Abstract
Aims
Next-generation sequencing is being implemented in the clinical laboratory environment for the purposes of candidate causal variant discovery in patients affected with a variety of genetic disorders. The successful implementation of this technology for diagnosing genetic disorders requires a rapid, user-friendly method to annotate variants and generate short lists of clinically relevant variants of interest. This report describes Omicia’s Opal platform, a new software tool designed for variant discovery and interpretation in a clinical laboratory environment. The software allows clinical scientists to process, analyze, interpret and report on personal genome files.
Materials & Methods
To demonstrate the software, the authors describe the interactive use of the system for the rapid discovery of disease-causing variants using three cases.
Results & Conclusion
Here, the authors show the features of the Opal system and their use in uncovering variants of clinical significance.
Keywords: analysis and selection tool, genome analysis, next-generation sequencing, variant annotation, variant workflows, variant annotation, whole-genome sequencing
Next-generation sequencing (NGS) for clinical research and diagnostics is expanding as technical complexity and costs decline. It is now possible to diagnose inherited disorders based on whole-genome or exome sequencing of affected and unaffected relatives or even single affected individuals. To facilitate the use of NGS as a diagnostic tool for identifying genetic causes of disease, novel informatics tools are needed to handle these large data sets with thousands to millions of detected variants from the reference sequence. NGS sequence analysis can be divided in two distinct separate steps: variant calling and variant analysis. Variant calling deals with the processing of raw data (BAM files or FASTQ files) as well as performing alignment, assembly and generation of variant call format (VCF) or genome variant format (GVF) variant files. Variant analysis utilizes a selection of tools that integrate the functional annotation of the variants generated from the variant calling pipeline. Here, the authors focus on variant analysis including causal variant discovery. A common method of causal variant discovery used primarily for research purposes and described in many recent publications is heuristic variant filtering [1–6]. This filtering method is based on assumptions about the attributes of the disease-causing variant(s), including the effect of the variant on the protein, the presumed absence of the variant in the Single Nucleotide Polymorphism database (dbSNP) or frequency cutoffs based on minor allele frequency from the 1000 Genomes Project. Typically, these filtering strategies are performed with software that requires knowledge of Linux or Unix command-line language and/or requires the user to learn complex programs. To analyze NGS data in the context of research projects, the authors' group at ARUP Laboratories performs variant calling utilizing Burrows–Wheeler Aligner (BWA) [7,8], Sequence Alignment/Map Tools (SAMTools) [9] and Genome Analysis Toolkit (GATK) [10,11] for sequence alignment and variant calling [12]. Sequence quality control steps and initial variant filtering is done with the SNP and Variation Suite (Golden Helix, Bozeman MT). Currently, variants are then analyzed by hand for the function of the gene as it relates to our particular clinical case, conservation of the base/amino acid change and whether the gene has previously been associated with the disorder of interest. While this method works well in the research setting, it is a time-consuming process that requires trained bioinformatics and scientific personnel.
Methods for analysis of NGS-based clinical tests, however, should be capable of fast and accurate clinical annotation, prioritization of detected variants by interactive data mining, and variant reporting capabilities. These methods should also be accessible to all clinical laboratory personnel involved in NGS test interpretation. These requirements for clinical use of NGS have contributed to the difficulty and longer analysis time for finding disease causing genes from human genome or exome sequencing data and have driven the development of software platforms for use in the clinical testing environment. Here the authors describe the most recently released version of Opal, the Omicia software platform, which addresses the need to quickly analyze, interpret and generate reports on personal genomes in a clinical setting. This platform is accessed using the Omicia Opal web interface and can be used to filter data consisting of millions of variants to a limited set of potential pathogenic candidate variants. Additional commercial software platforms for identifying causative variants for clinical analysis include Variant Analysis (Ingenuity), Silicon Valley Biosystems and Knome. A review of variant calling and analysis tools can be found at Pabinger et al. [13]. Here, the authors focus on Omicia’s unique approach to variant annotation and heuristic filtering as well as the integration of the Variant Annotation, Analysis and Selection Tool (VAAST) prioritization tool that allows variant prioritization without heuristic filtering [14]. The software is described in detail here and we demonstrate its utility with three cases of NGS data sets as examples of scenarios seen by clinical geneticists.
Materials & methods
Software architecture
Omicia Opal is implemented in a software-as-a-service model. All user interactions take place through web browsers using the secure https protocol. Users log in with a username and password (which can be specific to each project) and can then upload variant files in one of the acceptable formats as described in the following section. These files are transferred to Omicia’s Linux-based servers, where they are validated, stored and analyzed by Omicia’s Opal Annotation Pipeline and Omicia’s VAAST analysis tool, using cloud-based servers. Computationally annotated variants are loaded into Opal’s relational database for further variant mining by users through the Opal Variant Miner web interface. As a multitier system, Opal utilizes a variety of technologies and programming languages. The system is currently accessible through a 128-bit encrypted connection and is Health Insurance Portability and Accountability Act compliant. Each user is given access to only their own data, or data that the user elects to share with others by explicitly giving them permission through the user interface.
Omicia’s Opal annotation pipeline
Variants are defined as a sequence change from either the GRCh37/hg19 or NCBI36/hg18 genomic reference sequence. The variant files can contain whole-genome, exome or targeted NGS data from various platforms. Acceptable formats are VCF, GVF and the Complete Genomics’ master Var format. Upon upload, the Omicia Opal system processes each variant list through a series of annotation programs called the Omicia Opal Annotation Pipeline. The Annotation Pipeline runs on Omicia’s servers and annotates variants using the following multistep workflow. First, the files are converted from their input format into a common internal representation due to the significant diversity encountered in the implementation of the VCF formats, particularly in the description of the number of sequence reads per variant allele. Next, the version of the genomic reference sequence used in variant file generation is verified by comparison with known polymorphisms in the National Center for Biotechnology Information dbSNP 135 database [101]. If more than 60% of the single-nucleotide variants in an exome or genome file have positions on a particular version of the genomic reference sequence identical to known dbSNP entries, then that version is assigned to the variant file. For smaller numbers of variants, as seen in targeted-sequence projects, the pipeline relies strictly on user-supplied genomic reference annotation. The pipeline then gathers a number of frequently-used summary statistics about the files, including the number of variants, median Phred-like quality [15] of variants, median reads per variant and transition/transversion ratio. Genomes and exomes for which these measures fall more than two standard deviations from the median value in previously observed genomes or exomes are flagged as being problematic. A score is derived from these parameters, and noted as the Omicia Genome Clinical Grade. The pipeline then classifies the function of each variant using the ANNOVAR tool [16]. Each variant is classified according to the location within and effect on the protein. Variants in protein coding regions are classified as synonymous, nonsynonymous, stopgained, stop-lost, frameshift insertion/deletion, nonframeshift insertion/deletion and splice-site variants. Variants outside of protein coding regions are classified as either 3´ untranslated region (UTR), 5´ UTR, intronic, intergenic or splice-site variants. The Omicia system uses a combination of the Ensembl Database release 62 [17] and RefSeq [18] databases as a basis for these classifications and presents the variant and protein change in Human Genome Variation Society nomenclature [19]. The annotated gene names use the official HUGO Gene Nomenclature Committee gene symbols [20]. If the variant is present in the NCBI dbSNP 135 database, then the variant is annotated with the dbSNP identifier (rs number). Variants found in the Online Mendelian Inheritance in Man (OMIM) database of disease-causing mutations have a specific OMIM hyperlink to the variant [102]. OMIM variants are typically described in coordinates relative to protein sequences as described at the time of their publication, and, over time, these variant coordinates can become invalid as the reference genome is updated. The Omicia pipeline uses a variant alignment algorithm in order to annotate the variants’ position on the hg19 version of the reference genome. Variant annotations (and if available, hyperlinks) are also given for the following databases: the Human Genome Mutation Database, version 7.2 (HGMD) [103], Phencode collection of locus-specific databases [21], the National Human Genome Research Institute Catalog of Published Genome-Wide Association Studies and the Pharmacogenetics Knowledge Base (PharmGKB) [104]. In addition, variants are annotated with the allele and genotype frequency information from the 1000 Genomes Project [105]. This information can be used to distinguish common polymorphisms from rare, possibly disease-causing mutations.
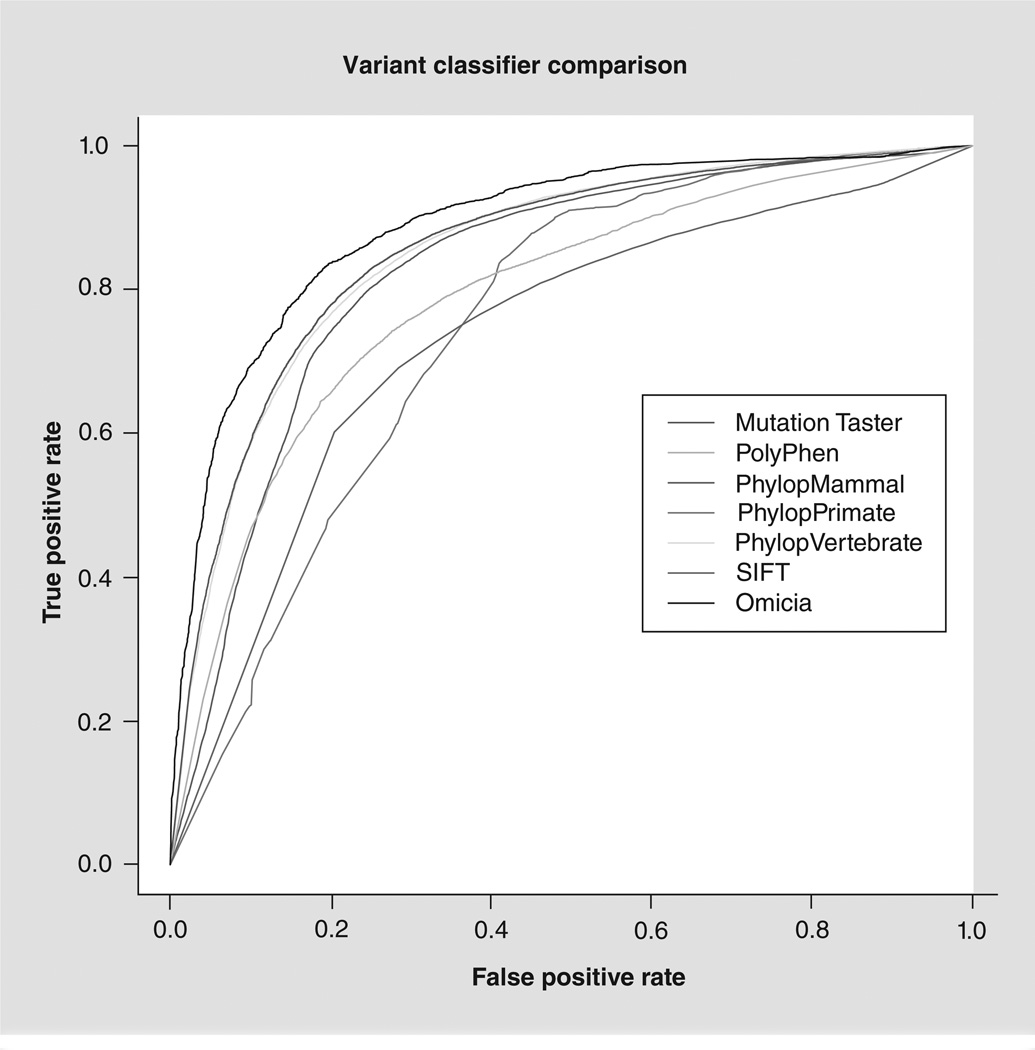
Next, scores predicting pathogenicity are generated for each protein-coding variant using the following programs: Sorting Intolerant from Tolerant (SIFT) [106], PolyPhen 2 [107], MutationTaster [108] and PhyloP [22]. The SIFT scores are a prediction of the tolerance for certain amino acid changes within the protein and are based on evolutionary conservation at that protein position. SIFT p-values below 0.05 indicate that the change is likely deleterious. PhyloP assesses the evolutionary conservation of each position. The PhyloP score is the −log(p-value) under a null hypothesis of neutral evolution, and a negative sign indicates faster than expected evolution, while positive values imply conservation. PolyPhen 2 is a tool that predicts possible impact of an amino acid substitution on the structure and function of a human protein using a number of structural, physical and comparative considerations. This algorithm produces one of three calls for nonsynonymous variants: benign, possibly damaging and probably damaging. MutationTaster predicts whether protein-coding variants are disease-causing or benign and assigns a p-value to these predictions. Each variant is further assessed for pathogenicity using a simple decision-tree algorithm, which generates the Omicia Variant Score, a random-forest classifier [23] trained using a selected set of mutations in the HGMD database marked as ‘disease-causing’, representing a highly reliable set of true disease-causing mutations. As a negative control, the authors developed a similarly-sized set of SNPs from the dbSNP132 database with a minor allele frequency above 5%, which therefore is assumed to be benign. The random-forest classifier was trained using the scores for variants that have a protein impact as attributes from the four programs SIFT, PolyPhen, MutationTaster and PhyloP. A randomly chosen subset of 10,000 variants was left out of the combined training set and used in validation. The underlying classifier assigns each variant a ‘benign’ or ‘pathogenic’ classification and a confidence value between 0 and 1 for that classification. To enable simple filtering strategies, these classifications are scaled to a single-valued 0–1 scale with 1 corresponding to variants that the Omicia Variant Score has highest confidence in being pathogenic and 0 corresponding to variants the Omicia Variant Score determines are most likely to be benign. A receiver operating characteristic curve, showing a performance comparison to the individual prediction programs, shows the improved performance of the integrated score (Figure 1). As can be seen in Figure 1, a score of 0.85 or higher generates a 1% false-positive prediction rate within our testing set. A variant score higher than 0.85 is considered to be likely pathogenic and a score between 0.5 and 0.85 is considered potentially pathogenic. Finally, the output of the Annotation Pipeline is loaded into a data repository utilized in the Variant Minerview. For further details on the method and the program, see the Omicia website [109].
Figure 1. Omicia score.
Receiver operating characteristic curve of the performance of different variant impact assessment algorithms on 10,000 test variants, including Human Gene Mutation Database disease-causing mutations and benign high frequency mutations from dbSNP.
Data for simulated case study
The simulated case study variant files with the spiked mutations were each loaded into the Omicia system in VCF format via its web interface [24]. The blinded study used the publicly available Complete Genomics variant sets from samples HG00731, HG00732 and HG00733, corresponding to the father, mother and daughter from a Puerto Rican family, respectively. The chosen mutations were added to each data set as appropriate for each disease inheritance scenario. The genome variant files were generated using Complete Genomics software version 2.0.0.26 and downloaded from the Complete Genomics website [110].
Case 1 simulated progressive familial intrahepatic cholestasis (OMIM211600), an autosomal recessive disorder caused by mutations in the ATP8B1 gene (RefSeq:NM_005603.4). To construct a compound heterozygous scenario in the daughter, the heterozygous chr18:55362420C>A (p.Gly308Val) [25] mutation was added into the mother’s variant file (HG00732). The heterozygous chr18:55342225C>T (p.Asp554Asn) [26] mutation was added into the father’s variant file (HG00731). Both heterozygous mutations were added to the daughter’s variant file (HG00733) to create the compound heterozygote.
Case 2 simulated a scenario of an autosomal dominant severe congenital neutropenia (OMIM202700), which is caused by variants in the ELANE gene (RefSeq:NM_001972.2), in two unrelated patients. For this scenario, the two mutations added were described by Dale et al. [27]. The heterozygous chr19:853338G>A mutation, p.Val72Met, was added into the variant file for sample HG00731 and the heterozygous chr19:855613C>T mutation, p.Pro110Leu was added to the variant file for sample HG00732.
Once all five of the simulated case study genomes had been processed through the Annotation Pipeline, a researcher was given the pedigrees and a short description of the symptoms and the Opal software was then used to find the causal variants and diseases. The researcher had no foreknowledge of the genes or mutations involved that had been added into the Complete Genomics variant files. To identify causative variants, default filters were used for each patient tested. These filters required variants to have protein impact (any mutation in a protein-coding region that is not synonymous) and required that variants be present in NCBI’s RefSeq gene database. The user also applied two basic quality filters to the data, requiring read coverage greater than or equal to 20, and a Complete Genomics quality score greater than or equal to 100 [106]. Finally, as the diseases were both rare, the user filtered variants to a set that had minor allele frequencies of less than 2% in the 1000 Genomes Project.
Case 3 used a single VCF file generated by a study looking for the causative variant for Ogden syndrome, a very rare X-linked disorder [28]. This variant file was generated by capture of the X chromosome of an affected infant male and subsequent NGS and is publically available on the ANNOVAR website [111]. The same default filters were used for Case 3 as described for Cases 1 and 2 with exceptions noted in the results section. More specific filtering steps for all cases are described below in the clinical test cases section.
Results
Omicia Opal web interface: features & layout
The home page is displayed after logging in to Opal using the secure login specific to each user. From here, the user can upload variant files, access tools for data analyses, access previously generated clinical reports and manage account settings. The upload page is where genomic data is uploaded into Opal. Opal accepts whole genome, exome or gene variant data sets in several common variant file formats: GVF [29], VCF, Complete Genomics master Var files and Illumina Clinical Service variant files. The data are then submitted to Opal’s Annotation Pipeline. The annotation process takes approximately 1 h for a typical whole genome variant file and approximately 20 min for an exome variant file. The variant data can be uploaded into folder-like projects. Each user can create her/his own personal workspace under the My Reports tab, and users with appropriate privileges can create other projects as desired, for example, granting access to colleagues as needed for a particular study or collaboration. Additionally, the Public Projects folder is publicly available to all Opal users and contains annotated data from several whole-genome NGS data sets, including those of James Watson, J. Craig Venter, and Stephen Quake. This Public Projects folder is provided for users without NGS data that want to familiarize themselves with the Omicia software and can be used free of charge.
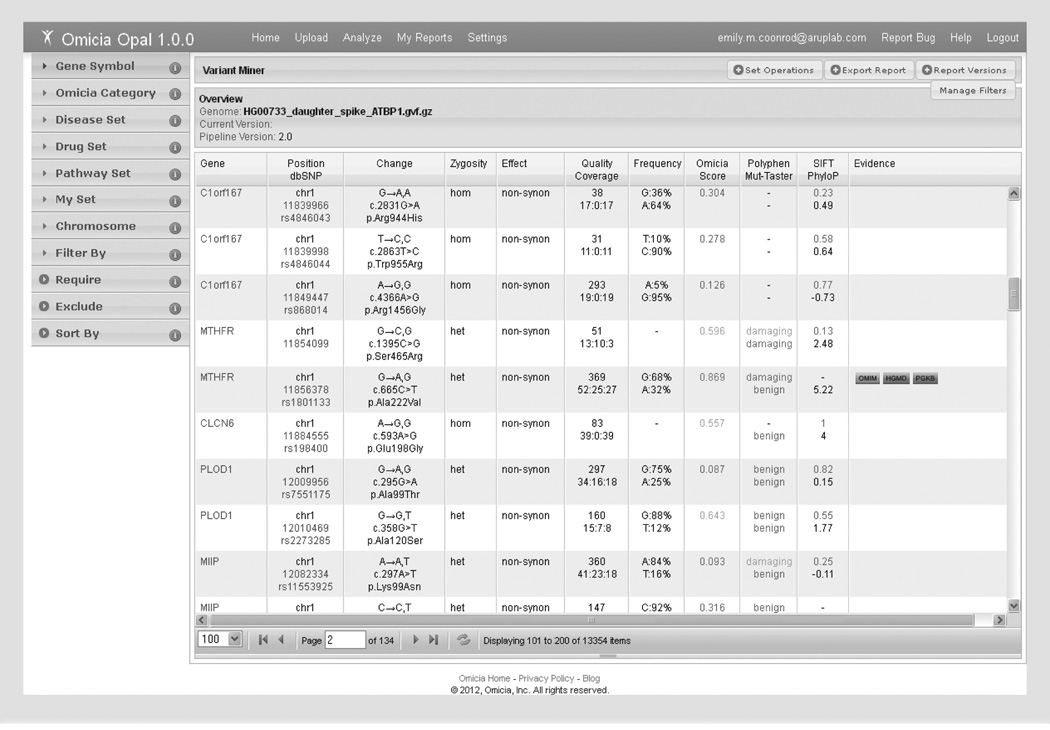
Clicking on the project’s folder in the My Reports tab transfers the user to the data sets in the project. The various report types are listed for each file and the Variant Miner Report is accessed from this window. The Variant Miner Report is the primary mechanism to identify variants of interest using a set of filtering criteria together with biological context that are accessed by clicking the Variant Miner button in the Variant Report. Figure 2 shows the Variant Report of a subset of variants from the Complete Genomics data set prior to applying any filters. The report is divided into two main panels: the variant annotation grid and filters and knowledge sets.
Figure 2. Opal Variant Miner webpage.
The Variant Miner consists of the variant annotation grid and filtering options by knowledge sets or variant properties. To the left of the table are the multiple available filtering options (in the collapsible windows). The bottom of the table lists the number of variants (items) left after each filtering step. Each variant is listed per table row, and ordered numerically by chromosome number and position. Hyperlinks to additional information are available for the Gene (in blue), Position/dbSNP (in blue), and Evidence (boxed) columns. Quality and Coverage information comes from the next-generation sequencing data file, if available. The allele frequency is from the 1000 Genomes frequency data. Red numbers and words in the Omicia/Polyphen/SIFT columns indicate predicted damaging variants, yellow indicates the prediction of a potentially damaging variant and green indicates a benign variant.
The Variant Miner Report displays the annotated variants in a table format referred to as the variant annotation grid. The variant annotation grid displays the following data columns: Gene, Position/dbSNP identifier, Change, Zygosity, Effect, Quality/Coverage, Frequency, Omicia score, MutationTaster (Mut-Taster)/Polyphen scores, SIFT/PhyloP scores and Evidence, but it is customizable and the user can select which annotations to display. The Gene column lists the gene containing the variant with HUGO Gene Nomenclature Committee [112] gene symbols serving as hyperlinks, which takes the user to a separate window with more information about the gene (described in detail in next paragraph). The Position/dbSNP column contains the variant’s chromosomal location with a link to the University of California Santa Cruz (UCSC) genome browser and the SNP identification number (rs number) if found in dbSNP, with an embedded URL link to dbSNP, if applicable. The Change column shows the reference nucleotide and the variant nucleotide as reported in the sample’s NGS data. In addition, the Human Genome Variation Society nomenclature is listed for the nucleotide (cDNA position) and protein change if the variant was present in the coding regions. The Zygosity column lists the genotype of the variant as either heterozygous (het) orhomozygous (hom). Effect refers to the impact of the variant on the gene and transcripts; that is, synonymous, nonsynonymous, stop gain/loss, indel/frameshift and splice variants. The Quality and Coverage is also uploaded into Opal if the NGS quality or read coverage data are available in the variant file. The Quality metric refers to the variant’s Phred-like quality score as generated by the user’s selected variant calling software. Below the quality score, the NGS read coverage depth is listed as ‘total reads: reference nucleotide reads (wild type reads): reads containing the variant’. In the Frequency column, the reference allele frequency is followed by the frequency of the variant, which is calculated from data generated by the 1000 Genomes Project and provided by dbSNP in the Global Minor Allele Frequency field. The Omicia score is an aggregation of scores from PolyPhen, MutationTaster, SIFT and PhyloP designed to indicate the probability that a variant is deleterious. The disease Evidence column information is specific to the variant listed in that row and shows hyperlinks to the databases or the literature references from which variant-specific information was mined by Opal. The literature and database evidence was gathered from OMIM, HGMD, the Phen code collection of Locus Specific Databases, the National Human Genome Research Institute Catalog of Published Genome-Wide Association Studies and PharmGKB.
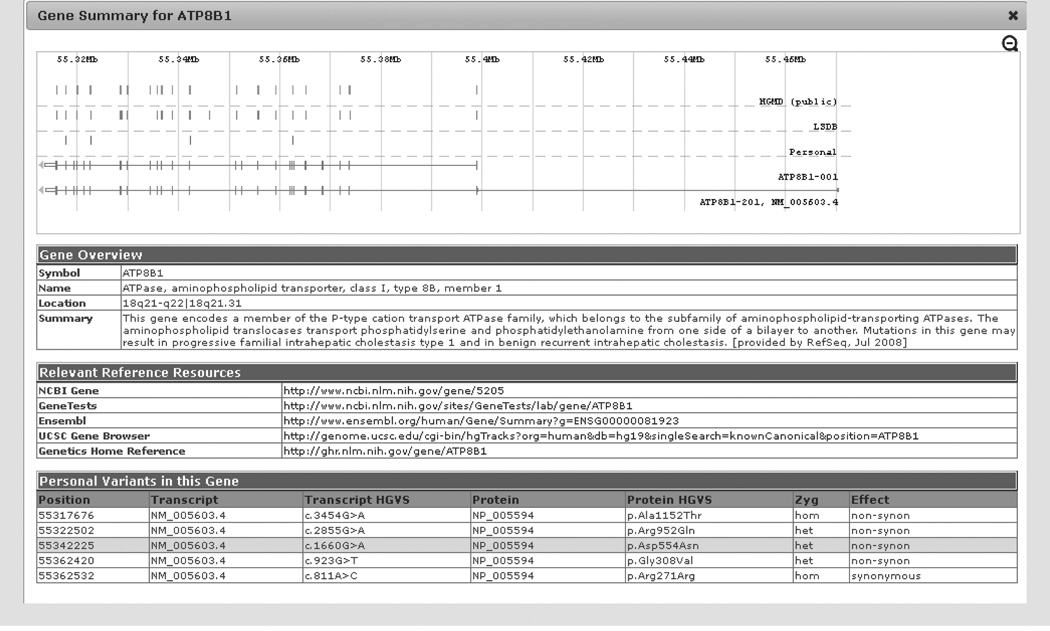
Each individual gene in the variant annotation grid is hyperlinked to a Gene Summary window (Figure 3). The Gene Summary window contains graphics that display the gene structure, the location of any personal variants from the sample, the location of any variants in the Locus Specific Databases and variants from HGMD specific to the gene. The gene symbol, full name, chromosomal cytoband location and summary of the NCBI listed gene function are found in the Gene Overview section of the Gene Summary window. The next section (Relevant Reference Resources) contains hyperlinks to the NCBI Gene, Gene Tests, Ensembl, UCSC Gene Browser and Genetics Home Reference web pages specific to the individual gene(if available). The Gene Tests link [113] provides information on the associated disease along with information on companies performing clinical testing and the types of clinical tests available for the gene. The genetics home reference webpage [114] has information on the gene’s involvement in human health. The last section shows the Personal Variants in this gene in the individual’s data set. Each personal variant will have the cDNA position and nucleotide change (e.g., c.1660G>A), the protein position and amino acid change (p.), along with the transcript number (NM_#) and protein number (NP_#) associated with the variant. Variant zygosity and protein effect is also listed. The highlighted row denotes the variant from which the Gene Summary was selected. If applicable, this window will also contain any Omicia Disease Categories and any information in the Associated Knowledge Sets, such as the Disease Set, Drug Set or Pathway Set related to the gene.
Figure 3. Gene summary window.
This window will open if the Gene symbol hyperlink is used from the variant annotation grid. The window contains the gene structure figure with the variant positions marked, a Gene Overview with the NCBI gene summary, and the Relevant Reference Resources has a number of hyperlinks. Any other variants found in the patient for this gene are listed under Personal Variants with the variant in the row where the gene link was instigated highlighted in yellow.
Searches & filters
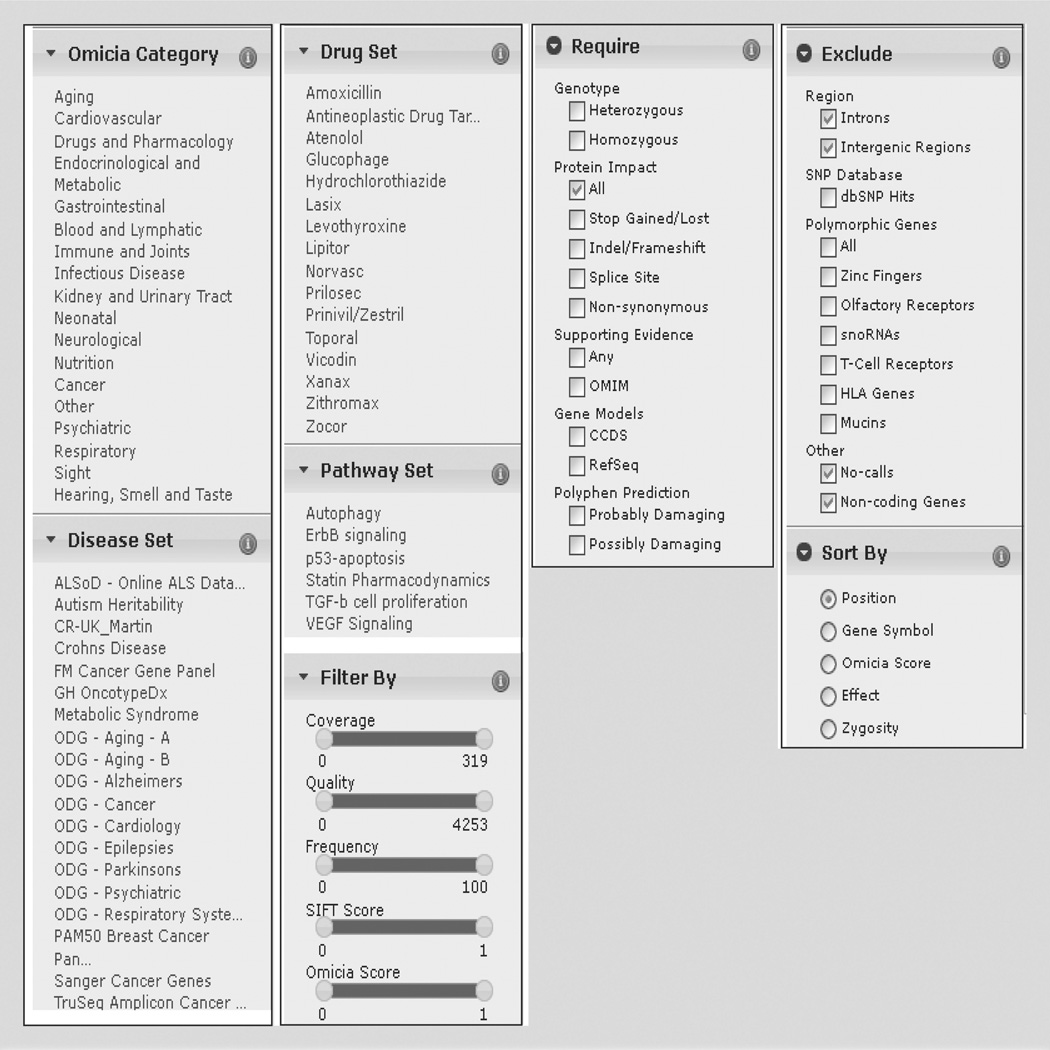
The Variant Miner view page also has a selection of searches and filtering methods, using evidence from scientific literature, variant properties and knowledge sets (Figures 2 & 4). The Filter By selection enables the user to filter variants using five numeric criteria generated in Opal’s Annotation Pipeline (Figure 4). Using interactive sliders, users can filter by NGS read coverage depth or variant quality, minor allele frequency of the variant in the 1000 Genomes Project data set, SIFT score and Omicia Variant Assessor Score. Users can also limit results based on the effect of the variant on the protein, for example, show only stop-gained or nonsynonymous variants, or require that variants have supporting disease evidence from any of the databases utilized in the Annotation Pipeline, for example, OMIM. Users can choose to exclude variants present in introns, intergenic regions, highly polymorphic genes and variants that are present in dbSNP 135. In addition, users can limit variants by chromosome number or even to specific genes by gene symbol.
Figure 4. Variant filtering windows.
The selections available in each of the various collapsible windows (found on the Variant Miner page) used for data filtering are displayed.
In addition, users can restrict variant lists to genes that are present in curated gene sets. Opal provides the following five groupings of gene sets: Omicia Categories, Disease Set, Drug Set, Pathway Set and My Set (Figures 2 & 4). The My Set filter contains a custom set of genes created by the user, and the other sets are populated and provide a convenient entry point into the genome for clinicians. The Harrison Category set contains genes that are associated with particular high-level disease areas, such as aging and cancer (Figure 4). Omicia curates the disease categories in collaboration with experts in each disease area. The category names are based on the section headers of the Harrison textbook Principles of Internal Medicine [30], which is used in the education of physicians. The Disease Set contains genes that are known to be related to particular diseases, for example, autism, Crohn’s disease and metabolic syndromes (Figure 4). Omicia compiles these disease-related gene sets in collaboration with disease experts. The Drug Set contains genes that are relevant to the safety and efficacy for a collection of top-prescribed drugs with examples including Lipitor®, Prilosec® and Xanax®. Omicia compiles these drug-related gene sets in collaboration with pharmacology experts. The Pathway Set contains sets of genes that are members of particular pathways, for example, the VEGF pathway. After changing a Filter or Knowledge Set, the variant annotation grid will update and the new resulting variant count is indicated at the bottom of the grid.
Genome operations: intersects & differences between data sets
The proband’s variants can also be filtered based on the presence or absence of variants in other data sets within the same workspace, for example, variants in the genomes of family members, unaffected control genomes or affected nonrelated genomes. After clicking the Set Operations button in the Variant Minerview, all other data sets in the same workspace are displayed. One or more data sets can be selected and used in the comparison. Once the background data set(s) is selected, there are four types of set operations available. The Variant Difference returns the variants that are different between the proband and the selected background data set(s). The Variant Intersect function returns the variants that are present both in the proband and selected other data sets (such as from an affected sibling). The Gene Difference returns proband variants that are present in the genes where the background data set(s) do not carry the same variants. The Gene Intersect returns variants that are present in genes where both the proband and the selected genome(s) carry variants. The gene intersect function is useful when testing several unrelated patients with the same disease because they may have mutations in the same gene but are unlikely to have variants at exactly the same positions.
If a set of filters is enabled in the proband genome, then a comparison is performed after the same filters are automatically applied to the background genomes. A list of variants or genes that meet the selected criteria are subsequently displayed and the data set can be filtered by the gene or variant list generated, by either eliminating variants or genes that are the same as the control or unaffected genomes (difference functions) or retaining only the variants or genes in common between data sets (intersect functions). Once the filtering and set operations are done, if desired, the variant view table can be exported as a text file using the Export button.
Clinical test case 1
To demonstrate the utility of the Omicia platform and filtering options, three test cases were performed. Case 1 is a study of an affected daughter and the unaffected parents. The researcher given the variant spiked study data was told that the affected daughter had pruritus and failure to thrive, and also that the inheritance pattern expected was recessive. Initially, the mother, daughter and father had a total of 3,840,652; 3,759,721 and 3,724,239 variants, respectively. After the default and basic filtering steps were performed on the proband (for read coverage, Complete Genomics quality score (100), and allele frequency) as described in the methods, a total of 901 variants remained. This variant set was intersected by gene with the parents using the set operations functions to look for either compound heterozygous or homozygous variants. After the genes were intersected with both parents, 227 genes remained. Removing variants with an Omicia Variant Assessor Score of 0.7 left 27 genes remaining. Of these 27 genes remaining five contained homozygous variants and 22 contained compound heterozygous variants. By requiring ‘supporting evidence’, every gene except ATP8B1 was removed. Variants in the ATP8B1 gene cause autosomal recessive progressive familial intrahepatic cholestasis. The patient was heterozygous for two known deleterious, nonsynonymous ATP8B1 variants (chr18:55342225C>T and chr18:55362420C>A).
Clinical test case 2
For Case 2, the researcher performing the blinded study was told that the genomes belonged to two unrelated patients with the same symptoms of recurrent bacterial infections in early childhood, and also that the expected inheritance pattern was autosomal dominant. The original Complete Genomics data for the mother and father in the previous case were spiked with the variants of interest and given to the researcher. After the default and basic filtering steps (for read coverage, quality (100) and allele frequency) were applied as described in the methods, the patients had 985 and 1024 variants left. These patients were intersected by gene using the set operations functions, (this intersect was done by gene rather than position because unrelated patients with the same symptoms may have the same causal gene, but different mutations within the gene). After applying the gene intersect, these patients had 307 genes in common. Then the ‘require supporting evidence’ filter was used, yielding only six candidate genes. When the Omicia Score was required to be higher than 0.7, only one gene was left: ELANE. The ELANE gene fits the phenotype of the patients and is known to cause autosomal dominant severe congenital neutropenia and cyclic neutropenia. This disease causes a deficiency of neutrophils which results in reoccurring infections. Each patient was heterozygous for a known deleterious nonsynonymous ELANE variant (chr19:853338G>A for one patient, and chr19:855613C>T for the other patient).
Clinical test case 3
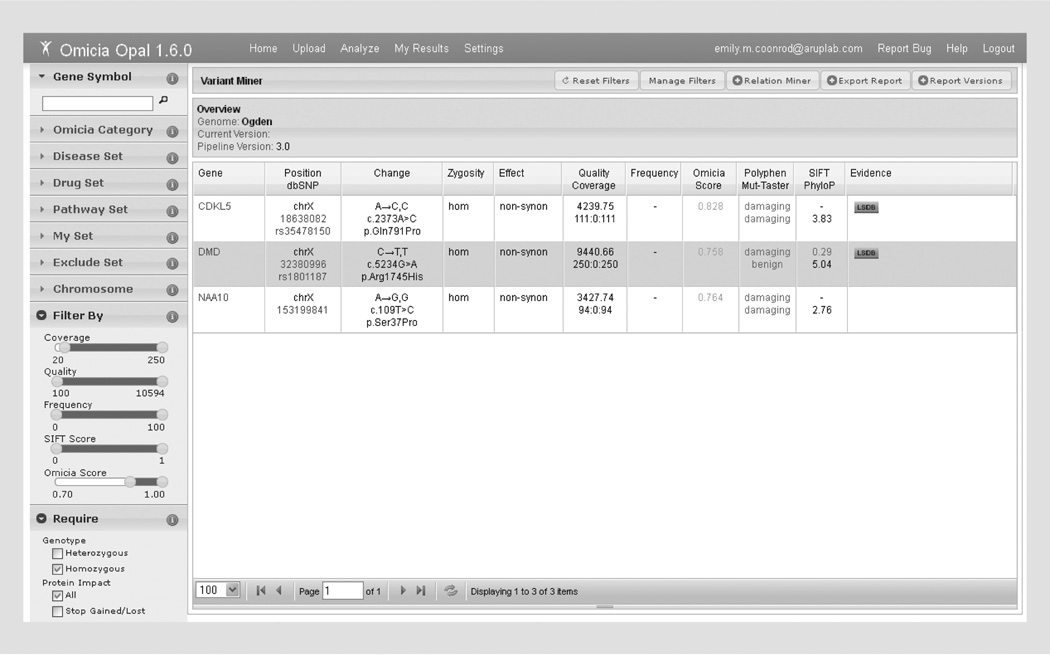
This test case used a VCF file generated by DNA capture of the X chromosome in an infant male affected with a very rare X-linked disorder, Ogden syndrome [28]. The VCF file from this single individual was retrieved from the ANNOVAR website [107] and uploaded into Opal. The default read (20) and quality (100) filters were applied, which decreased the number of candidates from 166 to 160. There was no allele frequency information associated with the variants, so no frequency filter was set. When the Omicia Score was required to be 0.7 or higher, seven variants remained. Requiring the variants to be homozygous (heterozygous variants on the X chromosome in a male are probably sequencing error) removed one variant for a total of six. Requiring the Polyphen prediction to be probably or possibly damaging dropped the number of candidates to the following three genes: CDKL5, DMD and NAA10 (Figure 5). Manual exploration of the variants using the Gene Summary tab showed that NAA10 is involved in Ogden syndrome, making the NAA10 c.109T>C variant the obvious choice for a candidate in this case.
Figure 5. Filtering results for clinical test case 3.
The three genes remaining after heuristic filtering in clinical test case 3 are shown in the variant miner view.
Integration of VAAST into Opal
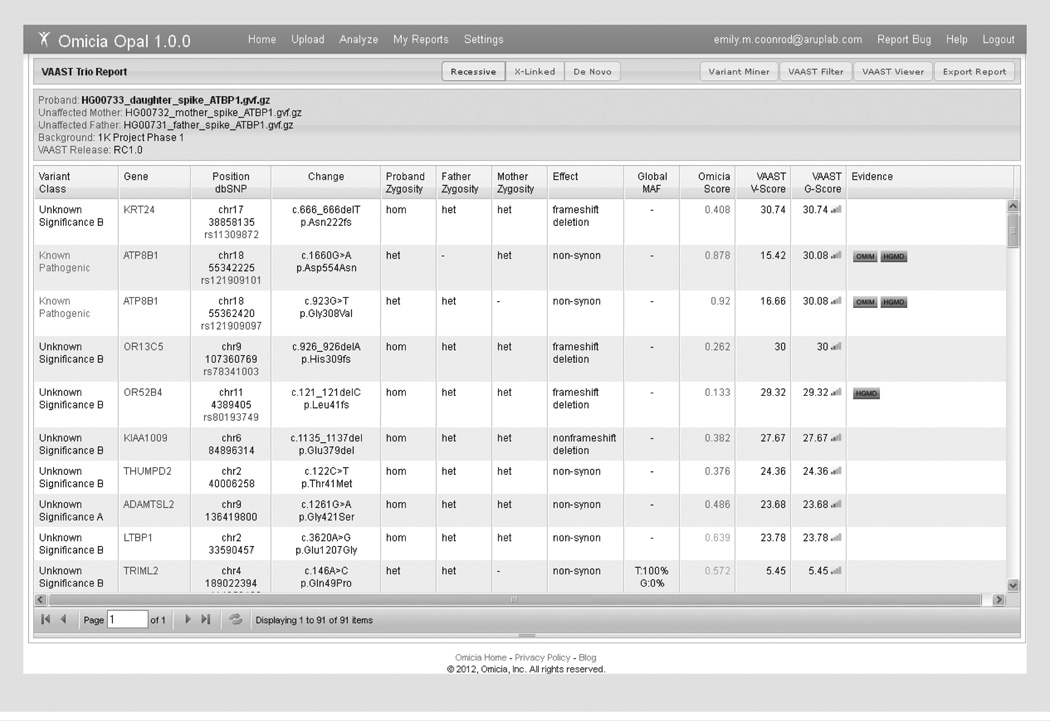
One of the main goals for further development of the Opal system was implementation of the VAAST for variant prioritization [115]. VAAST uses the predicted severity of a non-synonymous amino acid change from the reference and the allele frequency of the case’s variant change as found in a control data set to generate a list of genes ranked by the likelihood that the variants in that gene lead to disease [14]. The implementation of VAAST allows for variant prioritization without heuristic filtering methods or threshold- setting, which are commonly used for gene identification as described in the test cases above. The VAAST output contains three scores, the variant (V) score describes the impact of a variant, the gene (G) score, describes the combined impacts of a set of variants on the gene in question, and a p-value, determined through a permutation-based approach, indicates the statistical significance of the gene score. These scores are unique to each experiment but significance can be estimated from the p-value associated with each VAAST score (for details, see [33]). This tool recently became available in Opal after the initial blinded study was performed, so VAAST analysis was performed on the spiked data set for the trio with intrahepatic cholestasis (described as Clinical Test Case 1) to test variant prioritization. Upon running VAAST on the spiked data set from clinical test case 1, 91 variants were ranked. Figure 6 shows the top ten variants with the causative variants uncovered in the blinded study ranked 2nd and 3rd by the VAAST G-score. The gene with the highest VAAST score, KRT24, had a lower Omicia score than the variants in ATP8B1 and also did not have any functional evidence to support it as a candidate. Implementation of VAAST into the Opal system creates a fast, user-friendly format for performing VAAST analysis. Outside of the Opal system, running VAAST requires Linux tools, Linux commands, and takes time to learn how to run properly but can be accessed by academic users free of charge [111]. It is important to note that VAAST is useful as a ranking tool but the top ranked variants will still require vetting by the end user.
Figure 6. Variant Annotation, Analysis and Selection Tool Trio report.
Shown is the Variant Annotation, Analysis and Selection Tool data report from the trio analysis performed with the simulated clinical data described in Case 1. The ATP8B1 compound heterozygous changes rank 2nd and 3rd in this report for the Variant Annotation, Analysis and Selection Tool G-score.
Comparison of other commercially available software packages for clinical analysis of NGS data
There are a few additional commercial software packages available for clinical analysis and interpretation of NGS data. These packages include Variant Analysis from Ingenuity and the Knome suite. Other companies such as Silicon Valley Biosystems and Personalis offer full integrated end-to-end sequencing and interpretation services, and currently no independent software packages were offered. Table 1 shows a comparison of the features available in Ingenuity, Knome and Omicia’s software, as determined by publically available information on their websites. The alignment, variant calling and variant annotation columns refer to whether the companies offer these services regardless of method. Omicia, Ingenuity and Knome offer the user the ability to filter variants in a customizable manner for purposes of heuristic filtering. Omicia provides the user with a variant ranking (from the VAAST tool) which here refers to prioritization of variants by any method. Clinical reporting here refers to reports generated with a focus on interpretation for clinical testing purposes. One of the difficulties of uncovering causative variants in exome and genome sequencing is in integrating the candidate variant list with all available knowledge bases such as HGMD, OMIM, locus-specific databases, protein–protein interaction networks and published literature. All three companies have made efforts to integrate various knowledge bases into their user interface and/or clinical reports to streamline the process of narrowing down a list of variants to a very short list of candidates specific to the particular clinical case. This feature is attractive in a clinical setting due to the time requirements for laboratory personnel to find this information if it is not in one central location.
Table 1.
Comparison of commercially available software platforms for next-generation sequencing data analysis.
| Alignment | Variant calling |
Variant annotation |
Variant filtering |
Variant ranking |
Clinical reporting |
Access to knowledge bases |
|
|---|---|---|---|---|---|---|---|
| Omicia Opal | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Ingenuity variant analysis | ✓ | ✓ | ✓ | ✓ | |||
| Knome | ✓ | ✓ | ✓ | ✓ | ✓ |
Conclusion
This report demonstrates the use of the Omicia platform for the identification of clinically important variant(s) in personal genomes, exomes or other NGS assays. Here, the authors show an example of the successful identification of disease-causing variants from whole genome data in a trio and in two unrelated individuals. All test cases discusses and analyzed here are made available through the Opal system for free access to the research community, and as an educational tool for genome analysis.
The web application interface allows for rapid and easy annotation, prioritization and navigation of large variant data sets from various NGS platforms. The intuitive design allows the end user to analyze large variant data sets directly through annotation, multiple sort and filter selections, intersect and difference functions and VAAST analysis without the inclusion of internal or external bioinformatics groups, which shifts the power of analysis to the user. This shift is critical in a clinical laboratory setting where turnaround time, speed of analysis, accuracy and reproducibility of results are paramount.
Expert commentary
The current bottleneck in implementing NGS as a platform for clinical diagnostics is in the analysis and reporting of causative variants. Systems such as Omicia’s Opal platform will aid in the integration of NGS-based tests into the clinical laboratory by reducing data analysis time and, therefore, test turnaround time.
Five-year view
There will probably be an integration of NGS-based clinical testing into routine diagnostic testing in the next 5 years. A key development to realize this goal will be tools to rapidly analyze NGS data and decrease turn-around-times and accuracy of results in a HIPAA-compliant environment.
Availability & requirements
Opal is a web-based software application, which runs on modern web browsers, including Apple Safari versions 5 and higher, Mozilla Firefox versions 10 and higher, and Google Chrome versions 18 and higher. As a multi-tier web application, Opal is implemented in multiple software technologies, including HTML, CSS and JavaScript for presentation logic, Python for application logic, relational databases and file systems for data storage and Perl and Java for back-end computation, running on a Linux platform. Opal access is available at [105]. Opal’s core functionality including the annotation pipeline is free to all users with agreement to Opal’s terms of use; to access advanced premium features such as premium genome processing, genome comparisons and VAAST runs, there are additional costs.
Key issues.
Omicia’s Opal system annotates next-generation sequencing data and allows the user to perform heuristic filtering for causative variant discovery.
Three clinical case studies show that heuristic filtering of a human genome resulted in discovery of the causal variant.
Variant annotation, analysis and selection tool analysis of one of the simulated case studies revealed the causal variant and shows the utility of variant annotation, analysis and selection tool for clinical applications.
Omicia’s Opal system could be used for data analysis of next-generation sequencing-based clinical testing.
Acknowledgments
This work was supported by NIH SBIR grant R44HG3667 to MG Reese & A Russell, NIH ARRA GO grant RC2HG005619 to MG Reese and NIH SBIR grant R44HG006579 to A Russell, KV Voelkerding and MG Reese, all administered by the National Human Genome Research Institute (NHGRI). A Russell is an employee of Omicia Inc. and MG Reese is the co-founder, president and chief scientific officer of Omicia Inc.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Bainbridge MN, Wiszniewski W, Murdock DR, et al. Whole-genome sequencing for optimized patient management. Sci. Transl. Med. 2011;3(87):87re3. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA. 2009;106(45):19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. N. Engl. J. Med. 2010;362(13):1181–1191. doi: 10.1056/NEJMoa0908094. • Landmark publication describing the discovery of a causative variant for a Mendelian disorder using whole-genome sequencing.
- 4. Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2010;42(1):30–35. doi: 10.1038/ng.499. • Landmark publication describing the discovery of a causative variant for a Mendelian disorder using exome sequencing.
- 5.Sobreira NL, Cirulli ET, Avramopoulos D, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6(6):e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet. Med. 2011;13(3):255–262. doi: 10.1097/GIM.0b013e3182088158. • One of the first published cases where discovery of a causative variant using exome sequencing led to a treatment decision.
- 7.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Handsaker B, Wysoker A, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coonrod EM, Durtschi JD, Margraf RL, Voelkerding KV. Developing genome and exome sequencing for candidate gene identification in inherited disorders: an integrated technical and bioinformatics approach. Arch. Pathol. Lab. Med. 2013;137(3):415–433. doi: 10.5858/arpa.2012-0107-RA. [DOI] [PubMed] [Google Scholar]
- 13. Pabinger S, Dander A, Fischer M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform. 2013 doi: 10.1093/bib/bbs086. (Epub ahead of print). •• Provides a description and comparison of many different tools for working with next-generation sequencing data.
- 14. Yandell M, Huff C, Hu H, et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21(9):1529–1542. doi: 10.1101/gr.123158.111. •• Describes details of the variant annotation, analysis and selection tool algorithm.
- 15.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–194. [PubMed] [Google Scholar]
- 16.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flicek P, Ahmed I, Amode MR, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000;15(1):7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 20.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giardine B, Riemer C, Hefferon T, et al. PhenCode: connecting ENCODE data with mutations and phenotype. Hum. Mutat. 2007;28(6):554–562. doi: 10.1002/humu.20484. [DOI] [PubMed] [Google Scholar]
- 22.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20(1):110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiman L. Random forests. Machine Learning. 2001;45(1):5–23. [Google Scholar]
- 24.Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bull LN, van Eijk MJ, Pawlikowska L, et al. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat. Genet. 1998;18(3):219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- 26.Klomp LW, Bull LN, Knisely AS, et al. A missense mutation in FIC1 is associated with greenland familial cholestasis. Hepatology. 2000;32(6):1337–1341. doi: 10.1053/jhep.2000.20520. [DOI] [PubMed] [Google Scholar]
- 27.Dale DC, Person RE, Bolyard AA, et al. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96(7):2317–2322. [PubMed] [Google Scholar]
- 28.Rope AF, Wang K, Evjenth R, et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Hum. Genet. 2011;89(1):28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reese MG, Moore B, Batchelor C, et al. A standard variation file format for human genome sequences. Genome Biol. 2010;11(8):R88. doi: 10.1186/gb-2010-11-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison TR, Wilson JD. Principles of Internal Medicine. McGraw-Hill: Health Profession Division, NY, USA; 1991. Harrison’s. [Google Scholar]
Websites
- 101.dbSNP. www.ncbi.nlm.nih.gov/projects/SNP.
- 102.OMIM. www.ncbi.nlm.nih.gov/omim.
- 103.HGMD. http://www.hgmd.cf.ac.uk/ac/index.php.
- 104.PharmGKB. http://www.pharmgkb.org/
- 105.1000 Genomes. www.1000genomes.org.
- 106.SIFT. http://sift.jcvi.org/
- 107.PolyPhen2. http://genetics.bwh.harvard.edu/pph2.
- 108.Mutation Taster. http://www.mutationtaster.org/
- 109.Omicia. www.omicia.com.
- 110.Complete Genomics. www.completegenomics.com.
- 111.ANNOVAR. http://wannovar.usc.edu/example.html.
- 112.HGNC. www.genenames.org.
- 113.Gene Tests. www.ncbi.nlm.nih.gov/sites/GeneTests.
- 114.Genetics Home Reference. http://ghr.nlm.nih.gov/
- 115.VAAST. http://www.yandell-lab.org/software/vaast.html.
- 116.Omicia Opal. https://app.omicia.com/login?came_from=%2F.
- 117.Ingenuity Variant Analysis. http://www.ingenuity.com/products/variant-analysis.
- 118.Knome. http://www.knome.com/