Abstract
Many bacteria organize themselves into structurally complex communities known as biofilms in which the cells are held together by an extracellular matrix. In general, the amount of extracellular matrix is related to the robustness of the biofilm. Yet, the specific signals that regulate the synthesis of matrix remain poorly understood. Here we show that the matrix itself can be a cue that regulates the expression of the genes involved in matrix synthesis in Bacillus subtilis. The presence of the exopolysaccharide component of the matrix causes an increase in osmotic pressure that leads to an inhibition of matrix gene expression. We further show that non-specific changes in osmotic pressure also inhibit matrix gene expression and do so by activating the histidine kinase KinD. KinD, in turn, directs the phosphorylation of the master regulatory protein Spo0A, which at high levels represses matrix gene expression. Sensing a physical cue such as osmotic pressure, in addition to chemical cues, could be a strategy to non-specifically coordinate the behavior of cells in communities composed of many different species.
Introduction
The ability to form structured communities of cells (biofilms) is a widespread feature of bacteria (Branda et al., 2005b; Kolter and Greenberg, 2006; O’Toole et al., 2000). Cells in biofilms are held together by an extracellular matrix that consists of polysaccharide and protein and sometimes DNA (Branda et al., 2005a; Lopez et al., 2010) but whose composition can vary greatly depending on the conditions and the species present (Branda et al., 2005b; O’Toole et al., 2000). Given the chemical diversity of the matrix, how might a mixed community regulate the amount of matrix made? We reasoned that, in addition to chemical cues, the physical properties of diverse matrices might provide a common cue for regulating matrix synthesis. In particular, we focused on osmotic pressure because the high concentration of exopolysaccharide (EPS) polymers (up to 90% of a biofilm’s dry mass (Flemming and Wingender, 2010) would be expected to increase the osmotic pressure and there is ample precedent that osmotic pressure can regulate gene expression (Russo and Silhavy, 1991; Ruzal and Sanchez-Rivas, 1998).
An attractive organism in which to study biofilm development is the spore-forming bacterium Bacillus subtilis. B. subtilis forms highly structured colonies on semi-solid surfaces and thick pellicles at the air-liquid interface of standing cultures as a consequence of extracellular matrix production (Branda et al., 2001a; Lemon et al., 2008). The main components of the B. subtilis extracellular matrix are EPS, which is synthesized by enzymes encoded by the epsA-O operon and amyloid-like fibers, which are encoded by tapA-sipW-tasA (Branda et al., 2006; Branda et al., 2004); Romero et al. 2010). Both operons are subject to negative regulation by the repressor SinR (Chu et al., 2006). Derepression of the matrix operons is brought about by the antirepressor SinI (Bai et al., 1993; Chai et al., 2008), whose synthesis is turned on by the phosphorylated form of the regulatory protein Spo0A, which also directs the transcription of genes that govern entry into sporulation (Fujita et al., 2005; Fujita and Losick, 2005). At least four histidine kinases contribute to the phosphorylation of Spo0A via a multicomponent phosphorelay (Burbulys et al., 1991; Jiang et al., 2000). Low levels of Spo0A~P turn on SinI synthesis but are insufficient to activate genes contributing to sporulation, which have weak binding sites for the phosphoprotein (Fujita et al., 2005). High levels of Spo0A~P, on the other hand, repress the gene for SinI while turning on sporulation gene expression (Chai et al., 2011).
In earlier work, an intriguing connection was observed between EPS production and the onset of spore formation during biofilm formation. Efficient entry into sporulation was found to depend on matrix production in a manner that was bypassed by a mutation in the gene for the histidine kinase KinD (Aguilar et al., 2010). Thus, KinD was inferred to be a checkpoint protein that linked sporulation to EPS production. Some histidine kinases are known to be bifunctional in that they exhibit competing phosphatase and kinase activities (Kostakioti et al., 2009; Zhu et al., 2000). It was proposed that KinD is such a bifunctional kinase and that at low EPS levels, KinD is principally a phosphatase that keeps Spo0A~P concentrations low and that at high EPS levels it becomes a kinase, allowing the phosphoprotein to accumulate to high concentrations (Aguilar et al., 2010).
But how EPS production effects such a switch was mysterious. Here we report that a non-specific physical cue, increasing osmotic pressure from the accumulation of EPS during biofilm formation, stimulates the kinase activity of KinD, causing Spo0A~P levels to rise. This rise represses the gene for SinI and hence matrix gene expression and instead turns on genes for entry into sporulation. Thus, a physical property of the matrix, osmotic pressure, coordinates the behavior of cells in the biofilm.
RESULTS AND DISCUSSION
Polymer addition inhibits biofilm robustness and matrix gene expression
As B. subtilis biofilms develop, the amount of wrinkling and overall thickness increase and there is an apparent concomitant increase in robustness. We have developed a quantitative physical assay for biofilm robustness that involves measuring a pellicle’s shear strength as it grows in small circular wells. Pellicles were grown at 30°C in a cylindrical cell on an Ares G2 strain-controlled rheometer (Figs. 1a). A cylindrical bob coupled to a torque sensor was placed at the center of the well. Tests of pellicle strength were conducted by periodically (0.1Hz) rotating the well around its symmetrical axis at small angles (≤0.6°) and measuring the torque on the cylinder as shown in Fig. 1a. The maximal torque measured per period is defined as the oscillatory torque. Under these experimental conditions, the oscillatory torque reflects the viscoelastic properties of the biofilm. Thus, measuring the oscillatory torque as pellicles form can be used as an assay to probe the strength and hence the robustness of biofilms. Under our conditions, the first measurable increase in robustness occurred about 26 hours after inoculation (red curve in Fig 1c). After that, the strength increased steadily for ~8 hours before leveling off. We wished to ask how a change in the physical environment influenced biofilm development. Therefore, we performed torque measurements as a pellicle was forming in the presence of 20% (by mass) of 20 kDa polyethylene glycol (PEG). Surprisingly, while the initial stages of pellicle formation did not change, there was a marked change at later times; the strengthening stopped earlier and the mature pellicle was ~40% weaker (blue curve in Fig. 1b). In fact, the pellicles weakened monotonically with the amount of PEG added to the medium (Fig. 1c). [A similar correlation between polymer concentration and pellicle weakening was obtained using the maximal torque (yield torque) a pellicle can withstand before it ruptures (data not shown).] Moreover, pellicles grown in the presence of PEG had a visible defect in robustness (Fig. 1d). As shown Fig. 1D even though rates of strengthening during the initial stages were similar, biofilms grown in the presence of PEG stopped strengthening earlier than those grown in medium without PEG.
Figure 1. Real time measurements of pellicle strength.
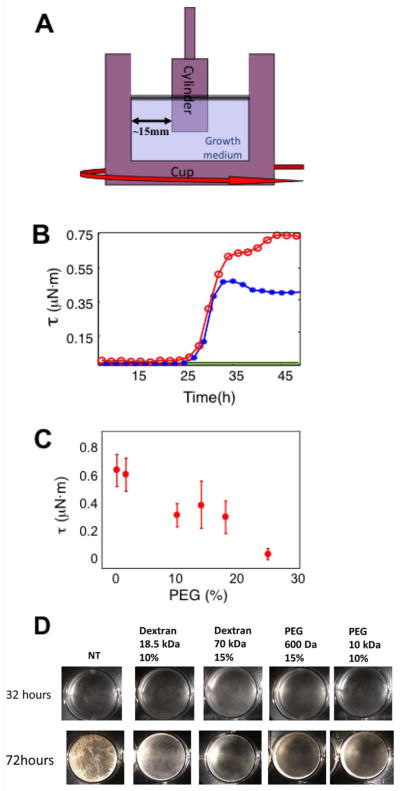
(a) Pellicles were grown inside a cuvette cell on top of an Ares G2 strain controlled rheometer. The diameter of the outside well was 34 mm and that of the inside cylinder was 15 mm. The setup was held at a constant temperature of 30°. ( (b) Oscillatory torque, τ, was measured at a strain of 0.5% approximately every two hours for wild type bacteria in MSgg (red), MSgg supplemented with 20% by mass of 20 kDa PEG (blue), MSgg with 15% by mass of PEG with no bacteria (green) and pure MSgg inoculated with a matrix mutant that was not capable of synthesizing exopolysaccharide (black). The pellicle began to show rigid behavior after ~30 hours for both pure and PEG supplemented medium. Note, however, that the supplemented medium saturated faster and did not reach the level of robustness of the pure medium. (c) Oscillatory torque, τ, for wild type pellicles grown in MSgg medium supplemented with 20 kDa PEG at 0 - 25% mass fractions. D) 3610 was grown in a liquid biofilm medium for 3 days with/without polymer addition. Pictures were taken using a SPOT camera (Diagnostic Instruments, USA).
Why did the addition of PEG impair the development of biofilm robustness? Since biofilm strength is associated with matrix, one possibility was that the addition of PEG led to a reduction in matrix gene expression and hence the production of matrix. To investigate this we asked whether the addition of PEG would inhibit the expression of the matrix operon tapA-sipW-tasA (henceforth simply tapA). Cells were grown in biofilm-inducing medium (MSgg) without shaking to facilitate pellicle formation. The medium was modified by addition of PEG or Dextran over a range of concentrations. After 48 hours we harvested the cells and measured the expression of tapA. The results show that increasing the concentration of polymer caused a marked reduction in matrix gene expression (Fig. 2a) and pellicle robustness (Fig. 1d), while having little or no effect on cell growth (Fig. S1).
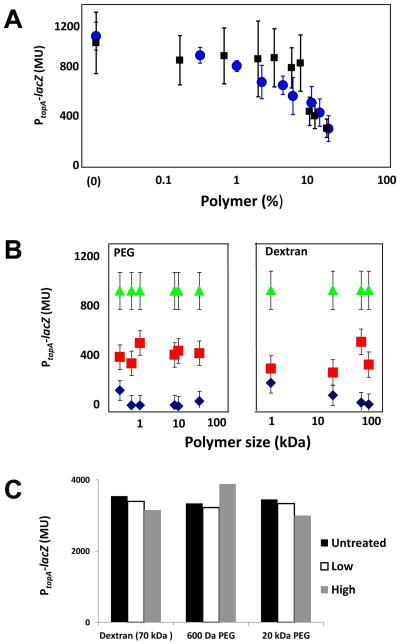
Figure 2. Osmotic pressure regulates matrix gene expression.
(a) PtapA activity in a wild type strain carrying PtapA-lacZ (RL4582) was measured as a function of the mass fraction of 20 kDa PEG (blue) and 70 kDa dextran (black) added to the medium. (b) PtapA activity in a wild type strain carrying PtapA-lacZ (RL4582) was measured medium supplemented with various concentrations of different-sized PEG (right) and Dextran (left) polymers prepared at set osmotic pressures: dark blue diamonds, magenta squares and green triangles corresponded respectively to 5-8,1-3 and 0 atm additional osmotic pressure exerted by the polymers. (c) PtapA-lacZ activity was measured in a sinR mutant (in which tapA is expressed constitutively) (RL4585). The curves represent 3 experiments, performed in duplicates, in which different polymers were used to increase the osmotic pressure; 70 kDa Dextran 600 Da PEG and 20 kDa PEG. Error bars are the standard deviation.
The effect of polymer correlates with changes in osmotic pressure
To investigate the basis for the polymer-induced decrease in the expression of tapA, we took advantage of the unique properties of polymeric solutions to differentiate between the effects of multiple physical parameters. To do this, we tested the effects of both PEG and dextran over a range of molecular sizes and concentrations as well as various concentrations of the dextran subunit dextrose. This enabled us to distinguish among the contributions of viscosity, osmotic pressure, diffusivity, ionic strength and chemical composition. Thus, for instance, solutions of 2.5% PEG of molecular weight 0.3 kD and 15% PEG of molecular weight 20 kDa exert similar osmotic pressures but their viscosities differ by more than one order of magnitude. Given that we could reproduce the effect for both small (dextrose or PEG 600 Da) and large (Dextran 110 kDa and PEG 35 kDa) molecules, we concluded that there was no correlation with size and diffusivity. The diffusivity of small molecules is almost completely unaltered by the addition of polymers and therefore cannot explain the effect we observed. In contrast, the diffusivity of large molecules depends strongly on the polymer size. As both PEG and dextran caused similar effects we also concluded that chemical composition of the polymeric additives was much less significant than the osmotic pressure. Additionally, the observed effects cannot be due to changes in ionic strength because PEG is neutral. We also ruled out any systematic dependence on viscosity of the polymeric solutions (Figure S2). Lastly and importantly, we found that the level of matrix gene expression correlated strongly with the osmotic pressure exerted by the polymers (Fig. 2b).
The tapA operon is under the negative control of the repressor SinR (Chai et al., 2008; Kearns et al., 2005). If osmotic pressure inhibits expression (blocks derepression) of the operon, then it should exert little or no effect in a sinR mutant in which tapA is expressed constitutively. As evidence that this is the case, pellicle morphology in a sinR mutant was largely unchanged in the presence of polymers (Figure S4). Also, tapA transcription was insensitive to osmotic pressure in a mutant lacking the repressor (Fig. 2C) (as compared to the wild type in Fig. 2C). [Note that the level of tapA expression is much higher in a sinR mutant than in the wild type) as seen from a comparison of the scales between panels B and C.] In toto, the results so far indicate that osmotic pressure acts at or upstream of SinR in the pathway governing tapA transcription.
Next, we asked whether in wild type cells increasing osmotic pressure would stimulate sporulation gene expression, which requires high levels of Spo0A~P (Fujita et al., 2005; Fujita and Losick, 2005). Transcription of a sporulation-specific promoter (spoIIA) significantly increased following the addition of various polymers (Fig. 3). Moreover, cells grown in biofilm-inducing medium supplemented with various polymers sporulated prematurely (Figure S5).
Figure 3. Polymer stimulates sporulation gene expression in a KinD-dependent manner.
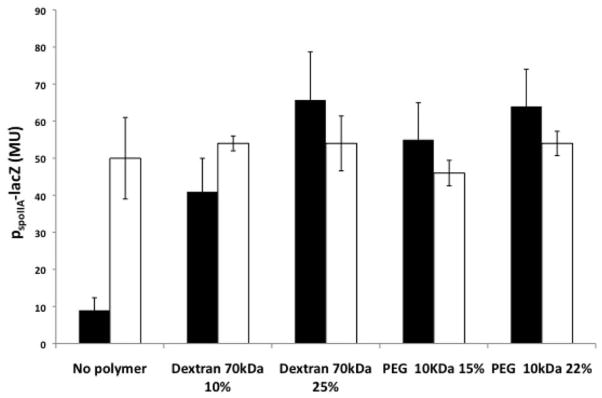
A wild type strain (IKG10; black columns) and its kinD mutant derivative (IKG603; white columns), both carrying a sporulation gene reporter PspoIIA-lacZ, were grown with the indicated polymers. The columns represent 3 experiments, performed in duplicates., Error bars are the standard deviation.
The effect of polymer is dependent on KinD
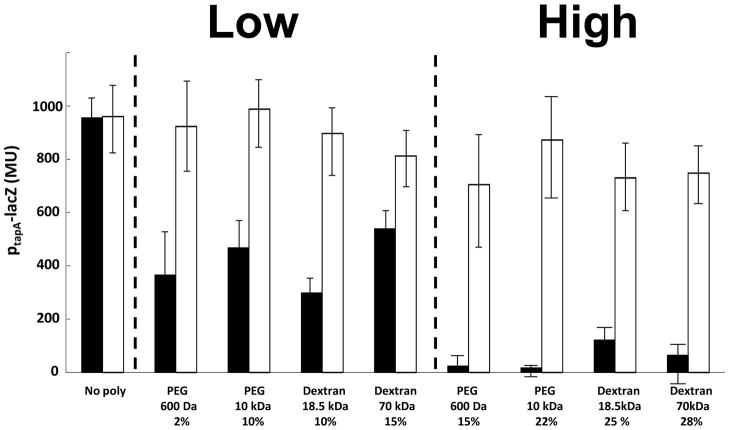
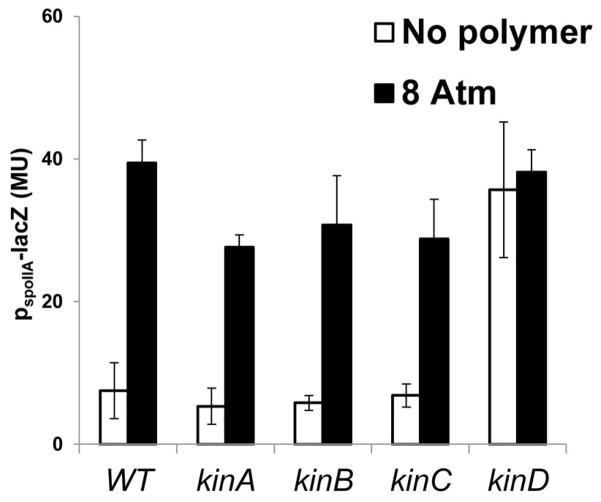
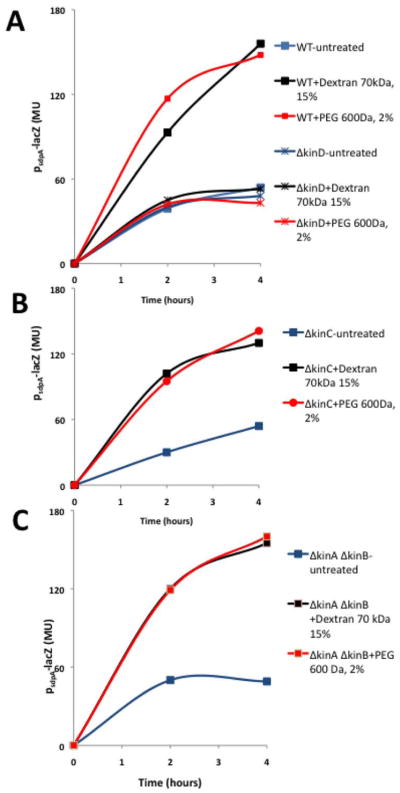
Previous work has indicated the histidine kinase KinD is a checkpoint protein that links the levels of Spo0A~P to the accumulation of extracellular matrix (Aguilar et al., 2010). As a result, KinD mutant cells exhibit a subtle biofilm defect, and sporulate prematurely (Vlamakis et al., 2008). It was proposed that KinD is principally a phosphatase at low levels of matrix, keeping Spo0A~P at low to moderate concentrations, and principally a kinase at high matrix levels, causing Spo0A~P to reach high concentrations. At low to moderate Spo0A~P concentrations, matrix genes would be ON and sporulation genes OFF whereas at high matrix levels, matrix genes would be OFF (via repression of the gene for SinI) and genes governing entry into sporulation would be ON (Chai et al., 2011). We therefore wondered whether the presence of the extracellular matrix is sensed by KinD as polymer-induced osmotic pressure. In contrast to the wild type, increasing osmotic pressure in a KinD mutant failed to inhibit expression of the matrix gene tapA (Fig. 4). In addition, a KinD mutant was largely unaltered in the pellicle morphology with or without added polymers (Figure S6). Also, transcription from the sporulation promoter spoIIA was higher in a KinD mutant than in the wild type in the absence of added polymer (in keeping with the idea that KinD is principally a phosphatase that keeps Spo0A~P levels low when unstimulated) (Fig. 3). This high level of sporulation transcription seen in the absence of KinD was not further increased by raising the osmotic pressure (Fig. 3). The effect was specific to KinD as mutants lacking KinA, KinB or KinC were unaltered in their response to changes in osmotic pressure (Fig. 5). This suggests that KinD is an osmosensor whose kinase activity increases and whose phosphatase activity decreases in response to increasing osmotic pressure, raising Spo0A~P levels.
Figure 4. Polymer inhibits matrix gene expression in a KinD-dependent manner.
A wild type strain (RL4582; black columns) and its kinD mutant derivative (IKG602; white columns), both carrying a matrix gene reporter PtapA-lacZ were grown with the indicated polymers. PtapA activity was measured under conditions of low (LOW) and high (HIGH) osmotic pressure. The columns represent 3 experiments, performed in duplicates, in which different polymers described in figure 2B were used to increase the osmotic pressure. Error bars are the standard deviation.
Figure 5. The uncoupling of sporulation gene expression from increasing osmotic pressure is specifically dependent on KinD.
Strain 3610 and its kinase mutant derivatives, kinA (IKG604), kinB (IKG605), kinC (IKG606), kinD (IKG603), all carrying PspoIIA-lacZ (a sporulation promoter), were grown in biofilm-inducing medium. PspoIIA activity is expressed in Miller units. The columns represent 3 experiments, performed in duplicates, in which different polymers described in figure 2B were used to increase the osmotic pressure. Error bars are the standard deviation.
A distinctive feature of KinD is that its activity depends on a lipoprotein called Med (Banse et al., 2011). We therefore wondered whether KinD activity would be dependent on Med in our osmosensing assay. We confirmed that this was indeed the case in experiments similar to those of Fig. 3 (data not shown).
If increasing osmotic pressure was elevating Spo0A~P levels, then in a rich medium (LB) in which the levels of the phosphoprotein are normally very low, polymer addition should turn on genes whose transcription requires low to moderate levels of Spo0A~P and are normally not expressed in LB medium. As shown in Fig. 6, the addition of either PEG or Dextran to LB medium significantly increased the transcription of sdp, an operon that is under the direct control of Spo0A~P and activated at low levels of the phosphoprotein (Fujita et al., 2005; Gonzalez-Pastor et al., 2003). This induction was dependent on KinD but not on KinA, KinB, or KinC (Fig. 6). In sum, our results are consistent with a model in which KinD is an osmosensor that detects increases in osmotic pressure from the synthesis of matrix and in response stimulates the phosphorylation of Spo0A.
Figure 6. Stimulatory effect of osmotic pressure on Spo0A-P-directed gene is specifically dependent on KinD.
Strain 3610 carrying PsdpA-lacZ (IKG609) and its derivatives with the kinase mutations ΔkinC (IKG611), ΔkinA ΔkinB (IKG612) and kinD (IKG610) were grown in LB (non-sporulation) medium in a shaking culture to early stationary phase. PsdpA activity is expressed in Miller units. The curves represent 3 experiments, performed in triplicates. Error bars are the standard deviation.
Finally, we investigated the idea that during biofilm formation KinD is responding to increasing osmotic pressure from the accumulation of matrix polymers. That is, whether the physical property of the matrix is an environmental signal that is detected by cells in the community and used to coordinate their behavior. If polymer-induced increases in osmotic pressure are indeed regulating matrix production and the onset of sporulation, then in biofilm-inducing medium a mutant blocked in EPS production (Δeps) (Branda et al., 2004; Chu et al., 2006) should have low levels of Spo0A~P. Indeed, as seen previously (Vlamakis et al., 2008), in the absence of EPS, matrix gene transcription was elevated (Fig. 7A) [recall that high levels of Spo0A~P inhibit matrix gene transcription (Chai et al., 2011)] and sporulation transcription was markedly decreased (Fig. 7B) (Aguilar et al., 2010). These effects were reversed by artificially raising the osmotic pressure to 2 atm and 5-8 atm (based on using several different polymers; Fig. 7A and 7B) in a manner that was dependent on KinD (Fig. 7C and 7D). In sum, these findings reinforce the view that KinD is an osmosensor whose kinase activity is stimulated and whose phosphatase activity decreased by the increase in osmotic pressure caused by EPS synthesis.
Figure 7. Increasing osmotic pressure mimics the effect of matrix production on matrix and sporulation gene expression in a matrix mutant.
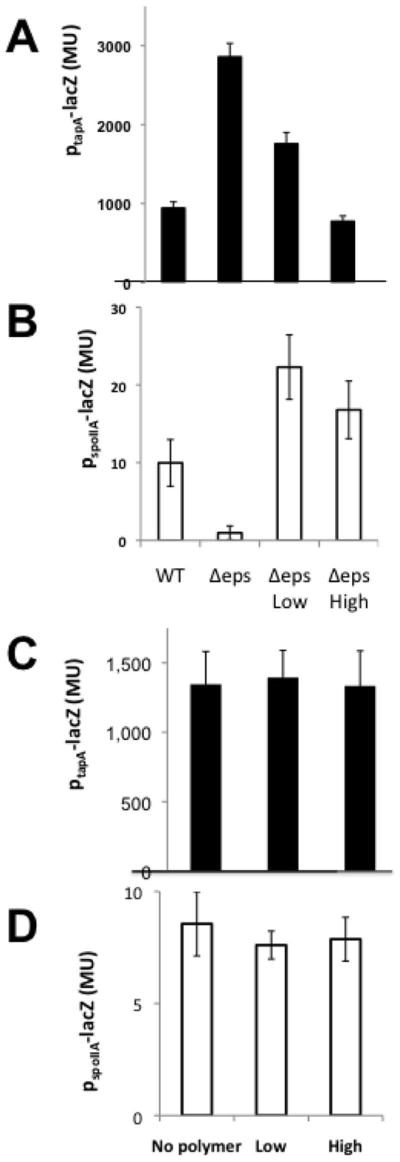
Panels (A) and (B): The activities of PtapA-lacZ in a wild type strain (RL4582) and its Δeps (IKG600) mutant derivative (A) and of PspoIIA-lacZ in a wild type strain (IKG10) and its Δeps derivative (IKG601) (B) were determined without added polymer and with osmolarity increased (using the polymers described in Fig. 2B) to 2 atm (LOW) and 5-8 atm (High). The results shown are the average of using multiple polymers in triplicate. Panels (c) and (d): The activities of PtapA-lacZ in a Δeps mutant lacking kinD (IKG607) (c) and of PspoIIA-lacZ in an Δeps mutant lacking kinD (IKG608) (d) were determined without added polymer and at 2 atm (LOW) and 5-8 atm (High) as indicated above. The columns represent 3 experiments, performed in duplicates, in which different polymers described in figure 2B were used to increase the osmotic pressure. Error bars are the standard deviation.
As a final test of the idea that KinD activity is controlled by osmotic pressure, we measured the osmotic pressure within the pellicle itself. Using a freezing-point osmometer (AI model 3300), we found that the osmotic pressure of the pellicle was higher than that of the medium by 3±1 atm.
Evidence that osmosensing depends on a transmembrane segment
Interestingly, EnvZ of Escherichia coli, a histidine kinase that also senses and responds to changes in osmotic pressure, is known to function as both a kinase and a phosphatase (Hsing et al., 1998; Russo and Silhavy, 1991). Earlier work had shown that the transmembrane segments of EnvZ play an important role in this osmosensing. (Tokishita et al., 1992). Intriguingly, we note a 16-residue long region of similarity between transmembrane segments of the proteins: IVtL--LFasLVttYL in EnvZ versus IVlLniLFi-LVlyYL in KinD (upper case letters indicate residues that are identical between the two sequences; the segment of similarity is highlighted in yellow in Figure 8A).
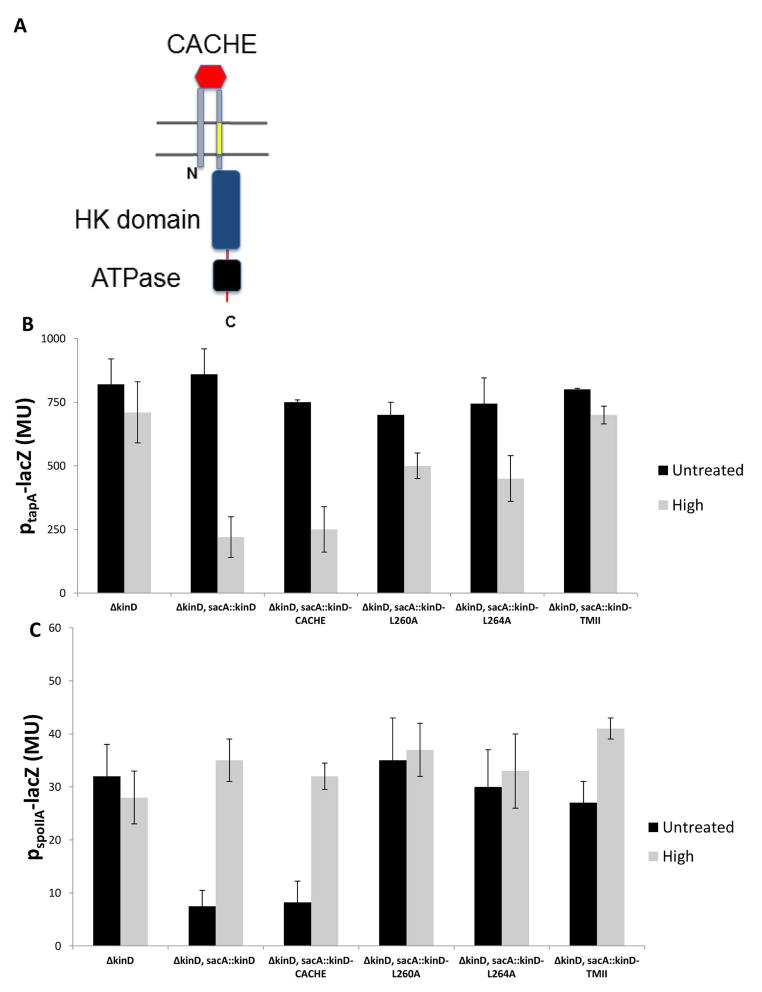
Figure 8. Amino acid substitutions in a transmembrane segment of KinD impair osmosening.
Panel A. Transmembrane segment II of KinD, which is similar to the transmembrane segment of EnvZ is highlighted in yellow. Panels B and C. The activities of PspoIIA-lacZ (b) and PtapA-lacZ (c) were measured at the indicated atms with derivatives of a mutant deleted for kinD (RL4552) and containing the indicated mutant and wild type alleles of kinD at sacA (IKG613). kinD-CACHE is mutant for the CACHE domain (Chen et al, 2012; (IKG614) and kinD-TMII contains the coding sequence for the transmembrane segment of KinB in place of the coding sequence for the second (II) transmembrane domain of KinD (IKG615). High osmolarity was as described for Fig. 7. The columns represent 3 experiments, performed in duplicates, in which different polymers described in figure 2B were used to increase the osmotic pressure. Error bars are the standard deviation.
We created alanine substitutions for several of the residues in the transmembrane segment, asking whether the mutants would be impaired in osmosensing. As shown in Figure 5, replacing two highly conserved leucine residues with alanine (L260A and in L264A) resulted in the partial loss of KinD’s ability to turn matrix genes OFF (Fig. 8B) and sporulation promoters OFF (Fig. 8C). Furthermore, a double mutant for L260 and L264 was conspicuously insensitive to osmotic pressure as judged by colony morphology (Figure S6) and transcription of tapA and spoIIA (Figure S7)
Substitution of the entire transmembrane segment with a transmembrane segment of KinB, a transmembrane kinase that is not required for osmosensing (Figures 8B and 8C), resulted in complete loss of osmosensing by KinD.
Interestingly, KinD has a CACHE domain that appears to respond to a small molecule released by plant roots and to glycerol in high concentration (Chen et al., 2012); M. Shemesh & R.L., unpublished results). KinD mutant for the CACHE domain was largely unimpaired in osmosensing (Figure 8B and 8C). Conversely, however, the L260A and L264A mutants of transmembrane domain II were not blocked in responding to glycerol (data not shown). That these mutants remained active in responding to glycerol in a CACHE-domain dependent manner indicates that the L260A and L264A substitutions had not completely inactivated KinD. Rather, the effect of the substitutions was specific to osmosensing. We therefore hypothesize that KinD is a bifunctional kinase that can independently sense a small molecule(s) via its CACHE domain and osmotic pressure via its transmembrane segment.
Our results suggest that B. subtilis uses the polymers it produces during biofilm formation as a physical cue to coordinate the behavior of cells in the community. Conceivably, the physical properties of matrix polymers are also exploited for synchronizing development in biofilms composed of multiple species. As an indication of the plausibility of this idea, EPS extracted from P. aeruginosa and E. coli pellicles was observed to substitute for B. subtilis EPS in inhibiting matrix gene expression in an eps mutant of B. subtilis (Fig. 9). It will be interesting to see whether the physical properties of biofilms are used as a developmental cue by diverse kinds of biofilm-forming bacteria, including those that form communities in consortia with other bacteria.
Figure 9. Heterologous EPS inhibits matrix gene expression.
An Δeps mutant containing PtapA-lacZ (IKG600) was grown in medium supplemented with either 20 or 30% EPS extracted from B. subtilis 3610, Pseudomonas aeruginosa PA14 and E. coli W3100 pellicles. Percent EPS was calculated by dry weight in the total solution. The assay was performed in 96 well plates.
Finally, we comment on the approach we devised to measure the rheological properties of the biofilm. Probing physical properties ordinarily requires applying physical forces: to measure the strength of a material, it needs to be stretched or squeezed. Thus, biofilm rheological assays usually involve measuring either local microscopic strength of submerged biofilms (Hohne et al., 2009) or impeding the growth of the biofilm (Jones et al., 2011). By allowing B. subtilis to form biofilms naturally around the force probe of the rheometer, we were able to probe the macroscopic robustness of the developing pellicle over time under conditions similar to those used in traditional, bench-top studies without impeding biofilm development. This approach is different and complementary to previous assays in which large biofilms are first grown and then transported to a rheometer (Jones et al., 2011) for mechanical testing or alternatively are grown within a confined microenvironment. Additionally, by using polymers as our osmotic agents rather than small molecules that are more commonly used (such as salts and sugars), we were able to avoid alternative physiological effects well as distinguish among various possible physical properties such as viscosity and ionic effects. We hope that our approach to studying rheology will prove to be of broad applicability in the biofilm field.
MATERIALS AND METHODS
Strains and media
Bacillus subtilis NCIB3610 and its derivatives were grown in Luria-Bertani (LB) medium at 37°C or MSgg medium (Branda et al., 2001a) at 23°C.
Strains used in this work
All B. subtilis strains are derivatives of NCIB 3610, a wild strain that forms robust biofilms (Branda et al., 2001b), and are listed in Table S1.
Strain construction
Strain were constructed using standard methods (Sambrook and Russell, 2001). SPP1 phage-mediated transduction was used to introduce antibiotic resistance-linked mutations from PY79 derivatives into NCIB3610 in different backgrounds (Yasbin and Young, 1974). For mutants of kinD were created using a QuikChange II XL site-directed mutagenesis kit (Stratagene).
Pellicle formation
For pellicle formation in liquid medium, cells were grown to exponential phase and 3μl of culture were mixed with 3 ml of biofilm-inducing medium in a 12-well plate (VWR). Plates were incubated at 22°C. Images of colonies and pellicles were taken using a SPOT camera (Diagnostic Instruments, USA).
β-Galactosidase assays
Pellicles were formed in MSgg medium (Branda et al., 2001b) with/without polymers as indicated in the legend of each figure. The pellicle were collected, washed twice with PBS and sonicated to remove the extra-cellular matrix. OD600 had been measured. 108-109 cells were taken for the assay. Cells were spun down and pellets were re-suspended in 1 ml Z buffer (40 mM NaH2PO4, 60 mM Na2HPO4, 1 mM MgSO4, 10 mM KCl and 38 mM β-mercaptoethanol) supplemented with 200 μg ml−1 freshly made lysozyme. Resuspensions were incubated at 30°C for 15 min. Reactions were started by adding 200 μl of 4 mg ml−1 ONPG (2-nitrophenyl β-D-galactopyranoside) and stopped by adding 500 μl of 1 M Na2CO3. The soluble fractions were transferred to cuvettes (VWR), and OD420 values of the samples were recorded using a Pharmacia ultraspectrometer 2000. The β-galactosidase-specific activity was calculated according to the equation [(OD420/(time × OD600)] × dilution factor × 1000. Assays were conducted in duplicate.
Osmotic pressure modification
The osmotic pressure in the experimental system was modified by the addition of polymers to the growth medium. Dexran and PEG samples were purchased from Sigma-Aldrich Co. Several weight fractions of polymer were dissolved in MSgg or LB and left to tumble slowly until the solution turned optically clear. The osmotic pressure of the modified medium was measure with an “Advanced Instruments Freezing Point Osmometer” (model 3300). The contribution of the polymer to the osmotic pressure was calculated by subtracting the measured osmotic pressure of the pure medium. In addition, by comparing these values with the measurements of the osmotic pressure of the identical mass fractions of polymer dissolved in deionized water we found that the contribution of the polymer to the osmotic pressure was largely identical for all our media. The viscosity of the modified medium was measure on an “Ares G2” using a double-walled Cuvette Geometry.
Pellicle rheology
For the time lapse measurements (Fig. 1c) pellicles were grown on top of an “Ares G2” strain controlled rheometer. 15 ml of MSgg (with or without PEG depending on the experimental intention) were poured into a stainless steel 34-mm well. A cylinder (15-mm diameter) was placed at the center of the well, 7-mm above the bottom so that the liquid/air interface was at approximately half the cylinders height. The cylinder was coupled to the torque sensor of the rheometer. Repetitive, low strain (≤1%) tests of the pellicles strength were conducted by measuring the oscillatory torque on the cylinder, τ, while rotating the well around it’s symmetry axis periodically at small amplitudes (≤0.6°). In this geometrical configuration the measurements were largely insensitive to the fluid and biomass on which the biofilm was formed. This is due to the existence of a large gap between the inner and outer walls. To verify this we used an identical setup and measurement parameters to measure the shear response of inoculated LB medium at an optical density (OD600) greater than 0.5. For this sample we did not resolve any signal above the noise level of the instrument. The whole setup was isolated and held under a controlled temperature of 30°C.
To compare the strength of pellicles grown under different osmotic pressures (Fig. 1d) 6μl of cells in exponential phase were mixed with 6.5 ml of MSgg modified with appropriate amounts of 20 kDa PEG in six well plates (VWR). Roughened PMMA cylinders (15mm diameter) were placed in the center of the wells 1 mm above the bottom. The top of these cylinders consisted of a smaller diameter cylinder (Fig. 1a) that could be mounted on a “Bohlin Gemini II” stress controlled rheometer without disturbing the sample. The plates were incubated in ambient conditions (23°C). The shear strength was measured after ~72 hours of incubation. The first step was to measure the oscillatory torque at low strains (<5%) (red circles in Fig. 1). Then, we continually sheared the pellicles at a strain rate of 1% per second. The torque increased monotonically until at high enough strains the pellicles began to rupture. (We disregarded tests where rupture initiated at one of the walls.) The maximal torque achieved before rupture is defined as the yield torque.
EPS extraction
E. coli strain W3100 was grown in static M9 glycerol for 4 days. B. subtilis strain 3610 was grown in static biofilm-inducing medium for 3 days. P. aeriginosa PA14 was grown in static M63 glycerol medium for 7 days. Pellicles were harvested, washed twice in PBS, and mildly sonicated. Cells were removed by centrifugation. The supernatant fluid was mixed with cold isopropanol in a 5:1 ratio and incubated in 4°C overnight. Samples were centrifuged at 8000 rpm, 4°C for 10 mins. Pellets were re-suspended in a digestion mix of: 0.1M MgCl2, 0.1 mg/mL DNase, 0.1mg/mL RNase solution, mildly sonicated and incubated for 4 hrs at 37°C. Samples were extracted twice with phenol- chloroform. The aquatic fraction was dialyzed for 48 hrs against DDW using Slide-A-Lyzer Dialysis with 3.5 MWCO (Thermo Fisher).
Supplementary Material
Acknowledgments
We thank M.P Brenner and A. Seminara for helpful discussions and J. Willking for assistance with the rheology experiments. This work was supported by the Harvard Materials Research Science and Engineering Center (DMR-0820484) and by grants from BASF to D.W., R.L. and R.K., from the NIH to R.L. (GM18568) and to R.K (GM58213), and from the NSF (DMR-1006546) to D.W.
References
- Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. KinD Is a Checkpoint Protein Linking Spore Formation to Extracellular-Matrix Production in Bacillus subtilis Biofilms. MBio. 2010;1 doi: 10.1128/mBio.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 1993;7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001a;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001b;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Vik A, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005a;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005b;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Norman T, Kolter R, Losick R. Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillussubtilis. EMBO J. 2011:1402–1413. doi: 10.1038/emboj.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cao S, Chai Y, Clardy J, Kolter R, Guo J-h, Losick R. A Bacillus subtilis Sensor Kinase Involved in Triggering Biofilm Formation on the Roots of Tomato Plants. Molecular Microbiology. 2012 doi: 10.1111/j.1365-2958.2012.08109.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Fujita M, Gonzalez-Pastor JE, Losick R. High- and Low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. Evidence that entry into sporulation in Bacillussubtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Hohne DN, Younger JG, Solomon MJ. Flexible Microfluidic Device for Mechanical Property Characterization of Soft Viscoelastic Solids Such as Bacterial Biofilms. Langmuir. 2009;25:7743–7751. doi: 10.1021/la803413x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing W, Russo FD, Bernd KK, Silhavy TJ. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol. 1998;180:4538–4546. doi: 10.1128/jb.180.17.4538-4546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- Jones WL, Sutton MP, McKittrick L, Stewart PS. Chemical and antimicrobial treatments change the viscoelastic properties of bacterial biofilms. Biofouling. 2011;27:207–215. doi: 10.1080/08927014.2011.554977. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Kolter R, Greenberg EP. Microbial sciences: the superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- Kostakioti M, Hadjifrangiskou M, Pinkner JS, Hultgren SJ. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol Microbiol. 2009;73:1020–1031. doi: 10.1111/j.1365-2958.2009.06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon KP, Earl AM, Vlamakis HC, Aguilar C, Kolter R. Biofilm development with an emphasis on Bacillus subtilis. Curr Top Microbiol Immunol. 2008;322:1–16. doi: 10.1007/978-3-540-75418-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Russo FD, Silhavy TJ. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- Ruzal SM, Sanchez-Rivas C. In Bacillus subtilis DegU-P is a positive regulator of the osmotic response. Curr Microbiol. 1998;37:368–372. doi: 10.1007/s002849900395. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Tokishita S, Kojima A, Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the transmembrane regions of membrane-located protein kinase, EnvZ. J Biochem. 1992;111:707–713. doi: 10.1093/oxfordjournals.jbchem.a123823. [DOI] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qin L, Yoshida T, Inouye M. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc Natl Acad Sci U S A. 2000;97:7808–7813. doi: 10.1073/pnas.97.14.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.