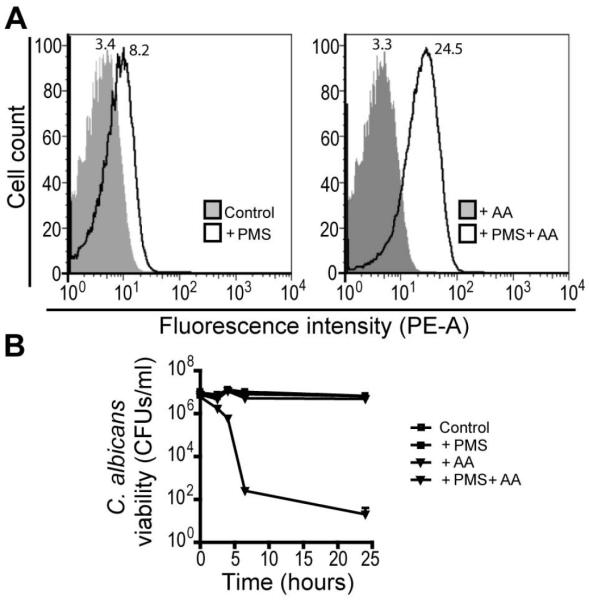
Reduction of PMS is required for its uptake and modification by C. albicans suspensions. C. albicans was grown on YNBA for 24 hours, and cells were then suspended in saline alone (control) or saline solution plus 100 mM ascorbic acid (AA), PMS (1mM) or PMS and AA (PMS+AA). A. Cells were analyzed by flow cytometry 4 hours post challenge to determine the levels of intracellular fluorescence. Mean fluorescence intensity (MFI) of each treatment is shown at the top of the FACS histogram. Data are representative of three independent experiments. B. C. albicans survival for each treatment as determined by CFU counts (n=3).
DISCUSSION
Our studies elucidate novel attributes that make P. aeruginosa 5MPCA potently toxic towards fungi, especially in environments such as bacterial-fungal biofilms. The striking antifungal properties of 5MPCA have remained poorly characterized since this phenazine is relatively unstable at neutral pH when chemically synthesized (Hansford et al., 1972) and is not found in P. aeruginosa monoculture supernatants (Byng et al., 1979). Here, we demonstrate that PMS is a powerful surrogate to study 5MPCA antifungal activity (Fig. 2). PMS can be modified by amino acids in vitro (Fig. 3A), and analysis of soluble fungal proteins extracted from C. albicans cultured either with PMS or P. aeruginosa indicate that these methylphenaziniums are covalently bound to proteins in vivo. The extracted red fungal soluble proteins showed similar spectral characteristics to those of the chemically synthesized red derivatives (Fig. 3C, D, S2A and B). Notably, the derivatives formed upon modification of PMS (Fig. 2B, 3A and B) or 5MPCA (Gibson et al., 2009) were fluorescent (Fig. S2C), accumulated in the fungal cell and participated in C. albicans killing (Fig. 5). In contrast, when modification occurred outside of C. albicans, the newly formed red compounds neither promoted the red pigmentation of fungal cells nor exhibited anticandicidal activity even in the presence of reducing agents (Fig S3) suggesting that the phenazines covalently bound to amino-containing compounds were not taken up by the fungus and that intracellular modification is an important requirement for toxicity and killing. Our proposed model summarizing these findings is presented in Fig. 7.
Fig. 7.
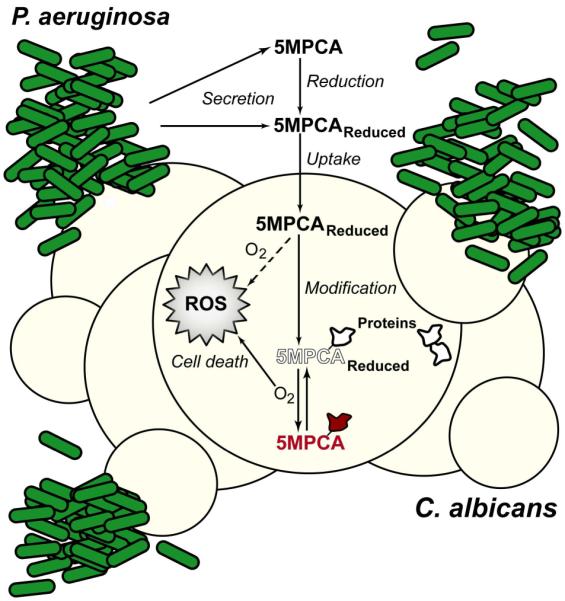
Proposed model for the mechanisms mediating 5MPCA antifungal properties, based on observations with the structurally similar methylphenazinium, PMS. In mixed species C. albicans–P. aeruginosa colony biofilms, 5MPCA is secreted by P. aeruginosa. This methylphenazinium is either released in its reduced form, or reduced extracellularly by bacterial or fungal factors within the metabolically active biofilm. Reduced 5MPCA enters to C. albicans cells, where it can be oxidized (dotted arrow) to generate toxic by-products such as superoxide and H2O2 (ROS). In addition, 5MPCA reacts with cellular amines within macromolecules, such as proteins leading to the accumulation of red, fluorescent methylphenazinium derivatives within C. albicans. As metabolic activity decreases, oxygen (O2) tension increases thereby causing more efficient redox cycling of 5MPCA itself (dotted arrow) and protein-bound 5MPCA derivatives. Oxidation of colorless, reduced 5MPCA and its derivatives (outline text) by O2 causes the generation and accumulation of toxic levels of ROS, resulting in fungal cell death. Metabolic reducing equivalents, such as NADH, can re-reduce 5MPCA and 5MPCA-derivatives (filled text), thus closing the redox cycle. As the metabolism of biofilm cells slows or stops, the proportion of the 5MPCA derivatives in the oxidized state increases, conferring the characteristic red color and high level fluorescence that is apparent in fungal cells from P. aeruginosa-C. albicans co-cultures.
The reduction of PMS and 5MPCA is important for both the uptake and modification of these methylphenazines. Within metabolically active fungal colonies, oxidized PMS was reduced (Fig. S7A), perhaps due to the fungal secretion of small molecules with low redox potentials such as reduced riboflavin (Ksenzhek and Petrova, 1983; Worst et al., 1998). These molecules may transfer electrons to extracellular PMS within the interior of the fungal colony. In co-culture biofilms, P. aeruginosa itself may also contribute to the reduction of its own excreted 5MPCA as it does with other secreted phenazines such as PYO (Price-Whelan et al., 2006, 2007). We demonstrated that PMS reduction promoted uptake by C. albicans cells (Fig. 6A and S7A), possibly due to an increase in its lipophilicity as the reduction of PMS makes it less soluble in aqueous solutions (Zaugg, 1964) and enhances its uptake across cell membranes (Bisschop et al., 1979; French et al., 1973). Our in vitro chemical studies showed that reduction of PMS is also required for spontaneous chemical conversion to red derivatives that are covalently linked to amino acids (Fig. 3A) and proteins under physiological pH conditions (see Supporting Information for experimental details). Further evidence for a requirement for reduction of the methylphenazinium by aspects of C. albicans metabolism was provided by our finding that no red pigmentation was developed when either P. aeruginosa or PMS was introduced onto lawns of heat-killed, and thus metabolically inactive, C. albicans cells (Fig. S7B). We cannot speculate whether the partially-reduced or fully-reduced form of PMS and 5MPCA is most important for C. albicans uptake and killing. However, the stability of the partially reduced semiquinoid form of PMS (Zaugg, 1964) is likely a crucial factor in this methylphenazinium toxicity.
Our data suggest that some of the toxic properties of 5MPCA are associated with its ability to generate ROS (Fig. 4 and S5). In fact, our results predict that reduced 5MPCA or 5MPCA derivatives form ROS when exposed to oxygen as has been previously demonstrated for PMS (Halaka et al., 1982). Similar redox activities have been shown to be important for the toxicity of other Pseudomonas phenazines, such as PYO, and likely play a major role in fungal killing (Hassan and Fridovich, 1980; Muller, 2002). We show evidence that the sequestered 5MPCA that is covalently bound to intracellular soluble proteins retains its ability to be reduced intracellularly and oxidized by oxygen (Fig 3D, S2A and B). The intracellular reducing power within the fungal cells, such as the accumulation of NADH, likely contributes to the redox cycling of these intracellular red methylphenazinium derivatives (Fig. 3D and S2B) and thus, to the generation of ROS. In addition, our findings suggest that another lethal mechanism independent of ROS production could be the covalent modification of vital intracellular amine targets by 5MPCA and PMS that may lead directly to their inactivation and fungal killing. Notably, PYO, a well studied P. aeruginosa toxin (Kerr et al., 1999; Thomashow et al., 1990), was not modified by amine-containing compounds in vitro, and we did not obtain any indication of intracellular modification in vivo (Fig. S4). Consistent with our model that methylphenazinium modification and intracellular accumulation increase effectiveness of these toxins, 5MPCA and PMS had greater lethal effects than PYO towards C. albicans (Fig. S4A and Gibson et al., 2009). Further studies will be developed to determine if 5MPCA also plays a role in P. aeruginosa infection of other species.
Our findings emphasizing a specific role for the reduced forms of 5MPCA may have implications for other aspects of phenazine biochemistry in P. aeruginosa. For instance, aeruginosins A and B (Hansford et al., 1972; Herbert and Holliman, 1969; Holliman, 1969), two 5MPCA-derived red soluble pigments (Fig. S1B and C) produced in very late P. aeruginosa mono-cultures, may be spontaneous products of the reduced form of this methylphenazinium with ammonia. Notably, as we demonstrated with PMS covalently modified by arginine, aeruginosin A also does not have any antifungal activity towards C. albicans (Gibson et al., 2009). Furthermore, we found that, when grown on agar with amino acids such as arginine as sole carbon and energy source, P. aeruginosa produced large amounts of soluble red pigments (data not shown). This suggests that, under the reducing conditions that prevail within the interior of mature colonies, secreted 5MPCA reacts with free aminoacids within the medium to yield these red-pigmented derivatives. Another role for reduced forms of 5MPCA may be suggested by in vitro enzymatic studies on the late steps of PYO biosynthesis. It has been reported that both PhzM and PhzS (Fig. 1A) are required for the conversion of PCA to 5MPCA, suggesting that this step is unfavorable in the absence of a reaction that removed the 5MPCA product (Parsons et al., 2007). We propose that close proximity of fungal cells within the biofilm environment allows for the continuous uptake and subsequent modification of reduced forms of 5MPCA by C. albicans thereby decreasing levels of free 5MPCA in P. aeruginosa and thus, driving the conversion of PCA to 5MPCA forward (Fig. 1A). Hence, under mono-culture conditions, free 5MPCA is not found to accumulate extracellulary, maybe due to its constant conversion to PYO and its extracellular modification by free amine-compounds, whereas under mixed-species biofilm conditions 5MPCA is taken-up and modified by the fungus.
In conclusion, in environments such as the lungs of cystic fibrosis patients, where P. aeruginosa and C. albicans are often found together (Chotirmall et al., 2010), molecules such as 5MPCA may play an important role controlling fungal populations. Previous work has shown that other fungi similarly accumulate and are killed by 5MPCA (Gibson et al., 2009), suggesting that this effective antimicrobial strategy maybe broadly applicable to several fungi or even other eukaryotes. Furthermore, it remains to be determined if 5MPCA also contributes to the damage of human tissues that occurs during P. aeruginosa infections. Thus, our research highlights the importance of studying mixed-species microbial communities to identify novel microbial toxins that may not be readily extractable or stable under mono-culture conditions and might play an important role in vivo.
EXPERIMENTAL PROCEDURES
Strains and culture conditions
The fungal and bacterial strains used in this study are described in Table S1. Fungal strains were grown on YPD (Gibson et al., 2009) or yeast nitrogen base (Hogan et al., 2004) synthetic agar (YNBA; pH 5.5) containing YNB salts with ammonium sulfate (RPI), 2 % glucose, 1.5% agar (Fisher bioReagents) and 0.2 % yeast synthetic Drop-out medium supplement without tryptophan (Sigma). P. aeruginosa PA14 wild type and mutant monocultures, and C. albicans–P. aeruginosa co-cultures were grown as previously described (Gibson et al., 2009) with minor modifications as follows. The fungal lawns were prepared by spreading 700 μl of an YPD-grown overnight C. albicans culture onto an agar plate. After 48 hours, P. aeruginosa strains were spot inoculated onto lawns, and co-cultures were incubated at 30°C for the specified time. Stock solutions of 500 mM rutin hydrate (Sigma), dissolved in DMSO; 1M PMS (Acros-organics) prepared in sterile distilled water; and 24 mM PYO (Cayman-chemicals) in ethanol were added to molten agar to the appropriate final concentrations. All assays performed with PMS were carried out in the dark.
C. albicans viability and pigmentation assays
C. albicans from exponential phase cultures grown to an absorbance (OD600) of 3.0 (107 cells/ml) in 5 ml YPD were spot inoculated (5-10 μl) onto plates of YNBA containing 1 mM of PMS or PYO and grown for the time specified. Where indicated, rutin (2 mM) was supplemented to the agar. The viability of yeast cells cultured with P. aeruginosa, or grown on YNBA containing PMS or PYO was assessed by taking agar core samples (Gibson et al., 2009) from the co-culture areas or by collecting the whole colonies formed on monoculture conditions, and suspending them on saline solution (0.85% NaCl). Each cell suspension was washed twice with saline followed by methylene blue staining as reported (Gibson et al., 2009). Where indicated, clonogenenicity assays were also performed after each treatment. The absorbance of treated cells suspended in saline was measured followed by diluting them down to 0.1 OD600. The cell numbers were counted using a hemocytometer and then 100 cells were plated on YPD medium. After 3 days of incubation the final number of colonies was counted. To visualize the redox activity of the red-pigmented cells, 15 PMS-grown (72 hours) colonies were collected and suspended in 1.5 ml of sterile saline solution and treated with either an oxidant or reducing agent.
ROS determination and oxidative stress responsive genes analysis
Intracellular ROS accumulation was examined using the fluorescent probe dichlorodihydrofluorescein diacetate (DCFH-DA) as previously described (Deveau et al., 2010) with minor modifications as follows. C. albicnas cultured on YNBA medium alone or with either 1 mM PMS or 10 mM H2O2 for 18 hours were washed and suspended in saline. Two aliquots from each sample were diluted 1:5 in saline and then treated, where indicated, with fresh DCFH-DA (50 μM) for 30 minutes. After washing with 500 μl of phosphate saline buffer (PBS), green fluorescence was determined by flow cytometry in a BD Canto instrument using the green fluorescent protein (GFP-A) channel.
The induction of ROS responsive genes was evaluated using C. albicans CAI-4 strains containing the yEGFP open reading frame fused to either CAT1, TTR1 or TRX1 promoter regions (Barelle et al., 2004; Enjalbert et al., 2007). To generate the TTR1 and TRX1 reporter strains, we linearized pTTR1-GFP and pTRX1-GFP plasmids with StuI and integrated at the RSP1 locus in the strain CAI-4 as previously described (Barelle et al., 2004; Enjalbert et al., 2007). The genotypes of transformants were confirmed by PCR (Barelle et al., 2004; Enjalbert et al., 2007), and their GFP fluorescence was evaluated by microscopy analysis of cells exposed to 10 mM H2O2 for 1 hour (Deveau et al., 2010). To examine the green fluorescence generated by pGFP- (a strain harboring the promoterless yEGFP fused to the ADH1 promoter, that only express basal fluorescence levels), CAT1-, TTR1- and TRX1-strains upon exposure to PMS, we cultured them on YNBA with or without methylphenazinium (1 mM) for 18 hours and measured the fluorescence levels by flow cytometry as described above.
In vitro red pigment formation and analysis
Analysis of the red pigment formed from the reactions of PMS with arginine and the redox properties of the product was done by monitoring the UV-Vis spectra of the reactions. See supporting information section for detailed experimental procedures used for the chemical reactions to form the red derivatives with amino acids. (Katritzky, 1985; Mclwain, 1937; Rajagopalan and Handler, 1962)
Extraction and analysis of C. albicans soluble proteins
For protein extraction, C. albicans was either grown on YNB medium with or without 1mM PMS or co-cultured with P. aeruginosa strains previously inoculated on the top of a polycarbonate filter (25 mm, GE water & Process technologies) placed on C. albicans pre-grown lawns. Fungal cells from colonies formed on YNBA medium or from C. albicans co-cultured with P. aeruginosa were collected and suspended in saline after 96 or 48 hours of incubation at 30°C, respectively. Each concentrated solution was then aliquoted and diluted down to an absorbance of ~30.0 (OD600) in fresh saline. After a short centrifugation time, the pellet was weighed and a specific volume (5 ml/ g of wet weight) of CelLytic yeast buffer (Sigma) was added. Cells exposed either to PMS or 5MPCA were incubated at 30°C under constant agitation for 18 or 6 hours, respectively. Subsequently, the final volume of each cell suspension was adjusted to equal densities using a previously reported hypertonic buffer (Sithanandam et al., 1998) without EDTA, and then, completely disrupted with one volume 0.5 mm zilica/sirconia beads for ~4 minutes (5 times × 50 seconds). Cellular debris was removed by centrifugation (15 minutes at 16,100xg) and the supernatant was then subjected to ultracentrifugation (100,000xg for 1 hour) to pellet membranes and organelles from the suspension. The soluble fraction was then treated with Benzonase Nuclease (Novagen) 50U/ml (5 minutes) to finally remove nucleic acids. Protein concentration was determined by Bradford assay following manufacturer recommendations (Biorad). The redox properties and fluorescence of the proteinaceous solutions were then analyzed by monitoring their UV-Vis and fluorescence emission spectra (Excitation 488 nm) using a SpectraMax M2 spectrophotometer. To test the covalent biding of the soluble proteins and methylphenaziniums, 800 mg of each red pigmented protein solution was treated with 8 M Urea (Sigma) and 6 M guanidinium chloride (Sigma) (Macheroux et al., 1998) for 1 hour. Treated and untreated soluble proteins were precipitated with 30% TCA and after centrifugation, pellet pigmentation was visually compared.
Analyses of PMS and 5MPCA induced intracellular fluorescence by flow cytometry
Samples of C. albicans colonies grown on YNBA with or without PMS were collected at indicated time points and suspended in saline. After two washes with 500 μl saline solution (as described above), 5 μl of the suspension were visualized by epifluorescence microscopy as described previously (Gibson et al., 2009). Intracellular fluorescence of 1 × 105 cells per treatment was also analyzed by flow cytometry in a BD Canto instrument using the R-phycoerythrin (PE-A) channel (excitation, 488 nm). 5 × 104 PMS-exposed cells from each population detected by flow cytometry were sorted according to their levels of fluorescence, and suspended into sterile saline solution (final volume 1 ml) prior to survival determination. Each analysis was performed on three independent co-cultures or YNBA colonies. Viability of sorted populations exhibiting different levels of fluorescence upon PMS exposure was assessed by plating a 10−2 dilution (~50 sorted cells/ml) on YNBA. Colony forming units (CFUs) were determined after 48 hours of incubation. Intracellular fluorescence of C. albicans grown with P. aeruginosa for 24 hours was also assessed by flow cytometry as described above. For experimental details of cell fluorescence analysis upon reduction or heat killed treatments see the Supporting Information section.
Effect of reduced PMS in cell suspensions
To assess the effect of redox state on PMS-mediated killing of C. albicans, a cell suspension from a 24 hours YNBA colony was diluted (~106 cells per ml) in 500 μl of saline solution alone (0.85 % NaCl) or saline containing either 100 mM AA at pH 3.0, PMS (1 mM) or PMS plus AA. The pH of the solutions was adjusted with 10 N HCl to pH 3.0 when it was required. The experiments were conducted under these conditions since we found that PMS was undesirably converted to its fully reduced, water insoluble form by these concentrations of AA at higher pH values or by the use of the strong reducing agent Na2S2O4 (Zaugg, 1964). Samples were removed at each time point and cells were washed with saline solution twice prior to analyses. CFUs of the collected samples were determined by serial dilutions followed by triplicate plate counts on YNBA medium at the beginning of the assay and after the specified time points.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Gordon W. Gribble for his important intellectual contributions to this project and to Ambrose Cheung for his comments and suggestions on this manuscript. We thank Anna K. Mapp for allowing us to perform the mass spectrometry analysis at her laboratory. We are thankful to Andres Lorente for his insightful ideas on protein isolation and to Aurélie Deveau for her help with yeast transformation and valuable suggestions. We thank Yoshiyuki Nakagawa for providing the C. albicans CU2 and the catalase deficient mutant cat1/cat1 strains. We are very grateful to Gary Ward for their help and assistance on cell sorting. This work was supported by the Pew Biomedical Scholars Program (D.A.H.) and the Cystic Fibrosis Foundation (D.A.H.)
REFERENCES
- Anjaiah V, Koedam N, Nowrk-Thompson B, Loper J, Höfte M, Tambong J, Cornelis P. Involvement of phenazines and anthranilate in the antagonism of Pseudomonas aeruginosa PNA1 and Tn5 derivatives Toward Fusarium spp. and Pythium spp. Molecular Plant-Microbe interactions. 1998;11:847–854. [Google Scholar]
- Barelle CJ, Manson CL, MacCallum DM, Odds FC, Gow NA, Brown AJ. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast. 2004;21:333–340. doi: 10.1002/yea.1099. [DOI] [PubMed] [Google Scholar]
- Baron SS, Rowe JJ. Antibiotic action of pyocyanin. Antimicrob Agents Chemother. 1981;20:814–820. doi: 10.1128/aac.20.6.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind A, Bertele RM, Harms K, Horl G, Jungwirth R, Petermuller C, Przyklenk B, Weisslein-Pfister C. Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection. 1987;15:270–277. doi: 10.1007/BF01644137. [DOI] [PubMed] [Google Scholar]
- Bisschop A, Bergsma J, Konings WN. Site of interaction between phenazine methosulphate and the respiratory chain of Bacillus subtilis. Eur J Biochem. 1979;93:369–374. doi: 10.1111/j.1432-1033.1979.tb12832.x. [DOI] [PubMed] [Google Scholar]
- Bolwerk A, Lagopodi AL, Wijfjes AH, Lamers GE, Chin AWTF, Lugtenberg BJ, Bloemberg GV. Interactions in the tomato rhizosphere of two Pseudomonas biocontrol strains with the phytopathogenic fungus Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant Microbe Interact. 2003;16:983–993. doi: 10.1094/MPMI.2003.16.11.983. [DOI] [PubMed] [Google Scholar]
- Brand A, Barnes JD, Mackenzie KS, Odds FC, Gow NA. Cell wall glycans and soluble factors determine the interactions between the hyphae of Candida albicans and Pseudomonas aeruginosa. FEMS Microbiol Lett. 2008;287:48–55. doi: 10.1111/j.1574-6968.2008.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byng GS, Eustice DC, Jensen RA. Biosynthesis of phenazine pigments in mutant and wild-type cultures of Pseudomonas aeruginosa. J Bacteriol. 1979;138:846–852. doi: 10.1128/jb.138.3.846-852.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayet C, Steele RH, Breckinridge BS. A chemiluminescence (CL) of phenazine methosulfate (PMS) in the presence of hydrogen peroxide (HOOH) induced by reductants including reduced nicotinamide adenine dinucleotide (NADH) and ascorbic acid (AA) Biochem Biophys Res Commun. 1963;10:390–395. doi: 10.1016/0006-291x(63)90543-6. [DOI] [PubMed] [Google Scholar]
- Chotirmall SH, O’Donoghue E, Bennett K, Gunaratnam C, O’Neill SJ, McElvaney NG. Sputum Candida Albicans presages Fev1 decline and hospitalized exacerbations in cystic fibrosis. Chest. 2010 doi: 10.1378/chest.09-2996. In press. [DOI] [PubMed] [Google Scholar]
- Cugini C, Calfee MW, Farrow JM, 3rd, Morales DK, Pesci EC, Hogan DA. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:896–906. doi: 10.1111/j.1365-2958.2007.05840.x. [DOI] [PubMed] [Google Scholar]
- Deveau A, Piispanen AE, Jackson AA, Hogan DA. Farnesol induces hydrogen peroxide resistance in Candida albicans yeast by inhibiting the Ras-cyclic AMP signaling pathway. Eukaryot Cell. 2010;9:569–577. doi: 10.1128/EC.00321-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11:30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell. 2006;17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, MacCallum DM, Odds FC, Brown AJ. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect Immun. 2007;75:2143–2151. doi: 10.1128/IAI.01680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SW, Palmer DS, Sim WA. Phenazine methosulfate uptake by rat liver mitochondria. Can J Biochem. 1973;51:235–240. doi: 10.1139/o73-030. [DOI] [PubMed] [Google Scholar]
- Garcia-Sancho J, Herreros B. Effects of redox agents on the Ca2+-activated K+ channel. Cell Calcium. 1983;4:493–497. doi: 10.1016/0143-4160(83)90024-6. [DOI] [PubMed] [Google Scholar]
- Gibson J, Sood A, Hogan DA. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl Environ Microbiol. 2009;75:504–513. doi: 10.1128/AEM.01037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Haque A, Mukhopadhyay G, Narayan RP, Prasad R. Interactions between bacteria and Candida in the burn wound. Burns. 2005;31:375–378. doi: 10.1016/j.burns.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Halaka FG, Babcock GT, Dye JL. Properties of 5-methylphenazinium methyl sulfate. Reaction of the oxidized form with NADH and of the reduced form with oxygen. J Biol Chem. 1982;257:1458–1461. [PubMed] [Google Scholar]
- Hansford GS, Holliman FG, Herbert RB. Pigments of Pseudomonas species. IV. In vitro and in vivo conversion of 5-methylphenazinium-1-carboxylate into aeruginosin A. J Chem Soc Perkin 1. 1972;1:103–105. doi: 10.1039/p19720000103. [DOI] [PubMed] [Google Scholar]
- Hassan HM, Fridovich I. Mechanism of the antibiotic action of pyocyanine. J Bacteriol. 1980;141:156–163. doi: 10.1128/jb.141.1.156-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert RB, Holliman FG. Pigments of pseudomonas species. II. Structure of aeruginosin B. J Chem Soc Perkin 1. 1969;18:2517–2520. doi: 10.1039/j39690002517. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 2002;296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- Holliman FG. Pigments of pseudomonas species. I. Structure and synthesis of aeruginosin A. J Chem Soc Perkin 1. 1969;18:2514–2516. doi: 10.1039/j39690002514. [DOI] [PubMed] [Google Scholar]
- Hughes WT, Kim HK. Mycoflora in cystic fibrosis: some ecologic aspects of Pseudomonas aeruginosa and Candida albicans. Mycopathol Mycol Appl. 1973;50:261–269. doi: 10.1007/BF02053377. [DOI] [PubMed] [Google Scholar]
- Katritzky A. Handbook of Heterocyclic Chemestry. Pergamon Press; Oxford, New York, Toronto, Sydney, Frankfurt: 1985. pp. 208–209. [Google Scholar]
- Kehrmann F, Havas E. On the acknowledgement of the phenazine. Berichte der Deutschen Chemischen Gessellschaft. 1913;46:341–352. [Google Scholar]
- Kerr JR. Suppression of fungal growth exhibited by Pseudomonas aeruginosa. J Clin Microbiol. 1994;32:525–527. doi: 10.1128/jcm.32.2.525-527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JR, Taylor GW, Rutman A, Hoiby N, Cole PJ, Wilson R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol. 1999;52:385–387. doi: 10.1136/jcp.52.5.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings WN, Barnes EM, Jr., Kaback HR. Mechanisms of active transport in isolated membrane vesicles. 2. The coupling of reduced phenazine methosulfate to the concentrative uptake of beta-galactosides and amino acids. J Biol Chem. 1971;246:5857–5861. [PubMed] [Google Scholar]
- Ksenzhek O, Petrova S. Electrochemical properties of flavins in aqueous solutions. Bioelectrochm Bioenerg. 1983;11:105–127. [Google Scholar]
- Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004a;10:599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun. 2004b;72:4275–4278. doi: 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch AS, Robertson GT. Bacterial and fungal biofilm infections. Annu Rev Med. 2008;59:415–428. doi: 10.1146/annurev.med.59.110106.132000. [DOI] [PubMed] [Google Scholar]
- Macheroux P, Hill S, Austin S, Eydmann T, Jones T, Kim SO, Poole R, Dixon R. Electron donation to the flavoprotein NifL, a redox-sensing transcriptional regulator. Biochem J. 1998;332(Pt 2):413–419. doi: 10.1042/bj3320413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- Maridonneau-Parini I, Mirabelli F, Richelmi P, Bellomo G. Cytotoxicity of phenazine methosulfate in isolated rat hepatocytes is associated with superoxide anion production, thiol oxidation and alterations in intracellular calcium ion homeostasis. Toxicol Lett. 1986;31:175–181. doi: 10.1016/0378-4274(86)90012-3. [DOI] [PubMed] [Google Scholar]
- Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol. 2006;44:417–445. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- Mclwain H. The phenazine series. Part IV. Reactions of alkyl phenazonium salts; the phenazyls. J Chem Soc. 1937:1704–1711. [Google Scholar]
- Morales DK, Hogan DA. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010;6:e1000886. doi: 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa Hassan H, Fridovich I. Mechanism of the antibiotic action of pyocyanine. J Bacteriol. 1980;141:156–163. doi: 10.1128/jb.141.1.156-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. Pyocyanin induces oxidative stress in human endothelial cells and modulates the glutathione redox cycle. Free Radic Biol Med. 2002;33:1527–1533. doi: 10.1016/s0891-5849(02)01087-0. [DOI] [PubMed] [Google Scholar]
- Muller M. Polyphenol cytotoxicity induced by the bacterial toxin pyocyanin: role of NQO1. Free Radic Biol Med. 2009;47:84–91. doi: 10.1016/j.freeradbiomed.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Nishikimi M, Appaji N, Yagi K. The occurrence of superoxide anion in the reaction of reduced phenazine methosulfate and molecular oxygen. Biochem Biophys Res Commun. 1972;46:849–854. doi: 10.1016/s0006-291x(72)80218-3. [DOI] [PubMed] [Google Scholar]
- Ottaway JH. Some rate constants for the phenazine methosulphate-catalysed oxidation of reduced nicotinamide-adenine dinucleotide. Biochem J. 1966;99:253–256. doi: 10.1042/bj0990253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JF, Greenhagen BT, Shi K, Calabrese K, Robinson H, Ladner JE. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry. 2007;46:1821–1828. doi: 10.1021/bi6024403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. 2010;8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- Price-Whelan A, Dietrich LE, Newman DK. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- Price-Whelan A, Dietrich LE, Newman DK. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan KV, Handler P. Oxidation of phenazine methosulfate by hepatic aldehyde oxidase. Biochem Biophys Res Commun. 1962;8:43–47. doi: 10.1016/0006-291x(62)90232-2. [DOI] [PubMed] [Google Scholar]
- Ran H, Hassett DJ, Lau GW. Human targets of Pseudomonas aeruginosa pyocyanin. Proc Natl Acad Sci U S A. 2003;100:14315–14320. doi: 10.1073/pnas.2332354100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbachenko IM, Lisovskaya IL, Tikhonov VP. Oxidation-induced calcium-dependent dehydration of normal human red blood cells. Free Radic Res. 2007;41:536–545. doi: 10.1080/10715760601161452. [DOI] [PubMed] [Google Scholar]
- Shirtliff ME, Peters BM, Jabra-Rizk MA. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett. 2009;299:1–8. doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sithanandam G, Ramakrishna G, Diwan BA, Anderson LM. Selective mutation of K-ras by N-ethylnitrosourea shifts from codon 12 to codon 61 during fetal mouse lung maturation. Oncogene. 1998;17:493–502. doi: 10.1038/sj.onc.1201958. [DOI] [PubMed] [Google Scholar]
- Thomashow LS, Weller DM. Role of phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. Journal of Bacteriology. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow LS, Weller DM, Bonsall RF, Pierson LS. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol. 1990;56:908–912. doi: 10.1128/aem.56.4.908-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worst DJ, Gerrits MM, Vandenbroucke-Grauls CM, Kusters JG. Helicobacter pylori ribBA-mediated riboflavin production is involved in iron acquisition. J Bacteriol. 1998;180:1473–1479. doi: 10.1128/jb.180.6.1473-1479.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaugg WS. Spectroscopic characteristics and some chemical properties of N-methylphenazinium methyl sulfate (phenazine methosulfate) and pyocyanine at the semiquinoid oxidation level. J Biol Chem. 1964;239:3964–3970. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.